Abstract
Leptospira interrogans differs from other spirochetes in that it contains homologs of all the Escherichia coli lpx genes required for the biosynthesis of the lipid A anchor of lipopolysaccharide (LPS). LPS from L. interrogans cells is unusual in that it activates TLR2 rather than TLR4. The structure of L. interrogans lipid A has now been determined by a combination of matrix-assisted laser desorption ionization time-of-flight mass spectrometry, NMR spectroscopy, and biochemical studies. Lipid A was released from LPS of L. interrogans serovar Pomona by 100 °C hydrolysis at pH 4.5 in the presence of SDS. Following purification by anion exchange and thin layer chromatography, the major component was shown to have a molecular weight of 1727. Mild hydrolysis with dilute NaOH reduced this to 1338, consistent with the presence of four N-linked and two O-linked acyl chains. The lipid A molecules of both the virulent and nonvirulent forms of L. interrogans serovar Icterohaemorrhagiae (strain Verdun) were identical to those of L. interrogans Pomona by the above criteria. Given the selectivity of L. interrogans LpxA for 3-hydroxylaurate, we propose that L. interrogans lipid A is acylated with R-3-hydroxylaurate at positions 3 and 3′ and with R-3-hydroxypalmitate at positions 2 and 2′. The hydroxyacyl chain composition was validated by gas chromatography and mass spectrometry of fatty acid methyl esters. Intact hexa-acylated lipid A of L. interrogans Pomona was also analyzed by NMR, confirming the presence a β-1′,6-linked disaccharide of 2,3-diamino-2,3-dideoxy-D-glucopyranose units. Two secondary unsaturated acyl chains are attached to the distal residue. The 1-position of the disaccharide is derivatized with an axial phosphate moiety, but the 4′-OH is unsubstituted. 1H and 31P NMR analyses revealed that the 1-phosphate group is methylated. Purified L. interrogans lipid A is inactive against human THP-1 cells but does stimulate tumor necrosis factor production by mouse RAW264.7 cells.
Nearly all of the diverse eubacteria that are enclosed by two membranes synthesize lipid A as the hydrophobic anchor of their outer membrane lipopolysaccharide (LPS)1 (1, 2). Several spirochetes of clinical importance, such as Treponema pallidum, Treponema denticola, and Borrelia burgdorferi, possess an outer membrane but do not make LPS (3–5). Accordingly, they lack the lpx genes (6, 7), which are required for lipid A assembly in Escherichia coli and other Gram-negative organisms (2). The absence of lipid A in the outer membranes of T. pallidum, T. denticola, and B. burgdorferi may be compensated for by alternative lipids (8), lipoproteins (9), or other complex glycoconjugates (4). Whatever the explanation, spirochetes lacking LPS are not easily cultivated outside of their hosts (10, 11).
Disease-causing serovars of Leptospira interrogans spp. are members of a distinct spirochete group (12, 13) that can survive or proliferate either in a mammal or in the environment, typically in fresh water contaminated by the urine of infected animals (14). L. interrogans causes a hemorrhagic fever in humans known as Weil’s disease, which may be fatal in untreated cases because of liver, kidney, or pulmonary damage (14, 15). Early biochemical, serologic, and genetic studies showed that LPS is present in Leptospira, but its covalent structure has not been characterized (12, 16–20). However, numerous studies have shown that leptospiral LPS possesses much lower endotoxic activity than typical Gram-negative LPS (21). Werts et al. (22) found that highly purified L. interrogans LPS is unusual because it activates TLR2 rather than TLR4. The latter is the classical signaling receptor of the innate immune system that detects the lipid A moiety of most other Gram-negative LPSs (23–26).
The recently completed sequencing of the L. interrogans serovar Lai genome strongly supports the idea that these spirochetes synthesize lipid A and LPS, because the genome encodes a complete set of Lpx orthologs and LPS-related glycosyl transferases (13). Likewise, the earlier studies of Adler and coworkers demonstrated the existence of typical O-antigen gene clusters in various strains of L. interrogans, indicating that some form of LPS must be present (12, 27).
