Abstract
We report here the functional characterization of an essential Saccharomyces cerevisiae gene, MPR1, coding for a regulatory proteasomal subunit for which the name Rpn11p has been proposed. For this study we made use of the mpr1-1 mutation that causes the following pleiotropic defects. At 24°C growth is delayed on glucose and impaired on glycerol, whereas no growth is seen at 36°C on either carbon source. Microscopic observation of cells growing on glucose at 24°C shows that most of them bear a large bud, whereas mitochondrial morphology is profoundly altered. A shift to the nonpermissive temperature produces aberrant elongated cell morphologies, whereas the nucleus fails to divide. Flow cytometry profiles after the shift to the nonpermissive temperature indicate overreplication of both nuclear and mitochondrial DNA. Consistently with the identification of Mpr1p with a proteasomal subunit, the mutation is complemented by the human POH1 proteasomal gene. Moreover, the mpr1-1 mutant grown to stationary phase accumulates ubiquitinated proteins. Localization of the Rpn11p/Mpr1p protein has been studied by green fluorescent protein fusion, and the fusion protein has been found to be mainly associated to cytoplasmic structures. For the first time, a proteasomal mutation has also revealed an associated mitochondrial phenotype. We actually showed, by the use of [rho°] cells derived from the mutant, that the increase in DNA content per cell is due in part to an increase in the amount of mitochondrial DNA. Moreover, microscopy of mpr1-1 cells grown on glucose showed that multiple punctate mitochondrial structures were present in place of the tubular network found in the wild-type strain. These data strongly suggest that mpr1-1 is a valuable tool with which to study the possible roles of proteasomal function in mitochondrial biogenesis.
INTRODUCTION
Nucleocytoplasmic interactions have often been studied by looking for nuclear suppressors of mitochondrial mutations. We have previously described the mpr1-1 mutation, which was isolated as a nuclear suppressor of a mitochondrial mutation that resulted in defective tRNA processing (Zennaro et al., 1989; Rinaldi et al., 1994). Although the mechanism of the mitochondrial suppression was unclear, the MPR1 gene was found to be essential (Rinaldi et al., 1995), and its temperature-sensitive allele, mpr1-1, caused a pleiotropic phenotype. The deduced amino acid sequence of Mpr1p (Rinaldi et al., 1995) clearly indicated that it belongs to a conserved gene family and exhibits high identity to the recently published sequence of the human proteasomal regulatory subunit Poh1p (Spataro et al., 1997).
Control of many cellular activities (such as metabolic adaptation, cell differentiation, cell cycle control, and stress response) requires degradation of regulatory proteins such as cyclins, transcriptional activators and repressors (for reviews see Nurse, 1990; Rechsteiner et al., 1993; Hochstrasser 1995; Murray, 1995; Hilt and Wolf 1996; King et al., 1996). The ATP- and ubiquitin-dependent 26S proteasome is a functional complex capable of recognizing and degrading such regulatory proteins when they are to be eliminated (Peters, 1994). The 26S proteasome is known to be composed of two subunits: the 19S regulatory particle recognizes and unfolds ubiquitinated proteins, which are then degraded by the 20S proteolytic subunit. In Saccharomyces cerevisiae, the structure of the latter subunit has been resolved by crystallography, and all the corresponding genes have been identified (Groll et al., 1997). Recently also the 19S regulatory particle has been isolated, and its 17 protein subunits have been isolated and sequenced. One of them, called Rpn11p, was found to be the product of the MPR1 gene we had studied (Glickman et al., 1998). We therefore will call this gene RPN11/MPR1.
In S. cerevisiae, the specific roles of several components of the proteasomal regulatory complex have been identified by the study of the effects of mutations in the corresponding genes. Some of the components are known to be involved in cell cycle progression. Ghislain et al. (1993) have described two thermosensitive mutations (cim3 and cim5), which are colethal with cdc28-1N. In these mutant strains, the cell cycle was arrested in G2-M upon shifting to the nonpermissive temperature (Ghislain et al., 1993). Mutations in other proteasomal genes resulted in a similar arrest of the cell cycle and/or a failure to replicate the spindle pole body (McDonald and Byers, 1997). Other mutations, such as one affecting the regulatory subunit Nin1p (Kominami et al., 1994), result in a block both in the G1-S and the G2-M transitions. In many cases, these mutations were accompanied by additional effects such as the accumulation of ubiquitinated proteins.
Despite our knowledge of the structure and subunits of the proteasome, little is known about the intracellular localization of the proteasome complex. Immunocytochemical studies have revealed the presence of proteasomes in the nucleus and in the cytoplasm of a variety of cells and tissues. However, the intracellular distribution of proteasomes varies during the cell cycle and during development in higher eukaryotes, and several observations point to a highly dynamic state of proteasomes in the cell (Amsterdam et al., 1993; Rivett 1993; Peters et al., 1994; Dawson et al., 1995).
In this report, we describe the function and localization of Mpr1p, and we analyze in detail the pleiotropic effects of a mutated allele (mpr1-1) of the RPN11/MPR1 gene. This is the first case in which a clear mitochondrial phenotype can be associated with a proteasomal mutation.
MATERIALS AND METHODS
Strains, Plasmids, and Media
The yeast strains and plasmids used in this study are listed in Table 1.
Table 1.
Yeast strains and plasmids
| Strain | Genotype | Source |
|---|---|---|
| W303 | MATaMATα, his3-11/his3-11, ade2-1/ade2-1, leu2-3,112/leu2-3,112, ura3-1/ura3-1,trp1-Δ2/trp1-Δ2, can1-100 [rho+] | Thomas and Rothstein, 1989 |
| W303-1B | MATα, his3-11, ade2-1, leu2-3,112, ura3-1, trp1-Δ2, can1-100 [rho+] | Thomas and Rothstein, 1989 |
| R117 | MATa/MATα, his1, ura1,2 MPR1/mpr1 [Ts932] | Rinaldi et al., 1994 |
| R117/a12 | MATa, his1, ade2, leu2, ura3, mpr1 [rho+] | Rinaldi et al., 1994 |
| W303/5 | MATa/MATα, his3-11/his3-11, ade2-1/ade2-1, leu2-3,112/leu2-3,112, ura3-1/ura3-1, trp1-Δ2/trp1-Δ2, can1-100, MPR1/mpr1∷URA3 [rho+] | This study |
| FY1679 | MATa/MATα, his3-Δ200/+, leu2-Δ1/+, ura3-52/ura3-52, trp1-Δ63/+, [rho+] | Winston et al., 1995 |
| FY1679/1 | MATa/MATα, his3-Δ200/+, leu2-Δ1/+, ura3-52/ura3-52, trp1-Δ63/+, MPR1/mpr1∷URA3, [rho+] | This study |
| W303-pI | MATα, his3-11, ade2-1, leu2-3,112, ura3-1, trp1-Δ2, mpr1∷pYI-mpr1, can1-100 [rho+] | This study |
| W303-MPR1 | W303-pI with the integrative plasmid pYI-mpr1 excised | This study |
| W303-mpr1 | W303-pI with the integrative plasmid pYI-MPR1 excised | This study |
| W303-mpr1 [rho°] | MATα, his3-11, ade2-1, leu2-3,112, ura3-1, trp1-Δ2, mpr1, can1-100 [rho°] | This study |
| W303-MPR1 [rho°] | MATα, his3-11, ade2-1, leu2-3,112, ura3-1, trp1-Δ2, MPR1, can1-100 [rho°] | This study |
| Plasmid | Insert | Vector | Reference |
|---|---|---|---|
| pYC | YCp50 | Rose et al., 1987 | |
| pYC31 | MPR1; 3100 bp | YCp50 | |
| YCpMPR1 | MPR1, 2400 bp | YCp50 | |
| YEpMPR1 | MPR1, 2400 bp | YEplac181 | Gietz and Sugino, 1988 |
| KSpMPR1 | MPR1, 2400 bp | pBluescript KS+ (Stratagene) | |
| pKSmpr1∷URA3 | mpr1∷URA3, 4000 bp | pBluescript KS+ | |
| pCRII-mpr1 | mpr1, 1625 bp | pCRII (Invitrogen) | |
| pYI-mpr1 | mpr1, 1625 bp | YIplac211 | Gietz and Sugino, 1988 |
| TU65 | GFP (green fluorescent protein), 800 bp | pBluescript KS+ (Stratagene) | Chalfie et al., 1994 |
| pRSETB8 | GFP (green fluorescent protein), 800 bp | pRSETB (Invitrogen) | |
| p100GFP | GFP under UASga11-10/CYC1 promoter | pEMBLYex4 | Baldari et al., 1987 |
| pKS-EcoRI | pKS without EcoRI restriction site | pBluescript KS+ | |
| KSpGFP | GFP, 800 bp | pBluescript KS+ | |
| pCRII-MPR1 | MPR1, 1452 bp | pCRII | |
| KSpMPR1-GFP | MPR1-GFP, 2250 bp | pBluescript KS+ | |
| YEpMPR1-GFP | MPR1-GFP, 2250 bp | YEplac181 | Gietz and Sugino, 1988 |
| YCpMPR1-GFP | MPR1-GFP, 2250 bp | YCplac111 | Gietz and Sugino, 1988 |
| pSK15 | H. sapiens POH1 cDNA, 1500 bp | pBluescript SK+ | |
| pYES-POH1 | H. sapiens POH1 cDNA, 1500 bp | pYES2 (Invitrogen) |
Yeast Culture Media.
