Abstract
Accumulation of myeloid-derived suppressor cells (MDSCs) associated with inhibition of dendritic cell (DC) differentiation is one of the major immunological abnormalities in cancer and leads to suppression of antitumor immune responses. The molecular mechanism of this phenomenon remains unclear. We report here that STAT3-inducible up-regulation of the myeloid-related protein S100A9 enhances MDSC production in cancer. Mice lacking this protein mounted potent antitumor immune responses and rejected implanted tumors. This effect was reversed by administration of wild-type MDSCs from tumor-bearing mice to S100A9-null mice. Overexpression of S100A9 in cultured embryonic stem cells or transgenic mice inhibited the differentiation of DCs and macrophages and induced accumulation of MDSCs. This study demonstrates that tumor-induced up-regulation of S100A9 protein is critically important for accumulation of MDSCs and reveals a novel molecular mechanism of immunological abnormalities in cancer.
Defective myeloid cell differentiation is one of the major factors underlying the immune nonresponsiveness of both solid tumors and hematological malignancies (1). Consequences of defective myeloid cell differentiation include decreased production of mature, functionally competent DCs and accumulation of myeloid-derived suppressor cells (MDSCs) (2–4). MDSCs are a mixed group of myeloid cells including immature granulocytes, macrophages, DCs, and myeloid progenitors. In mice, MDSCs with the phenotype of Gr-1+CD11b+ cells were detected in practically all tested tumor models. Significant accumulation of these cells was found in patients with various types of cancer (5–9). Numerous studies have demonstrated a critical role of MDSCs in the suppression of T cell responses and induction of T cell tolerance in cancer (10–13). MDSCs may account in large part for the limited effectiveness of cancer vaccines and other therapies such as anti–vascular endothelial growth factor (VEGF) treatment (8, 14, 15). Recent studies have also demonstrated an important function of these cells in infection, inflammation, and prevention of graft rejection (16–20).
Despite the wealth of information regarding the functional importance of MDSCs, the mechanism responsible for their accumulation remains unknown. Various tumor-derived factors (VEGF, IL-6, IL-10, M-CSF, and GM-CSF) can inhibit DC differentiation from hematopoietic progenitor cells (HPCs) in vitro and in vivo (1). In search of specific genes responsible for abnormal myeloid cell differentiation in cancer, we evaluated the effect of VEGF on HPC gene expression by differential display analysis and consistently found up-regulation of the mouse S100A9 gene. S100A9, also referred to as myeloid-related protein 14 (MRP14) or calgranulin B, is a member of the large family of S100 proteins. It is expressed together with its dimerization partner S100A8 (MRP8 or calgranulin A) in circulating neutrophils and monocytes, but not in resting tissue macrophages (21). S100A9 protein contains two helix–loop–helix motifs (EF-hand), which have high affinity for Ca2+ (22). Upon elevation of intracellular calcium, S100A8 and S100A9 translocate from the cytosol to the cytoskeleton and the plasma membrane of myeloid cells (23) and are released as a heterodimer to recruit leukocytes to sites of inflammation or tumors (24). Until now, their role in myeloid cell differentiation remained unknown. We report here that up-regulation of S100A9 in myeloid precursors in cancer inhibits DC and macrophage differentiation and induces accumulation of MDSCs. This may represent a universal molecular mechanism of tumor-induced abnormalities in myeloid cells in cancer, directly linking inflammation and immune suppression.
RESULTS
S100A9 expression is associated with accumulation of MDSCs in cancer
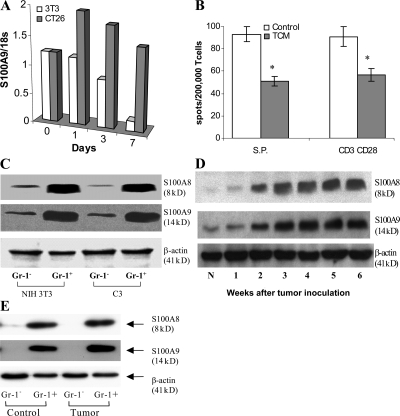
S100A9 and S100A8 mRNAs and proteins were readily detectable in enriched bone marrow HPCs, but gradually decreased during culture with GM-CSF and IL-4 and were almost undetectable by day 7, when >70% of cells in culture were DCs (Fig. 1 A and Fig. S1, A and B, available at http://www.jem.org/cgi/content/full/jem.20080132/DC1). Thus, under physiological conditions, DC differentiation was associated with marked down-regulation of S100A8 and S100A9. To evaluate the effect of tumor-derived factors on S100A9 expression, HPCs were cultured for 7 d with GM-CSF and IL-4 in CT-26 tumor cell–conditioned medium (TCM) or 3T3 fibroblast-conditioned medium (FCM). TCM prevented the down-regulation of S100A9 mRNA expression during differentiation in vitro (Fig. 1 A). Previous studies have shown that in contrast to FCM, TCM inhibits the differentiation of DCs from HPCs and induces the accumulation of Gr-1+CD11b+ MDSCs (25–27). To verify the suppressive activity of these MDSCs, HPCs were cultured for 5 d with GM-CSF, IL-4, and TCM. Consistent with previous results, TCM substantially decreased the proportion of CD11c+ DCs and increased the proportion of Gr-1+CD11b+ MDSCs (Fig. S2). Gr-1+CD11b+ cells were then sorted and added to splenocytes from C57BL/6 mice stimulated with anti-CD3/CD28 antibody or to OT-1 splenocytes (CD8+ T cells from these mice have a TCR that recognizes the OVA-derived peptide SIINFEKL) stimulated with control or specific peptide. Gr-1+CD11b+ cells generated in TCM significantly suppressed T cell responses (Fig. 1 B).
Figure 1.
Expression of S100A9 and A8 in myeloid cells in tumor-bearing mice. (A) HPCs from bone marrow of naive mice were cultured with GM-CSF and IL-4 for 7 d in the presence of 25% (vol/vol) conditioned media from NIH 3T3 fibroblasts (3T3) or CT26 colon cancer cells (CT26). RNA was extracted on days 0, 1, 3, and 7, and qRT-PCR was performed. The level of expression of S100A9 was normalized to 18S rRNA. Two experiments yielded the same results. (B) HPCs from naive C57BL/6 mice were cultured for 5 d with GM-CSF and IL-4 in complete medium alone (control) or conditioned media from EL-4 cells (TCM). Gr-1+ cells were isolated at the end of 5-d culture and added at 1:4 ratio to splenocytes from OT-1 mice. The number of IFN-γ–producing cells in response to stimulation by the specific or control peptides was evaluated in an ELISPOT assay and calculated per 2 × 105 splenocytes. S.P. represents the number of spots in cells stimulated with control peptide subtracted from the number of spots in cells stimulated with specific peptide. In parallel, Gr-1+ cells were added at 1:4 ratio to splenocytes from naive C57BL/6 mice and stimulated with 0.5 μg/ml anti-CD3 and 5 μg/ml anti-CD28 antibodies. The number of IFN-γ cells was evaluated in ELISPOT assay. Two experiments were performed. * represents the statistically significant (P < 0.05) differences between control and TCM groups. (C) HPCs from C57BL/6 mice were cultured for 5 d with GM-CSF and IL-4 in the presence of conditioned media (25% vol/vol) from NIH 3T3 fibroblasts (NIH 3T3) or C3 fibrosarcoma cells (C3). Gr-1+ or Gr-1− cells were isolated using magnetic beads by two rounds of isolation. Proteins extracted from cells were subjected to Western blotting using indicated antibodies. Two experiments with the same results were performed. (D) Expression of S100A8 and A9 proteins in whole cell lysates of splenocytes from CT26 tumor-bearing mice. N, naive tumor-free mice. (E) Gr-1+ and Gr-1− cells were isolated from spleens of naive mice (control) or mice bearing 3-wk-old CT26 tumor (tumor) as described above. The levels of proteins were evaluated by Western blotting. Two experiments with the same results were performed.
We asked whether the observed up-regulation of S100A8 and S100A9 by TCM was associated with MDSC accumulation. Gr-1+ cells isolated after a 5-d culture of HPCs with GM-CSF and IL-4 in TCM expressed substantially more S100A8 and S100A9 proteins than did Gr-1− cells (Fig. 1 C). No differences in the levels of S100A8 and S100A9 were found between Gr-1+ cells isolated from populations cultured with FCM versus TCM.
The correlation between MDSC accumulation and S100A8/A9 levels was investigated in CT26 colon carcinoma-bearing mice. Consistent with previous observations (28, 29), the proportion of MDSCs in the spleens of these mice increased gradually during tumor growth, and by 3 wk after inoculation was more than fivefold higher than in the spleens of control mice (unpublished data). Amounts of S100A8 and S100A9 increased in spleens of tumor-bearing mice in parallel with MDSC accumulation (Fig. 1 D). Similar to the in vitro experiments, S100A8/A9 proteins were detected in the Gr-1+, but not the Gr-1− population, and no differences in protein levels were noted between Gr-1+ cells from naive and tumor-bearing mice (Fig. 1 E). Thus, the increased expression of S100A8 and S100A9 in spleens of tumor-bearing mice and the TCM-inducible up-regulation of S100A8 and S100A9 in vitro are associated exclusively with the accumulation of MDSCs.
S100A9 is essential for MDSC accumulation in cancer
To determine whether MDSC accumulation requires S100A9, we examined S100A9-deficient (S100A9KO) mice (32). No differences were found in the proportions of the populations of myeloid cells in spleens (Fig. S3 A, available at http://www.jem.org/cgi/content/full/jem.20080132/DC1), blood, and bone marrow (not depicted) between wild-type and S100A9KO mice. Likewise, no significant differences in the presence of the myeloid populations were observed after in vitro differentiation of HPCs (Fig. S3 B).
To evaluate differentiation of myeloid cells in the presence of tumor-derived factors, enriched HPCs isolated from S100A9KO mice and their wild-type littermates were cultured with GM-CSF for 5 d in TCM. Consistent with previous observations (for review see [1]), tumor-derived factors significantly reduced the differentiation of DCs and macrophages and substantially increased the production of Gr-1+CD11b+ MDSCs in wild-type populations. In contrast, TCM did not appreciably inhibit the differentiation of myeloid cells from S100A9KO mice (Fig. 2 A). Thus, loss of S100A9 prevents the defective differentiation of TCM-treated myeloid cells.
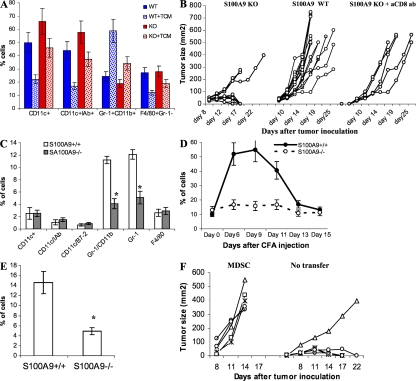
Figure 2.
