Abstract
The pH low-insertion peptide (pHLIP) serves as a model system for peptide insertion and folding across a lipid bilayer. It has three general states: (I) soluble in water or (II) bound to the surface of a lipid bilayer as an unstructured monomer, and (III) inserted across the bilayer as a monomeric α-helix. We used fluorescence spectroscopy and isothermal titration calorimetry to study the interactions of pHLIP with a palmitoyloleoylphosphatidylcholine (POPC) lipid bilayer and to calculate the transition energies between states. We found that the Gibbs free energy of binding to a POPC surface at low pHLIP concentration (state I–state II transition) at 37°C is approximately −7 kcal/mol near neutral pH and that the free energy of insertion and folding across a lipid bilayer at low pH (state II–state III transition) is nearly −2 kcal/mol. We discuss a number of related thermodynamic parameters from our measurements. Besides its fundamental interest as a model system for the study of membrane protein folding, pHLIP has utility as an agent to target diseased tissues and translocate molecules through the membrane into the cytoplasm of cells in environments with elevated levels of extracellular acidity, as in cancer and inflammation. The results give the amount of energy that might be used to move cargo molecules across a membrane.
Keywords: thermodynamics, drug delivery, imaging, membrane protein
The folding of helical membrane proteins can be conceptualized in terms of distinct steps: (i) peptide insertion and folding to form independently stable helices across a lipid bilayer, (ii) helix association to form a helix bundle intermediate, and (iii) further rearrangements of protein structure and/or binding of prosthetic groups to achieve a functional state (1). Insertion of most membrane proteins is facilitated in vivo by complex molecular machines, such as the translocon, which assist in placing hydrophobic sequences across the bilayer (2). In contrast to more hydrophobic sequences, moderately polar transmembrane domains of C-terminally-anchored proteins can postranslationally translocate themselves into membranes in a translocon-defective yeast strain (3, 4), seeking the free-energy minimum (5). Thus, insertion of a disordered peptide in aqueous solution to form a transbilayer α-helix is accompanied by a release of energy, so thermodynamic studies of the interaction of proteins and peptides with a lipid bilayer can provide insights regarding folding. However, such studies are significantly complicated by the poor solubility of membrane peptides, which has made it difficult to separate the contributions of peptide interactions with a lipid bilayer from self-interactions and aggregation.
We have been studying a useful system for measurement of the thermodynamics and kinetics of peptide insertion and folding across a lipid bilayer. It is based on the pH low-insertion peptide (pHLIP), which has three major states: (I) soluble in water in an unstructured, monomeric state, (II) bound to the surface of a lipid bilayer in an unstructured, monomeric state, and (III) inserted across the bilayer as an α-helix (6–9). The existence of three distinct equilibrium states makes it possible to separate the process of peptide attachment to a lipid bilayer from the process of peptide insertion/folding. Besides its interest for the study of membrane protein folding, pHLIP is a promising imaging and drug-delivery agent that can target acidic tissues as in cancer, stroke, ischemia, and others. Here, we report the results of a thermodynamic study of pHLIP interactions with a phospholipid bilayer and discuss general aspects of peptide interactions with a lipid bilayer and the implications of the results for the translocation of cargo molecules across a membrane by pHLIP.
Results
To study the interaction of pHLIP (AEQNPIYWARYADWLFTTPLLLLDLALLVDADEGT) with lipid bilayers at high (pH 8.0) and low pHs, we used fluorescence spectroscopy methods and isothermal titration calorimetry (ITC) [details in supporting information (SI) Text]. pHLIP contains two tryptophan residues having fluorescence that reports the attachment of the peptide to the membrane surface and the insertion of the peptide across the lipid bilayer as an α-helix (6,8,9). We chose 100-nm large unilamellar vesicles (LUV) containing palmitoyloleoylphosphatidylcholine (POPC) lipids with zwitterionic head groups to minimize any electrostatic component of the pHLIP interaction and to study peptide-membrane interactions over a wider range of temperatures than was possible in earlier work with dimyristoylphosphatidylcholine (DMPC) (6).
pHLIP Adsorption to the Bilayer Surface.
Experiments at different temperatures followed fluorescence spectra of pHLIP (Pt = 7.4 μM initial concentration) with progressive addition of POPC vesicles at pH 8.0. Fig. S1a presents the curves obtained. Fluorescence was measured from 330–360 nm when excited at 295 nm (short-wavelength emission was excluded to minimize the contribution of scattered light).
ITC was used to determine enthalpy changes of the pHLIP interaction with the lipid bilayer at various temperatures. Each injection of a small aliquot of peptide was accompanied by the release of the same amount of heat, calculated by the integration of the titration peaks (Fig. S1c). This is a steady-state experiment, because the lipid is in great excess. Complete binding of the peptide to lipids was seen, even at the end of the titration, where the lipid: peptide ratio exceeded 600:1. The average heat values of the reaction (ΔH) (calculated per peptide molecule, not per lipid) measured at 15°C, 22°C, and 37°C temperatures were −25.5, −21.2, and −17.6 kcal/mol, respectively (Fig. S1d). In a titration experiment, the peptide solution was first loaded in the calorimetric cell, and then vesicles were injected in a sequence of steps (Fig. S1e). Progressive injections produced decreasing exothermic heats of reaction. The cumulative heat of reaction after the ith injection (the titration curve) is presented in Fig. S1f.