Rich medium was YP (1% bactopeptone and 1% yeast extract), containing 2% glucose (YPD), 2% glycerol (YPG), or 2% galactose (YPGal). Minimal medium was WO (0.17% yeast nitrogen base, 0.5% ammonium sulfate, and 2% glucose). All media were supplemented with 2.3% bacto agar (Difco, Detroit, MI) for solid media, and WO was supplemented with the appropriate nutritional requirements according to the phenotype of the strains.
Isolation of the RPN11/MPR1 Gene.
To isolate the gene that complements the growth defect of the mpr1-1 mutant, the mutant strain R117/a12 was transformed with a nuclear DNA library constructed in the YCp50 centromeric vector (Rose et al., 1987). One plasmid (pYC31), containing an insert of 3.1 kb, complemented the thermosensitive phenotype of the mutant strain. This insert contained a truncated NIC96 gene and two unknown ORFs. The ORF responsible for the suppression was present in an EcoRI–BamHI fragment of 2.4 kb (YCpMPR1). The gene was called MPR1 (GenBank accession number X79561).
Construction of W303-mpr1 Strain.
Isogenic strains in a W303 genetic context have been constructed as follows. The W303-1B haploid strain was transformed with the circular integrative plasmid YIplac211 (Gietz and Sugino, 1988) containing mpr1-1 and URA3 as selectable markers (pYI-mpr1). Twenty-five URA3 transformants of the resulting strain W303-pI were cured with 5FOA; among the URA3 clones, four showed the same growth defect as the strain R117/a12, indicating that the RPN11/MPR1 gene had been excised with the URA3 marker: the substitution of RPN11/MPR1 with mpr1-1 in the W303-1B strain results in the same phenotype as the one observed in R117/a12. The W303-1B strain containing mpr1-1 was called W303-mpr1, whereas the same strain having retained the wild-type MPR1 gene was called W303-MPR1. Correct integration of the mutant allele was demonstrated by PCR analysis.
Allelism Test between the Isolated MPR1 Gene and the mpr1-1 Mutation.
The first step was to construct a haploid strain deleted for MPR1 and made viable by the presence of the episomal plasmid YEpMPR1 (LEU2). This was done by sporulating the W303/5 diploid strain (MPR1/mpr1::URA3) transformed with the same plasmid YEpMPR1. A viable spore was then crossed with the mutant W303-mpr1, the corresponding diploid was sporulated, and tetrad analysis was performed. In case of allelism two types of spores should be obtained: 1) W303-mpr1::URA3 + YEpMPR1 (which cannot lose the plasmid); and 2) W303-mpr1 + YEpMPR1 (which can lose the plasmid). This is was what we obtained.
Construction of GFP Plasmids.
To construct the p100GFP plasmid, the wild-type GFP was isolated as a KpnI–PstI fragment from plasmid TU65 (a kind gift from Dr. M. Chalfie, Department of Biological Sciences, Columbia University, New York, NY) (Chalfie et al., 1994). After blunting the KpnI overhang with T7 polymerase, GFP was ligated into SmaI–PstI-restricted pEMBLYex4 (episomal plasmid; Baldari et al., 1987) under the control of the UASgal1-10/CYC1 promoter. The fusion RPN11/MPR1-GFP was constructed as follows. The GFP gene from the pRSETB8 plasmid was cloned in pBluescript KS+ (Stratagene, La Jolla, CA) lacking the EcoRI site of the polylinker (pKS-EcoRI). This plasmid was obtained by ligation of pBluescript KS+ digested with the restriction enzymes EcoRV and SmaI (resulting plasmid, KSpGFP). Two oligonucleotides with an EcoRI site at the extremities were used to amplify the RPN11/MPR1 promoter region and the full gene except the stop codon. This fragment of 1452 bp was first cloned in the pCRII plasmid (resulting plasmid, pCRII-MPR1) and then cloned in KSpGFP (resulting plasmid, KSpMPR1-GFP). This construction was also cloned in a centromeric and multicopy plasmid (YEpMPR1-GFP and YCpMPR1-GFP).
Construction of the Yeast Plasmid Containing the POH1 Gene.
The human cDNA (pSK15) corresponding to the POH1 gene (GenBank accession number U86782) was purchased from the Research Genetics/IMAGE Consortium (Genome Systems, St. Louis, MO) (GenBank accession number AA084170). This cDNA was cloned in the plasmid pYES2 containing the galactose-activated GAL1 promoter (resulting plasmid, pYES-POH1).
Transformation Procedures
Transformation of the wild-type strain was done with a standard lithium chloride procedure (Ito et al., 1983). The mutant strain has a low-efficiency transformation phenotype and had to be transformed by the protoplast procedure: a culture of 50 ml in exponential growth phase was harvested and resuspended in 10 ml of 1 M sorbitol, 20 mM dithiothreitol, and 10 mM EDTA (pH 8) and incubated for 10 min at room temperature. After centrifugation cells were resuspended in 10 ml of 1 M sorbitol containing 0.5 mg of cytohelicase (Sigma, St. Louis, MO). After incubation for 10 min at room temperature, protoplasts were harvested by centrifugation at 1500 rpm for 10 min, washed with 10 ml of 1 M sorbitol, resuspended in YPD containing 1 M sorbitol, and incubated for 30 min at 24°C. Protoplasts were then centrifuged and resuspended in 1 M sorbitol, 10 mM Tris-HCl (pH 7.4), and 10 mM CaCl2. Plasmid DNA was added to 100 μl of this protoplast suspension for each transformation, and after 10 min at room temperature, 1 ml of 20% polyethylene glycol 4000 in a 10 mM Tris-HCl (pH 7.4)/10 mM CaCl2 solution was added. After 10 min at room temperature and 10 min on ice, protoplasts were mixed to 10 ml of soft agar (1% agar) containing 1 M sorbitol supplemented with the appropriate amino acids and spread onto minimal sorbitol plates.
DNA Techniques
DNA techniques were performed as described by Sambrook et al. (1995). The oligonucleotides used for mpr1-1 amplification (bold in Figure 2) are the following: IX13, 5′-CCCTACGGTCTGTTGTTGTTCTGATTCCC-3′; IX14, 5′-CCACAGATGGAAACGCATTTAATGGTGATG-3′. For GFP fusion (underlined in Figure 2): 5′-EcoRIMPR, 5′-CGGAATTCGAATGATGGTTGCACTC-3′; 3′-EcoRIMPR, 5′-GCCTTAAGTTTAATTGCCACTGAAT-3′. Induction of [rho°] with ethidium bromide was obtained by growing the mpr1-1 mutant and RPN11/MPR1 strains for 24 h in the presence of 25 μg/ml ethidium bromide. The [rho°] mutants were selected by their inability to grow on respiratory medium followed by observation of 4,6-diamidino-2-phenylindole (DAPI) staining.