Lack of S100A9 affects generation of MDSCs under pathological conditions. (A) Enriched HPCs were isolated from bone marrow of S100A9 KO mice (KO) and their wild-type littermates (WT). Cells were cultured with GM-CSF and IL-4 for 5 d in complete culture medium or in the presence of EL-4 TCM. The cell phenotypes were evaluated by flow cytometry. Mean ± SD of the proportions of indicated cell populations from three mice are shown. (B) EL-4 cells (3 × 105) were injected s.c. into wild-type (WT) or S100A9 knockout (KO) mice and tumor size was measured. Anti-CD8 antibody (200 μg) was injected i.p. into KO mice 3 and 1 d before injection of tumor cells. The S100A9KO group included 12 mice, WT group 15 mice, and KO mice treated with anti-CD8 antibody 5 mice. Tumor size for each mouse is shown. (C) Phenotypes of splenocytes from wild-type (S100A9+/+) and knockout (S100A9−/−) mice 15 d after EL-4 cell injection were evaluated using multicolor flow cytometry. * represents statistically significant differences between the groups (P < 0.05). (D) Wild-type and S100A9 knockout mice were injected s.c. with 200 μl of CFA. The presence of Gr-1+CD11b+ cells was monitored in peripheral blood. Each group included 4 mice. Mean ± SD are shown. (E) Gr-1+CD11b+ cells in spleens of wild-type and S100A9 knockout mice on day 12 after CFA injection. Four mice per genotype were analyzed. * represents statistically significant differences between the groups (P < 0.05). (F) S100A9−/− mice were injected s.c. with 2 × 105 EL-4 tumor cells and then split into 2 groups of 5 mice. One group was left untreated (no transfer) and the other group was treated i.v. with 3 × 106 Gr-1+CD11b+ cells isolated from tumor-bearing wild-type mice (MDSC) on days 1, 3, 5, and 7 after tumor inoculation. Tumor size for each mouse is shown.
To evaluate the effect of S100A9 deficiency on tumor growth, control and S100A9KO mice were injected s.c. with 5 × 105 EL-4 lymphoma cells. By days 8–10 after inoculation, tumor growth was evident in all mice (Fig. 2 B). Tumors continued to grow aggressively in all 15 wild-type mice. In sharp contrast, tumors were rejected in 9 out of 12 S100A9KO mice (P = 0.0002). Mean tumor size in the remaining three S100A9KO mice was significantly smaller than in the wild-type mice (238.7 ± 51.8 vs. 521.5 ± 199.3, P = 0.008). Similar results were obtained in the C3 sarcoma tumor model (Fig. S4, available at http://www.jem.org/cgi/content/full/jem.20080132/DC1).
To investigate whether immunological mechanisms were involved in tumor rejection, S100A9KO mice were injected i.p. with 200 μg of anti-CD8 antibody 3 d and 1 d before tumor injection. This treatment reduced the level of CD8+ T cells in peripheral blood by more than fivefold (unpublished data). None of the S100A9KO mice treated with anti-CD8 antibody rejected their tumors. Tumor growth in these mice was similar to that in S100A9 wild-type mice (Fig. 2 B). Splenocytes were obtained from S100A9KO and wild-type littermates 12–13 d after injection of EL-4 cells and restimulated for 6 d with irradiated EL-4 tumor cells. T cells were collected and used as effectors in cytotoxicity assays against either EL-4 cells, unrelated B16 melanoma cells, or peritoneal macrophages obtained from naive C57BL/6 mice. T cells from S100A9KO mice showed substantially higher cytotoxicity against EL-4 cells than did T cells from wild-type mice. However, no differences were found in cytotoxicity against macrophages or B16 cells (Fig. S5, available at http://www.jem.org/cgi/content/full/jem.20080132/DC1). Tumors removed from wild-type mice 12 d after tumor cell injection had clearly visible infiltration of S100A9+ cells and Gr-1+ cells, whereas these cells were absent from tumors from S100A9KO mice (Fig. S6, A and B). Tumors from S100A9 KO mice had visible infiltration of CD8+ and CD4+ T cells, which was absent in tumor from S100A9 wild-type mice (Fig. S6, C and D) Tumors from S100A9 wild-type mice had viable cells and infiltrated adjacent adipose tissue. In contrast, tumors from S100A9KO mice had extensive necrosis and apoptosis at the periphery (Fig. S6 E). By day 15 after tumor cell injection, tumor-bearing wild-type mice expressed substantially more Gr-1+CD11b+ MDSCs than did tumor-free mice (11% in Fig. 2 C vs. 4% in Fig. 2 A). In contrast, the proportion of these cells in tumor-bearing S100A9KO mice was the same as in tumor-free S100A9KO mice and significantly (P < 0.05) lower than in tumor-bearing wild-type mice (Fig. 2 C). Thus, MDSCs do not accumulate in tumors in the absence of S100A9.
To address the need for S100A9 for MDSC generation in a tumor-independent model, we injected mice with CFA to mimic the conditions of infection and inflammation, which are known to stimulate production of MDSCs. The level of MDSCs was monitored in peripheral blood by flow cytometry. CFA induced a more than fivefold increase in the proportion of circulating MDSCs in wild-type mice, which peaked on days 6–9 after injection and returned to control level by day 13. In contrast, the number of circulating MDSCs did not increase in S100A9KO mice at any time point (Fig. 2 D). A similar pattern was observed in spleens on day 13 after CFA injection (Fig. 2 E). Thus, the absence of S100A9 protein prevents accumulation of MDSCs in response to CFA, as well as to tumor challenge.
To address the potential contribution of MDSC accumulation to tumor rejection, we performed adoptive transfers of MDSCs. S100A9KO mice were injected with EL-4 lymphoma cells, and then split into two groups of five mice each. One group was untreated and the other group was injected with MDSCs isolated from EL-4 tumor-bearing wild-type mice. 4 out of 5 untreated S100A9KO mice rejected their tumors by day 17, whereas all 5 S100A9KO mice injected with MDSCs developed rapidly growing tumors (Fig. 2 F). Collectively, these results suggest that the absence of MDSC accumulation in S100A9 KO mice is responsible for their ability to reject tumors.
S100A9 overexpression impairs differentiation of DCs and induces accumulation of MDSCs
We asked whether overexpression of S100A9 and A8 could directly regulate the differentiation of DCs and other myeloid cells. To address this question, we generated stable clones of R1 embryonic stem (ES) cells that overexpress S100A8, S100A9, or both (S100A8/9). R1 ES cells transfected with empty vector or another member of S100 family, S100A1, served as controls. Cells were subjected to in vitro differentiation of DCs, as previously described (33). The expression of S100A8 and S100A9 was verified by qRT-PCR, as well as by Western blotting (Figs. 3, A and B). Endogenous S100A8 and S100A9 genes were expressed in the control ES cells, but were shut down in embryoid bodies (EBs). The expression of these genes in transfected ES cells was continuously maintained during cell culture (Fig. S7, available at http://www.jem.org/cgi/content/full/jem.20080132/DC1).
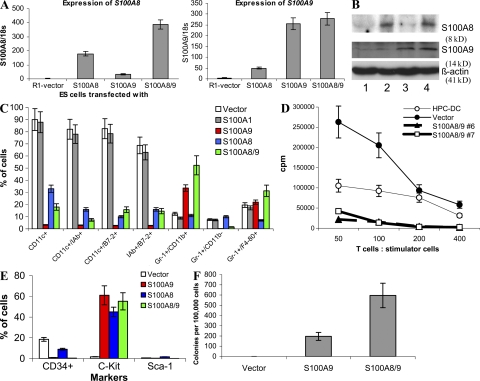
Figure 3.
Effect of S100A9 overexpression on DC differentiation from ES cells. ES cells were transfected with empty vector (vector), S100A1, S100A8, S100A9, or a combination of S100A8 and 9 (S100A8/9). (A) Expression of S100A8 and A9 was evaluated in transfected ES cells using qRT-PCR. (B) Western blot assay for S100A9 and A8 in ES cells transfected with pcDNA-S100A9 and pcDNA-S100A8. Lane 1, empty vector; lane 2, S100A8; lane 3, S100A9; and lane 4, S100A8/9. (C) Phenotypes of cells generated from ES cells cultured for 35 d using the combination of cytokines described in Materials and methods. Expression of different surface molecules was evaluated by flow cytometry. Mean and SD from four different experiments are shown. (D) Effector T cells were isolated from allogeneic BALB/c mice and 105 cells per well were mixed in triplicates of U-bottomed 96-well plates with different numbers of irradiated cells generated from ES cells. Vector, ES cells transfected with empty vector; S100A8/9, ES cells transfected with S100A8 and A9. Two different clones (#6 and #7) of ES cells are shown. HPC-DC, DCs generated from bone marrow progenitor cells by 5-d culture with GM-CSF and IL-4. T cell proliferation was measured by 3[H]thymidine uptake and expressed as counts per minute (CPM). Three experiments were performed. Mean ± SD is shown. T cells alone showed <500 CPM count (not depicted). (E and F) Cells were generated from ES cells and either used for analysis of phenotype by flow cytometry (E) or for colony formation in semisolid medium supporting the growth of myeloid colonies (StemCell Technologies). (F) Colony formation was evaluated in duplicates, and the number of colonies was calculated per 105 cells.
On day 35 after the start of the cultures, cells generated from EBs were collected to evaluate their phenotype by flow cytometry. The total numbers of these cells were similar in all groups (unpublished data). More than 80% of cells generated from control- or S100A1-transfected ES cells had a phenotype of mature DCs (CD11c+IAb+ or CD11c+B7-2+; Fig. 3 C). In sharp contrast, ES cells transfected with S100A9 or S100A8/A9 displayed a dramatically reduced ability to generate DCs. Instead of DCs, these ES cells produced predominantly Gr-1+ cells. The effect of S100A8 on DC differentiation was similar, although much less pronounced. S100A8 did not increase the level of Gr-1+CD11b+ cells (Fig. 3 C). Reduced DC generation from S100A8/9-transfected ES cells was verified in a functional test using the allogeneic mixed leukocyte reaction, a hallmark of DC activity. Cells differentiated from S100A8/9 ES cells had a much lower ability to stimulate the proliferation of allogeneic T cells than did cells differentiated from control ES cells (Fig. 3 D).
S100A9-, A8-, or 8/9-transfected progeny did not express CD34 or Sca-1 markers. However, more than half of these cells expressed c-kit (Fig. 4 E), suggesting that S100A8/9 might cause accumulation of myeloid progenitor cells. This hypothesis was tested in a colony formation assay. Cells differentiated from control ES cells did not form colonies, whereas a significant number of colonies were formed by cells differentiated from S100A9 or S100A8/9 ES cells (Figs. 3 F). Most of the colonies morphologically resembled granulocyte-macrophage CFU-GMs (Fig. S8, available at http://www.jem.org/cgi/content/full/jem.20080132/DC1).
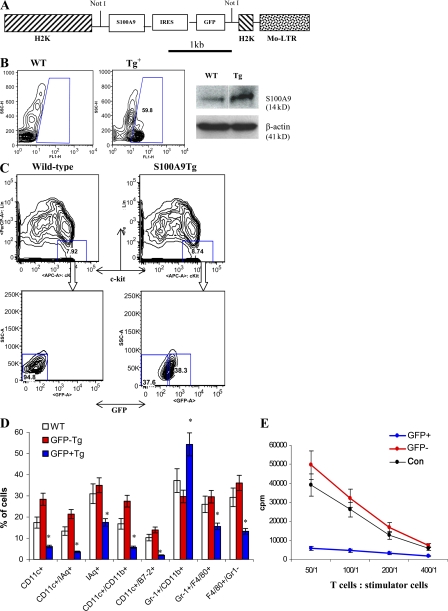
Figure 4.
