Abstract
Membrane fusion is an essential step during entry of enveloped viruses into cells. Conventional fusion assays are generally limited to observation of ensembles of multiple fusion events, confounding more detailed analysis of the sequence of the molecular steps involved. We have developed an in vitro, two-color fluorescence assay to monitor kinetics of single virus particles fusing with a target bilayer on an essentially fluid support. Analysis of lipid- and content-mixing trajectories on a particle-by-particle basis provides evidence for multiple, long-lived kinetic intermediates leading to hemifusion, followed by a single, rate-limiting step to pore formation. We interpret the series of intermediates preceding hemifusion as a result of the requirement that multiple copies of the trimeric hemagglutinin fusion protein be activated to initiate the fusion process.
Keywords: enveloped viruses, lipid bilayer, single molecule, virus entry
Enveloped viruses contain a lipid-bilayer membrane surrounding internal protein and nucleic-acid components. During entry, the membrane of the virus fuses with a membrane of the host cell and releases the viral genome—a process that requires specific, virally encoded envelope proteins. Much of our current understanding of viral membrane fusion comes from structural and mechanistic studies of the influenza virus hemagglutinin (HA), a homotrimeric envelope protein anchored by a C-terminal transmembrane segment (1–3). Processing of the HA precursor polypeptide, HA0, into HA1 and HA2 is necessary for subsequent fusion to occur. Like many other enveloped viruses, influenza virus enters cells by clathrin-mediated endocytic pathways, which deliver it to endosomes (4). Acidification of the endosomal lumen triggers fusogenic conformational changes in HA. Fig. 1 illustrates the sequence of low-pH-induced molecular events by which HA is believed to drive membrane fusion.
Fig. 1.
Proposed mechanism for viral fusion proteins. (A) In the prefusion state, the protein, anchored in the viral membrane by a C-terminal transmembrane segment, folds so that the fusion peptide (green) is sequestered. (B) A ligand-dependent trigger (e.g., proton binding, for HA and many other viral fusion proteins) induces a conformational change in which the fusion peptide projects toward the target membrane, forming an extended intermediate that bridges the two membranes. (C) The intermediate collapses, by zipping up of the C-terminal part of the ectodomain (blue) alongside the trimer-clustered N-terminal part (red). (D) The collapse pulls the two membranes together, leading to formation of a hemifusion stalk. (E) A fusion pore opens up, and snapping into place of the membrane-proximal and transmembrane segments of the protein completes the conformational transition and stabilizes the fusion pore. (Figure adapted from ref. 40.)
The structures of HA0, of the (HA1-HA2)3 ectodomain before acidification, and of the (HA2)3 ectodomain after fusion provide the outline of the fusion mechanism (5, 6). The initial large-scale event after proton binding appears to be release of the hydrophobic, N-terminal segment of HA2, from the pocket in which it lodges in the neutral pH form of (HA1-HA2)3. This “fusion peptide” is projected toward the target-cell membrane by a conformational transition that converts a loop in HA2 into an α-helix and creates a long, central coiled-coil. Insertion of the fusion peptide into the cell membrane connects the two lipid bilayers through a postulated, extended intermediate. There is good evidence for such a “prehairpin intermediate” in the case of the envelope protein of HIV-1, (gp120-gp41)3 (7–9). Collapse or “fold-back” of this intermediate can then force the two bilayers together.
Theoretical, computational, and physicochemical studies of lipid-bilayer fusion suggest multiple kinetic barriers (1). A so-called “hydration force” hinders the approach of two parallel bilayers to separations closer than 10–15 Å (10). Distortion of the two bilayers into a “hemifusion stalk” provides a further barrier (1). The hemifusion stalk (in which only the proximal leaflets of the two bilayers have merged) may proceed directly toward nascent fusion pores or toward a longer-lived hemifusion diaphragm (11). Formation of a committed, expanding pore from a hemifusion structure appears to be an additional, kinetically disfavored step (12).
Conventional fusion assays provide some kinetic information by detecting ion-conducting pores or redistribution of fluorescent dye between pairs of fusing cells or between viruses and liposomes in suspension (13–17). The data are generally limited to averaged observations of a large number of fusion events, and information about short-lived or rare intermediate states is lost, as the underlying individual fusion events dephase over time. The characterization of hemifusion and pore formation by individual retrovirus particles demonstrates the strength of single-particle observations for extracting detailed kinetic information on fusion intermediates (18). Similar approaches have enabled visualization of hemifusion by individual influenza virions (19, 20), but direct observation of the entire influenza fusion process and its kinetic characterization at the single-particle level has been difficult to achieve. We report here the development of a method for real-time detection of both hemifusion and pore formation for individual, intact influenza virus particles fusing with a target lipid bilayer. We use a planar bilayer on a glass coverslip, with a thin dextran cushion to provide an essentially liquid support. This approach allows the use of total internal reflection fluorescence (TIRF) microscopy to image the fusion kinetics of fluorescently labeled particles, with high sensitivity and time resolution. A lipophilic fluorophore is incorporated into the viral lipid envelope; its escape into the target bilayer reports hemifusion. Formation of a pore between the viral and target membranes is measured by diffusion of aqueous dye from the interior of the virus particle to the space below the supported bilayer. We demonstrate the use of this method to obtain detailed kinetic information for the hemifusion and pore-formation processes at a level hitherto unavailable from use of ensemble-averaged techniques.