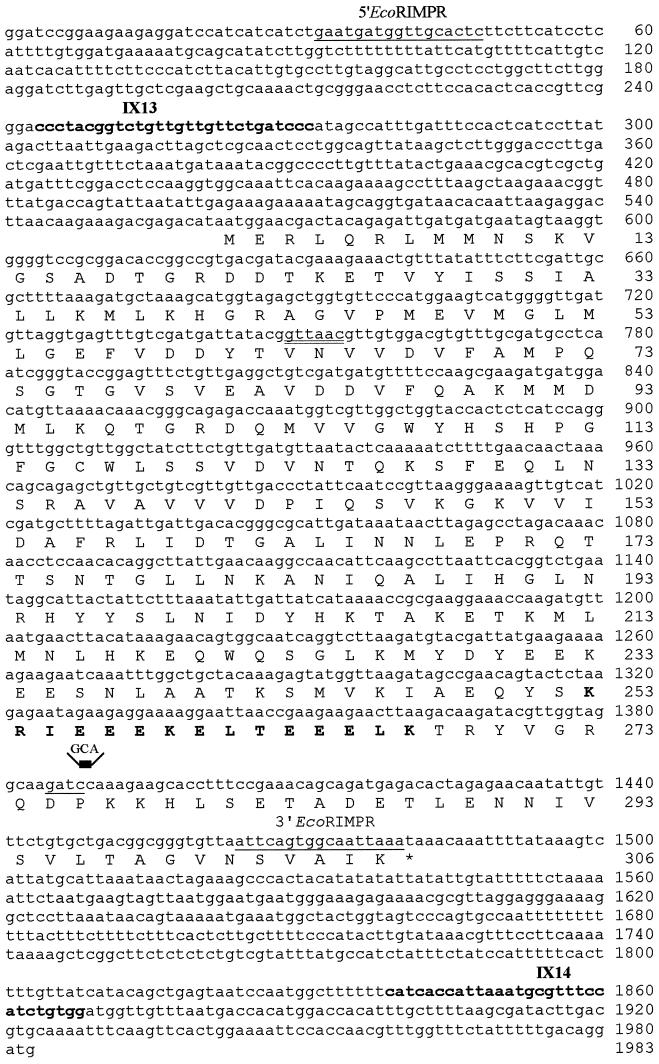
Figure 2.
RPN11/MPR1 sequence (GenBank accession number X79561), its deduced amino acid sequence, and sequence of the mpr1-1 allele. The HpaI site (double underlined) was used to insert the URA3 gene to interrupt the RPN11/MPR1 gene. The oligonucleotides IX13 and IX14 (bold) were used to amplify the mutant allele mpr1-1, which contained a CC to GCA change at nucleotides 1388–1389 (the Sau3A site is underlined); the mutated sequence is indicated above the line. The oligonucleotides 5′EcoRIMPR and 3′EcoRIMPR (underlined) were used to amplify the wild-type allele to fuse RPN11/MPR1 to GFP (see MATERIALS AND METHODS). The amino acid sequence that forms a putative coiled-coils domain is shown in bold type.
Microscopy
For DAPI staining, cells were harvested during the exponential phase on YPD and fixed with 1% formaldehyde for 30 min. DAPI was added at the concentration of 1 μg/ml, and cells were observed by fluorescence microscopy. The vital dye 2-(4-dimetylaminostyryl)-N-methylpyridinium iodide (DASPMI), at the final concentration of 10−6 M, was used to stain the wild-type and mutant strains grown at 24°C on YPD. Cells were then observed by confocal microscopy. Cells containing the MPR1-GFP fusion were cultured in YPD at 24°C and harvested in logarithmic phase. Green fluorescent protein (Gfp) was detected by confocal microscopy, and cells were photographed directly from the culture.
Flow Cytometric Analyses
DNA content distributions were determined after staining with propidium iodide. Propidium iodide fluorescence signal intensities were acquired from a FACStarplus (Becton Dickinson, Mountain View, CA) equipped with an argon ion laser (excitation wavelength, 488 nm; laser power, 200 mW). The sample flow rate during analysis did not exceed 500–600 cells per second. Typically, 40,000 cells were analyzed per sample. Only raw data have been used to prepare the experimental figures.
Protein Analysis
For anti-ubiquitin immunoblot, yeast cell extracts were prepared as follows. Cells were allowed to grow to stationary phase on YPD at 24°C; half of these cultures were shifted at 36°C for 5 h. Cells were than harvested by centrifugation and resuspended in water. Samples were heated 10 min at 95°C; an equal amount of glass beads was added, and cells were broken by vortexing for 30 s six times with intermitted heating (1 min at 95°C). Equal volumes of 4.5% SDS and 2.25 mM EDTA were added, and samples were vortexed and heated for 10 min at 95°C (Gerlinger et al., 1997). After centrifugation, protein concentration was determined, and the same quantity of proteins (40 μg) was loaded on an SDS gel (10%). Ubiquitination of total cellular proteins was assessed by immunoblotting with rabbit anti-ubiquitin antibodies (Sigma).
Growth Curves
Growth curves were performed at 24°C in YPD medium using a stationary inoculum of 104 cells from cultures 2 wk old. Every 2 h cells were counted with the Burker chamber (Fortuna, West Germany). Routinely, viable cell count was determined and found to correspond. The percentage of [rho−] was determined by plating on YP plus glucose, followed by replica plating on YP plus glycerol.
Computer Analysis
Analysis of the Mpr1p was performed with the program COILS (Lupas’s method; Lupas et al., 1991; Lupas, 1996) and with the Protein Sequence Analysis (PSA) server (BioMolecular Engineering Research Center [BMERC], Boston University, Boston, MA) (Stultz et al., 1993; White et al., 1994).
RESULTS
Isolation of RPN11/MPR1 Gene
The mpr1-1 mutation was initially isolated in the diploid strain R117 as a nuclear suppressor of a mitochondrial tRNA mutation (ts932), resulting in defective processing of tRNAAsp (Zennaro et al., 1989; Rinaldi et al., 1994). R117 harbors a nuclear mutation that, in a heterozygous context, can suppress the defect of mitochondrial tRNA processing in ts932 and allows the formation of detectable amounts of mature tRNAAsp (Rinaldi et al., 1994). We then constructed a haploid strain (R117/a12) containing the suppressor allele in a wild-type mitochondrial context and found that it caused a growth defect in rich media containing either glucose or glycerol. This phenotype allowed us to clone the wild-type allele RPN11/MPR1 by complementation. The isolated RPN11/MPR1 gene was confirmed to be allelic to mpr1-1 by standard genetic analysis (see MATERIALS AND METHODS).
As shown in Figure 1, the RPN11/MPR1 gene complemented the thermosensitive phenotype of the mutant strain. RPN11/MPR1 codes for a protein of 306 amino acids (Mpr1p) (Figure 2) (Rinaldi et al., 1995). RPN11/MPR1 sequence (GenBank accession number X79561) was found to be identical to the ORF YFR004W, identified in sequence but not in function by Murakami et al. (1995) in the framework of the European Biotechnology Programme on Yeast Genome Sequencing.
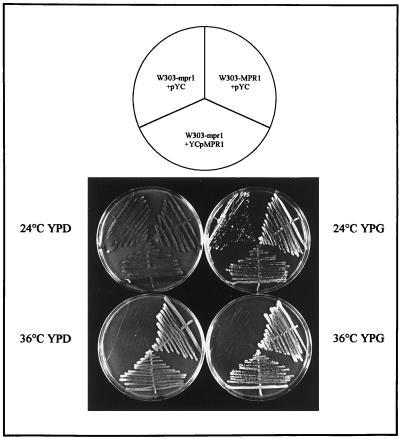
Figure 1.
The RPN11/MPR1 gene complements the thermosensitive phenotype of the mpr1-1 mutant strain. The plasmid YCpMPR1 and the control empty centromeric plasmid pYC (see MATERIALS AND METHODS) were transferred into the wild-type W303-MPR1 and mutant (W303-mpr1) strains. One transformant from each experiment was streaked on YPD and YPG and incubated at 24 and 36°C for 3 d.
The RPN11/MPR1 Gene Is Essential for Viability
To construct RPN11/MPR1-deleted cells, a one-step gene replacement method (Rothstein et al., 1991) was used. The URA3 gene marker was inserted into the coding region of RPN11/MPR1 at the HpaI site (located after amino acid 63) in the plasmid KSpMPR1 (resulting plasmid, pKSmpr1::URA3). This construction was used to transform two different wild-type diploid strains (W303 and FY1679, yielding strains W303/5 and FY1679/1). Ten tetrads from each transformed strain were dissected; only two spores were found to grow for each tetrad, and both were auxotrophic for uracil, indicating that the zpn11/MPR1 gene disruption was lethal. The two nongrowing spores of each tetrad, supposed to contain RPN11/mpr1::URA3 disruption, were examined under the microscope during incubation after dissection; they stopped growing after two to three divisions with elongated buds.
Identification of the Mutation in the mpr1-1 Gene
To determine the nucleotide change in the mpr1-1 allele, two oligonucleotides (IX13 and IX14; see MATERIALS AND METHODS) were used to amplify the mutated form of the gene (Figure 2). Total DNA was extracted from the mutant strain, and two independent PCR products were directly sequenced. The mpr1-1 allele was also cloned from a third independent amplification in the pCRII vector (pCRII-mpr1) and sequenced; in all cases, the same nucleotide changes were found; namely, a dinucleotide CC was replaced by a trinucleotide GCA causing a change of proline 276 to alanine (P276A), followed by a frame shift leading to the production of a truncated protein of 285 amino acids (Figure 2). This change resulted in the loss of the unique Sau3A restriction site in the gene. Although the presence of the wild-type gene RPN11/MPR1 on centromeric or multicopy plasmids complemented the mpr1-1 mutation, a centromeric plasmid bearing the RPN11/MPR1 gene truncated at the Sau3A restriction site was not capable of restoring the correct growth of the mutant. We conclude that the 31 C-terminal amino acids of Mpr1p are necessary for its function in cell growth.