We analyze the titration data using a binding equilibrium (Eq. 1) model rather than a partition model, because we think that it is more accurate to consider lipids as individual molecules than as a phase.
where P is the peptide, L is the lipid molecule, and n is the coordination number. Our goal was to measure the adsorption constant and to estimate the approximate number of lipids interacting with one peptide molecule (coordination number). We analyze the titration data using Scatchard plots, where Pb is the amount of peptide bound to lipids, Pf is the amount of unbound peptide (free peptide in solution), equal to Pt − Pb (Pt is the total concentration of the peptide), and Lt is the total amount of lipid at each step of the titration experiment. Pb was calculated from the titration data:
where cp is the peptide concentration at each step of titration, and S, S0, and Smax are the current, initial, and maximum values of the signals measured over the course of a titration experiment, respectively.
Mathematical Analysis of Scatchard Plots.
The titration data are straightforward to interpret and analyze in a Scatchard plot, because distinct types of interactions are represented by straight lines described by the equation:
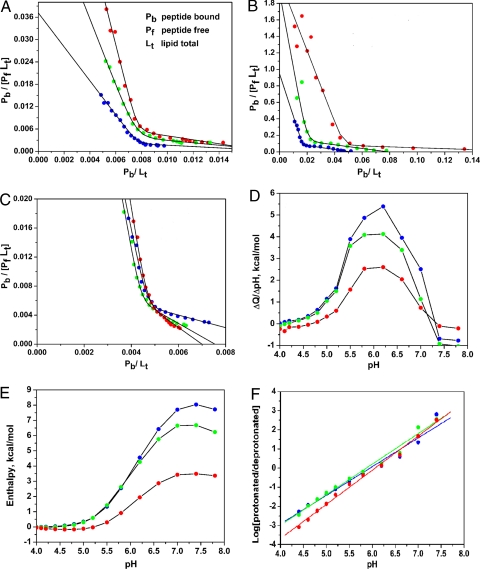
where y is Pb/PfLt, x is Pb/Lt, K is the adsorption constant, and n is the number of lipids. Thus, intersections of the line with the x and y axes give values of 1/n and K, respectively. The Scatchard plots constructed from both fluorescence and ITC titration data are presented in Fig. 1 A–C. The existence of two different types of pHLIP interaction with the bilayer at low and high lipid/peptide ratios is evident from the dramatic change of slope between two linear regions. These two types of binding can be described by adsorption constants K1 and K2, and coordination numbers n1 and n2 at low and high peptide/lipid ratios, respectively, and the slopes of the linear parts of the curves are then given by −K1n1 and −K2n2, respectively. Both in the fluorescence and ITC data, increasing the availability of bilayer surface area for peptide binding results in a sharp transition near the transition point xt.
To fit the experimental data over all x values, we need to describe the transition from one type of binding to the other. The changes of slope (Kn versus x) during the transition follow an S-type transition (possibly a first-order phase transition), and the slope changes are well approximated by using an empirical function:
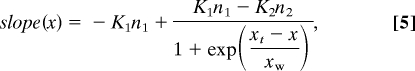 |
where xw is the width of transition. Then, an expression for the total transition curve, y(x), is obtained by integration of equation (5):
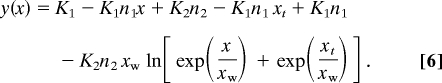 |
Eq. 6 was fit to the experimental data from the fluorescence and ITC measurements. The proposed model describes the experimental data with high precision and allows calculation of the adsorption constants and coordination numbers at low and high lipid/peptide ratios. Changes of Gibbs free energy associated with pHLIP binding to the bilayer surface (energy per peptide, not per lipid) were calculated from the standard equation:
where K is the adsorption constant. Tables S1 and S2 contain the thermodynamic parameters obtained from fitting the experimental fluorescence data. The variation of the adsorption constants, coordination numbers and free energy during the titrations are presented in Fig. S2 a and b, c and d, and g–i. Large, positive changes in heat capacity (Δcp) were determined from the temperature dependence of ΔH.
Fig. 1.
Thermodynamic data on pHLIP interaction with POPC vesicles. (A–C) Scatchard plots constructed from the titration of pHLIP with POPC vesicles at pH 8.0 (A) and pH 5.0 (B) monitored by changes of tryptophan fluorescence and (C) by changes of heat release measured by ITC (titration data presented in Figs. S1 a, b, and f, respectively). (D) The pH-dependence of the heat effects induced by the pH changes of pHLIP-POPC. (E) The changes of enthalpy calculated from data in D over the course of the pHLIP transition from the denatured to the native state in lipid bilayers. (F) The pH-dependence of the logarithmic ratio of protonated to deprotonated forms of pHLIP. These plots were used to calculate the number of protons necessary to induce the transition. Red, 15°C; green, 22°C; blue, 37°C.
pHLIP Insertion to Form a Transbilayer Helix.