Results
Fluid Supported Bilayers.
Two requirements for single-particle observation using fluorescence microscopy are a planar geometry and a fluid support. Conventional liposome-based assays are not easily adapted to single-particle detection, and frictional coupling with glass constrains the bilayer and can interfere with fusion assays. To provide a support that allows both the membrane components and the soluble components beneath it to diffuse freely, we functionalized the glass with a thin (≈1 nm), hydrophilic film of dextran polymer. In aqueous buffers, the dextran film hydrates and swells into a hydrophilic cushion, ≈100 nm thick (21). Because the bilayer can move freely over such a lubricating layer, it exhibits self-healing properties, preventing the formation of defects.
Supported lipid bilayers were formed on the dextran cushion from lipid vesicles, which adsorb to the hydrophilic surface and fuse among themselves until they reach critical size and rupture into a planar bilayer (22, 23). In the experiments described here, liposomes containing 80% phosphatidylcholine and 20% cholesterol were made by extrusion through filters with pores of 100 nm. The liposomes were doped with 1% bovine ganglioside GD1a, to provide the sialic acid receptor for influenza virus. We tested the fluidity of the supported bilayer by fluorescence recovery after photobleaching (FRAP) [supporting information (SI) Fig. S1]. In these experiments, the measured diffusion coefficient for the lipid was between 0.5 and 2 μm2/s, indicating unhindered diffusion that is indistinguishable from fluid bilayers supported on glass (21, 23, 24). We have thus generated a fully fluid, planar bilayer with an aqueous reservoir in the dextran, into which the contents of a fusing virion or vesicle can be released.
Hemifusion.
Hemifusion can be detected by observing dequenching of a fluorescent probe embedded in the membrane of the impinging virion—a method also used in various bulk-phase fusion assays (13–15, 17). Quenching of fluorescence from dye incorporated at high concentration is alleviated when the membrane in which it resides merges with a larger bilayer. The fatty-acyl linked dyes we have used incorporate into the viral membrane and hence dequench completely when that leaflet merges with the apposing leaflet of the supported bilayer (hemifusion). To allow simultaneous detection with the red fluorophore with which we label the virion interior (see below), we synthesized octadecyl-rhodamine 110 (Rh110C18), a green-emitting molecule (see SI Methods). Particles were labeled by incubating them with 30 μM Rh110C18; excess dye was removed by gel filtration.
A diagram of the experimental setup appears in Fig. 2A. The coverslip bearing the supported bilayer is mounted in a flow cell on the stage of a TIRF microscope. Virus particles labeled with Rh110C18 are injected into the flow cell, and attachment to the ganglioside receptors is monitored by the appearance of diffraction-limited spots in the image. Fig. 2B Lower Left shows an image of the Rh110C18 fluorescence from a 140 × 70-μm2 area of the target membrane. Each bright spot represents a single virus particle bound to the membrane. Omission of ganglioside from the supported bilayer resulted in a 100-fold decrease in the number of docked particles, confirming the specificity of attachment through sialic-acid receptors. We triggered fusion by lowering the pH in the flow cell from 7.4 to 4.6. Release of Rh110C18 into the supported bilayer could be detected by a sudden increase in fluorescence (green trace in Fig. 2C). The dequenching spike was followed by an intensity decrease as dye molecules diffused out of the observation area surrounding each particle. Release and diffusion of Rh110C18 could also be detected in time-lapse images as an outward expanding fluorescent “cloud” (Fig. 2B Lower Right and Movie S1).
Fig. 2.
Experimental design. (A) Virus particles are labeled with two fluorescent dyes to monitor the kinetics of hemifusion and fusion pore formation. Fluorescence is collected by a high-NA microscope objective and imaged onto a CCD. (B) Fluorescence images before (Left) and during (Right) the fusion of individual viral particles. B Upper and Lower correspond to the red and green fluorescence, respectively, of the same ≈70 × 140-μm2 area of the supported bilayer. Dequenching of the Rh110C18 membrane dye upon hemifusion gives rise to the transient brightening of individual particles. (C) The fluorescence intensity of the red SRB viral content tracer (upper trace), the green Rh110C18 membrane dye (middle trace), and the fluorescein pH sensor (lower trace) provide exact times elapsed between pH drop, hemifusion, and fusion.
For precise determination of the time at which the pH drop reached any particular virion, we introduced a low concentration of biotinylated lipid into the bilayer (10−3 mol %), to capture fluoresceinated streptavidin. The low background of fluorescein thus created served as a detector for the shift in pH, because fluorescein adopts a nonfluorescent configuration below pH ≈ 6.4 (25). The sudden decrease in the green background fluorescence that corresponds to the pH change is clearly visualized in Fig. 2 B and C. The time to hemifusion of any single particle can thus be determined by measuring the elapsed time between the drop in fluorescein background and the dequenching of Rh110C18.
Fusion-Pore Formation.
To label the interior of the influenza virions, we incubated the virus preparation overnight in a concentrated (10 mM) solution of sulforhodamine B (SRB) and then removed excess dye by gel filtration. SRB penetrates the virion bilayer and accumulates in the viral interior. Dye-loaded virus was used within a few hours, to avoid loss by back diffusion. We combined the interior labeling procedure with Rh110C18 membrane labeling to produce doubly labeled particles. Colocalization analysis showed that among the membrane-docked, fluorescent particles, 90% contained Rh110C18, 40% contained SRB, and 30% contained both dyes. The efficiency of docking to the surface, as measured by the particle density, was not affected by either of the two dyes.