The putative sequence of Mpr1p allowed us to predict a secondary structure of nine repeat strand–turn–helix domains and reveals the presence of a hypothetical “KEKE motif” (Realini et al., 1994). This motif can form a coiled-coils structure in the C-terminal part of the protein (Figure 2, bold type), a feature found in most known regulatory subunits of the proteasome. The mutant form of Mpr1p retains this domain. An other conserved domain (MPN domain), possibly involved in the interaction with the 20S particle, has been identified in the N-terminal part of the protein (Hofmann and Bucher, 1998).
Growth Phenotype Associated with the mpr1-1 Mutation
The growth defect of the original mpr1-1 mutant strain R117/a12 consisted of thermosensitivity (on glucose- or glycerol-containing media) at 36°C and delayed growth at the permissive temperature of 24°C on YP plus glucose. Figure 3 shows the growth curves at 24°C on YPD of isogenic W303-MPR1 wild-type and W303-mpr1 mutant strains. The generation time of the mutant was the same as that of the wild type, and the delayed growth was due to an increased length of the lag phase. The curve was repeated several times, and the length of the lag phase was the same. In contrast, on the same rich medium containing glycerol, at 24°C growth started only after repeated transfers on YPG. The mechanism of this adaptation is presently under study but is not due to selection of revertants.
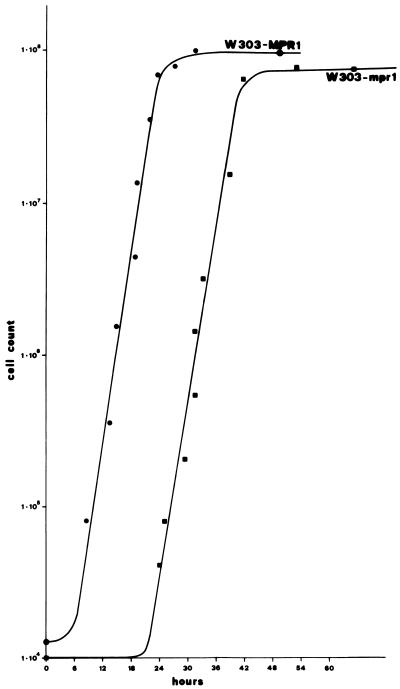
Figure 3.
Growth curves at 24°C on rich glucose-containing medium of the wild-type strain W303-MPR1 and of the mutant strain W303-mpr1. Growth was monitored by counting cells with a Burker chamber, and a viable cell count was found to correspond; the percentage of [zho−] did not vary significantly along the curve. •, W303-MPR1; ▪, W303-mpr1.
Pleiotropic Phenotypes of the mpr1-1 Mutant
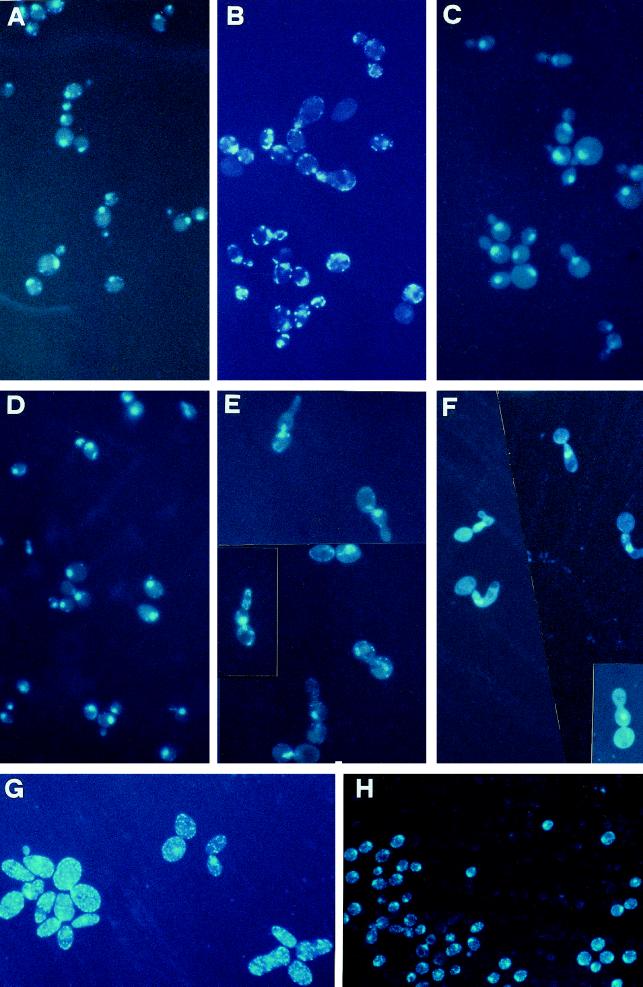
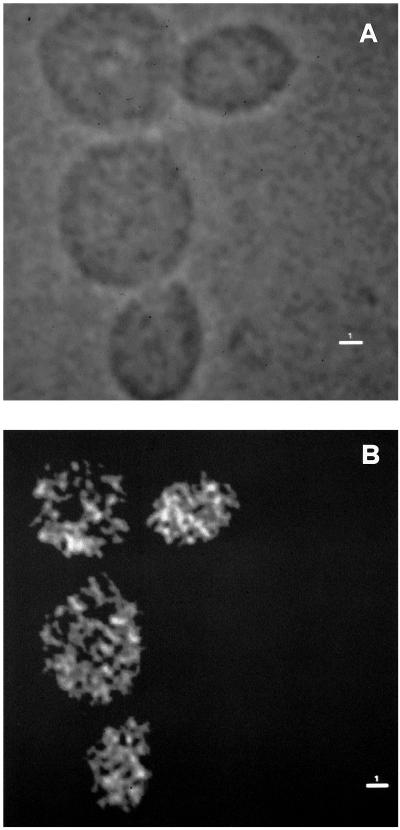
Microscopic observation after DAPI staining of the mutant cells growing at 24°C on YP plus glucose showed that cells were enlarged and a high proportion of the yeast population exhibited very large buds compared with the wild-type strain growing in the same condition (Figure 4, A and B). Moreover, the cytoplasm of mutant cells showed the presence of abundant punctate structures not present in the isogenic wild-type strain growing on YP plus glucose. To see whether they corresponded to mitochondria, we extended the DAPI-staining analysis to a [rho°] mpr1-1 derivative (obtained by ethidium bromide treatment; see MATERIALS AND METHODS). We observed a complete absence of the above-mentioned structures in the [rho°] cells (Figure 4C). We also compared the DAPI-stained mutant and wild-type cells after a shift for 5 h at the nonpermissive temperature. In this condition, mpr1-1 cells exhibited an aberrant morphology characterized by elongated daughter cells unable to perform a further division, whereas the nucleus was mislocalized and often only present in the first bud (Figure 4E). This is better seen in the [rho°] mpr1-1 derivative (Figure 4F). Figure 4G shows the original mutant strain R117/a12 growing on YP plus glucose at 24°C and (Figure 4H) the same strain, transformed with the plasmid YCpMPR1, growing in the same condition and stained with DAPI. In the R117/a12 strain, the multiplicity of the punctate structures is even higher than in the W303-mpr1 derivative.
Figure 4.
Morphological defects of strains containing the mutated allele mpr1-1. Cells were grown on rich glucose-containing medium at 24°C to exponential growth phase and then shifted at 36°C for 5 h and stained with DAPI (see MATERIALS AND METHODS). (A–C) W303-MPR1, W303-mpr1, and the corresponding W303-mpr1-1 [rho°] derivative grown at 24°C and stained with DAPI. (D–F) The same strains after a shift of 5 h at 36°C. (G and H) The original mutant strain R117/a12 and the same strain transformed with the RPN11/MPR1 allele grown at 24°C to exponential phase and stained with DAPI. Magnification was the same in all panels.
