Abstract
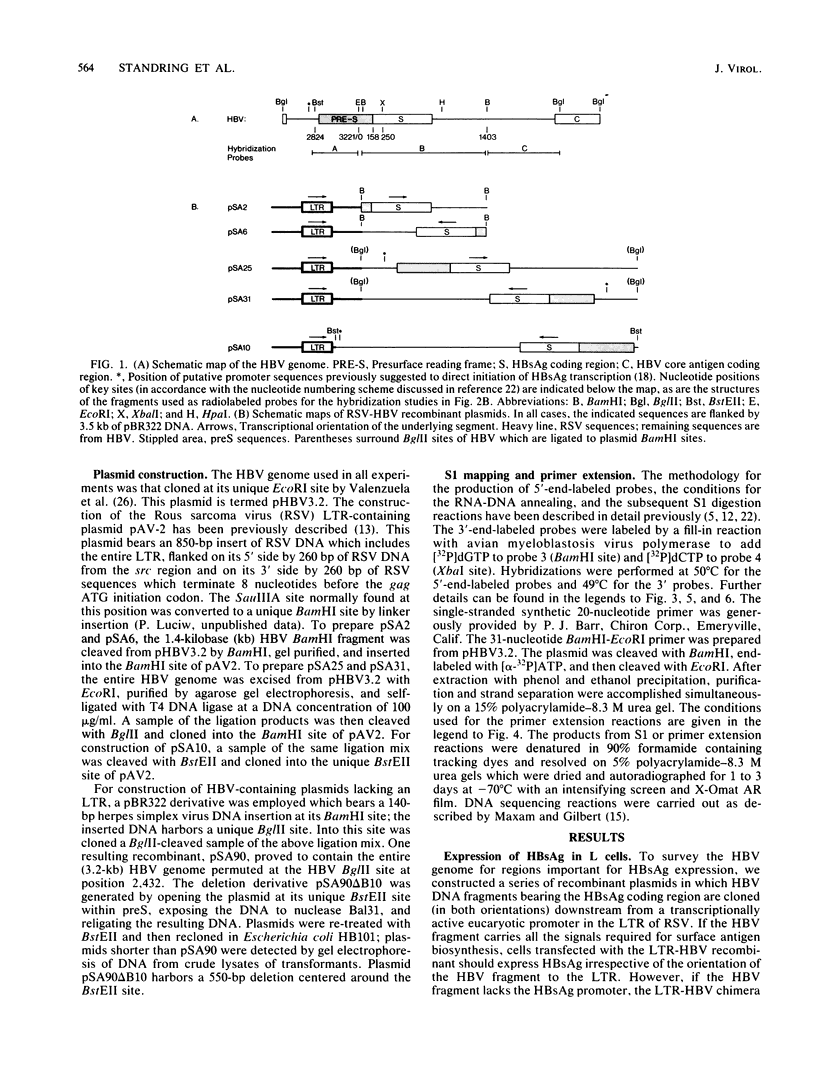
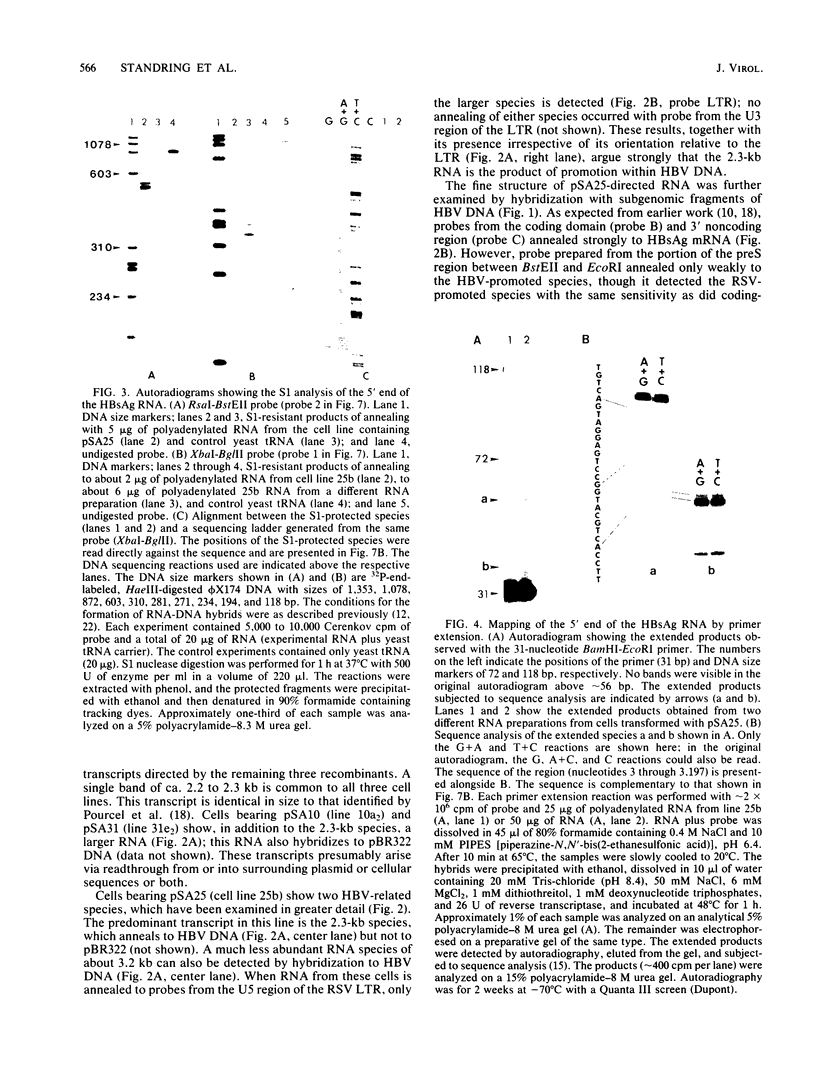
Cloned hepatitis B virus (HBV) DNA directs the synthesis of the viral surface antigen (HBsAg) when introduced into mouse L cells by DNA transformation. We have used recombinants between the Rous sarcoma virus long terminal repeat and subgenomic fragments of HBV DNA to localize regions of the HBV genome required for HBsAg expression. Examination of HBV-specific RNA from such transformants indicates that transcription initiates at three distinct sites (153, 163, and 183 nucleotides upstream from the translation initiation codon for mature HBsAg). Thus in these cells, a large segment of the presurface reading frame is not represented in HBsAg mRNA. The termination site of this RNA lies within the coding sequences for the viral core antigen, some 1,094 +/- 10 base pairs downstream from the TAA stop codon for HBsAg. Two additional open reading frames are present in the resultant unspliced HBsAg RNA.
Full text
PDF

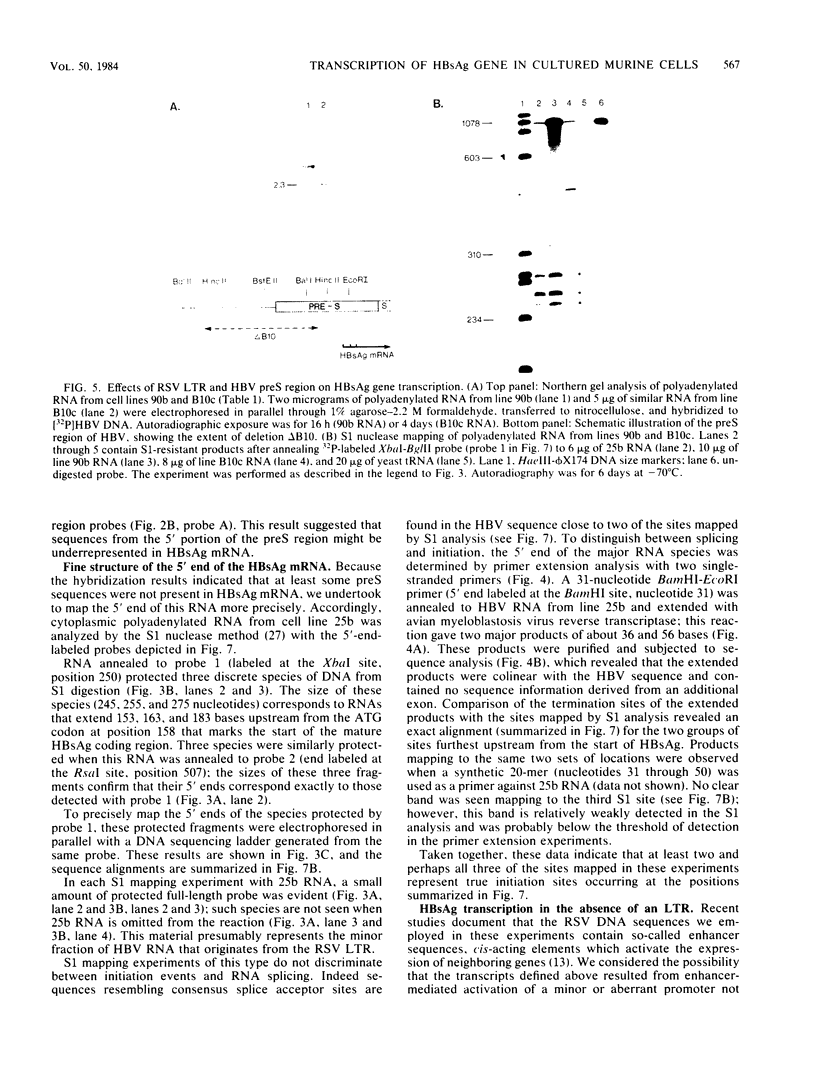




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Birnboim H. C., Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979 Nov 24;7(6):1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cattaneo R., Will H., Darai G., Pfaff E., Schaller H. Detection of an element of the SV40 late promoter in vectors used for expression studies in COS cells. EMBO J. 1983;2(4):511–514. doi: 10.1002/j.1460-2075.1983.tb01455.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cattaneo R., Will H., Hernandez N., Schaller H. Signals regulating hepatitis B surface antigen transcription. Nature. 1983 Sep 22;305(5932):336–338. doi: 10.1038/305336a0. [DOI] [PubMed] [Google Scholar]