Fig. 2B Upper shows the fluorescence of the SRB from the same 140 × 70-μm2 area of the target membrane as depicted in Fig. 2B Lower. The red trace in Fig. 2B shows rapid decay of the SRB signal of the same particle for which the hemifusion trace is shown. Loss of red content signal starts several seconds after the Rh110C18 dequenching burst. The decay reports loss of SRB from the virion after fusion-pore formation, as the dye diffuses into the fluid support of the bilayer. After hemifusion, a redistribution of lipophilic dye from the inner to the outer leaflet of the viral membrane, called “flip-flopping” (26), results in rapid depletion of fluorescent probe from the inner leaflet and the absence of a second dequenching signal upon pore formation. A reliable measure for the time elapsed between hemifusion and fusion-pore formation can readily be obtained for each particle by determining the time between initial dequenching of the green Rh110C18 fluorescence and the decrease in red SRB intensity. Hemifusion kinetics were not affected by the addition of the interior dye, nor were kinetics of fusion-pore formation altered by the presence of SRB in the viral membrane (Fig. S2).
Intermediate States.
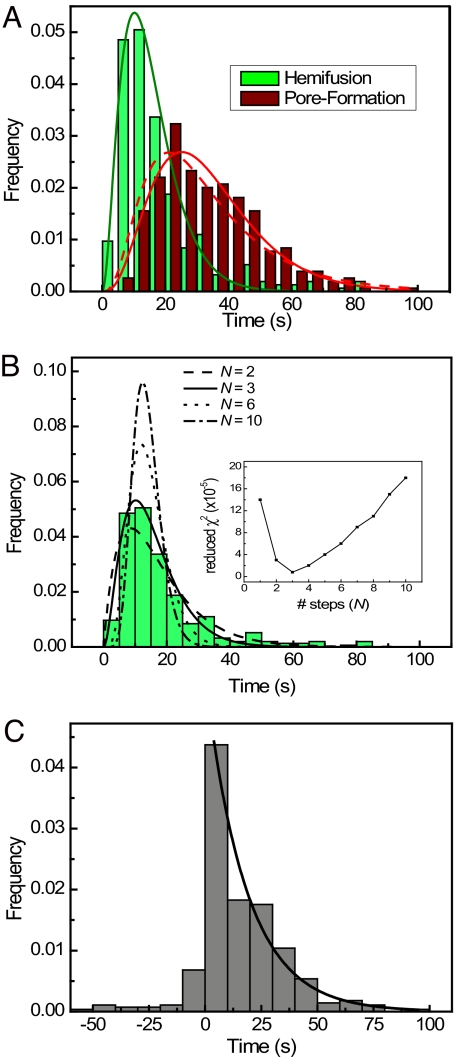
We determined the times elapsed between pH drop and hemifusion and between pH drop and fusion, by locating the maximum and minimum slopes in the single-particle traces for Rh110C18 and SRB (Fig. 2C), respectively. Fig. 3A shows the distribution of lag times for hemifusion and pore formation (n = 309) compiled from experiments conducted at 23°C and pH 4.6. Both histograms show a rise and decay in the frequency of events, indicating intermediate states. Thus, the rate-limiting step of the first event (hemifusion) cannot be a single, one-step transition, because we would then have observed an exponentially distributed lag time. A simple kinetic model describes a series of N transitions between initial and final states, with a single rate constant, k1, for each transition:
where A is the initial configuration at t = 0, the time of the pH drop, and H is the hemifused state at time t. The same scheme also represents N independent events, with the requirement that all N must take place to induce hemifusion (see SI Methods). The probability density for this scheme is a gamma distribution (27):
Use of this expression to fit the hemifusion lag times (Fig. 3A) yields k1 = 0.20 ± 0.02 s−1 and N = 3.1 ± 0.2. We also show fits with k1 as the only free parameter and N fixed at 2, 3, 6, or 10 (Fig. 3B). Fig. 3B Inset shows the χ2 goodness-of-fit as a function of N, with a clear minimum at N = 3.
Fig. 3.
Fusion kinetics of fluorescently labeled influenza virus. (A) Time elapsed between pH decrease and hemifusion (green) and pore formation (red) of individual particles. The presence of intermediate states before hemifusion is clearly visible as a rise and decay in the histograms. Solid lines are best fits to a gamma function with N transitions [N = 3 for hemifusion (green); N = 4 for pore formation (red)]. The dashed line represents a convolution of the N = 3 gamma distribution of hemifusion times with the experimentally observed single-exponential transition between hemifusion and pore formation. (B) Hemifusion histogram from A compared with gamma distribution fits with varying numbers of steps. (Inset) Fitting error for fits with 1–10 transitions. (C) Distribution of lag times between hemifusion and pore formation of individual particles. The solid line represents a single-exponential fit with a rate constant of 0.55 ± 0.04 s−1. See also Figs. S2 and S3.