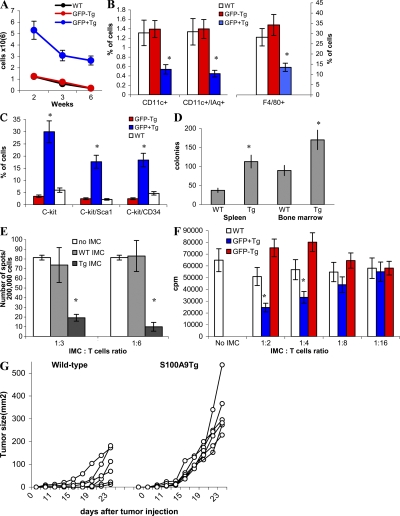
Effect of S100A9 overexpression on myeloid cell differentiation in transgenic mice. (A) Vector used for generation of transgenic mice. (B, left) GFP+ cells in peripheral blood of wild-type (WT) or transgenic mice (Tg) that express S100A9. (right) The level of S100A9 protein in spleens of wild-type (WT) and S100A9 transgenic mice (Tg). (C) Bone marrow progenitor cells were enriched by lineage depletion kit (MACS), and then stained with a cocktail of lineage-specific antibodies. Lin−c-kit+ GFP− or GFP+ populations were sorted by flow cytometry. (D) Sorted cells were cultured for 5 d with cocktail of cytokines as described in Materials and methods. After that time, media was replaced and 0.5 × 106 cells were plated in 24-well plates and cultured with GM-CSF alone for additional 7 d. LPS was added for the last 24 h of culture. The phenotype of cells was evaluated using multicolor flow cytometry. WT, wild-type FVB/N mice; GFP− and GFP+, S100A9 Tg mice. Three experiments were performed. (E) Sorted cells were used as stimulators of allogeneic (BALB/c) T cells. Cell proliferation was measured in triplicate 4-d cultures by 3[H]thymidine uptake. Mean ± SD from two experiments are shown. T cells alone showed count <500 CPM.
To investigate the role of S100A9 in myeloid cell differentiation in vivo, we generated transgenic mice (S100A9Tg) that overexpress S100A9 in hematopoietic cells under control of the H2K-promoter/enhancer and Moloney MuLV enhancer/poly(A) site (34). This promoter previously demonstrated high-level expression of targeted cDNA in hematopoietic cells from spleens, lymph nodes, and thymus (34), as well as in HPCs and stem cells in bone marrow (35). To better trace the expression of the transgene transcript, GFP was linked to the S100A9 cDNA behind an IRES sequence to permit independent translation (Fig. 4 A). FVB/N mice derived from the zygotes injected with this H2K-S100A9-GFP transgenic construct were genotyped by slot blot hybridization to a GFP probe and phenotyped by FACS analysis for GFP expression in peripheral blood cells. Nine founder mice stably integrated the transgene. Expression of GFP protein in peripheral blood cells was significant in one founder mouse (Fig. 4 B, left). Transgene-positive offspring from this founder line exhibited a consistent level of GFP expression during breeding for several generations to wild-type mice. The level of GFP expression was stable during the lifetime of these mice for at least 32 wk (the period of observation). The level of S100A9 protein in spleens obtained from transgenic mice was substantially higher than in wild-type mice (Fig. 4 B, right).
To investigate the effect of S100A9 on the differentiation of myeloid cells, we isolated lineage-negative, c-kit–positive HPCs from bone marrow of control (wild-type) FVB/N mice and S100A9Tg mice. Lin−c-kit+GFP− and Lin−c-kit+GFP+ cells were sorted from S100A9Tg mice (Fig. 4 C). HPCs were cultured for 5 d with a cocktail of cytokines (Flt3, stem cell factor [SCF], IL-11, GM-CSF, and IL-3), followed by a 7-d incubation with GM-CSF; LPS was present in the culture medium for the last 24 h. As shown in Fig. 4 D, the cells generated from wild-type HPCs were CD11c+IAq+ DCs (20%), F4/80+Gr-1− macrophages (30%), and Gr-1+CD11b+ immature myeloid cells (IMCs; 35%). No statistically significant differences were found between the percentages of cell types generated from wild-type HPCs and GFP− HPCs from S100A9Tg mice. GFP+ HPCs from S100A9Tg mice produced few DCs and macrophages, whereas >50% of the total cell population consisted of Gr-1+CD11b+ IMCs (Fig. 4 D). In contrast to cells generated from wild-type and GFP− transgenic HPCs, cells generated from GFP+ transgenic HPCs failed to stimulate allogeneic T cells (Fig. 4 E), confirming the very low presence of DCs.
The total number of GFP−Gr-1+CD11b+ IMCs in S100A9Tg mice was the same as in wild-type mice. In contrast, the number of GFP+ IMCs was fivefold higher than in wild-type mice. Although the total number of GFP+ IMC cells decreased in older mice, the difference between S100A9Tg and wild-type mice was maintained (Fig. 5 A). The proportions of DCs and macrophages were evaluated in spleens of 3-wk-old wild-type and S100A9Tg mice. In S100A9Tg mice, GFP+ and GFP− populations were analyzed separately. No differences were found in the proportions of DCs and macrophages between splenocytes from wild-type mice and GFP− splenocytes from S100A9Tg mice. In the population of GFP+ splenocytes, the percentages of DCs and macrophages were significantly lower than in GFP− splenocytes (Fig. 5 B).
Figure 5.
Phenotype and functional activity of myeloid cells in S100A9 transgenic mice. (A) S100A9Tg mice and their wild-type littermates were killed at indicated times after birth. The number of Gr-1+CD11b+ cells in spleens of wild-type (control) and transgenic mice were evaluated. GFP− and GFP+ splenocytes were counted separately. Each group included three mice. Mean ± SD is shown. (B) The proportion CD11c+ DCs and F4/80+ macrophages in spleens of 3-wk-old wild-type and transgenic mice. (C) The proportion of indicated populations of cells in bone marrow of wild-type and S100A9Tg mice. In transgenic mice, the proportion of cells was calculated separately for GFP+ and GFP− cells. Each group included three mice. Mean ± SD. * represents statistically significant differences between GFP+ and GFP− cells (P < 0.05). (D) Colony formation assay of splenocytes and bone marrow cells isolated from wild-type or S100A9Tg mice. Total myeloid colonies were calculated per 2 × 105 cells in spleen and per 2 × 104 cells in bone marrow. * represents statistically significant differences between WT and Tg mice (P < 0.05). (E) Gr-1+CD11b+ IMC were sorted from spleens of wild-type (WT) or S100A9Tg mice (Tg) and added at a 1:3 or 1:6 ratio to splenocytes from FVB/N mice immunized with specific MHC class I–restricted PDSLRDLSVF peptide. Cells were stimulated with control (RAHYNIVTF) or specific peptides, and the number of IFN-γ–producing cells was evaluated in quadruplicates in ELISPOT assay. The numbers of spots in cells stimulated with control peptide were subtracted from values in cells stimulated with specific peptide. Mean ± SD from three experiments are shown. * represents statistically significant differences from splenocytes cultured without IMC (P < 0.05). (F) Gr-1+CD11b+ IMCs were sorted from WT or S100A9Tg mice (Tg). In transgenic mice, these cells were separated into GFP+ and GFP− cells. T cells were isolated from naive FVB/N mice and DCs were generated from bone marrow of naive BALB/c mice. DCs were mixed with T cells at 1:50 ratio and IMC were added at the indicated ratio. Cells were cultured in triplicates in round-bottomed 96-well plates for 4 d. 3[H]thymidine uptake was measured. Mean ± SD from three experiments are shown. * represents statistically significant differences from splenocytes cultured without IMC (P < 0.05). (G) AVN cells (5 × 105) were injected s.c. into wild-type or S100A9Tg mice and tumor size was monitored. Each group included six mice.
Next, we evaluated the level of myeloid precursors in wild-type and S100A9Tg mice. Percentages of c-kit+ cells in bone marrow were sixfold higher in GFP+ populations of S100A9Tg mice than in either GFP− populations or wild-type mice (Fig. 5 C). Differences in c-kit expression in spleens were less dramatic but statistically significant (P < 0.05; unpublished data). The ability of bone marrow cells and splenocytes from S100A9Tg mice to form myeloid colonies was significantly (P < 0.05) greater than that of cells from wild-type mice (Fig. 5 D and Fig. S9, available at http://www.jem.org/cgi/content/full/jem.20080132/DC1). Collectively, these data indicate that overexpression of S100A9 in hematopoietic cells results in accumulation of myeloid progenitors and IMC at the expense of DC and macrophage differentiation.
To evaluate the possible immunosuppressive activity of IMCs generated in S100A9Tg mice, Gr-1+CD11b+ cells were sorted from wild-type and transgenic tumor-free mice. Two experimental systems were used: peptide-specific CD8+ T cell response and allogenic mixed leukocyte reaction. In the first experimental system, peptide-specific CD8+ T cells were generated by immunizing naive FVB/N mice with DCs loaded with MHC class I (H2-Dq)–restricted rat HER-2/neu–derived peptide PDSLRDLSVF. Splenocytes were isolated and stimulated with control or specific peptide in the presence of IMCs. The number of IFN-γ–producing cells was evaluated by ELISPOT assay. IMCs from S100A9Tg mice significantly reduced the number of antigen-specific CD8+ T cells, whereas IMCs from wild-type mice did not (Fig. 5 E). In the second experimental system, T cells isolated from naive FVB/N mice were stimulated with allogeneic DCs from BALB/c mice in the presence of IMCs. IMCs from S100A9Tg, but not wild-type mice, significantly suppressed T cell proliferation at 1:2 and 1:4 ratios (Fig. 5 F).
To assess possible effects of S100A9 overexpression on tumor growth, wild-type and S100A9Tg mice were injected s.c. with 5 × 105 AVN tumor cells. AVN cells were established from a spontaneous, relatively immunogenic, slow-growing tumor arising in FVB/N MMTV-Neu Tg mice (36). Injection of tumor cells into S100A9Tg mice resulted in rapid tumor growth (Fig. 5 G). Tumors were clearly detectable in all S100A9Tg mice by week 2 and were >200 mm2 in size 1 wk later (mean size 286.5 ± 153.9 mm2). In contrast, only a few tumors were visible by week 2 in wild-type mice. By day 25, the mean tumor size of wild-type mice was only 84.6 ± 71.9 mm2 (P = 0.001).
STAT3 regulates expression of S100A9
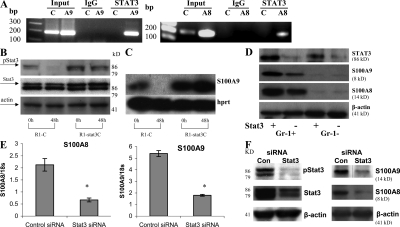
Previous studies have shown that the effect of tumors on DCs and MDSCs is mediated via hyperactivation of the Jak2–STAT3 pathway, which is triggered by most of the tumor-derived factors implicated in abnormal myeloid cell differentiation in cancer (27, 30). The promoter regions of S100A8 and S100A9 contain several potential STAT3 binding sites (TTCCC G/A G/T AA). In chromatin immunoprecipitation assays of 32D myeloid cells, STAT3 interacted with the promoter regions of both S100A8 and S100A9 (Fig. 6 A). Thus, it is possible that STAT3 regulates the transcription of S100A8 and S100A9.