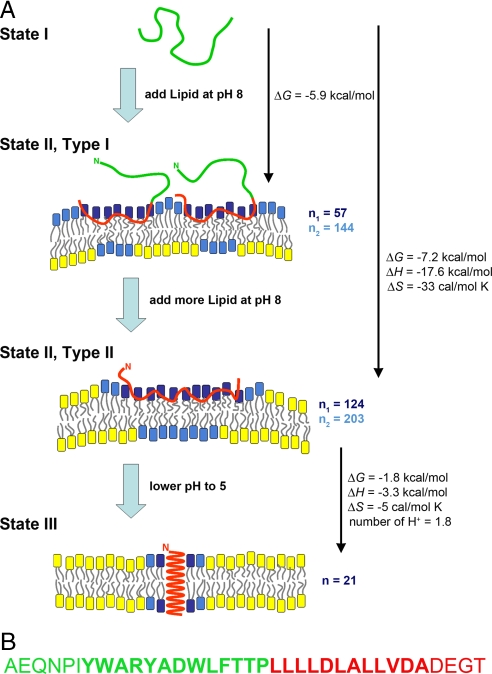
Thermodynamic parameters for pHLIP insertion and folding across the lipid bilayer were obtained from fluorescence spectroscopy and ITC. Fig. S1b presents titration curves obtained from pHLIP fluorescence at pH 5.0, varying POPC concentration at each of three temperatures. The data are rendered in Scatchard plots In Fig. 1b and analyzed as above. Fig. S2 g–i shows the changes of adsorption constant, coordination number and free energy. Table S2 has the parameters found in the analysis of the fluorescence Scatchard plots at pH 5.0. These results report both processes: peptide attachment (state I to state II) and pHLIP insertion/folding into the bilayer (state II to state III). An interesting observation is that the number of lipids seen to interact with the peptide is only half of that in the peptide attachment to the surface and that it decreases with temperature (Table 1).
Table 1.
Summarized thermodynamic parameters of pHLIP attachment and insertion/folding in a lipid bilayer
| Type | Parameter | 15°C | 22°C | 37°C |
|---|---|---|---|---|
| Transition from state I to state II (attachment to membrane) | ||||
| Type I (crowded pHLIP) | K (M−1) | 6.3 × 103 | 10.1 × 103 | 12.7 × 103 |
| nfluor | 56 | 52 | 57 | |
| nITC | 94 | 132 | 144 | |
| ΔG (kcal mol−1) | −5.0 | −5.4 | −5.9 | |
| Type II (dispersed pHLIP) | K (M−1) | 5.8 × 104 | 7.8 × 104 | 20.9 × 104 |
| nfluor | 123 | 125 | 124 | |
| nITC | 200 | 212 | 202 | |
| ΔG (kcal mol−1) | −6.3 | −6.7 | −7.2 | |
| ΔH (kcal mol−1) | −25.5 | −21.2 | −17.6 | |
| ΔS (kcal mol−1K−1) | −0.067 | −0.072 | −0.033 | |
| Δcp = 340 cal mol−1K−1 | ||||
| Transition from state I to state III via state II | ||||
| Type I (crowded pHLIP) | K (M−1) | 1.3 × 105 | 1.7 × 105 | 1.1 × 105 |
| nfluor | 19 | 13 | 6 | |
| ΔG (kcal mol−1) | −6.8 | −7.1 | −7.2 | |
| Type II (dispersed pHLIP) | K (M−1) | 9.6 × 105 | 19.3 × 105 | 20.9 × 105 |
| nfluor | 54 | 47 | 21 | |
| ΔG (kcal mol−1) | −7.9 | −8.5 | −9.0 | |
| Transition from state II to state III (folding across bilayer) | ||||
| Type II (dispersed pHLIP) | ΔG (kcal mol−1) | −1.6 | −1.8 | −1.8 |
| ΔH (kcal mol−1) | −8.04 | −6.68 | −3.33 | |
| ΔS (kcal mol−1K−1) | −0.022 | −0.017 | −0.005 | |
| No. of H+ | 1.5 | 1.6 | 1.8 | |
| Δcp = 216 cal mol−1K−1 | ||||
K is the adsorption constant, n is the coordination number (number of lipids), ΔG, ΔH, and ΔS are changes of free energy, enthalpy, and entropy of peptide–membrane interaction, and Δcp is the heat capacity.