The pore-formation lag-time distribution can be fit by a gamma distribution with N = 4 (solid red curve in Fig. 3A), suggesting a single step from hemifusion to fusion. A more direct analysis takes advantage of our determination of hemifusion and fusion from the same particle, thus allowing us to determine the distribution of time intervals between the two events. Among the particles that contained both lipid and content dyes, ≈10% showed both hemifusion and fusion signals. For these particles, the distribution of hemifusion decay times (the time between hemifusion and pore formation) is shown in Fig. 3C. In 90% of the traces, dequenching of Rh110C18 (hemifusion) preceded loss of SRB signal (fusion), consistent with our assumption that a hemifused membrane is an essential intermediate, rather than an abortive, off-pathway state (28, 29). A single exponential decay (k2 = 0.55 ± 0.004 s−1) fits the positive lag times, supporting our conclusion that the transition from hemifusion to pore formation involves a single, rate-limiting step. Furthermore, the pore-formation lag-time distribution can be described by a convolution of the N = 3 gamma distribution of hemifusion times with the experimentally observed single-exponential transition between hemifusion and pore formation (Fig. 3A, dashed red curve; and see SI Methods and Fig. S4). These results all indicate that three events must occur before hemifusion and that there is a single, rate-limiting step between hemifusion and pore formation.
pH Dependence of Hemifusion.
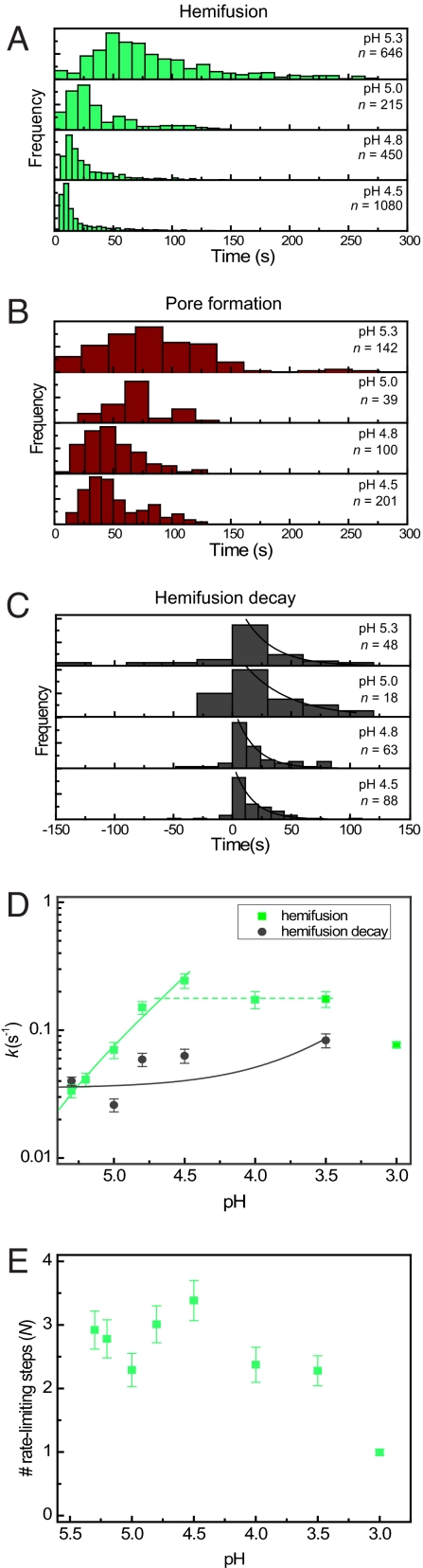
Proton binding to HA initiates the fusion process (30, 31). To explore the mechanism of pH activation, we varied the pH of the activating buffer. As expected, the lag time between the pH drop and hemifusion increased with increasing pH (Fig. 4 A and B). The hemifusion decay time was relatively independent of pH, however, and the rate constant for the fusion step changed by less than 3-fold between pH 4.5 and 5.3 (Fig. 4 C and D). The pH insensitivity suggests that virus particles are already committed to fusion once they have reached the hemifusion intermediate. The observation that disappearance of content dye followed hemifusion in a single transition with simple, first-order kinetics confirms that loss of SRB signal occurred through formation of a fusion pore, rather than through fusion-independent leakage of the dye.
Fig. 4.
Fusion kinetics under varying pH conditions. (A–C) Hemifusion (A), pore-formation (B), and hemifusion decay (C) histograms for events recorded at varying acidic pH conditions. (D) Kinetic rate constants for transitions between prehemifusion intermediates (green squares) and decay of hemifusion to formation of fusion pores (gray circles) plotted as a function of proton concentration. The solid lines are plotted from least-squares fit (y = a + bx) of the rate constants as a function of proton concentration (green line: a = −0.01135, b = 8114, R = 0.987; gray line: a = 0.03522, b = 160.6, R = 0.635). A horizontal dotted green line has been included to emphasize the plateau in the hemifusion rate constants below pH 4.8. The solid lines appear curved in the log–log plot as a result of the nonzero y-intercepts. (E) Number of transitions N preceding hemifusion as a function of proton concentration. Values for N are obtained by fitting the hemifusion histograms with gamma distributions.