Figure 6.
Regulation of S100A9 expression by STAT3 (A) ChIP assay. Nuclear extracts from 32D cells were sonicated and ChIP with either anti-STAT3 antibody (STAT3) or control rabbit IgG (IgG) was performed. PCR was performed with primers specific for promoter regions of the S100A9 (A9), S100A8 (A8), or β-actin (C) genes. Input, PCR reaction performed with DNA isolated from nuclear extract without immunoprecipitation. (B and C) R1 ES cells were transfected with either control plasmid (R1-C) or Stat3C plasmid (R1-Stat3C) (56) and cultured with complete medium containing LIF. (B) The level of Stat3 protein was determined by Western blotting prior (0 h) and 48 h (48 h) after LIF withdrawal from ES cells. (C) The expression of S100A9 was measured by Southern blotting of RT-PCR products. (D) Deletion of STAT3 was achieved in STAT3loxP/loxP x Mx-Cre transgenic mice by two successive i.p. injections of 100 μg poly(I:C) 7 d apart. Undeleted STAT3loxP/loxP mice were used as a control. Gr-1+ and Gr-1− cells were isolated from spleens of control (STAT3 +) and STAT3 deficient mice (STAT3 -), and the levels of STAT3, S100A8, and S100A9 proteins were evaluated by Western blotting. Two experiments with the same results were performed. (E and F) 32D cells were transfected with STAT3 siRNA. As a control, nontargeting siRNA pool was used (siGenome). (E) Expression of S100A8 and A9 mRNA was measured in quadruplicates by qPCR. Mean ± SD are shown. * represents statistically significant difference from control (P < 0.05). (F) The level of indicated proteins was evaluated in Western blotting. Two experiments with the same results were performed.
In support of this hypothesis, ES cells cultured in the presence of leukemia inhibitory factor (LIF) displayed functionally active phospho-STAT3 and high S100A9 mRNA expression, whereas LIF withdrawal substantially reduced amounts of both pY705STAT3 and S100A9 mRNA within 48 h (Fig. 6, B and C). Expression of the constitutively active STAT3 mutant STAT3C in ES cells maintained a high level of S100A9 expression after LIF withdrawal (Fig. 6, B and C).
To examine the role of STAT3 in the regulation of S100A9 expression in vivo, we used Mx1-Cre/STAT3loxP bitransgenic mice, which have an inducible deletion of the STAT3 gene (31). In these mice, ablation of STAT3 in hematopoietic cells is achieved after injection of poly(I:C). STAT3loxP/loxP mice (without Mx1-Cre) were used as controls. Splenocytes were collected 10 d after injection of poly(I:C). Poly(I:C) treatment substantially reduced amounts of STAT3 and S100A8/A9 in both Gr-1+ and Gr-1− cells of Mx1-Cre/Stat3loxP mice, but not of control mice (Fig. 6 D).
Finally, we depleted STAT3 from 32D cells by RNA interference. 32D cells were transfected with either control siRNA or STAT3 siRNA, and down-regulation of STAT3 by STAT3 siRNA was confirmed by Western blotting (Fig. 6 F). STAT3 depletion resulted in the down-regulation of S100A8 and S100A9 gene expression (Fig. 6 E), as well as the levels of both proteins (Fig. 6 F). Collectively, these data demonstrate that STAT3 up-regulates the expression of S100A8 and S100A9 both in vitro and in vivo.
S100A9 regulates myeloid cell differentiation via reactive oxygen species (ROS)
It is possible that overexpression of S100A9 in myeloid cells results in increased secretion of the protein, which in turn inhibits HPC differentiation. To investigate this possibility, three independent assays were performed. First, splenocytes (106/ml) from control and S100A9Tg mice were cultured for 48 h, and extracellular levels of S100A8 and S100A9 proteins were measured by ELISA. The concentrations of the proteins were similar in both groups (∼150 ng/ml).
Second, Lin−c-kit+ myeloid progenitors derived from the bone marrow of naive FVB/N mice were placed in the bottom chambers of 24-well Transwell plates. Splenocytes (1.5 × 106 cells per ml) from wild-type or S100A9Tg mice were placed in the top chambers (which are separated from the bottom chambers by a semipermeable membrane) and were cultured with a cocktail of cytokines supporting myeloid differentiation. Splenocytes in the top chambers were replaced every 3 d. The progenitor cells differentiated into mature myeloid cells regardless of whether the splenocytes in the top chambers overexpressed S100A9 or not (unpublished data).
Third, Lin−c-kit+ progenitors were cultured in the presence of different concentrations of recombinant S100A8, S100A9, or S100A8/A9 heterodimer. None of these proteins at concentrations of 1 or 5 μg/ml significantly reduced the proportion of mature myeloid cells generated from myeloid progenitors. Collectively, the results suggest that overexpression of S100A9 inhibits myeloid differentiation primarily via intracellular mechanisms.
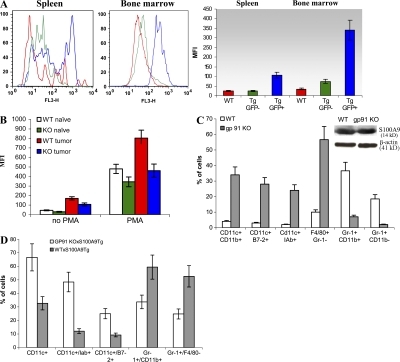
We and others have previously shown that one of the main characteristics of MDSCs from tumor-bearing mice is high production of ROS (28, 29, 37). Recent studies have implicated S100A9 protein in the regulation of ROS generation by NAPDH oxidase (23, 38–41). We hypothesized that tumor-induced up-regulation of S100A9 in progenitors might inhibit myeloid differentiation by increasing production of ROS. Consistent with this hypothesis, amounts of ROS in the spleen and bone marrow were four- to fivefold greater in Gr-1+GFP+ cells from S100A9Tg mice than in Gr-1+GFP− cells from wild-type and S100A9T mice (Fig. 7 A).
Figure 7.
S100A9 affects myeloid cell differentiation via up-regulation of ROS. (A) ROS levels in S100A9Tg mice. Splenocytes and bone marrow cells were collected from 3-wk-old wild-type and S100A9Tg FVB/N mice. Cells were stimulated with PMA, labeled with APC-conjugated anti–Gr-1 antibody, and loaded with the oxidation-sensitive dye hydroethidine. The level of ROS production was evaluated in GFP+ or GFP−Gr-1+ cells. (left) fluorescence histograms from one typical experiment. (right) Graphs representing mean fluorescence intensity (MFI) in all performed experiments. Red, Gr-1+ cells from wild-type mice; green, GFP− Gr-1+ cells from S100A9 Tg mice; blue, GFP+Gr-1+ cells from S100A9 Tg mice. Three experiments with the same results were performed. (B) EL-4 tumors were established in wild-type and S100A9KO mice as described in Fig. 3. Splenocytes were collected 12 d after tumor inoculation. Cells were stimulated with PMA and labeled with APC-conjugated anti–Gr-1 antibody and PE-conjugated anti-CD11b antibody. ROS were measured in Gr-1+CD11b+ cells using DCFDA. Two experiments with similar results were performed. (C) EL-4 tumors were established in wild-type C57BL/6 or gp91−/− mice. 3 wk later, when tumors reached 1.5 cm in diameter, MDSCs were isolated from spleens using magnetic beads and cultured for 7 d in vitro, as described in Fig. 3 B. Proportions of different cell populations were evaluated. Cumulative results from three experiments are shown. (inset) The level of S100A9 protein in freshly isolated MDSCs. (D) Phenotypes of cells in F1 offspring from crosses between S100A9Tg and gp91phox KO mice or wild-type C57BL/6 mice. Lin−c-kit+ progenitor cells were sorted and cultured with cocktail of cytokines to generate myeloid cells. Proportions of different cell populations were evaluated by flow cytometry. Mean ± SD of cumulative results of two experiments is shown.
We determined whether tumor burden affected ROS levels in MDSCs in S100A9KO mice. EL-4 tumors were established in wild-type and S100A9KO mice, and ROS levels were evaluated in Gr-1+CD11b+ MDSCs in spleens at a time when all mice had similar tumor sizes (12 d after tumor inoculation). Consistent with previous reports, MDSCs from wild-type, tumor-bearing mice produced substantially more ROS than did Gr-1+CD11b+ IMCs from naive mice and (Fig. 7 B). ROS levels in IMCs from naive S100A9KO mice were only slightly lower than in IMCs from wild-type mice. However, ROS levels in MDSCs from tumor-bearing S100A9KO mice were substantially lower than in tumor-bearing wild-type mice and were equivalent to ROS levels in naive wild-type mice (Fig. 7 B).
We asked whether ROS affected the differentiation of MDSCs from tumor-bearing mice. EL-4 tumors were established in wild-type and gp91−/− mice, which lack a critical component of the NADPH complex and produce very little ROS. MDSCs (90–95% pure) were isolated from spleens 3 wk after tumor inoculation and cultured in the presence of GM-CSF and TCM for 7 d. MDSCs from tumor-bearing wild-type and gp91−/− mice expressed S100A9 (Fig. 7 C, inset). However, the lack of ROS in gp91−/− mice dramatically enhanced MDSC differentiation. Despite the presence of S100A9, <5% of gp91−/− cells retained an immature phenotype (Gr-1+CD11b+) after 7 d in culture, compared with >30% of wild-type cells. Less than 10% of wild-type cells were F4/80+Gr-1− macrophages or CD11c+ DCs. In sharp contrast, the majority of cells generated from gp91−/− MDSCs were macrophages and DCs (Fig. 7 C). These findings suggest that ROS mediate the inhibitory effects of S100A9 on myeloid cell differentiation.
To further explore this premise, F1 mice were generated by crossing S100A9Tg and gp91phox KO mice. As a control, we used the F1 cross of S100A9Tg mice with wild-type C57BL/6 mice. Mice that contained GFP-positive cells and exhibited suboptimal ROS production in response to PMA stimulation were selected for experiments. Lin−c-kit+ progenitor cells were sorted and cultured with a cocktail of cytokines to generate myeloid cells. Progenitor cells from gp91phoxKO x S100A9Tg F1 mice produced significantly more CD11c+ DCs and significantly less Gr-1+CD11b+ cells than did progenitors from C57BL/6 x S100A9Tg mice (Fig. 7 D). Collectively, these diverse data suggest that S100A9 may suppress myeloid cell differentiation via persistent up-regulation of ROS in progenitor cells.
DISCUSSION
This study demonstrates a novel role for the Ca2+ binding myeloid-related proteins S100A8 and S100A9 in myeloid cell differentiation. It suggests that up-regulation of these proteins in HPCs is primarily responsible for defective myeloid cell differentiation in cancer. S100A8 and S100A9 were previously implicated in trafficking of granulocytes and monocytes, especially during inflammation and metabolism of arachidonic acid (23). Here, we show that up-regulation of these proteins results in inhibition of DC differentiation and accumulation of MDSCs. The STAT3 transcription factor up-regulates the expression of S100A8 and S100A9, which is consistent with the previous observation that different tumor-derived factors mediate their immunosuppressive effects via hyperactivation of Jak2–STAT3 signaling in myeloid cells (27). Two of the cytokines (IL-10 and M-CSF) that were directly implicated in tumor-induced abnormalities of DCs (42, 43) were previously found to up-regulate S100A9 (44, 45). Consistent with these results, a direct connection between STAT3 activity and S100A9 expression in breast cancer cells was recently reported (46).