- Dubois M. F., Pourcel C., Rousset S., Chany C., Tiollais P. Excretion of hepatitis B surface antigen particles from mouse cells transformed with cloned viral DNA. Proc Natl Acad Sci U S A. 1980 Aug;77(8):4549–4553. doi: 10.1073/pnas.77.8.4549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Favaloro J., Treisman R., Kamen R. Transcription maps of polyoma virus-specific RNA: analysis by two-dimensional nuclease S1 gel mapping. Methods Enzymol. 1980;65(1):718–749. doi: 10.1016/s0076-6879(80)65070-8. [DOI] [PubMed] [Google Scholar]
- Fitzgerald M., Shenk T. The sequence 5'-AAUAAA-3'forms parts of the recognition site for polyadenylation of late SV40 mRNAs. Cell. 1981 Apr;24(1):251–260. doi: 10.1016/0092-8674(81)90521-3. [DOI] [PubMed] [Google Scholar]
- Galibert F., Chen T. N., Mandart E. Nucleotide sequence of a cloned woodchuck hepatitis virus genome: comparison with the hepatitis B virus sequence. J Virol. 1982 Jan;41(1):51–65. doi: 10.1128/jvi.41.1.51-65.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galibert F., Mandart E., Fitoussi F., Tiollais P., Charnay P. Nucleotide sequence of the hepatitis B virus genome (subtype ayw) cloned in E. coli. Nature. 1979 Oct 25;281(5733):646–650. doi: 10.1038/281646a0. [DOI] [PubMed] [Google Scholar]
- Ganem D. Persistent infection of humans with hepatitis B virus: mechanisms and consequences. Rev Infect Dis. 1982 Sep-Oct;4(5):1026–1047. doi: 10.1093/clinids/4.5.1026. [DOI] [PubMed] [Google Scholar]
- Gough N. M. Core and E antigen synthesis in rodent cells transformed with hepatitis B virus DNA is associated with greater than genome length viral messenger RNAs. J Mol Biol. 1983 Apr 25;165(4):683–699. doi: 10.1016/s0022-2836(83)80274-5. [DOI] [PubMed] [Google Scholar]
- Gough N. M., Murray K. Expression of the hepatitis B virus surface, core and E antigen genes by stable rat and mouse cell lines. J Mol Biol. 1982 Nov 25;162(1):43–67. doi: 10.1016/0022-2836(82)90161-9. [DOI] [PubMed] [Google Scholar]