Gamma-distribution analysis of hemifusion kinetics in the pH range between 4.5 and 5.3 shows that the number of steps remains constant at approximately N = 3 and that the individual rate constants vary in parallel (Fig. 4 D and E). If proton binding were part of each of these (apparently identical) three steps, then they should continue to depend smoothly on proton concentration as the pH drops. But instead, the rate constant levels off, suggesting that the extent of proton binding might determine the effective concentration of a species, which then undergoes the actual, rate-limiting rearrangement. Below pH 4.5 or so, the concentration of proton-bound HA species is no longer an issue, and in that regime the actual rearrangement rate might determine the apparent kinetic constant. Both k and N drop sharply below pH 3.5, probably reflecting low-pH-induced inactivation or denaturation of HA.
Discussion
Observation of individual influenza A particles fusing with a target membrane has allowed us to obtain detailed information on the kinetics of both hemifusion and pore formation. Analysis of hemifusion reveals about three intermediate steps preceding the formation of a hemifusion stalk. In previous studies of hemifusion between red blood cells and HA-expressing cells, a time lag was found between acidification and initial detection of fluorescent dye redistribution; this result was interpreted in terms of the accumulation of hidden intermediates (14). Subsequent analyses have usually relied on a similar experimental format (32–34). The large numbers of individual fusion events that make up the observed signal in such an experiment quickly lose coherence, essentially blurring the kinetic information. The single-particle studies made possible by the experimental design we describe give a more direct view of the kinetics, without any dephasing, and thus permit estimates of the number of intermediates and of the rates of the transitions between them.
We do not yet have direct evidence for a molecular description of the multiple rate-limiting intermediates preceding hemifusion. The current picture of fusion as mediated by HA and other class I viral fusogens involves the formation of an extended or “prehairpin” intermediate—an extended trimer with its fusion peptides inserted into the target-cell membrane and its TM segments anchored in the viral bilayer (Fig. 1, second panel). The conformation of HIV-1 gp41 that is sensitive to peptide inhibitors such as T-20 is generally agreed to have such an extended structure; comparable data are not yet available for HA. We would expect the initial step of proton binding to result in the rapid establishment of a pre-equilibrium, determining the effective concentration of a fusogenic species and thus the probability of formation of the extended intermediate. In the higher pH range we have examined (i.e., above pH ≈ 4.5), the pH dependence of hemifusion suggests that the rate-limiting step is the formation (or clustering) of enough proton-bound, activated timers within the area of contact of virus and target bilayer to promote approach of the two membranes. Below pH ≈ 4.5, the local concentration of proton-bound HA is maximized and the relatively slow conformational rearrangements (extension followed by collapse) determine the overall hemifusion rate. The pH at which activation is no longer limiting agrees well with the pH at which the measured rate of HA conformational change becomes very rapid, as measured by fluorescence changes in detergent-solubilized HA from the same, X31, influenza strain (35).
The model we have used to analyze hemifusion kinetics assumes N steps, each with the same rate constant. The value of N is thus a formal rather than literal parameter, because various simplifying assumptions apply. We can envision two ways to account for the consistent best fit of N = 3. One relates N to the number of molecular transitions an HA trimer, or a synchronized group of HA trimers, must undergo in sequence. For example, proton-triggered extension, accompanied by dissociation of the HA1 domains and extraction of the fusion peptide, subsequent collapse of the extended intermediate, and merging of the proximal membrane leaflets might each be a relatively slow process. Progression through these transitions in sequence would result in a kinetic pathway with several rate-limiting steps, for which hemifusion times would then be described by a gamma distribution with N equal to the number of sequential rate-limiting transitions. If one of these steps were slowed significantly, N would become equal to 1, because this single, rate-limiting transition would now dominate the kinetics of product formation. One experimental realization of such a condition is the hemifusion experiment at high pH, at which the first pH-dependent step should be rate-limiting. Hemifusion is indeed retarded at the highest pH for which we have measured fusion events (Fig. 4). N remains at 3, however, even at the lowest proton concentrations, indicating either that each of the sequential molecular steps is equally sensitive to pH, an unlikely possibility, or that an explanation for a constant N throughout a large range of proton concentrations must be found elsewhere.
An alternative explanation for N > 1 invokes only one rate-limiting transition at any proton concentration but requires that N of these rate-limiting transitions take place in parallel before hemifusion can occur. In the most straightforward version of this model, N would correspond to the number of HA trimers needed to facilitate hemifusion. Even if the particular rate-limiting step were different at high pH than at low (for example, exposure and activation of HA2 versus postactivation collapse), the requirement that a minimum number of trimers must proceed through the same series of molecular steps leads to a description of the hemifusion kinetics as a set of parallel rate-limiting transitions. If the parallel transitions are stochastic, this description yields hemifusion kinetics that obeys a gamma distribution with N equal to the number of participating trimers (see SI Methods). It is kinetically indistinguishable from the case with N sequential, rate-limiting steps, but it does not require the unrealistic assumption that N sequential steps remain equally rate-limiting over a 100-fold range of proton concentrations. Therefore, our observations suggest that stochastic transitions of three neighboring HA trimers mediate hemifusion. This three-HA minimal fusion model is consistent with the conclusions of Danielli et al. (14), who found that the lag time between pH drop and redistribution of R18 from HA-expressing cells has a power-law dependence (with N = 3) on the average number of HA trimers per cell. Others have attempted to differentiate between the number of HA molecules in a fusion aggregate and the number actually activated by proton binding, one estimate being ≈8 for the former number and 2 for the latter (33).