Several lines of evidence presented here support the conclusion that up-regulation of S100A9 causes inhibition of DC and macrophage differentiation and accumulation of MDSCs.
First, S100A9 was crucial for the in vivo increase in the number of MDSCs in response to inoculation of mice with tumor cells or CFA. S100A9 was also critical for the inhibitory effects of tumor-derived factors on myeloid cell differentiation in vitro. Our data showing normal myeloid cell differentiation in naive S100A9KO mice are consistent with previous studies in which abnormalities in myeloid cells of S100A9KO mice were not detected (32, 47). Apparently, under physiological conditions, S100A9 is not essential for myeloid cell differentiation. However, in the pathological response to challenge with tumor-derived factors or infection, S100A9 is required for both hyperproduction of MDSCs and blockade of DC differentiation.
Second, overexpression of S100A9 in ES cells dramatically compromised their ability to differentiate into mature myeloid cells, particularly DCs and macrophages. The model of DC generation from ES cells has now been validated in several studies (33, 48, 49), and it was chosen for our studies because precursors of granulocytes and monocytes normally express high levels of S100A9 (23). Therefore, we hypothesized that if S100A9 is important for myeloid differentiation, it would most likely affect the differentiation of early progenitor cells. Our data, indeed, demonstrate that S100A9 overexpression blocks normal DC differentiation. This is consistent with the observation that mature DCs have very low or undetectable levels of this protein. Our results concur with those of Hashimoto et al., who performed serial analysis of gene expression in human DCs and found that, in contrast to monocytes, DCs did not express transcripts for S100A8 or S100A9 (50). Apparently, to progress toward macrophage and DC differentiation, myeloid cells must down-regulate S100A9 expression. If such down-regulation is prevented, DC and macrophage differentiation is halted. Instead, ES cells overexpressing S100A8/A9 produced Gr-1+ cells and myeloid progenitors able to form myeloid colonies, which is reminiscent of the phenomenon observed in tumor-bearing mice (1). Because S100A8 and S100A9 form heterodimers, our initial hypothesis was that overexpression of both proteins would have a stronger effect on DC differentiation than would S100A9 alone. However, although double-transfected ES cells formed myeloid colonies more readily than did S100A9 single transfectants, they affected myeloid cell differentiation equally. This may suggest that up-regulation of S100A9 homodimers is sufficient to block DC differentiation. In line with these findings was the observation that the effect of S100A8 overexpression on DC differentiation was less pronounced than that of S100A9. In addition, S100A8 failed to induce the accumulation of Gr-1+CD11b+ cells. Although different functions of S100A9 and S100A8 have been described in several experimental systems (51), more studies are needed to clarify the mechanism of this phenomenon.
Third, overexpression of S100A9 in myeloid cells of transgenic mice inhibited differentiation of macrophages and DCs and induced the accumulation of IMCs. Cells overexpressing S100A9 had a potent suppressive effect on T cell activity similar to that seen in tumor-associated MDSCs. Moreover, S100A9Tg mice were more susceptible to challenge with immunogenic tumors than wild-type mice. It is important to point out that this effect may not necessarily be caused entirely by accumulation of MDSCs or loss of DCs. Although our preliminary experiments indicate that T cells from S100A9Tg mice do not express S100A9 and exhibit normal function, we cannot yet exclude a possible contribution of lymphoid cells to immune suppression in this mouse model.
Use of the S100A9 expression vector containing the GFP reporter allowed us to isolate GFP-positive cells with S100A9 overexpression and to compare them to GFP-negative cells from the same mouse as an internal control to rule out contributions of systemic or microenvironment effects. Our data strongly suggest that S100A9 exerts its effect on myeloid cell differentiation via intracellular mechanisms rather than through extracellular secretion of protein. GFP− myeloid cells in S100A9Tg mice displayed a normal phenotype despite the fact that they were located in close proximity to GFP+ cells that were dramatically affected. In addition, splenocytes from control and Tg mice secreted similar levels of S100A9, as well as S100A8. Finally, soluble factors from S100A9Tg splenocytes, as well as recombinant proteins, did not affect the differentiation of wild-type myeloid cells.
The molecular mechanism of S100A8/A9 effects on myeloid cell differentiation is currently not clear and requires further investigation. Several potential mechanisms could be explored.
These proteins may affect DC differentiation via their Ca2+ binding activity or by inhibiting casein kinase I and II activity, which may be associated with myeloid cell maturation and function. Up-regulation of S100A9 may sensitize myeloid cells to the effects of tumor-derived factors. For instance, we have shown that overexpression of S100A9 results in up-regulation of c-kit on the surface of myeloid cells. Tumor-derived SCF, the c-kit ligand, has been implicated in the accumulation of MDSCs in cancer (52). However, the fact that S100A8 overexpression induced up-regulation of c-kit, but did not affect production of Gr-1+CD11b+ cells argues against a c-kit–dependent mechanism. Recently, a novel role of S100A9 as an endogenous ligand for Toll-like receptor 4 (TLR4) was revealed (53). We have previously demonstrated that MDSC expansion requires signaling through the downstream target TLR MyD88 (16). It is possible that hyperactivation of TLR signaling may interfere with the normal process of myeloid cell differentiation, and S100A9 may be a major mediator of this effect.
Another possible mechanism by which S100A8/A9 might inhibit myeloid cell differentiation is linked to the ability of these proteins to facilitate ROS production by myeloid cells. S100A8/A9 is actively involved in the regulation of ROS generation by NAPDH oxidase (23). Studies demonstrated that S100A8 and S100A9 directly bind to p67phox and p47phox, critical components of the NADPH complex (38, 39). This binding potentiates NADPH oxidase activation in neutrophils. Furthermore, S100A9 inhibits the expression of a substantial number of genes in myeloid cells, including gp91phox (50), suggesting that differences in ROS production in the absence of S100A9 may reflect a general role of this protein during myeloid differentiation. ROS, in turn, are the major factors inhibiting differentiation of DCs in tumor-bearing mice (29). Our data ROS play an important role in S100A9-mediated effects on myeloid cell differentiation.
In recent years, it has become increasingly evident that inflammation plays a critical role in tumor progression (54). This study not only demonstrates a novel molecular mechanism responsible for the suppression of immune responses in cancer via accumulation of MDSCs but also provides a potential direct molecular link between immune suppression, MDSCs, and proinflammatory factors. Together with present and previous observations that lack of S100A9 or STAT3 function promotes tumor rejection via immunological mechanisms (31), these cumulative findings reveal a pathway that negatively regulates the immune response in cancer.
MATERIALS AND METHODS
Reagents.
The following antibody-producing hybridomas were purchased from the American Type Culture Collection (ATCC) and used as supernatants: anti-CD4 (L3T4, TIB-207), anti-CD8 (Lyt-2.2, TIB-210), and anti-MHC II (TIB-120). Anti-IAb, IAd/IEd, Gr-1, CD4, CD8, CD11b, CD86 (B7-2), CD45, CD11c, CD34, Sca-1, c-Kit, isotype control antibodies, and mouse cell lineage depletion kit were obtained from BD Biosciences. Anti-CD3 and anti–Ter-119 antibodies were obtained from BioLegend, anti-F4/80 antibody from Serotec, streptavidin microbeads and mouse lineage cell depletion kit from Miltenyi Biotec, anti–mouse S100A8, and anti-S100A9 from Santa Cruz Biotechnology. For immunohistochemical staining, anti-S100A9, anti–goat, anti–rat conjugated with biotin antibodies, and Vectastain ABC kit were purchased from Vector Laboratories. Myeloid Long-Term Culture Medium for Primitive Mouse Hematopoietic Cells (MyeloCult media) and hydrocortisone were purchased from StemCell Technologies. Low-Tox rabbit complement and Lympholyte M were obtained from Cedarlane Laboratories. Recombinant murine GM-CSF, IL-4, TNF-α, IL-3, IL-11, Flt3L, and SCF were obtained from R&D Systems. LPS was obtained from Sigma-Aldrich. For depletion of CD8+ cells anti-CD8 Lyt 2.2 antibody (ATCC) were generated as ascitis (provided by E. Celis, H. Lee Moffitt Cancer Center, Tampa, FL). Recombinant S100A9 and A8 proteins were previously described (53). AVN mammary carcinoma cell line was provided by K. Knutson (Mayo Clinic, Rochester, MN).
Animals.
6–8-wk-old female BALB/c and C57BL/6 mice were purchased from Harlan. FVB/N mice were purchased from Charles River Laboratories. The gp91phox KO and OT-1 mice were obtained from The Jackson Laboratory.
S100A9-deficient mice were previously described (32) and housed in pathogen-free units of the Division of Comparative Medicine vivarium at H. Lee Moffitt Cancer Center, University of South Florida. These mice were backcrossed for six generations to C57BL/6 mice. Control groups included mice with wild-type genotype (S100A9+/+) from the same generation of backcross.
All animal experiments were approved by the Institutional Animal Care and Use Committee and performed in accordance with U.S. Public Health Service policy and National Research Council guidelines.
Preparation of cells.
Spleens were collected from control or tumor-bearing mice. Single-cell suspensions of splenocytes were treated with ACK buffer to remove red blood cells. Bone marrow cells were harvested from the femurs and tibias of mice and enriched for HPCs by depletion of lineage-specific cells, as previously described (55). In brief, bone marrow cells were incubated with a mixture of antibodies (TIB-207, TIB-210, TIB-120, anti-TER-119, anti-Gr-1, and anti-B220) for 30 min on ice, washed, and treated with complement for 1 h at 37°C. Dead cells were then removed by centrifugation over a Lympholyte M gradient. The resulting fraction contained <20% of lineage-positive cells as detected by flow cytometry. 500,000 enriched HPCs were placed into each well of 24-well plates in 2-ml of RPMI supplemented with 10% FBS, 20 ng/ml GM-CSF, and 10 ng/ml IL-4. This complete medium was replaced every 3 d, and cells were collected for further analysis at indicated time points. To assess the effects of tumor-derived factors on DC differentiation, HPCs were fed with complete medium supplemented with 25% medium conditioned for 48 h by subconfluent cultures of NIH 3T3 fibroblasts as a control, or the C3, CT26, or EL4 tumor cell lines. For DC activation, 5 ng/ml TNF-α was added to RPMI complete medium for 48 h or 1 μg/ml LPS for 24 h before cell analysis.
CTL assay.
Splenocytes from EL4 tumor-bearing mice were cultured for 7 d with irradiated EL4 cells, in 10% FBS RPMI, supplemented with 20 ng/ml IL-15 and 20 ng/ml IL-21. On day 7, cytotoxicity was performed in a standard 6-h 51Cr-release CTL assay. The target cells were EL-4 incubated in duplicates at a serials effector/target ratio.
Real-time qPCR.