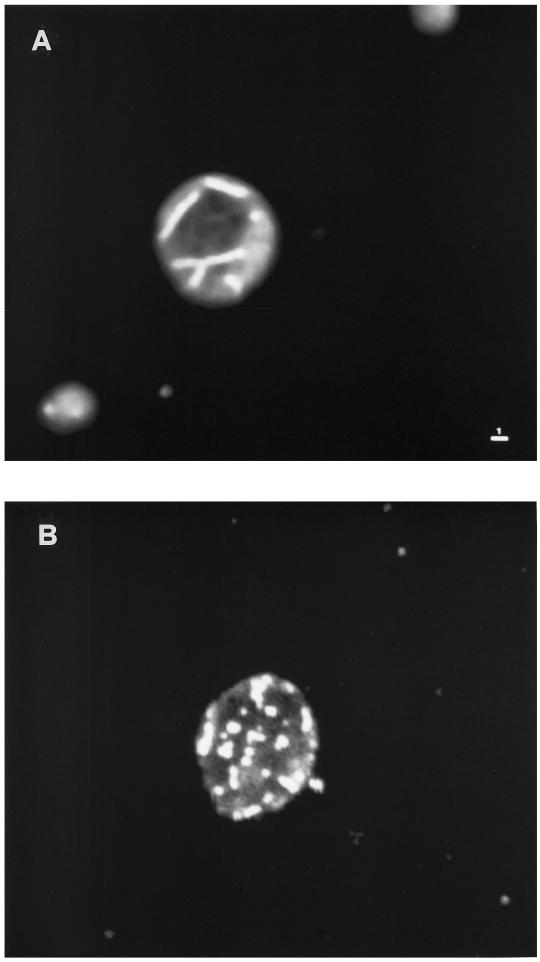
We then used the mitochondrial vital dye DASPMI, which stains functional mitochondrial membranes, to directly visualize the mitochondria. Observation of stained mutant cells by confocal microscopy, with spatial reconstruction of 0.2-μm sections, confirmed the identity of the punctate structures with mitochondria, whereas in the wild-type strain grown at 24°C, thread-like structures were present (Figure 5).
Figure 5.
Mitochondrial morphology of the W303-MPR1 (A) and W303-mpr1 strains (B). Wild-type and mutant strains were stained with DASPMI after growth on rich glucose-containing medium at 24°C and observed by confocal microscopy. The photographs represent the reconstruction of 0.2-μm sections. Bar, 1 μm.
Flow Cytometry Analysis of the mpr1-1 Mutant
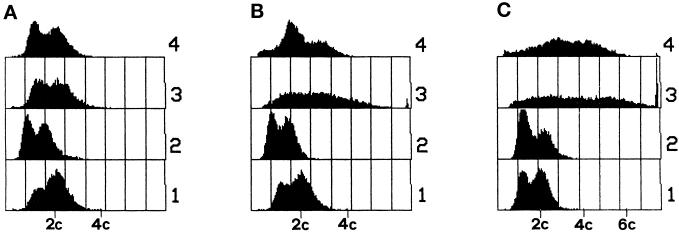
The total DNA content of cells was estimated by flow cytometry after growth on YPD to midexponential phase at 24°C and after a subsequent shift for 3 or 5 h to the nonpermissive temperature. To evaluate the contribution of mitochondrial DNA to the observed profiles, we performed the experiments also in the [rho°] derivatives of the W303-MPR1 and W303-mpr1 strains. Results are reported in Figure 6. Although at 24°C the four flow cytometry profiles were substantially similar except for a slightly lower DNA content in the two [rho°] strains, the shift to 36°C for 5 h produced a dramatic effect in the mpr1-1 strains, with amounts of DNA per cell in a region extending up to >7C in the [rho+] and to 6C in the [rho°] derivative. The same result was obtained with the original mutant R117/a12.
Figure 6.
Quantitative analysis of DNA content by flow cytometry. (A) The wild-type haploid strain W303-MPR1 (1), its [rho°] derivative (2), the haploid W303-mpr1 (3), and its [rho°] derivative (4) were cultured in YPD at 24°C to midexponential phase. Part of the same cultures was split in two parts and shifted at 36°C for 3 h (B) and 5 h (C).
In other words, the increase in DNA content per cell is not restricted to an increase of nuclear DNA but reaches higher values in the [rho+] mpr1-1 strain than in the [rho°] derivative, suggesting an important contribution of mitochondrial DNA to the aberrant flow cytometry profile. This is consistent with the high mitochondrial multiplicity observed by DAPI staining.
Mpr1p Belongs to a Conserved Protein Family
The sequence of Rpn11p/Mpr1p was compared with similar proteins present in databases. Figure 7 shows the alignment of Mpr1p and its similar proteins. Some of these protein sequences had been deduced from cDNA data. The Mpr1p sequence, in particular in the N-terminal part, is well conserved from yeast to human, whereas the C-terminal part, where the mpr1-1 mutation is localized, is less conserved. It is worth noting that Mpr1p exhibits 68.4% identity and 74.7% similarity with the human proteasomal Poh1p protein (Spataro et al., 1997), and, in fact, Glickman et al. (1998) have demonstrated that Rpn11p/Mpr1p is one of the subunits of the proteasomal regulatory particle.
Figure 7.
Alignment of the predicted amino acid sequences of Mpr1p and its homologues (BOXSHADE program, Genetics Computer Group [GCG], Madison, WI). ∼, cDNA sequences not present in databases. The accession numbers are as follows: Caenorhabditis elegans Ypt5p, U00032; S. pombe Pad1p, D31731; S. cerevisiae Mpr1p, X79561; Arabidopsis thaliana cDNA, T43507; Oryza sativa cDNA, D41810; Drosophila melanogaster cDNA, AA141347; Homo sapiens Poh1p, U86782; Mus musculus PAD1, Y13071.
The Schizosaccharomyces pombe Pad1p, which has 64% of identity with Rpn11p/Mpr1p, has been implicated in stress response and drug resistance and reported to be a coactivator of the transcriptional factor Pap1p. We therefore examined some aspects of stress response in the mutant strain. Preliminary data showed that the mutant strain is more sensitive than the wild-type strain to the presence of cadmium and vanadate but not to H2O2 (our unpublished results). This effect is under investigation.
The Homologous Gene HsPOH1 Suppresses the Mutant Phenotype
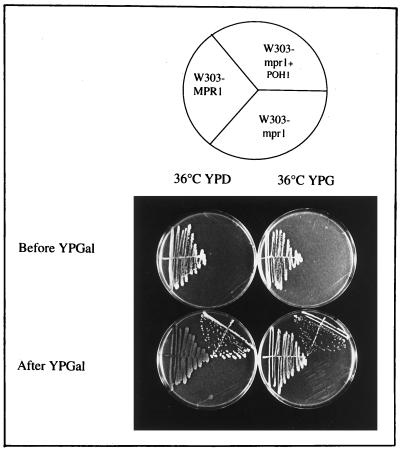
To determine whether the function of RPN11/MPR1 is conserved, we tested the ability of the human gene to complement the mpr1-1 mutation in S. cerevisiae. The plasmid pYES-POH1 was used to transform W303-mpr1. The transformants were tested for growth at 36°C before and after promoter activation with galactose. Figure 8 shows that the POH1 gene suppressed thermosensitivity on glucose and on glycerol.
Figure 8.
The growth defect of the mutant strain mpr1-1 is suppressed by the human gene POH1. The mutant strain mpr1-1 transformed with the human gene POH1 (pYES-POH1) under the GAL1 promoter is unable to grow at 36°C when the GAL1 promoter is repressed; after the induction of the promoter in galactose medium, the same transformed mutant strain grows either on glucose or glycerol media at the nonpermissive temperature. Plates were incubated 4 d at 36°C.
Localization of Mpr1p
We fused the Mpr1p protein with the Gfp from Aequorea victoria to localize the Mpr1p protein (Chalfie et al., 1994; Niedenthal et al., 1996). The construction in which the GFP gene is fused at the 3′ end of RPN11/MPR1 is described in detail in MATERIALS AND METHODS. The mutant W303-mpr1 and the wild-type W303-MPR1 strains were transformed both with centromeric and multicopy plasmids bearing RPN11/MPR1-GFP fusion (YCpMPR1-GFP and YEpMPR1-GFP respectively). The transformed W303-mpr1 mutant strain grew on glucose- and on glycerol-containing media at 36°C, thus indicating that the Mpr1p-Gfp fusion was functional. Transformants were analyzed by confocal microscopy. The localization of Mpr1p in W303-MPR1 is shown in Figure 9: in the tested conditions, the protein was essentially localized in the cytoplasm, but the localization was not uniform, suggesting that the protein might be associated with cytoplasmic structures. No difference was observed between the mutant and wild-type strains transformed with centromeric and multicopy plasmids bearing Mpr1p-Gfp fusion (our unpublished results). To verify that this particular localization was not due to the Gfp alone, we observed the localization of the Gfp expressed in the W303-MPR1 strain. The p100GFP plasmid, containing the GFP gene under the control of a galactose-inducible yeast promoter (see MATERIALS AND METHODS), was used to transform the W303-MPR1 strain. After the induction of the promoter with galactose, transformants were observed by confocal microscopy, and the localization of Gfp was uniform in the cytoplasm, showing that the observed result is not obtained with the Gfp alone.