If we assume that at pH 3.5–4, the rate of hemifusion reflects a lower limit for the rate of extended intermediate collapse, then the lifetime of this intermediate is maximally 15–20 s. The prehairpin intermediate of gp41 has a much longer lifetime (many minutes), as indicated by the “window of opportunity” for T-20 inhibition after association of gp120 and CD4 (36). The balance between the membrane tension against which collapse of the extended intermediate must act and the free energy gained by zipping up of the outer layer structure against the inner-layer coiled-coil may determine these lifetimes.
The types of observations we have made so far limit the detail with which we can trace the fusion pathway, but the advantages of observing fusion of individual virions with a defined and homogeneous membrane are evident. The defined contact area probably includes a uniform patch of 20–30 HA trimers, depending on the deformability of the fluid supported bilayer. Gangliosides can serve as authentic influenza virus receptors (37). The ganglioside density in our supported bilayers is approximately 1 per 60 nm2, within the range of concentration for glycosphingolipids in various cell types. Gangliosides will probably tend to cluster through local phase separations (38, 39), and rapid lateral diffusion is in any case likely to ensure that most HA1 subunits within the contact region are attached to the membrane. Thus, the way the virus “sees” the bilayer in these experiments probably resembles the situation within an acidifying endosome more closely than in previously available assays. We therefore believe that we have developed a useful format in which to dissect the mechanism of viral membrane fusion.
Methods
Flow-Cell Fabrication.
Glass microscope cover slips (25 × 25 mm, No. 1.5; VWR Scientific) were cleaned and functionalized with (3-glycidooxypropyl)tri methoxysilane (adapted from ref. 21). The resultant surface-exposed epoxy groups were allowed to covalently couple to dextran (Dextran 500; GE Healthcare; mean molecular weight, 5 × 105). Flow-cell channels were constructed by sandwiching 100-μm-thick double-stick tape (Grace Bio-Labs), with a 15 × 2-mm channel cut out, between the dextran-functionalized coverslip and a fused quartz microscope slide with holes drilled at either end of the channel. Polyethylene tubing (Intramedic; I.D. = 0.38 mm) was inserted into the holes to allow exchange of buffer solutions, and the flow cell was sealed with epoxy glue.
Supported Bilayer Preparation.
A solution was prepared with a 4:4:2:0.1:5 × 10−5 ratio of 1,2,dioleoyl-sn-glycero-3-phosphocholine (DOPC) (Avanti Polar Lipids), 1-oleoyl-2-palmitoyl-sn-glycero-3-phosphocholine (POPC) (Avanti Polar Lipids), cholesterol (Avanti Polar Lipids), bovine brain disialoganglioside GD1a (Sigma), and N-((6-(biotinoyl)amino)hexanoyl)-1,2-dihexadecanoyl-sn-glycero-3-phosphoethanolamine (biotin-X DHPE) (Molecular Probes) in HNE (5 mM Hepes, 145 mM NaCl, 0.2 mM EDTA) buffer at a concentration of ≈10 g/liter. Liposomes were extruded through a polycarbonate membrane filter with a pore size of 100 nm. Planar supported bilayers were formed over hydrated dextran surfaces by the vesicle-spreading method (23).
Labeling and Purification of Viral Particles.
Influenza particles were labeled with sulforhodamine B (SRB) (Aldrich) and rhodamine 110 octadecyl ester (Rh110C18; for synthesis and purification details, see SI Methods). Ten microliters of influenza virus (≈10 μg of viral protein) were mixed with 20 μl of 20 mM SRB in HNE buffer and left at room temperature for 16–20 h. Unincorporated dye was separated from the virus with a gel-filtration column (PD-10 desalting column; GE Healthcare) in a total volume of 0.8 ml. A 2 mM solution of Rh110C18 was prepared in dimethylformamide, and 13 μl was added to the SRB-labeled virus particles and agitated for 3 h. A second PD-10 column was used to separate the virus from free Rh110C18. Both dyes remained stably associated with the virus for several hours.
Fluorescence Imaging of Fusion.
Single-particle fusion assays were conducted on an inverted fluorescence microscope (Nikon TE-2000U) with a high numerical aperture objective (N.A. = 1.45). Viral particles were illuminated with the 488- and 568-nm lines from an argon/krypton laser (Coherent Innova 70C). Using 500- to 540-nm and 600- to 640-nm dual band-pass emission filters (Chroma Technology), the emission was split into separate green and red channels and focused onto separate regions of an electron multiplying CCD camera (Andor Technology DV 887-BI).
Labeled virus (≈50 ng/ml) and fluorescein-labeled streptavidin (Molecular Probes; 2 μg/ml) were introduced into the flow cell by using a syringe pump (VWR Scientific). The fusion reaction was initiated by flowing an acidic buffer containing 10 mM citric or acetic acid, 140 mM NaCl, 0.1 mM EDTA, and 0.01% NaN3. Time-lapsed fluorescence images were recorded at 10 Hz for 200–400 s by using Andor iQ imaging software (Andor Technology).
Data Analysis.
The position of each particle in the red and green fluorescence images was determined by particle tracking and cross-correlation analysis. Fluorescence trajectories were from calculated by integrating the intensities from a 4 × 4-pixel region around each particle. All data were analyzed with software written in MATLAB (The MathWorks).
Further experimental details and a discussion of the various kinetic models used in the data analysis can be found in SI Methods.
Supplementary Material
Acknowledgments.