Total RNA was extracted from cells with Trizol (Invitrogen). Traces of DNA were removed by treatment with DNase I. The cDNA was synthesized from 1 μg of total RNA using random hexamers and Superscript II reverse transcription (Invitrogen) according to the manufacturer's protocol. PCR was performed with 2.5 μl cDNA, TaqMan Universal PCR Master Mix (Applied Biosystems), and target gene assay mix containing sequence-specific primers for S100A8 or S100A9 and 6-carboxyfluorescein (6-FAM) dye-labeled TaqMan minor groove binder (MGB) probe (Applied Biosystems). Amplification with 18S endogenous control assay mix was used for controls. Data quantitation was performed using the relative standard curve method. Expression levels of the genes were normalized by 18S rRNA.
RT-PCR and Southern blots.
Total RNA was extracted from cells with Trizol. Traces of DNA were removed by treatment with DNase I. The cDNA was synthesized from 1 μg of total RNA using random hexamers and Superscript II reverse transcription according to the manufacturer's protocol. Samples were subjected to initial denaturation at 94°C for 3 min and 24 cycles (for S100A8 and S100A9) or 28 cycles (for hprt) of PCR (94°C for 30 s, 54°C for 30 s, and 72°C for 45 s) with final extension for 7 min at 72°C. The number of cycles was selected after preliminary experiments to avoid saturation of the PCR products. PCR primer pairs used in this study include the following: S100A8 forward for HPC and DC cultures: 5′-ACAATGCCGTCTGAACTGG; reverse, 5′-CTCTGCTACTCCTTGTGGCTGTCT-3′, or S100A8 forward for ES transfectants, 5′-GGAAATCACCATGCCCTCTACAA-3′; reverse, 5′-ATGCCACACCCACTTTTATCACC-3′. S100A9 forward for HPC and DC cultures, 5′-GGAGCGCAGCATAACCAC-3′; reverse, 5′-GCCATTGAGTAAGCCATTCC-3′, or S100A9 forward for ES transfectants, 5′-CATCGACACCTTCAATCAATACTC-3′; reverse, 5′-GCCATCAGCATCATACACTCCTC-3′. hprt: forward, 5′-GATTCAACTTGCGCTCATCTTAGGC-3′; reverse, 5′-GTTGGATACAGGCCAGACTTTGTTG-3′. The PCR products were separated on 1% agarose gels. The sizes of PCR products were: 279 bp for S100A8 in HPC and DCs and 174 bp for ES transfectants; 378 bp for S100A9 in HPC and DC cultures and 206 bp for ES transfectants; and 164 bp for hprt. PCR products were transferred in an alkaline buffer (0.4 N NaOH, 1 M NaCl) onto Hybond N+ nylon transfer membranes (GE Healthcare), and probed with 32P-labeled oligonucleotide probes: S100A8, 5′-GGAGTTCCTTGCGATGGTGATA-3′; S100A9, 5′-ACATCATGGAGGACCTGGACACA-3′; hprt, 5′-GTTGTTGGATATGCCTTGAC-3′.
Western blot analyses were performed as previously described (27).
Chromatin immunoprecipitation assay (ChIP).
32D cells were cultured in 10% FBS RPMI 1640 supplemented with IL3. Preparation of chromatin-DNA and ChIP assay were performed with kit (Millipore) per the manufacturer's instructions, using antibodies against STAT3 (Cell Signaling Technology), normal rabbit IgG (Santa Cruz Biotechnology), and protein A agarose/salmon sperm DNA (Millipore). Sonication was performed using a Branson Sonifier (model 450; VWR Scientific). After reversal of cross-linking, purified DNA was subjected to PCR with the following primers spanning the potential STAT3 binding site in the S100A9 promoter: forward 5′-ACTTCTGGAAATCAGTCTGGCGGT-3′; reverse, 5′-TGGGTTCTTTCCAGCTTCTGGCTA-3′. Primers for S100A8 promoter: forward, 5′-ACACCTCGTACAACTGGAACCACA-3′; reverse, 5′-TCAGCATCATCCAAGAGAGCCCAA-3′. Primers for β-actin: forward, 5′-TAGGGTGTAGACTCTTTGCAGCCA-3′; reverse, 5′-AGCGTCTGGTTCCCAATACTGTGT-3′.
Experiments with ES cells.
S100A1, S100A8, and S100A9 were amplified from mRNA of HPC cells isolated from BALB/c bone marrow using RT-PCR. The pairs of primers were as follows: for S100A1 forward, 5′-GCTCGAGGCCACCATGGGCTCTGAG-3′ containing XhoI site; reverse, 5′-AGAATTCGTGCTCAACTGGTCTCCCA-3′; containing EcoRI site; S100A8 forward, 5′-CTCGAGCCACCATGCCGTCTGAACTG-3′, containing XhoI site at the 5′ end and modified Kozak sequence; reverse, 5′-CAGGAATTCAGCCCTAGGCCAGAAG-3′, containing EcoRI site at 5′ end; S100A9 forward, 5′-AGATATCAGATCTGCCACCATGGCCAACAAAGCA-3′, containing EcoRV and BglII sites at 5′ end and modified Kozak sequence; reverse, 5′-AGTCTCGAGGCTGACCTCTTAATTA-3′, containing XhoI site at 5′ end. For S100A8, the amplified 311-bp fragment was digested using XhoI and EcoRI, and subsequently inserted into pcDNA3.1(−). For S100A9, the amplified 375-bp fragment was digested using EcoRV and XhoI, and inserted into pcDNA3.1(+). S100A1 fragment was cloned into pcDNA3.1(−). The vectors were verified by sequencing. To create cell lines expressing both S100A8 and S100A9, the latter gene was recloned using EcoRV and XhoI into pcDNA3-Hygro. R1 ES cells were transfected with Lipofectamine (Invitrogen) mixed with empty pcDNA3.1 vector; pcDNA3.1 containing S100A1, S100A9, or both S100A8 and S100A9 (S100A8/9); or the STAT3c plasmid (R1-Stat3C, (56) or its vector control RcCMV-Neo (R1-C). ES cells were grown in knockout DMEM containing 2 mM l-glutamine, 0.1 mM nonessential amino acids, 100 μM 2-mercaptoethenol, 15% ES cell certified FBS (Invitrogen), 1,000 U/ml LIF (ESGRO; Chemicon International, Inc.), and 200 μg/ml G418 or both G418 and 100 μg/ml hygromycin.
DC differentiation from S100A9-overexpressing ES cells.
Differentiation of DCs from ES cells was performed as previously described (33). In brief, ES cells were grown on gelatinized plates in complete knockout DMEM, as described above. To induce formation of embryoid bodies, ES cells were transferred to bacterial-grade Petri dishes in medium without LIF and cultured for 14 d; they were fed every other day. On day 14, 1 EB was transferred to each well of a 24-well plate and incubated for 30 d in 2 ml/well RPMI medium containing 15% FBS, 20 ng/ml GM-CSF, and 10 ng/ml IL-3. Half of the medium was changed every 3 d. 5 ng/ml TNF-α was added for the last 5 d before analysis.
Assessment of colony formation.
Colony formation by cells derived from in vitro differentiation was measured using semisolid 1% methylcellulose medium supplemented with recombinant cytokines supporting the optimal growth of myeloid colonies (MethoCult M3534; Stem Cell Technologies). Cells generated from ES cells on day 35 were collected and seeded at 25,000 cells/well in 6-well plates. Colonies were scored on day 10.
S100A9 transgenic mice.
The 375-bp BglII–XhoI S100A9 fragment from pcDNA-S100A9 was inserted into pMIGR1 upstream of an IRES-GFP. A 1.65-kb BglII–SalI fragment containing S100A9-IRES-GFP was end-filled and ligated to NotI linkers and cloned downstream of the H2K promoter at the NotI site of pH2K-i-LTR that had been previously modified to convert a 5′ EcoRI site to SalI (provided by P. Persons, St. Jude Children's Hospital, Memphis, TN). After verification of construct orientation and sequence, the 5.15-kb H2K-S100A9-IRES-GFP transgene was isolated by SalI digestion and electroelution from 1% agarose gel, followed by ethanol precipitation. To generate H2K-S100A9 transgenic mice, the Moffitt Mouse Models Core facility microinjected the purified DNA at 3 ng/μl into pronuclei of 0.5 dpc fertilized zygotes from the FVB/N strain and implanted surviving embryos into the oviducts of pseudopregnant CD1 foster recipients per standard procedures. Transgenic founders and their offspring were genotyped by slot-blot hybridization of tail DNA to a radiolabeled GFP probe.
Generation of myeloid cells from sorted HPCs in transgenic mice.
Bone marrow cells obtained from wild-type or S100A9Tg mice were depleted of lineage-positive cells using mouse lineage cell depletion kit (Miltenyi Biotec), and stained with the following antibodies: pacific blue–conjugated CD3; Percp-conjugated CD4, CD8, Ter-119, CD11b, Gr-1, and B220; and APC-conjugated c-kit. Lin−c-Kit+ GFP+ and Lin−c-Kit+ GFP− stem cells were sorted using a FACSAria. To differentiate myeloid progenitor cells, 35,000 Lin−c-Kit+ GFP+ or Lin−c-Kit+ GFP− cells were placed in each well of 24-well plates and cultured for 5 d in Myeloid Long-Term Culture Medium for Primitive Mouse Hematopoietic Cells, supplemented with 20 ng/ml of IL-3, IL-11, SCF, Flt3L and GM-CSF. 500,000 cells were cultured in RPMI supplemented with 10% FBS and 20 ng/ml of GM-CSF for another 7 d. 24 h before cell analysis, 1 μg/ml LPS was added to activate DCs.
Isolation of Gr-1+ cells.
Gr-1+ cell isolation was performed using MiniMACS microbeads according to the manufacturer's protocol (Miltenyi Biotec). The purity of Gr-1+ cells was consistently >95% in all samples. The depletion of Gr-1+ cell was achieved by two rounds of isolation using magnetic beads. Negative fractions were collected and used as Gr-1− cells. They contained <5% Gr-1+ cells. In some experiments, Gr-1+CD11b+ cells were sorted using FACSAria cell sorter (BD Biosciences).
IFN-γ ELISPOT assay.
MDSCs were isolated from spleens of wild-type and S100A9Tg mice using cell sorting on FACSAria cell sorter. The purity of cell populations was >99%. As responder cells, we used splenocytes from FVB/N mice immunized twice with s.c. injection of 5 × 105 DCs loaded with MHC class I (H2-Dq) restricted rat HER-2/neu–derived peptide PDSLRDLSVF. MDSCs were mixed with splenocytes at 1:3 and 1:6 ratios. The number of IFN-γ–producing cells in response to stimulation with the specific (PDSLRDLSVF) or control (RAHYNIVTF) peptides (10 μg/ml) was evaluated in a 42-h ELISPOT assay performed as previously described (57). The numbers of spots were counted in triplicates and calculated using an automatic ELISPOT counter (Cellular Technology, Ltd).
Allogeneic mixed leukocyte reaction.
T cells were isolated from naive FVB/N mice using a T cell enrichment column (R&D Systems), and 105 cells were placed in each well of round-bottomed 96-well plates. DCs were generated from BALB/c mice using GM-CSF and IL-4. DCs were mixed with T cells at a 1:50 ratio. MDSCs isolated from wild-type and S100A9Tg mice were added to cultures at different MDSC/T cells ratios (from 1:2 to 1:16). Cell proliferation was evaluated in triplicates after a 96-h incubation using 3[H]thymidine incorporation.