The transition from state II to state III is like the transition from a set of denatured (D) states to the native (N) state, in other words, the process of folding of the protein in the membrane environment, assuming that the “native state” is like that found in bacteriorhodopsin—a transmembrane helix. Here, the denatured state is unlikely to be the state on the folding pathway in vivo, but may be informative to examine. To find changes in the heat associated with the D–N transition and to estimate the number of protons involved in the process, we used isothermal acid-titration calorimetry (IATC) (10, 11). pHLIP was preequilibrated with vesicles at pH 8.0, placing most of the peptide in state II, and then aliquots of acid were injected to induce peptide insertion across the bilayer (state III). Each injection of acid into the peptide-POPC solution was accompanied by the release of heat (Fig. S1g). In separate experiments, acid was injected into a solution of POPC at various temperatures to measure the heat of dilution (Fig. S1h). Separately, a microelectrode was used to measure stepwise pH values. By subtracting the dilution heat measured at the same pH, the heat effects of peptide insertion and folding were evaluated. Fig. 1d shows the pH dependence of the released heat induced by pH changes at various temperatures. The enthalpy change decreased with increasing temperature (Fig. 1e). The pKa was found to be ≈6.0, correlating well with earlier data (6). The change in heat capacity (Δcp) was about 216 cal mol−1 K−1 based on the temperature dependence of ΔH. The apparent number of protons (h) involved in the process of pHLIP folding can be estimated from the plots on Fig. 1f according to the equation:
The number of protons inducing the insertion and folding of pHLIP is equal to 1.5–1.8 from these measurements.
Discussion
pHLIP is a peptide that lives in three worlds: soluble as an unstructured monomer, bound to a bilayer as an unstructured monomer, and inserted across a bilayer as a monomeric helix. In contrast to previously studied amphipathic peptides, it does not aggregate, form helical structure on the surface of a membrane, or induce pore formation (6–9, 12). Folding (formation of a transmembrane helix) accompanies insertion of the peptide into the lipid bilayer. It is triggered by a drop of pH, which leads to the protonation of Asp residues and the enhancement of peptide hydrophobicity (8). The pHLIP interaction with the lipid bilayer allows its attachment to the membrane surface (state II) and its insertion/folding (state III) to be distinguished, giving an opportunity for thermodynamic studies of membrane protein folding and stability, and motivating the experiments we report here.
Changes of pHLIP tryptophan fluorescence were used to monitor interaction with large unilamellar POPC vesicles. We previously demonstrated that the emission of tryptophan residues of pHLIP is distinct in each of the three states. In state I (in solution), the tryptophan residues of pHLIP are completely exposed to the polar and flexible water environment, resulting in long wavelength fluorescence, (Class III according to the Burstein and Reshetnyak classification (13, 14). In state II (adsorbed to the bilayer surface), tryptophan residues are partially buried in the hydrophobic core of the lipid bilayer, leading to a blue shift of the emission and an increase of intensity. And in state III (transbilayer α-helix), one of the tryptophan residues is completely buried in the hydrophobic core and gives a more pronounced blue shift and an additional increase in intensity. Thus, using tryptophan fluorescence spectroscopy, we can probe the location of pHLIP in the bilayer; however, no information about internal changes in the bilayer is obtained. Therefore, we also used ITC to monitor heat changes during the interaction states, which include changes in both peptide–lipid and lipid–lipid interactions.
We analyzed the titration data using a binding equilibrium model rather than a partition model, since it seems more accurate to consider lipids as individual molecules rather than as a phase. Our goal was to estimate not only the adsorption constant but also the number of lipids interacting with pHLIP. Scatchard plots of the data show the existence of two distinct types of peptide interaction with the lipid bilayer. The first type of interaction (type I, Fig. 2a) occurs at low lipid/peptide ratio and is associated with a “parking problem,” where the total accessible membrane surface is not enough to accommodate the full lengths of peptides on it. Type I binding energetically favors partial adsorption of 2 peptides instead of the whole length of a single peptide (see free energy values in Table 1). The second type of interaction (type II, Fig. 2a) occurs at high lipid/peptide ratios, when there is enough accessible space on the bilayer for peptides to occupy surface area without competing with each other. Fluorescence and ITC data for the adsorption constants give similar values for the free energy changes for both types of interactions of pHLIP with the bilayer at pH8.0. However, the coordination numbers from the ITC titration data are 1.5–2.5 times higher than values obtained from the fluorescence data. Therefore, the coordination numbers are presented separately in Table 1, while other parameters were averaged.
Fig. 2.
A schematic representation of pHLIP interaction with a lipid bilayer is shown. (A) In state I, the peptide is in solution at neutral and basic pHs. By addition of vesicles, the unstructured peptide is adsorbed on the membrane surface. State II, type I interactions occur at a low lipid/peptide ratio, when the total accessible membrane surface is not enough to accommodate the full length of peptide on it. We reason that the hydrophobic motif near the C terminus is adsorbed first (red sequence in B). By addition of more vesicles, the transition to the type II (state II) interaction is seen, at a high lipid/peptide ratio when there is enough accessible space on a membrane for peptides to freely occupy lipid surface area without competing with each other. In state III, a drop of pH leads to the protonation of Asp residues, increasing peptide hydrophobicity, and resulting in the insertion and formation of a transmembrane α-helix. Our thermodynamic measurements suggest the existence of three major populations of lipids: (i) lipids interacting with the peptide directly (lipids with blue head groups); (ii) lipids not interacting with the peptide directly but influenced by the interaction (lipids with cyan head groups, possibly in both leaflets); and (iii) lipids that are not involved in the interaction with pHLIP (lipids with yellow head groups). The values for energy changes are taken from Table 1 (for 37°C). (B) The pHLIP sequence is presented, with the presumed transmembrane part in bold (based on the helix seen in bacteriorhodopsin). Red is used to denote the hydrophobic sequence and the C terminus, which are expected to initially interact with the bilayer.