We thank David Stevens and Rose Gonsalves for assistance. The work was supported by a pilot grant from the Harvard Medical School Department of Biological Chemistry and Molecular Pharmacology, by NIH grants AI57159 (to S.C.H.) and AI72346 (to A.M.v.O.), and by the Medical Research Council (J.J.S.). S.C.H. is a Howard Hughes Medical Institute Investigator.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0807771105/DCSupplemental.
References
- 1.Chernomordik LV, Kozlov MM. Protein-lipid interplay in fusion and fission of biological membranes. Annu Rev Biochem. 2003;72:175–207. doi: 10.1146/annurev.biochem.72.121801.161504. [DOI] [PubMed] [Google Scholar]
- 2.Harrison SC. Mechanism of membrane fusion by viral envelope proteins. Adv Virus Res. 2005;64:231–261. doi: 10.1016/S0065-3527(05)64007-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Skehel JJ, Wiley DC. Receptor binding and membrane fusion in virus entry: The influenza hemagglutinin. Annu Rev Biochem. 2000;69:531–569. doi: 10.1146/annurev.biochem.69.1.531. [DOI] [PubMed] [Google Scholar]
- 4.Matlin KS, Reggio H, Helenius A, Simons K. Infectious entry pathway of influenza virus in a canine kidney cell line. J Cell Biol. 1981;91:601–613. doi: 10.1083/jcb.91.3.601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wilson IA, Skehel JJ, Wiley DC. Structure of the haemagglutinin membrane glycoprotein of influenza virus at 3 Å resolution. Nature. 1981;289:366–373. doi: 10.1038/289366a0. [DOI] [PubMed] [Google Scholar]
- 6.Bullough PA, Hughson FM, Skehel JJ, Wiley DC. Structure of influenza haemagglutinin at the pH of membrane fusion. Nature. 1994;371:37–43. doi: 10.1038/371037a0. [DOI] [PubMed] [Google Scholar]
- 7.Furuta RA, Wild CT, Weng Y, Weiss CD. Capture of an early fusion-active conformation of HIV-1 gp41. Nat Struct Biol. 1998;5:276–279. doi: 10.1038/nsb0498-276. [DOI] [PubMed] [Google Scholar]
- 8.Rimsky LT, Shugars DC, Matthews TJ. Determinants of human immunodeficiency virus type 1 resistance to gp41-derived inhibitory peptides. J Virol. 1998;72:986–993. doi: 10.1128/jvi.72.2.986-993.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Frey G, et al. A fusion-intermediate state of HIV-1 gp41 targeted by broadly neutralizing antibodies. Proc Natl Acad Sci USA. 2008;105:3739–3744. doi: 10.1073/pnas.0800255105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rand RP, Parsegian VA. Physical force considerations in model and biological membranes. Can J Biochem Cell Biol. 1984;62:752–759. doi: 10.1139/o84-097. [DOI] [PubMed] [Google Scholar]
- 11.Kasson PM, et al. Ensemble molecular dynamics yields submillisecond kinetics and intermediates of membrane fusion. Proc Natl Acad Sci USA. 2006;103:11916–11921. doi: 10.1073/pnas.0601597103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chanturiya A, Chernomordik LV, Zimmerberg J. Flickering fusion pores comparable with initial exocytotic pores occur in protein-free phospholipid bilayers. Proc Natl Acad Sci USA. 1997;94:14423–14428. doi: 10.1073/pnas.94.26.14423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chernomordik LV, Leikina E, Frolov V, Bronk P, Zimmerberg J. An early stage of membrane fusion mediated by the low pH conformation of influenza hemagglutinin depends upon membrane lipids. J Cell Biol. 1997;136:81–93. doi: 10.1083/jcb.136.1.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Danieli T, Pelletier SL, Henis YI, White JM. Membrane fusion mediated by the influenza virus hemagglutinin requires the concerted action of at least three hemagglutinin trimers. J Cell Biol. 1996;133:559–569. doi: 10.1083/jcb.133.3.559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kemble GW, Danieli T, White JM. Lipid-anchored influenza hemagglutinin promotes hemifusion, not complete fusion. Cell. 1994;76:383–391. doi: 10.1016/0092-8674(94)90344-1. [DOI] [PubMed] [Google Scholar]
- 16.Stegmann T, Hoekstra D, Scherphof G, Wilschut J. Kinetics of pH- dependent fusion between influenza virus and liposomes. Biochemistry. 1985;24:3107–3113. doi: 10.1021/bi00334a006. [DOI] [PubMed] [Google Scholar]
- 17.Hoekstra D, Klappe K, de Boer T, Wilschut J. Characterization of the fusogenic properties of sendai virus: Kinetics of fusion with erythrocyte membranes. Biochemistry. 1985;24:739–4745. doi: 10.1021/bi00339a005. [DOI] [PubMed] [Google Scholar]
- 18.Melikyan GB, Barnard RJO, Abrahamyan LG, Mothes W, Young JAT. Imaging individual retroviral fusion events: From hemifusion to pore formation and growth. Proc Natl Acad Sci USA. 2005;102:8728–8733. doi: 10.1073/pnas.0501864102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Imai M, Mizuno T, Kawasaki K. Membrane fusion by single influenza hemagglutinin trimers: Kinetic evidence from image analysis of hemagglutinin-reconstituted vesicles. J Biol Chem. 2006;281:12729–12735. doi: 10.1074/jbc.M600902200. [DOI] [PubMed] [Google Scholar]