- Laub O., Rall L. B., Truett M., Shaul Y., Standring D. N., Valenzuela P., Rutter W. J. Synthesis of hepatitis B surface antigen in mammalian cells: expression of the entire gene and the coding region. J Virol. 1983 Oct;48(1):271–280. doi: 10.1128/jvi.48.1.271-280.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luciw P. A., Bishop J. M., Varmus H. E., Capecchi M. R. Location and function of retroviral and SV40 sequences that enhance biochemical transformation after microinjection of DNA. Cell. 1983 Jul;33(3):705–716. doi: 10.1016/0092-8674(83)90013-2. [DOI] [PubMed] [Google Scholar]
- Machida A., Kishimoto S., Ohnuma H., Miyamoto H., Baba K., Oda K., Nakamura T., Miyakawa Y., Mayumi M. A hepatitis B surface antigen polypeptide (P31) with the receptor for polymerized human as well as chimpanzee albumins. Gastroenterology. 1983 Aug;85(2):268–274. [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Pasek M., Goto T., Gilbert W., Zink B., Schaller H., MacKay P., Leadbetter G., Murray K. Hepatitis B virus genes and their expression in E. coli. Nature. 1979 Dec 6;282(5739):575–579. doi: 10.1038/282575a0. [DOI] [PubMed] [Google Scholar]
- Payne G. S., Courtneidge S. A., Crittenden L. B., Fadly A. M., Bishop J. M., Varmus H. E. Analysis of avian leukosis virus DNA and RNA in bursal tumours: viral gene expression is not required for maintenance of the tumor state. Cell. 1981 Feb;23(2):311–322. doi: 10.1016/0092-8674(81)90127-6. [DOI] [PubMed] [Google Scholar]
- Pourcel C., Louise A., Gervais M., Chenciner N., Dubois M. F., Tiollais P. Transcription of the hepatitis B surface antigen gene in mouse cells transformed with cloned viral DNA. J Virol. 1982 Apr;42(1):100–105. doi: 10.1128/jvi.42.1.100-105.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proudfoot N. J., Brownlee G. G. 3' non-coding region sequences in eukaryotic messenger RNA. Nature. 1976 Sep 16;263(5574):211–214. doi: 10.1038/263211a0. [DOI] [PubMed] [Google Scholar]