To understand the coordination number difference, we consider what is measured in the titration experiments: fluorescence measures how many peptides bind to the membrane surface; calorimetry measures the total heat released during the interaction. Thus, it is reasonable to suppose that the peptide adsorbed to l lipid molecules (detectable by fluorescence spectroscopy) may also induce the perturbation of additional m lipid molecules. The ITC signal reports both processes, fluorescence reports only the first process. Combined ITC-fluorescence was previously used to study amphipathic peptides interacting with lipid bilayers (15, 16). However, in one case a partition model was used to analyze the titration data (15), and in the other case a simple binding model (with a single binding site) was used (16). In all cases (including our study) fluorescence and ITC methods gave similar values for free energy changes. Only through our use of a binding model with multiple coordination centers is it possible to see the lipid binding difference.
For both types I and II interactions, the adsorption constants increase with temperature, showing that binding of pHLIP to the bilayer surface is stronger at high temperatures. But, the coordination number does not change much with temperature, so stronger binding cannot be attributed to an increase of contacts between peptide and lipid bilayer. We assume that adsorption is partly driven by hydrophobic interactions due to the partition of hydrophobic residues (poly Leu motif in pHLIP, see 2b) into the lipid bilayer: an increase of temperature enhances hydrophobic interactions, and we observe stronger binding. However, we cannot conclude that pHLIP adsorption is from hydrophobic interactions alone, because the interaction is accompanied by a release of heat that decreases with temperature, giving a large positive value of 340 cal mol−1 K−1 for the heat capacity. A positive heat capacity was also observed for magainin interactions with bilayers and was interpreted as the exposure of hydrophobic surfaces of lipid to the aqueous environment (17).
We posit that several processes could contribute to the observed heat capacity changes during pHLIP interactions with the lipid bilayer. Partitioning of hydrophobic residues into the membrane should lead to a negative heat capacity change. However, pHLIP is not a fully hydrophobic peptide; it contains many polar and even potentially charged residues (see Fig. 2B). Thus we cannot exclude partial insertion of these residues into the lipid bilayer, which should result in a positive heat capacity change. Also, we have assumed that the peptides do not interact significantly with each other. Although this is reasonable at low surface concentration, where our previous data indicate that the peptide is monomeric (9), we have no data to show this at high concentration. And pHLIP interaction with the outer leaflet of a bilayer creates strain and distortion, because anisotropic area occupancy induces bilayer distortions (18,19). We previously reported such a distortion induced by pHLIP interaction with the outer leaflet of the red blood cell membrane (8) as well as distortion in a liposome system (12). The distortion is released when the peptide inserts and occupies equal areas in the two bilayer leaflets. The molecular mechanism of distortion of the membrane might be associated with the exposure of hydrophobic parts of lipids to the polar water environment and resulting in positive heat-capacity changes. Furthermore, the hydrophobic region of the lipid bilayer does not have a sharp boundary —there is a continuous gradient of water activity (20, 21), so that the hydrophobic interactions we see pertain to a continuum of environments.
We propose a distinction among three populations of lipids: (i) lipids interacting directly with the pHLIP peptide (blue head groups in Fig. 2A); (ii) lipids not interacting with the peptide directly, but perturbed by the interaction (cyan head groups in Fig. 2A; such lipids could be present in both outer and inner leaflets); and (iii) lipids not significantly perturbed (yellow head groups in Fig. 2A). It seems reasonable that fluorescence titration experiments report on the process of pHLIP interaction with the first type of lipid (blue), because this method follows peptide binding. However, ITC experiments reflect both processes: peptide interaction with lipids and lipid distortions, and the coordination numbers revealed by ITC are always higher than those numbers calculated from the fluorescence data. Thus, the peptide directly interacting with one lipid (probed by fluorescence and ITC) induces distortion of another lipid (probed by ITC). In state II, the numbers of lipids directly interacting with pHLIP are 55 and 124 for type I and II interactions, respectively. Type I interaction is the first event of pHLIP binding, and it seems reasonable that the polyLeu hydrophobic motif near the C terminus of pHLIP (green residues) would be involved. The C terminus inserts across the membrane and emerges inside the vesicle (or membrane, in cells) after a pH drop (8, 9). Our data clearly indicate that the peptide does not adopt any well defined secondary structure upon membrane adsorption. We estimated the total surface area of the unstructured polyLeu and the C terminus of pHLIP (red residues) by summing the surface areas for each amino acid taken from Table 2 in ref. 22. The total surface area of the “green” part of pHLIP (see Fig. 2B) is ≈3,500–4,000 Å2. For comparison, a cluster of 55 lipids with headgroup areas of ≈60–65 Å2 (23, 24) will occupy ≈3,300–3,600 Å2 as well. The transition from type I to type II is accompanied by the adsorption of the whole peptide on the membrane surface (the red residues on Fig. 2B), and pHLIP might occupy a total area of ≈8,000 Å2, which correlates well with the total surface area of about 124 lipids. The I-II transition is rather sharp (possibly first order), which is probably dictated by the requirement to maintain a balance between the energy gained because of the full-length peptide interaction with lipids and, at the same time, the lipid distortion induced by this interaction. For Type II there would be enough available membrane surface to complete this transition.