Figure 9.
Localization of the fusion protein Mpr1p-Gfp in wild-type cells detected by confocal microscopy. The haploid wild-type strain W303-MPR1 transformed with YCpMPR1-GFP was grown in rich medium at 24°C to exponential growth phase. (A) Phase image; (B) Gfp fluorescence. Bars, 1 μm.
The Mutant Strain Accumulates Ubiquitinated Proteins
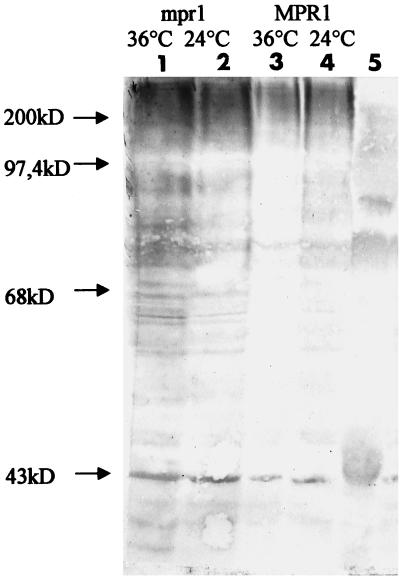
Yeast strains with functionally attenuated proteasomes accumulate ubiquitinated proteins (Ghislain et al., 1993; Gordon et al., 1993; Yokota et al., 1996; Kominami et al., 1997). We examined whether the mpr1-1 strain presented the same accumulation as described for the mutated form of other proteasomal subunits. Total proteins were extracted from the wild-type and mutant strains, both grown at 24°C to stationary phase; these strains were also shifted 5 h at the nonpermissive temperature of 36°C. As shown in Figure 10, accumulation of ubiquitinated proteins after the shift at the nonpermissive temperature in the mpr1-1 strain can indeed be observed.
Figure 10.
The mpr1-1 mutant strain accumulates ubiquitinated proteins. The wild-type strain containing RPN11/MPR1 and the mutant strain containing mpr1-1 were grown at 24°C to stationary phase, after which one-half of the culture was shifted for 5 h to 36°C. Proteins were then extracted, electrophoresed, and detected as indicated in MATERIALS AND METHODS. Lines 1 and 2, ubiquitinated proteins from the W303-mpr1 strain before (2) and after (1) incubation at 36°C. Lines 2 and 3, the same from the W303-MPR1 strain. Line 5, Protein molecular weight standards.
DISCUSSION
RPN11/MPR1 Encodes a Proteasomal Subunit
Our results are fully consistent with those recently reported by Glickman et al. (1998) showing that the sequence of the proteasomal protein Rpn11p corresponds to the deduced sequence of MPR1 (Rinaldi et al., 1995). In particular the identification of Mpr1p with Rpn11p is consistent with the similarity of the cell cycle defects to those observed in mutants of other proteasomal subunits such as Nin1p, Sug2/Pcs1p, Cim3p/Sug1p, and Cim5p (Ghislain et al., 1993; Kominami et al., 1995; McDonald and Byers, 1997) and by the pleiotropy of the defects observed in the mpr1-1 mutant, including the accumulation of ubiquitinated proteins. Moreover, the mpr1-1 mutation is complemented by human proteasomal subunit Poh1p.
RPN11/MPR1 is an essential gene, as are most previously studied 19S proteasomal genes. The mpr1-1 mutation is a missense mutation followed by a frame shift producing premature termination. Hence the mutated protein is altered and truncated in its C-terminal part, which has been shown to be necessary to rescue the growth defect.
Pleiotropic Effects of the mpr1-1 Mutation
The main phenotypes of the mpr1-1 mutant can be summarized as follows: 1) Microscopic analysis of DAPI-stained cells grown on YP plus glucose at the permissive temperature shows enlarged cells, mostly bearing large buds and containing multiple punctate mitochondrial structures. 2) In YP medium containing glucose, the growth rate at 24°C is not decreased compared with the wild type but only delayed by a substantially longer lag phase. One might think that some factor(s) accumulated in the stationary phase must be proteolytically degraded before growth starts, or, alternatively, that a factor abnormally degraded during the stationary phase must be newly synthesized. 3) Growth on the same medium containing glycerol is much more severely impaired even at the permissive temperature. 4) At 36°C, no growth is observed, and flow cytometry profiles show that after a shift to this temperature, the DNA content per cell is strongly increased in the [rho+] cells and, to a lesser extent, in the [rho°] cells, implying an overreplication of both nuclear and mitochondrial DNA. Microscopic analysis after the shift to 36°C shows aberrant morphologies with elongated buds. Only one nucleus seems to be present, and it is often localized in the first bud. 5) The mpr1-1 mutant accumulates ubiquitinated proteins in the stationary phase of growth at 24°C and after a shift to 36°C.
Although some of these phenotypes are caused by other proteasomal mutations, the close connection between cell cycle defects and the mitochondrial phenotype had not been observed before.
Only in one other case has a relationship been observed between a proteasomal mutation and a mitochondrial phenotype: a mutation in the YNT1 gene, coding for a proteasomal subunit, suppresses the mitochondrial defects (among which is a punctate mitochondrial morphology) caused by a mutated allele of YME1, a nuclear gene coding for a mitochondrial zinc-dependent protease (Thorsness et al., 1993; Campbell et al., 1994).
Mitochondria contain their own protein degradation system (possibly involved in proofreading of mitochondrial protein synthesis products), and a relationship between the two proteolytic systems might actually exist (Rep and Grivell, 1996; Suzuki et al., 1997).
Mitochondrial Morphology
Mitochondrial morphology in S. cerevisiae is highly variable depending on growth and physiological conditions. For example, a shift from fermentation to respiration conditions results in a change from tubular to punctate structure (Visser et al., 1995). During mating, mitochondria form a single dynamic network with a number of fission and fusion events (Nunnari et al., 1997). During meiosis, mitochondria are seen as highly organized thread-like structures, probably required for transmission to the four spores (Smith et al., 1995).
In the mpr1-1 mutant, this dynamic situation seems to be affected, because we do not observe the mitochondrial tubular structure usually observed in cells growing in YP plus glucose (Figure 5). The punctate mitochondrial morphology we observe in the mpr1-1 mutant growing on rich medium containing glucose has also been observed in the yme1 mutant altered in a mitochondrial protease. Analogous “disorganization” of the mitochondrial tubular network has also been seen in different mutants. Mutations in the actin-encoding ACT1 gene result in an altered mitochondrial morphology and movement during sporulation (Smith et al., 1995). Similar observations have been reported for mutants of the MDM10 and MMM1 genes, which encode mitochondrial proteins involved in mitochondrial inheritance during mitosis (Burgess et al., 1994; Sogo and Yaffe, 1994). In all these cases, round mitochondria are observed instead of tubular structures (Fisk and Yaffe, 1997).
Cell Cycle Progression in the mpr1-1 Mutant
The results of flow cytometry indicate that, after the shift to 36°C, a high proportion of the population has a very high DNA content and that both nuclear and mitochondrial DNA are involved in this increase. This is consistent with the results of DAPI staining. Several genes have been implicated in restricting DNA replication to once per cell cycle. Among these, cdc16 and cdc27 mutants have been shown to overreplicate DNA by multiple rounds of replication, without completing the cell cycle, but no effect of the [rho°] condition had been detected (Heichman and Roberts, 1996). Similar observations have been carried out by Moreno and Nurse (1994) in S. pombe.
In the mpr1-1 [rho°] mutant, we do not observe multiple rounds of replication of nuclear DNA but, rather, a situation that is more reminiscent of the doa4 mutant phenotype; in fact, highly increased amounts of nuclear DNA and similar flow cytometry profiles have been observed in mutants of the DOA4 gene, which codes for a deubiquitinating enzyme bearing similarities to the human tre-2 oncogene (Papa and Hochstrasser, 1993; Singer et al., 1996).
In addition to this overreplication of nuclear DNA, the mpr1-1 mutant also presents increased amounts of mitochondrial DNA. This increase might explain the suppression of the mitochondrial tRNA-processing defect initially observed in the mitochondrial mutant ts932.