Measurement of ROS.
ROS was measured by labeling cells either with the oxidation-sensitive dye DCFDA (28) for experiments with S100A9 KO mice and ES cells or hydroethidine (58) for experiments with S100A9 Tg mice.
Hematoxylin-eosin and immunohistochemical staining of tumor tissues.
EL4 tumor tissues were either fixed in paraformaldehyde or frozen with OCT. Hematoxylin-eosin staining was performed on paraffin embedding tissues. 5-μm-thick sections were stained with hematoxylin-eosin and examined under light microscopy. Immunohistochemical staining was performed on frozen tissue sections using goat anti–mouse S100A9 antibody (R&D Systems), followed by staining with anti–goat IgG conjugated with biotin and ABC kit (Vector Laboratories) and counterstained with hematoxylin or stained with rat anti–mouse Gr-1 antibody (BD Biosciences), followed by staining with anti-rat IgG conjugated with biotin and ABC kit (Vector Laboratories) and counterstained with hematoxylin.
Down-regulation of Stat3 by siRNA in 32D cells.
32D cells (106) were mixed with 100 nM Stat3 siRNA (Dharmacon) in 100 μl 32D nucleofector solution, and transfection was performed using Nucleofector I (Amaxa). As a control, nontargeting siRNA pool was used (siGenome). The gene expression was evaluated 36 h later using qRT-PCR, and protein levels were measured after 72 h by Western blotting.
Statistics.
Statistical analysis was performed using parametric and nonparametric methods on JMP software (SAS Institute). In all cases, P values were calculated using two-sided t test.
Online supplemental material.
Fig. S1 shows the expression of S100A9 and A8 in myeloid cells in tumor-bearing mice. Fig. S2 demonstrates that tumor cell-conditioned medium inhibits differentiation of DCs and induces accumulation of MDSCs in vitro. Fig. S3 shows that differentiation of myeloid cells in S100A9-deficient naive mice is not impaired. Fig. S4 shows that C3 tumor is rejected by most of S100A9 KO mice. Fig. S5 provides the analysis of cytotoxicity by T cells from S100A9 KO mice. Fig. S6 shows the immunohistochemical evaluation of tumors from S100A9 KO mice. Fig. S7 demonstrates the expression of S100A9 and A8 in transfected ES cells. Fig. S8 shows the phenotype of myeloid cell colonies in cells transfected with S100A9/A8. Fig. S9 shows the colony formation by spleen and bone marrow cells from S100A9Tg mice. The online supplemental material is available at http://www.jem.org/cgi/content/full/jem.20080132/DC1.
Acknowledgments
We thank Dr. D. Carbone for his support during initial phase of this study, Dr. H. Yu for providing STAT3-deficient mice, Dr. K. Knutson for providing ANV tumor cell line, Dr. E. Celis for providing anti-CD8 antibody, and Dr. D. Persons for providing pH2K-i-LTR plasmid, which was originally made by Dr. I. Weisman. We thank Dr. N. Olashaw for invaluable help in preparation of the manuscript.
This research was funded by National Institutes of Health grants CA84488 and CA100062 to D.I. Gabrilovich and facilitated by the Flow Cytometry, Molecular Biology, Mouse Models, and Tissue Core Facilities supported by National Cancer Institute Cancer Center Support Grant P30-CA76292.
The authors have no conflicting financial interests.
Abbreviations used: ChIP, chromatin immunoprecipitation assay; EB, embryoid body; ES, embryonic stem; FCM, fibroblast-conditioned medium; HPC, hematopoietic progenitor cell; IMC, immature myeloid cell; LIF, leukemia inhibitory factor; MDSC, myeloid-derived suppressor cell; ROS, reactive oxygen species; SCF, stem cell factor; TCM, tumor cell–conditioned medium; TLR, Toll-like receptor; VEGF, vascular endothelial growth factor.
References
- 1.Gabrilovich, D.I. 2004. The mechanisms and functional significance of tumour-induced dendritic-cell defects. Nat. Rev. Immunol. 4:941–952. [DOI] [PubMed] [Google Scholar]
- 2.Serafini, P., I. Borrello, and V. Bronte. 2006. Myeloid suppressor cells in cancer: recruitment, phenotype, properties, and mechanisms of immune suppression. Semin. Cancer Biol. 16:53–65. [DOI] [PubMed] [Google Scholar]
- 3.Kusmartsev, S., and D.I. Gabrilovich. 2006. Role of immature myeloid cells in mechanisms of immune evasion in cancer. Cancer Immunol. Immunother. 55:237–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gabrilovich, D., V. Bronte, S.-H. Chen, M.P. Colombo, A. Ochoa, S. Ostrand-Rosenberg, and H. Schreiber. 2007. The terminology issue for myeloid-derived suppressor cells. Cancer Res. 67:425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ochoa, A.C., A.H. Zea, C. Hernandez, and P.C. Rodriguez. 2007. Arginase, prostaglandins, and myeloid-derived suppressor cells in renal cell carcinoma. Clin. Cancer Res. 13:721s–726s. [DOI] [PubMed] [Google Scholar]
- 6.Almand, B., J.I. Clark, E. Nikitina, N.R. English, S.C. Knight, D.P. Carbone, and D.I. Gabrilovich. 2001. Increased production of immature myeloid cells in cancer patients. A mechanism of immunosuppression in cancer. J. Immunol. 166:678–689. [DOI] [PubMed] [Google Scholar]
- 7.Mirza, N., M. Fishman, I. Fricke, M. Dunn, A. Neuger, T. Frost, R. Lush, S. Antonia, and D. Gabrilovich. 2006. All-trans-retinoic acid improves differentiation of myeloid cells and immune response in cancer patients. Cancer Res. 66:9299–9307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fricke, I., N. Mirza, J. Dupont, G. Lockhart, A. Jackson, J.-H. Lee, J.A. Sosman, and D.I. Gabrilovich. 2007. Treatment of cancer patients with VEGF-Trap overcomes defects in DC differentiation but is insufficient to improve antigen-specific immune responses. Clin. Cancer Res. 13:4840–4848. [DOI] [PubMed] [Google Scholar]
- 9.Zea, A.H., P.C. Rodriguez, M.B. Atkins, C. Hernandez, S. Signoretti, J. Zabaleta, D. McDermott, D. Quiceno, A. Youmans, A. O'Neill, et al. 2005. Arginase-producing myeloid suppressor cells in renal cell carcinoma patients: a mechanism of tumor evasion. Cancer Res. 65:3044–3048. [DOI] [PubMed] [Google Scholar]
- 10.Sinha, P., V.K. Clements, A.M. Fulton, and S. Ostrand-Rosenberg. 2007. Prostaglandin E2 promotes tumor progression by inducing myeloid-derived suppressor cells. Cancer Res. 67:4507–4513. [DOI] [PubMed] [Google Scholar]
- 11.Huang, B., P.Y. Pan, Q. Li, A.I. Sato, D.E. Levy, J. Bromberg, C.M. Divino, and S.H. Chen. 2006. Gr-1+CD115+ immature myeloid suppressor cells mediate the development of tumor-induced T regulatory cells and T-cell anergy in tumor-bearing host. Cancer Res. 66:1123–1131. [DOI] [PubMed] [Google Scholar]
- 12.Nagaraj, S., K. Gupta, V. Pisarev, L. Kinarsky, S. Sherman, L. Kang, D. Herber, J. Schneck, and D. Gabrilovich. 2007. Altered recognition of antigen is a novel mechanism of CD8+ T cell tolerance in cancer. Nat. Med. 13:828–835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kusmartsev, S., S. Nagaraj, and D.I. Gabrilovich. 2005. Tumor-associated CD8+ T cell tolerance induced by bone marrow-derived immature myeloid cells. J. Immunol. 175:4583–4592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Antonia, S.J., N. Mirza, I. Fricke, A. Chiappori, P. Thompson, N. Williams, G. Bepler, G. Simon, W. Janssen, J.H. Lee, et al. 2006. Combination of p53 cancer vaccine with chemotherapy in patients with extensive stage small cell lung cancer. Clin. Cancer Res. 12:878–887. [DOI] [PubMed] [Google Scholar]
- 15.Shojaei, F., X. Wu, A.K. Malik, C. Zhong, M.E. Baldwin, S. Schanz, G. Fuh, H.P. Gerber, and N. Ferrara. 2007. Tumor refractoriness to anti-VEGF treatment is mediated by CD11b(+)Gr1(+) myeloid cells. Nat. Biotechnol. 25:911–920. [DOI] [PubMed] [Google Scholar]
- 16.Delano, M.J., P.O. Scumpia, J.S. Weinstein, D. Coco, S. Nagaraj, K.M. Kelly-Scumpia, K.A. O'Malley, J.L. Wynn, S. Antonenko, S.Z. Al-Quran, et al. 2007. MyD88-dependent expansion of an immature GR-1+CD11b+ population induces T cell suppression and Th2 polarization in sepsis. J. Exp. Med. 204:1463–1474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gomez-Garcia, L., L.M. Lopez-Marin, R. Saavedra, J.L. Reyes, M. Rodriguez-Sosa, and L.I. Terrazas. 2005. Intact glycans from cestode antigens are involved in innate activation of myeloid suppressor cells. Parasite Immunol. 27:395–405. [DOI] [PubMed] [Google Scholar]
- 18.Ezernitchi, A.V., I. Vaknin, L. Cohen-Daniel, O. Levy, E. Manaster, A. Halabi, E. Pikarsky, L. Shapira, and M. Baniyash. 2006. TCR zeta down-regulation under chronic inflammation is mediated by myeloid suppressor cells differentially distributed between various lymphatic organs. J. Immunol. 177:4763–4772. [DOI] [PubMed] [Google Scholar]
- 19.MacDonald, K.P., V. Rowe, A.D. Clouston, J.K. Welply, R.D. Kuns, J.L. Ferrara, R. Thomas, and G.R. Hill. 2005. Cytokine expanded myeloid precursors function as regulatory antigen-presenting cells and promote tolerance through IL-10-producing regulatory T cells. J. Immunol. 174:1841–1850. [DOI] [PubMed] [Google Scholar]
- 20.Paraiso, K.H., T. Ghansah, A. Costello, R.W. Engelman, and W.G. Kerr. 2007. Induced SHIP deficiency expands myeloid regulatory cells and abrogates graft-versus-host disease. J. Immunol. 178:2893–2900. [DOI] [PubMed] [Google Scholar]
- 21.Foell, D., and J. Roth. 2004. Proinflammatory S100-proteins in arthritis and autoimmune disease. Arthritis Rheum. 50:3762–3771. [DOI] [PubMed] [Google Scholar]