The experiments performed at pH 5.0 follow the transitions from state I (at pH 5.0) to state III (transmembrane alpha-helix) via state II. The affinity of pHLIP for the bilayer at pH 5.0 is 10–20 times higher than at pH 8.0 because of the formation of the transmembrane helix. The strength of interaction increases with temperature, possibly because of the better attachment of pHLIP to the bilayer surface (discussed above). A significant difference between the process of peptide adsorption and insertion/folding is in the number of lipids interacting with pHLIP in state II vs. state III. Two layers of lipids surrounding the α-helix would account for the state III interaction at low lipid/peptide ratio (Fig. 2A), and the number of perturbed lipids decreases with an increase of temperature as thermal fluctuations increase. The minimum number of bilayer lipids with headgroup areas of 65 Å2 needed to surround a transmembrane helix 12 Å in diameter is ≈14. If a second shell forms around the helix, the total number of lipids involved is about 40–44. These numbers correlate with the coordination number obtained from the analysis of titration data at low pHs. At low lipid/peptide ratios the values of coordination number are lower and change from 19 to 6 with increasing temperature. We assume that each peptide helix might be surrounded by the same number of lipids (14) as in the case of high lipid/peptide ratios, but the lipids might be shared between helices (one lipid interacting with two different helices at the same time). However, we cannot exclude the possibility of some helix aggregation at low lipid/peptide ratios.
The isothermal acid titration calorimetry (IATC) experiments measure the heat released during folding in the lipid environment (D–N transition), that is the conversion from state II to state III, due, in part, to the formation of hydrogen bonds in the transmembrane helix, where these bonds are only partial in the partly hydrophobic environment of state II. At higher temperatures, less heat is released than at low temperatures, which might be explained by the destabilization of hydrogen bonds because of increased molecular fluctuations, resulting in a positive heat capacity (216 cal mol−1 K−1). By subtracting the values of free-energy changes for the state I–state II transition from the values of free-energy changes calculated for the state I to state III (via state II) transition, we estimated the free energy associated with the process of folding (state II–state III transition) to be approximately −1.6 to -−1.8 kcal/mol or approximately −0.1 kcal/mol per residue. This energy includes the net stabilization of hydrogen bonds due to helix formation. Previously, in studies of amphipathic peptides, it was found that −0.2 kcal/mol per residue is attributed to the changes of free energy of helix formation at the surface of a lipid bilayer (15). The number of protons involved in the process of protonation of pHLIP is ≈1.5–1.8. Previously, we demonstrated by mutation that the two Asp residues located in the transmembrane part of pHLIP are involved in protonation and peptide insertion/folding (8). Recognizing the nature of the hydrophobic gradient, partial titration of one of the Asp groups could account for the fractional number.
It is of interest to compare pHLIP with another class of well studied peptides, the amphiphatic membrane peptides. Despite a superficial similarity between pHLIP and amphiphatic peptides, insertion events appear to be driven by different mechanisms. Amphiphatic peptides form α-helices on the surfaces of bilayers (25). As with any anisotropic inclusions they occupy only one leaflet of a bilayer and induce distortion, which increases with peptide concentration and finally results in peptide aggregation and cooperative peptide insertion into the membrane, relieving the distortion and accompanied by the formation of a pore (26). The process of pHLIP insertion/folding also begins with adsorption of the peptide on a lipid bilayer surface. This adsorption might occur in two distinct steps, with a sharp transition between them. The peptide adsorption includes burial of hydrophobic and polar residues in the bilayer and exposure of hydrophobic surfaces of lipids to the water environment resulting from bilayer distortion. However, the pHLIP membrane distortion may be qualitatively different from that of the amphiphilic peptides and is not enough to induce peptide insertion and transmembrane helix formation, possibly because of the high flexibility of the peptide on the membrane surface, where it is in an extended and unstructured configuration (in contrast to inclusion of rigid α-helices in the case of amphipathic peptides). The subsequent, pH-driven process of adsorbed peptide insertion and folding across the bilayer is driven by the titration of carboxyl groups and enhanced formation of hydrogen bonds, accompanied by a reduction of lipid distortion. We have no evidence that pHLIP folding requires the step of α-helix formation on the bilayer surface before insertion. We assume that insertion and folding may occur cooperatively and simultaneously, in contrast to β-barrel membrane proteins, which fold via distinct intermediates (27). Molecular dynamics calculations support a cooperative model: a straight α-helix was not observed as a stable interface-bound conformation (28). However, detailed kinetic studies might provide more mechanistic insights in the future.
From its physical chemical properties, pHLIP is an example of a moderately polar transmembrane peptide. Such peptides are found in the tail-anchored proteins, which typically have a large N-terminal domain, a transmembrane part, and a short C terminus. The translocation of the C-terminal domain across all target membranes occurs posttranslationally (3, 4, 29). The study of such peptides and proteins is most interesting, because, like pHLIP, they can exhibit spontaneous insertion and folding across a membrane.