- Rall L. B., Standring D. N., Laub O., Rutter W. J. Transcription of hepatitis B virus by RNA polymerase II. Mol Cell Biol. 1983 Oct;3(10):1766–1773. doi: 10.1128/mcb.3.10.1766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw M. W., Choppin P. W., Lamb R. A. A previously unrecognized influenza B virus glycoprotein from a bicistronic mRNA that also encodes the viral neuraminidase. Proc Natl Acad Sci U S A. 1983 Aug;80(16):4879–4883. doi: 10.1073/pnas.80.16.4879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Standring D. N., Rall L. B., Laub O., Rutter W. J. Hepatitis B virus encodes an RNA polymerase III transcript. Mol Cell Biol. 1983 Oct;3(10):1774–1782. doi: 10.1128/mcb.3.10.1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stibbe W., Gerlich W. H. Structural relationships between minor and major proteins of hepatitis B surface antigen. J Virol. 1983 May;46(2):626–628. doi: 10.1128/jvi.46.2.626-628.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas P. S. Hybridization of denatured RNA and small DNA fragments transferred to nitrocellulose. Proc Natl Acad Sci U S A. 1980 Sep;77(9):5201–5205. doi: 10.1073/pnas.77.9.5201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiollais P., Charnay P., Vyas G. N. Biology of hepatitis B virus. Science. 1981 Jul 24;213(4506):406–411. doi: 10.1126/science.6264599. [DOI] [PubMed] [Google Scholar]
- Weaver R. F., Weissmann C. Mapping of RNA by a modification of the Berk-Sharp procedure: the 5' termini of 15 S beta-globin mRNA precursor and mature 10 s beta-globin mRNA have identical map coordinates. Nucleic Acids Res. 1979 Nov 10;7(5):1175–1193. doi: 10.1093/nar/7.5.1175. [DOI] [PMC free article] [PubMed] [Google Scholar]