- 22.Leukert, N., T. Vogl, K. Struppat, R. Reichelt, C. Sorg, and J. Roth. 2006. Calcium-dependent tetramer formation of S100A8 and S100A9 is essential for biological activity. J. Mol. Biol. 359:961–972. [DOI] [PubMed] [Google Scholar]
- 23.Nacken, W., J. Roth, C. Sorg, and C. Kerkhoff. 2003. S100A9/S100A8: myeloid representatives of the S100 protein family as prominent players in innate immunity. Microsc. Res. Tech. 60:569–580. [DOI] [PubMed] [Google Scholar]
- 24.Hiratsuka, S., A. Watanabe, H. Aburatani, and Y. Maru. 2006. Tumour-mediated upregulation of chemoattractants and recruitment of myeloid cells predetermines lung metastasis. Nat. Cell Biol. 8:1369–1375. [DOI] [PubMed] [Google Scholar]
- 25.Menetrier-Caux, C., M.C. Thomachot, L. Alberti, G. Montmain, and J.Y. Blay. 2001. IL-4 prevents the blockade of dendritic cell differentiation induced by tumor cells. Cancer Res. 61:3096–3104. [PubMed] [Google Scholar]
- 26.Gabrilovich, D.I., P. Cheng, Y. Fan, B. Yu, E. Nikitina, A. Sirotkin, M. Shurin, T. Oyama, Y. Adachi, S. Nadaf, et al. 2002. H1o histone and differentiation of dendritic cells. a molecular target for tumor-derived factors. J. Leukoc. Biol. 72:285–296. [PubMed] [Google Scholar]
- 27.Nefedova, Y., M. Huang, S. Kusmartsev, R. Bhattacharya, P. Cheng, R. Salup, R. Jove, and D. Gabrilovich. 2004. Hyperactivation of STAT3 is involved in abnormal differentiation of dendritic cells in cancer. J. Immunol. 172:464–474. [DOI] [PubMed] [Google Scholar]
- 28.Kusmartsev, S., Y. Nefedova, D. Yoder, and D.I. Gabrilovich. 2004. Antigen-specific inhibition of CD8+ T cell response by immature myeloid cells in cancer is mediated by reactive oxygen species. J. Immunol. 172:989–999. [DOI] [PubMed] [Google Scholar]
- 29.Kusmartsev, S., and D.I. Gabrilovich. 2003. Inhibition of myeloid cell differentiation in cancer: The role of reactive oxygen species. J. Leukoc. Biol. 74:186–196. [DOI] [PubMed] [Google Scholar]
- 30.Wang, T., G. Niu, M. Kortylewski, L. Burdelya, K. Shain, S. Zhang, R. Bhattacharya, D. Gabrilovich, R. Heller, D. Coppola, et al. 2004. Regulation of the innate and adaptive immune responses by Stat-3 signaling in tumor cells. Nat. Med. 10:48–54. [DOI] [PubMed] [Google Scholar]
- 31.Kortylewski, M., M. Kujawski, T. Wang, S. Wei, S. Zhang, S. Pilon-Thomas, G. Niu, H. Kay, J. Mule, W.G. Kerr, et al. 2005. Inhibiting Stat3 signaling in the hematopoietic system elicits multicomponent antitumor immunity. Nat. Med. 11:1314–1321. [DOI] [PubMed] [Google Scholar]
- 32.Manitz, M.P., B. Horst, S. Seeliger, A. Strey, B.V. Skryabin, M. Gunzer, W. Frings, F. Schonlau, J. Roth, C. Sorg, and W. Nacken. 2003. Loss of S100A9 (MRP14) results in reduced interleukin-8-induced CD11b surface expression, a polarized microfilament system, and diminished responsiveness to chemoattractants in vitro. Mol. Cell. Biol. 23:1034–1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cheng, P., Y. Nefedova, L. Miele, B.A. Osborne, and D. Gabrilovich. 2003. Notch signaling is necessary but not sufficient for differentiation of dendritic cells. Blood. 102:3980–3988. [DOI] [PubMed] [Google Scholar]
- 34.Domen, J., K.L. Gandy, and I.L. Weissman. 1998. Systemic overexpression of BCL-2 in the hematopoietic system protects transgenic mice from the consequences of lethal irradiation. Blood. 91:2272–2282. [PubMed] [Google Scholar]
- 35.Dominici, M., M. Tadjali, S. Kepes, E.R. Allay, K. Boyd, P.A. Ney, E. Horwitz, and D.A. Persons. 2005. Transgenic mice with pancellular enhanced green fluorescent protein expression in primitive hematopoietic cells and all blood cell progeny. Genesis. 42:17–22. [DOI] [PubMed] [Google Scholar]
- 36.Knutson, K.L., H. Lu, B. Stone, J.M. Reiman, M.D. Behrens, C.M. Prosperi, E.A. Gad, A. Smorlesi, and M.L. Disis. 2006. Immunoediting of cancers may lead to epithelial to mesenchymal transition. J. Immunol. 177:1526–1533. [DOI] [PubMed] [Google Scholar]
- 37.Sinha, P., V.K. Clements, and S. Ostrand-Rosenberg. 2005. Reduction of myeloid-derived suppressor cells and induction of M1 macrophages facilitate the rejection of established metastatic disease. J. Immunol. 174:636–645. [DOI] [PubMed] [Google Scholar]
- 38.Doussiere, J., F. Bouzidi, and P.V. Vignais. 2002. The S100A8/A9 protein as a partner for the cytosolic factors of NADPH oxidase activation in neutrophils. Eur. J. Biochem. 269:3246–3255. [DOI] [PubMed] [Google Scholar]
- 39.Doussiere, J., F. Bouzidi, and P.V. Vignais. 2001. A phenylarsine oxide-binding protein of neutrophil cytosol, which belongs to the S100 family, potentiates NADPH oxidase activation. Biochem. Biophys. Res. Commun. 285:1317–1320. [DOI] [PubMed] [Google Scholar]
- 40.Kerkhoff, C., W. Nacken, M. Benedyk, M. Dagher, C. Sopalla, and J. Doussiere. 2005. The arachidonic acid-binding protein S100A8/A9 promotes NADPH oxidase activation by interaction with p67phox and Rac-2. FASEB J. 19:467–469. [DOI] [PubMed] [Google Scholar]
- 41.Benedyk, M., C. Sopalla, W. Nacken, G. Bode, H. Melkonyan, B. Banfi, and C. Kerkhoff. 2007. HaCaT keratinocytes overexpressing the S100 proteins S100A8 and S100A9 show increased NADPH oxidase and NF-kappaB activities. J. Invest. Dermatol. 127:2001–2011. [DOI] [PubMed] [Google Scholar]
- 42.Menetrier-Caux, C., G. Montmain, M. Dieu, C. Bain, M. Favrot, C. Caux, and J. Blay. 1998. Inhibition of the differentiation of dendritic cells from CD34(+) progenitors by tumor cells: role of interleukin-6 and macrophage-colony-stimulating factor. Blood. 92:4778. [PubMed] [Google Scholar]
- 43.Steinbrink, K., M. Wolfl, H. Jonuleit, J. Knop, and A.H. Enk. 1997. Induction of tolerance by IL-10-treated dendritic cells. J. Immunol. 159:4772–4780. [PubMed] [Google Scholar]
- 44.Kumar, A., A. Steinkasserer, and S. Berchtold. 2003. Interleukin-10 influences the expression of MRP8 and MRP14 in human dendritic cells. Int. Arch. Allergy Immunol. 132:40–47. [DOI] [PubMed] [Google Scholar]
- 45.Henkel, G.W., S.R. McKercher, and R.A. Maki. 2002. Identification of three genes up-regulated in PU.1 rescued monocytic precursor cells. Int. Immunol. 14:723–732. [DOI] [PubMed] [Google Scholar]
- 46.Li, C., F. Zhang, M. Lin, and J. Liu. 2004. Induction of S100A9 Gene Expression by Cytokine Oncostatin M in Breast Cancer Cells Through the STAT3 Signaling Cascade. Breast Cancer Res. Treat. 87:123–134. [DOI] [PubMed] [Google Scholar]
- 47.Hobbs, J.A., R. May, K. Tanousis, E. McNeill, M. Mathies, C. Gebhardt, R. Henderson, M.J. Robinson, and N. Hogg. 2003. Myeloid cell function in MRP-14 (S100A9) null mice. Mol. Cell. Biol. 23:2564–2576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fairchild, P.J., F.A. Brook, R.L. Gardner, L. Graca, V. Strong, Y. Tone, M. Tone, K.F. Nolan, and H. Waldmann. 2000. Directed differentiation of dendritic cells from mouse embryonic stem cells. Curr. Biol. 10:1515–1518. [DOI] [PubMed] [Google Scholar]
- 49.Senju, S., S. Hirata, H. Matsuyoshi, M. Masuda, Y. Uemura, K. Araki, K. Yamamura, and Y. Nishimura. 2003. Generation and genetic modification of dendritic cells derived from mouse embryonic stem cells. Blood. 101:3501–3508. [DOI] [PubMed] [Google Scholar]
- 50.Hashimoto, S., T. Suzuki, H.-Y. Dong, S. Nagai, N. Yamazaki, and K. Matsushima. 1999. Serial analysis of gene expression in human monocyte-derived dendritic cells. Blood. 94:845–852. [PubMed] [Google Scholar]
- 51.Roth, J., T. Vogl, C. Sorg, and C. Sunderkotter. 2003. Phagocyte-specific S100 proteins: a novel group of proinflammatory molecules. Trends Immunol. 24:155–158. [DOI] [PubMed] [Google Scholar]
- 52.Pan, P.Y., G.X. Wang, B. Yin, J. Ozao, T. Ku, C.M. Divino, and S.H. Chen. 2007. Reversion of immune tolerance in advanced malignancy: modulation of myeloid derived suppressor cell development by blockade of SCF function. Blood. 111:219–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Vogl, T., K. Tenbrock, S. Ludwig, N. Leukert, C. Ehrhardt, M.A. van Zoelen, W. Nacken, D. Foell, T. van der Poll, C. Sorg, and J. Roth. 2007. Mrp8 and Mrp14 are endogenous activators of Toll-like receptor 4, promoting lethal, endotoxin-induced shock. Nat. Med. 13:1042–1049. [DOI] [PubMed] [Google Scholar]
- 54.van Kempen, L.C., K.E. de Visser, and L.M. Coussens. 2006. Inflammation, proteases and cancer. Eur. J. Cancer. 42:728–734. [DOI] [PubMed] [Google Scholar]
- 55.Cheng, P., A. Zlobin, V. Volgina, S. Gottipati, B. Osborne, L. Miele, and D.I. Gabrilovich. 2001. Notch-1 regulates NF-kappa B activity in hematopoietic progenitor cells. J. Immunol. 167:4458–4467. [DOI] [PubMed] [Google Scholar]
- 56.Bromberg, J.F., M.H. Wrzeszczynska, G. Devgan, Y. Zhao, R.G. Pestell, C. Albanese, and J.E. Darnell. 1999. Stat3 as an oncogene. Cell. 98:295–303. [DOI] [PubMed] [Google Scholar]
- 57.Gabrilovich, D.I., M. Velders, E. Sotomayor, and W.M. Kast. 2001. Mechanism of immune dysfunction in cancer mediated by immature Gr-1+ myeloid cells. J. Immunol. 166:5398–5406. [DOI] [PubMed] [Google Scholar]
- 58.Guthrie, H.D., and G.R. Welch. 2006. Determination of intracellular reactive oxygen species and high mitochondrial membrane potential in Percoll-treated viable boar sperm using fluorescence-activated flow cytometry. J. Anim. Sci. 84:2089–2100. [DOI] [PubMed] [Google Scholar]