We have shown that pHLIP can be used for targeting and translocation of cargo molecules across the membranes of cells in diseased tissues, such as tumor cells in acidic environments (7). Our thermodynamic study provides data on the energy of peptide insertion/folding that can contribute to the translocation of cargo molecules. We have also found that the pHLIP peptide can target acidic tumors or sites of inflammation in mice and rats, respectively (8). A puzzle in these experiments is why the peptide is not purged more rapidly from the circulation (which usually happens with low-molecular-weight compounds and peptides), but persists for hours to find its target. The present measurements offer a plausible explanation: The surface binding is strong, so that the peptide is mostly bound to membranes, and hops from one location to another through association/dissociation until it finds itself in an acidic environment, where it is stabilized by the insertion step.
Methods
For more detailed descriptions of methods used, see SI Text.
pHLIP Peptide.
The pHLIP sequence: AEQNPIYWARYADWLFTTPLLLLDLALLVDADEGT was prepared by solid-phase peptide synthesis using standard Fmoc (9-fluorenylmethyloxycarbonyl) chemistry.
Vesicle Preparations.
Large unilamellar POPC vesicles were prepared by extrusion.
Fluorescence Measurements.
Tryptophan fluorescence measurements were carried out on a PC1 spectrofluorometer (ISS) with regulated temperature-control units.
ITC Measurements.
ITC was performed by using a VP-ITC ultrasensitive microcalorimeter (MicroCal). The experimental data fitting was performed in Mathematica 5 (Wolfram Research), Statistica (StatSoft 2006), and Origin7.0 (Original Lab Corp.).
Supplementary Material
Acknowledgments.
We thank Dr. Ewa Folta-Stogniew, (Howard Hughes Medical Institute Biopolymer Facility) and Janet Crawford (the W. M. Keck Foundation Biotechnology Resource Laboratory, Yale University), for assistance with ITC measurements and peptide synthesis/purification, respectively. This work was supported in part by a University of Rhode Island (URI) Undergraduate Research Grant (to M.S.), Department of Defense (DOD) Prostate Cancer Research Program Congressionally Directed Medical Research Programs (CDMRP) Grant PC050351 (to Y.K.R.), URI Research Awards (to Y.K.R. and O.A.A.), DOD Breast Cancer Research Program CDMRP Grant BC061356, and National Institutes of Health Grants RCA125280A (to O.A.A.) and GM073857 and GM070895 (to D.M.E.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0804746105/DCSupplemental.
References
- 1.Engelman DM, et al. Membrane protein folding: beyond the two stage model. FEBS Lett. 2003;555:122–125. doi: 10.1016/s0014-5793(03)01106-2. [DOI] [PubMed] [Google Scholar]
- 2.Osborne AR, Rapoport TA. Protein translocation is mediated by oligomers of the SecY complex with one SecY copy forming the channel. Cell. 2007;129:97–110. doi: 10.1016/j.cell.2007.02.036. [DOI] [PubMed] [Google Scholar]
- 3.Brambillasca SM, Yabal M, Makarow M, Borgese N. Unassisted translocation of large polypeptide domains across phospholipid bilayers. J Cell Biol. 2006;175:767–777. doi: 10.1083/jcb.200608101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Borgese N, Brambillasca S, Colombo S. How tails guide tail-anchored proteins to their destinations. Curr Opin Cell Biol. 2007;19:368–375. doi: 10.1016/j.ceb.2007.04.019. [DOI] [PubMed] [Google Scholar]
- 5.Engelman DM, Steitz TA. The spontaneous insertion of proteins into and across membranes: The helical hairpin hypothesis. Cell. 1981;23:411–422. doi: 10.1016/0092-8674(81)90136-7. [DOI] [PubMed] [Google Scholar]
- 6.Hunt JF, Rath P, Rothschild KJ, Engelman DM. Spontaneous, pH-dependent membrane insertion of a transbilayer alpha-helix. Biochemistry. 1997;36:15177–151792. doi: 10.1021/bi970147b. [DOI] [PubMed] [Google Scholar]
- 7.Reshetnyak YK, Andreev OA, Lehnert U, Engelman DM. Translocation of molecules into cells by pH-dependent insertion of a transmembrane helix. Proc Natl Acad Sci USA. 2006;103:6460–6465. doi: 10.1073/pnas.0601463103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Andreev OA, et al. Mechanism and uses of a peptide that targets tumors and other acidic tissues. Proc Natl Acad Sci USA. 2007;104:7893–7898. doi: 10.1073/pnas.0702439104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Reshetnyak YK, Segala M, Andreev OA, Engelman DM. A monomeric membrane peptide that lives in three worlds: In solution, attached to and inserted across lipid bilayers. Biophys J. 2007;93:2363–2672. doi: 10.1529/biophysj.107.109967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nakamura S, Kidokoro S. Isothermal acid-titration calorimetry for evaluating the pH dependence of protein stability. Biophys Chem. 2004;109:229–249. doi: 10.1016/j.bpc.2003.11.006. [DOI] [PubMed] [Google Scholar]