Homology with Genes Involved in AP-1-dependent Transcription
Another aspect worth noting is the high degree of similarity (64.6% identity) of Mpr1p with Pad1p of S. pombe, which has been reported to act as a positive regulator of Pap1p-dependent transcription. Pap1p belongs to the AP-1 protein family, which controls the expression of genes involved in drug resistance and stress response (Kim and Struhl, 1995). It may be interesting to note that the PAD1 gene, isolated in a high-copy-number plasmid conferring pleiotropic drug resistance in fission yeast, is truncated (Shimanuki et al., 1995). This truncated form of PAD1 stops at the Sau3A site, where we identified the mpr1-1 mutation. Therefore, one could envisage that the C-terminal part of the protein might not be necessary for drug resistance.
On the other hand, the mutations present in the mpr1-1 allele are localized downstream of the above-mentioned site in the C-terminal region of the gene, which we have shown to be necessary for the suppression of the growth defect present in the mpr1-1 mutant. If the high similarity between Mpr1p and Pad1p is to be taken into account, this might suggest that different regions of the protein might be involved in the effects on growth and stress response. The possible involvement of Mpr1p in stress response has yet to be thoroughly investigated.
Localization of Mpr1p
The cytoplasmic localization of the Mpr1p-Gfp fusion is somewhat surprising, because most proteasomal proteins have been shown to have a nuclear localization (Nelson et al., 1993; McDonald and Byers, 1997). In our experiment no nuclear localization was evident, even if a presence of the protein in the nucleus cannot be ruled out. This is similar to the case of the proteasomal subunit Nin1p, which has been found to be essentially present in the cytoplasm in the form of dotted structures (Kominami and Toh-e, 1994).
Conclusion
Rpn11p/Mpr1p is a proteasomal regulatory protein that belongs to a highly conserved family. The mpr1-1 mutation is the first reported case of a mutation in this gene family. Its highly pleiotropic phenotype reflects multiple functions, including mitochondrial ones. This indicates a possible involvement of the proteasome in mitochondrial biogenesis. The mpr1-1 mutation will thus provide a unique opportunity to investigate this new aspect of yeast cell biology.
ACKNOWLEDGMENTS
We are deeply indebted to Andrea Onetti for confocal microscopy. Thanks are due to Hiroshi Fukuhara for helpful discussions and to Daniel Finley for critical reading of the manuscript. We are also indebted to Dr. Martin Chalfie for providing the GFP gene. We also thank Francesco Castelli for skillful technical assistance. This work was supported by the Commission of European Community, Human Capital and Mobility contract ERB CHRX-CT94-0520, and by Ministero Università e Ricerca Scientifica e Tecnologica-University La Sapienza Cofin 1997.
REFERENCES
- Amsterdam A, Pitzer F, Baumeister W. Changes in intracellular localization of proteasomes in immortalized ovarian granulosa cells during mitosis associated with a role in cell cycle control. Proc Natl Acad Sci USA. 1993;90:99–103. doi: 10.1073/pnas.90.1.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldari C, Murray JA, Ghiara P, Cesareni G, Galeotti CL. A novel leader peptide which allows efficient secretion of a fragment of human interleukin 1 beta in Saccharomyces cerevisiae. EMBO J. 1987;6:229–234. doi: 10.1002/j.1460-2075.1987.tb04743.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgess SM, Delannoy M, Jensen RE. MMM1 encodes a mitochondrial outer membrane protein essential for establishing and maintaining the structure of yeast mitochondria. J Cell Biol. 1994;126:1375–1391. doi: 10.1083/jcb.126.6.1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell CL, Tanaka N, White KH, Thorsness PE. Mitochondrial morphological and functional defects in yeast caused by yme1 are suppressed by mutation of a 26S protease subunit homologue. Mol Biol Cell. 1994;5:899–905. doi: 10.1091/mbc.5.8.899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalfie M, Tu Y, Euskirchen G, Ward WW, Prasher DC. Green fluorescent protein as a marker for gene expression. Science. 1994;263:802–805. doi: 10.1126/science.8303295. [DOI] [PubMed] [Google Scholar]
- Dawson SP, Arnold JE, Mayer NJ, Reynolds SE, Billett MA, Gordon C, Colleaux L, Kloetzel PM, Tanaka K, Mayer RJ. Developmental changes of the 26S proteasome in abdominal intersegmental muscles of Manduca sexta. J Biol Chem. 1995;270:1850–1858. doi: 10.1074/jbc.270.4.1850. [DOI] [PubMed] [Google Scholar]
- Fisk HA, Yaffe MP. Mutational analysis of Mdm1p function in nuclear and mitochondrial inheritance. J Cell Biol. 1997;138:485–494. doi: 10.1083/jcb.138.3.485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerlinger UM, Guckel R, Hoffmann M, Wolf DH, Hilt W. Yeast cycloheximide-resistant crl mutants defective in protein degradation. Mol Biol Cell. 1997;8:2487–2477. doi: 10.1091/mbc.8.12.2487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghislain M, Udvardy A, Mann C. S. cerevisiae 26S protease mutants arrest cell division in G2/metaphase. Nature. 1993;366:358–362. doi: 10.1038/366358a0. [DOI] [PubMed] [Google Scholar]
- Gietz RD, Sugino A. New yeast-Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six-base pair restriction sites. Gene. 1988;74:527–534. doi: 10.1016/0378-1119(88)90185-0. [DOI] [PubMed] [Google Scholar]
- Glickman MH, Rubin DM, Fried VA, Finley D. The regulatory particle of the Saccharomyces cerevisiae proteasome. Mol Cell Biol. 1998;18:3149–3162. doi: 10.1128/mcb.18.6.3149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon C, McGurk G, Dillon P, Rosen C, Hastie D. Defective mitosis due to a mutation in the gene for a fission yeast 26S protease subunit. Nature. 1993;366:355–357. doi: 10.1038/366355a0. [DOI] [PubMed] [Google Scholar]
- Groll M, Ditzel L, Lowe J, Stock D, Bochtler M, Bartunik HD. Structure of 20S proteasome from yeast at 2,4A resolution. Nature. 1997;386:463–471. doi: 10.1038/386463a0. [DOI] [PubMed] [Google Scholar]
- Heichman KA, Roberts JM. The yeast CDC16 and CDC27 genes restrict DNA replication to once per cell cycle. Cell. 1996;85:39–48. doi: 10.1016/s0092-8674(00)81080-6. [DOI] [PubMed] [Google Scholar]
- Hilt W, Wolf DH. Proteasomes: destruction as a programme. Trends Biochem Sci. 1996;21:96–102. [PubMed] [Google Scholar]
- Hochstrasser M. Ubiquitin, proteasomes, and the regulation of intracellular protein degradation. Curr Opin Cell Biol. 1995;7:215–223. doi: 10.1016/0955-0674(95)80031-x. [DOI] [PubMed] [Google Scholar]
- Hofmann K, Bucher P. The PCI domain: a common theme in three multiprotein complexes. Trends Biochem Sci. 1998;23:204–205. doi: 10.1016/s0968-0004(98)01217-1. [DOI] [PubMed] [Google Scholar]
- Ito H, Fukuda Y, Murata K, Kimura A. Trasformation of intact yeast cells treated with alkali cations. J Bacteriol. 1983;153:163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Struhl K. Determinants of half-site spacing preferences that distinguish AP-1 and ATF/CREB bZIP domains. Nucleic Acids Res. 1995;23:2531–2537. doi: 10.1093/nar/23.13.2531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King RW, Deshaies RJ, Peters JM, Kirschner MW. How proteolysis drives the cell cycle. Science. 1996;274:1652–1658. doi: 10.1126/science.274.5293.1652. [DOI] [PubMed] [Google Scholar]
- Kominami K, et al. Nin1p, a regulatory subunit of the 26S proteasome, is necessary for activation of Cdc28p kinase of Saccharomyces cerevisiae. EMBO J. 1995;14:3105–3115. doi: 10.1002/j.1460-2075.1995.tb07313.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kominami K, et al. Yeast counterparts of subunits S5a and p58 (S3) of the human 26S proteasome are encoded by two multicopy suppressors of nin1-1. Mol Biol Cell. 1997;8:171–187. doi: 10.1091/mbc.8.1.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kominami K, Toh-e A. Characterization of the function of the NIN1 gene product of Saccharomyces cerevisiae. Exp Cell Res. 1994;211:203–211. doi: 10.1006/excr.1994.1079. [DOI] [PubMed] [Google Scholar]