- 20.Wessels L, Elting MW, Scimeca D, Weninger K. Rapid membrane fusion of individual virus particles with supported lipid bilayers. Biophys J. 2007;93:526–538. doi: 10.1529/biophysj.106.097485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Elender G, Kuhner M, Sackmann E. Functionalisation of si/sio2 and glass surfaces with ultrathin dextran films and deposition of lipid bilayers. Biosens Bioelectron. 1996;11:565–577. doi: 10.1016/0956-5663(96)83292-1. [DOI] [PubMed] [Google Scholar]
- 22.Johnson JM, Ha T, Chu S, Boxer SG. Early steps of supported bilayer formation probed by single vesicle fluorescence assays. Biophys J. 2002;83:3371–3379. doi: 10.1016/S0006-3495(02)75337-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nollert P, Kiefer H, Jahnig F. Lipid vesicle adsorption versus formation of planar bilayers on solid surfaces. Biophys J. 1995;69:1447–1455. doi: 10.1016/S0006-3495(95)80014-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hovis JS, Boxer SG. Patterning and composition arrays of supported lipid bilayers by microcontact printing. Langmuir. 2001;17:3400–3405. [Google Scholar]
- 25.Klonis N, Clayton AH, Voss EW, Jr, Sawyer WH. Spectral properties of fluorescein in solvent-water mixtures: Applications as a probe of hydrogen bonding environments in biological systems. Photochem Photobiol. 1998;67:500–510. [PubMed] [Google Scholar]
- 26.Melikyan GB, Deriy BN, Ok DC, Cohen FS. Voltage-dependent translocation of r18 and dii across lipid bilayers leads to fluorescence changes. Biophys J. 1996;71:2680–2691. doi: 10.1016/S0006-3495(96)79459-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Feller W. An Introduction to Probability Theory and Its Applications. New York: Wiley; 1968. [Google Scholar]
- 28.Zimmerberg J, Blumenthal R, Sarkar D, Curran M, Morris S. Restricted movement of lipid and aqueous dyes through pores formed by influenza hemagglutinin during cell fusion. J Cell Biol. 1994;127:1885–1894. doi: 10.1083/jcb.127.6.1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chernomordik LV, Frolov VA, Leikina E, Bronk P, Zimmerberg J. The pathway of membrane fusion catalyzed by influenza hemagglutinin: Restriction of lipids, hemifusion, and lipidic fusion pore formation. J Cell Biol. 1998;140:1369–1382. doi: 10.1083/jcb.140.6.1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.White J, Kartenbeck J, Helenius A. Membrane fusion activity of influenza virus. EMBO J. 1982;1:217–222. doi: 10.1002/j.1460-2075.1982.tb01150.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Skehel JJ, et al. Changes in the conformation of influenza virus hemagglutinin at the pH optimum of virus-mediated membrane fusion. Proc Natl Acad Sci USA. 1982;79:968–972. doi: 10.1073/pnas.79.4.968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Leikina E, Ramos C, Markovic I, Zimmerberg J, Chernomordik LV. Reversible stages of the low-ph-triggered conformational change in influenza virus hemagglutinin. EMBO J. 2002;21:5701–5710. doi: 10.1093/emboj/cdf559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mittal A, Shangguan T, Bentz J. Measuring pKa of activation and pKi of inactivation for influenza hemagglutinin from kinetics of membrane fusion of virions and of HA expressing cells. Biophys J. 2002;83:2652–2666. doi: 10.1016/S0006-3495(02)75275-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mittal A, Leikina E, Chernomordik LV, Bentz J. Kinetically differentiating influenza hemagglutinin fusion and hemifusion machines. Biophys. J. 2003;85:1713–1724. doi: 10.1016/S0006-3495(03)74601-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Krumbiegel M, Herrmann A, Blumenthal R. Kinetics of the low pH-induced conformational changes and fusogenic activity of influenza hemagglutinin. Biophys J. 1994;67:2355–2360. doi: 10.1016/S0006-3495(94)80721-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chan DC, Kim P. HIV entry and its inhibition. Cell. 1998;89:681–684. doi: 10.1016/s0092-8674(00)81430-0. [DOI] [PubMed] [Google Scholar]
- 37.Herrler G, Klenk HD. The surface receptor is a major determinant of the cell tropism of influenza C virus. Virology. 1987;159:102–108. doi: 10.1016/0042-6822(87)90352-7. [DOI] [PubMed] [Google Scholar]
- 38.Ferraretto A, Pitto M, Palestini P, Masserini M. Lipid domains in the membrane: Thermotropic properties of sphingomyelin vesicles containing gm1 ganglioside and cholesterol. Biochemistry. 1997;36:9232–9236. doi: 10.1021/bi970428j. [DOI] [PubMed] [Google Scholar]
- 39.Menke M, Kunneke S, Janshoff A. Lateral organization of GM1 in phase-separated monolayers visualized by scanning force microscopy. Eur Biophys J. 2002;31:317–322. doi: 10.1007/s00249-002-0232-4. [DOI] [PubMed] [Google Scholar]
- 40.Harrison SC. Viral membrane fusion. Nat Struct Mol Biol. 2008;15:690–698. doi: 10.1038/nsmb.1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.