- 11.Nakamura S, Kidokoro S. Direct observation of the enthalpy change accompanying the native to molten-globule transition of cytochrome c by using isothermal acid-titration calorimetry. Biophys Chem. 2005;113:161–168. doi: 10.1016/j.bpc.2004.09.002. [DOI] [PubMed] [Google Scholar]
- 12.Zoonens M, Reshetnyak YK, Engelman DM . Bilayer interactions of pHLIP, a peptide that can deliver drugs and target tumors. Biophys J. 2008;95:225–235. doi: 10.1529/biophysj.107.124156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Reshetnyak YK, Koshevnik Yu, Burstein EA. Decomposition of protein tryptophan fluorescence spectra into log-normal components: III. Correlation between fluorescence and microenvironment parameters of individual tryptophan residues. Biophys J. 2001;81:1735–1758. doi: 10.1016/S0006-3495(01)75825-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shen C, et al. The Protein Fluorescence And Structural Toolkit (PFAST): Database and programs for the analysis of protein fluorescence and structural data. Protein Struct Funct Bioinf. 2008;71:1744–1754. doi: 10.1002/prot.21857. [DOI] [PubMed] [Google Scholar]
- 15.Wieprecht T, Apostolov O, Beyermann M, Seelig J. Interaction of a mitochondrial presequence with lipid membranes: Role of helix formation for membrane binding and perturbation. Biochemistry. 2000;39:15297–15305. doi: 10.1021/bi001774v. [DOI] [PubMed] [Google Scholar]
- 16.Andrushchenko VV, et al. Thermodynamics of the interactions of tryptophan-rich cathelicidin antimicrobial peptides with model and natural membranes. Biochim Biophys Acta. 2008;1778:1004–1014. doi: 10.1016/j.bbamem.2007.12.022. [DOI] [PubMed] [Google Scholar]
- 17.Wieprecht T, Beyermann M, Seelig J. Binding of antibacterial magainin peptides to electrically neutral membranes: Thermodynamics and structure. Biochemistry. 1999;38:10377–10387. doi: 10.1021/bi990913+. [DOI] [PubMed] [Google Scholar]
- 18.Markin VS. Lateral organization of membranes and cell shapes. Biophys J. 1981;36:1–19. doi: 10.1016/S0006-3495(81)84713-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bohinc K, et al. Shape variation of bilayer membrane daughter vesicles induced by anisotropic membrane inclusions. Cell Mol Biol Lett. 2006;11:90–101. doi: 10.2478/s11658-006-0009-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Engelman DM. Membranes are more mosaic than fluid. Nature. 2005;438:578–580. doi: 10.1038/nature04394. [DOI] [PubMed] [Google Scholar]
- 21.White SH. Membrane protein insertion: The biology-physics nexus. J Gen Physiol. 2007;129:363–369. doi: 10.1085/jgp.200709741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pacios LF. Distinct molecular surfaces and hydrophobicity of amino acid residues in proteins. J Chem Inf Comput Sci. 2001;41:1427–1431. doi: 10.1021/ci010369n. [DOI] [PubMed] [Google Scholar]
- 23.Engelman DM. Surface area per lipid molecule in the intact membrane of the human red cell. Nature. 1969;223:1279–1280. doi: 10.1038/2231279a0. [DOI] [PubMed] [Google Scholar]
- 24.Kucerka N, Kiselev MA, Balgavý P. Determination of bilayer thickness and lipid surface area in unilamellar dimyristoylphosphatidylcholine vesicles from small-angle neutron scattering curves: A comparison of evaluation methods. Eur Biophys J. 2004;33:328–334. doi: 10.1007/s00249-003-0349-0. [DOI] [PubMed] [Google Scholar]
- 25.Fernández-Vidal M, Jayasinghe S, Ladokhin AS, White SH. Folding amphipathic helices into membranes: Amphiphilicity trumps hydrophobicity. Mol Biol. 2007;370:459–470. doi: 10.1016/j.jmb.2007.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zemel A, Ben-Shaul A, May S. Perturbation of a lipid membrane by amphipathic peptides and its role in pore formation. Eur Biophys J. 2005;34:230–242. doi: 10.1007/s00249-004-0445-9. [DOI] [PubMed] [Google Scholar]
- 27.Tamm LK, Hong H, Liang B. Folding and assembly of beta-barrel membrane proteins. Biochim Biophys Acta. 2004;1666:250–263. doi: 10.1016/j.bbamem.2004.06.011. [DOI] [PubMed] [Google Scholar]
- 28.Im W, Brooks CL. Chemical theory and computation special feature: Interfacial folding and membrane insertion of designed peptides studied by molecular dynamics simulations. Proc Natl Acad Sci USA. 2005;102:6771–6776. doi: 10.1073/pnas.0408135102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kutay U, Hartmann E, Rapoport TA. A class of membrane proteins with C-terminal anchor. Trends Cell Biol. 1993;3:72–75. doi: 10.1016/0962-8924(93)90066-a. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.