- Lupas A. Coiled coils: new structures and new functions. Trends Biochem Sci. 1996;21:375–382. [PubMed] [Google Scholar]
- Lupas A, VanDyke M, Stock J. Predicting coiled coils from protein sequences. Science. 1991;252:1162–1164. doi: 10.1126/science.252.5009.1162. [DOI] [PubMed] [Google Scholar]
- McDonald HB, Byers B. A proteasome cap subunit required for spindle pole body duplication in yeast. J Cell Biol. 1997;137:539–553. doi: 10.1083/jcb.137.3.539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno S, Nurse P. Regulation of progression through the G1 phase of the cell cycle by the rum1+ gene. Nature. 1994;367:236–242. doi: 10.1038/367236a0. [DOI] [PubMed] [Google Scholar]
- Murakami Y, et al. Analysis of the nucleotide sequence of chromosome VI from Saccharomyces cerevisiae. Nat Genet. 1995;10:261–268. doi: 10.1038/ng0795-261. [DOI] [PubMed] [Google Scholar]
- Murray A. Cyclin ubiquitination: the destructive end of mitosis. Cell. 1995;81:149–152. doi: 10.1016/0092-8674(95)90322-4. [DOI] [PubMed] [Google Scholar]
- Nelson MK, Kurihara T, Silver PA. Extragenic suppressor of mutations in the cytoplasmic C terminus of SEC63 define five genes in Saccharomyces cerevisiae. Genetics. 1993;134:159–173. doi: 10.1093/genetics/134.1.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niedenthal RK, Riles L, Johnston M, Hegemann JH. Green fluorescent protein as marker for gene expression and subcellular localization in budding yeast. Yeast. 1996;12:773–786. doi: 10.1002/(SICI)1097-0061(19960630)12:8%3C773::AID-YEA972%3E3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- Nunnari J, Marshall WF, Straight A, Murray A, Sedat JW, Walter P. Mitochondrial transmission during mating in Saccharomyces cerevisiae is determined by mitochondrial fusion and fission and the intramitochondrial segregation of mitochondrial DNA. Mol Biol Cell. 1997;8:1233–1242. doi: 10.1091/mbc.8.7.1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nurse P. Universal control mechanism regulation onset of M-phase. Nature. 1990;344:503–508. doi: 10.1038/344503a0. [DOI] [PubMed] [Google Scholar]
- Papa FR, Hochstrasser M. The yeast DOA4 gene encodes a deubiquitinating enzyme related to a product of the human tre-2 oncogene. Nature. 1993;366:313–319. doi: 10.1038/366313a0. [DOI] [PubMed] [Google Scholar]
- Peters JM. Proteasomes: protein degradation machines of the cell. Trends Biochem Sci. 1994;19:377–382. doi: 10.1016/0968-0004(94)90115-5. [DOI] [PubMed] [Google Scholar]
- Peters JM, Franke WW, Kleinschmidt JA. Distinct 19S and 20S subcomplexes of the 26S proteasome and their distribution in the nucleus and the cytoplasm. J Biol Chem. 1994;269:7709–7718. [PubMed] [Google Scholar]
- Realini C, Rogers SW, Rechsteiner M. Proposed roles in protein-protein association and presentation of peptides by MHC Class I receptor. FEBS Lett. 1994;348:109–113. doi: 10.1016/0014-5793(94)00569-9. [DOI] [PubMed] [Google Scholar]
- Rechsteiner M, Hoffman L, Dubiel W. The multicatalytic and 26S proteases. J Biol Chem. 1993;268:6065–6068. [PubMed] [Google Scholar]
- Rep M, Grivell LA. The role of protein degradation in mitochondrial function and biogenesis. Curr Genet. 1996;30:367–380. doi: 10.1007/s002940050145. [DOI] [PubMed] [Google Scholar]
- Rinaldi T, Bolotin-Fukuhara M, Frontali L. A Saccharomyces cerevisiae gene essential for viability has been conserved in evolution. Gene. 1995;160:135–136. doi: 10.1016/0378-1119(95)00212-o. [DOI] [PubMed] [Google Scholar]
- Rinaldi T, Francisci S, Zennaro E, Frontali L, Bolotin-Fukuhara M. Suppression of a mitochondrial point mutation in a tRNA gene can cast light on the mechanisms of 3′ end-processing. Curr Genet. 1994;25:451–455. doi: 10.1007/BF00351785. [DOI] [PubMed] [Google Scholar]
- Rivett AJ. Proteasomes: multicatalytic proteinase complexes. Biochem J. 1993;291:1–10. doi: 10.1042/bj2910001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose MD, Novick P, Thomas JH, Botstein D, Fink GR. A Saccharomyces cerevisiae genomic plasmid bank based on a centromere-containing shuttle vector. Gene. 1987;60:237–243. doi: 10.1016/0378-1119(87)90232-0. [DOI] [PubMed] [Google Scholar]
- Rothstein R. Targeting, disruption, replacement and allele rescue: integrative DNA transformation in yeast. Methods Enzymol. 1991;194:281–301. doi: 10.1016/0076-6879(91)94022-5. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd ed. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1995. [Google Scholar]
- Shimanuki M, Saka Y, Yanagida M, Toda T. A novel essential fission yeast gene pad1+ positively regulates pap1+-dependent transcription and is implicated in the maintenance of chromosome structure. J Cell Sci. 1995;108:569–579. doi: 10.1242/jcs.108.2.569. [DOI] [PubMed] [Google Scholar]
- Singer JD, Manning BM, Formosa T. Coordinating DNA replication to produce one copy of the genome requires genes that act in ubiquitin metabolism. Mol Cell Biol. 1996;16:1356–1366. doi: 10.1128/mcb.16.4.1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith MG, Simon VR, O’Sullivan H, Pon LA. Organelle-cytoskeletal interactions: actin mutations inhibit meiosis-dependent mitochondrial rearrangements in the budding yeast. Mol Biol Cell. 1995;6:1381–1396. doi: 10.1091/mbc.6.10.1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sogo LF, Yaffe MP. Regulation of mitochondrial morphology and inheritance by Mdm10p, a protein of mitochondrial outer membrane. J Cell Biol. 1994;126:1361–1373. doi: 10.1083/jcb.126.6.1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spataro V, Toda T, Craig R, Seeger M, Dubiel W, Harris AL, Norbury C. Resistance to diverse drugs and ultraviolet light conferred by overexpression of a novel human 26S proteasome subunit. J Biol Chem. 1997;272:30470–30475. doi: 10.1074/jbc.272.48.30470. [DOI] [PubMed] [Google Scholar]
- Stultz CM, White JV, Smith TF. Structural analysis based on state-space modeling. Protein Sci. 1993;2:305–314. doi: 10.1002/pro.5560020302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki CK, Rep M, van Dijl JM, Suda K, Grivell LA, Schatz G. ATP-dependent proteases that also chaperone protein biogenesis. Trends Biochem Sci. 1997;22:118–123. doi: 10.1016/s0968-0004(97)01020-7. [DOI] [PubMed] [Google Scholar]
- Thorsness PE, White KH, Fox TD. Inactivation of YME1, a member of the ftsH-sec18-PAS1-CDC48 family of putative ATPase-encoding genes, causes increased escape of DNA from mitochondria in Saccharomyces cerevisiae. Mol Cell Biol. 1993;13:5418–5426. doi: 10.1128/mcb.13.9.5418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visser W, van Spronsen EA, Nanninga N, Pronk JT, Kuenen JG, van Dijken JP. Effects of growth conditions on mitochondrial morphology in Saccharomyces cerevisiae. Anthonie van Leeuwenhoek. 1995;67:243–253. doi: 10.1007/BF00873688. [DOI] [PubMed] [Google Scholar]
- White JV, Stultz CM, Smith TF. Protein classification by stochastic modeling on optimal filtering of amino-acid sequences. Math Biosci. 1994;119:35–75. doi: 10.1016/0025-5564(94)90004-3. [DOI] [PubMed] [Google Scholar]
- Winston F, Dollard C, Ricupero-Hovasse SL. Construction of a set of convenient Saccharomyces cerevisiae strains that are isogenic to S288C. Yeast. 1995;11:53–55. doi: 10.1002/yea.320110107. [DOI] [PubMed] [Google Scholar]
- Yokota K, et al. cDNA cloning of p112, the largest regulatory subunit of the human 26S proteasome, and functional analysis of its yeast homologue, Sen3p. Mol Biol Cell. 1996;7:853–870. doi: 10.1091/mbc.7.6.853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zennaro E, Francisci S, Ragnini A, Frontali L, Bolotin-Fukuhara M. A point mutation in a mitochondrial tRNA gene abolishes its 3′ end processing. Nucleic Acids Res. 1989;17:5751–5764. doi: 10.1093/nar/17.14.5751. [DOI] [PMC free article] [PubMed] [Google Scholar]