Given the unusual bioactivity of L. interrogans LPS toward TLR2 (22) and the lack of structural studies, we now report methods for the purification and characterization of L. interrogans lipid A. A combination of mass spectrometry, NMR spectroscopy, bioinformatics, and enzymology was used to show that L. interrogans makes lipid A molecules in which the usual glucosamine units are replaced with the analog 2,3-diamino-2,3-dideoxy-D-glucopyranose (GlcN3N). As anticipated from studies with Acidithiobacillus ferrooxidans, described in the preceding manuscripts (28, 29), the L. interrogans genome (13) contains significant full-length orthologs of the enzymes GnnA and GnnB. These proteins are needed to convert UDP-GlcNAc to UDP 2-acetamido-3-amino-2,3-dideoxy-α-D-glucopyranose (UDP-GlcNAc3N), a novel sugar nucleotide that is the key precursor of lipid A molecules containing GlcN3N units (28, 29). The lipid A of L. interrogans contains a novel 1-phosphate residue that is capped with a methyl group. Methylated phosphate residues are uncommon in biology (30–32) and without precedent in lipid A biochemistry (2, 33). L. interrogans lipid A lacks a 4′-phosphate moiety. Two unsaturated ester-linked secondary acyl chains are present on the distal unit. L. interrogans lipid A is inactive in the limulus lysate assay and against human THP-1 cells, but it does stimulate mouse macrophage tumor cells with about one-tenth the potency of E. coli lipid A. Our purification and proposed structure for L. interrogans lipid A should facilitate further pharmacological studies. A preliminary communication of our structure has appeared in abstract form (34).
EXPERIMENTAL PROCEDURES
Materials
Glass-backed 0.25-mm silica gel 60 TLC plates were obtained from Merck. Chloroform, ammonium acetate, and sodium acetate were from EM Sciences, whereas pyridine, methanol, and formic acid were from Mallinckrodt. Deuterated solvents (CD3OD, CDCl3 containing 0.1% tetramethylsilane, and D2O) and 5-mm NMR tubes were from Sigma-Aldrich. Triton X-100 and the bicinchoninic assay kit were from Pierce.
Bacterial Strains
L. interrogans serovar Pomona (strain L170) (12, 19) was grown in a special medium with added pyruvate at 30 °C (35). The cells from stationary phase cultures were harvested and lyophilized. L. interrogans serovar Icterohaemorrhagiae (strain Verdun) was from the Centre de Reference des Leptospires (Institut Pasteur, Paris, France). Both the virulent and the avirulent variants of the Verdun strain were cultured as described previously (22). The bacteria were grown in EMJH medium at 30 °C under aerobic conditions to a density of ~5 × 108 cells/ml (36, 37). The preliminary sequence data for T. denticola were obtained from the Baylor College of Medicine (www.hgsc.bcm.tmc.edu).
Purification of LPS
The method described by Westphal and Jann (38) was used to extract LPS from lyophilized cells of L. interrogans serovar Pomona (5 g dry weight) (19). LPS from L. interrogans serovar Icterohaemorrhagiae (strain Verdun) was extracted using a modification of the hot phenol-water method (22, 38). Typically, 1 mg of LPS was recovered from 1012 bacteria. The LPS in the phenol phase was subjected to extensive dialysis against hot water (70 °C). The dialyzed material was clarified twice by low speed centrifugation (3000 × g for 15 min at 10 °C), and the LPS was collected by ultracentrifugation (3 h at 100,000 × g at 10 °C). The pellet was resuspended in endotoxin-free water. The ultracentrifugation step was repeated two or three times until the resuspended LPS showed no absorbance at 260 and 280 nm, after which it was lyophilized and weighed. Because phenol-extracted LPS may retain some impurities that result in TLR2-dependent activation of cells, LPS preparations intended for biological studies are further purified using a procedure that removes LPS-associated proteins (39–41). However, these steps were omitted for the LPS used in the present work.
Purification of Lipid A Released from L. interrogans LPS
Lipid A was released from the L. interrogans LPS by 100 °C hydrolysis at pH 4.5 in the presence of SDS (42) as described previously, followed by Bligh-Dyer extraction (43). In a typical preparation, 150 mg of crude LPS was resuspended in 20 ml of 12.5 mM sodium acetate, pH 4.5, containing 1% SDS in a 150-ml Corex glass centrifuge bottle. The mixture was then placed in a boiling water bath for 30 min (42). The crude lipid A was extracted and then purified on a 2-ml DEAE-cellulose column (Whatman DE-52), equilibrated in the acetate form in CHCl3/MeOH/H2O (2:3:1, v/v/v) (42, 44–47). The entire lipid A sample was dissolved in 9 ml of CHCl3/MeOH/H2O (2:3:1, v/v/v) and loaded onto the column. Most of the putative L. interrogans lipid A eluted with CHCl3, MeOH, 30 mM NH4Ac (2:3:1, v/v/v), suggesting that it is not strongly anionic (42, 45, 48). Fractions from the 30 mM NH4Ac wash were pooled and, following removal of the solvents (42), stored at −20 °C.
Further purification of the L. interrogans lipid A by preparative TLC was carried out as described by Que et al. (42, 48) for Rhizobium etli lipid A. Briefly, the lipid A was redissolved in 2 ml of CHCl3, MeOH (4:1, v/v), and a 0.2–0.5-mg sample was applied in 10-μl spots along a line at the origin of four 20 × 20-cm Silica Gel 60 analytical TLC plates (0.25-mm thickness), which were developed with the solvent CHCl3, pyridine, 88% formic acid, MeOH, H2O (60:35:10:5:2, v/v/v/v/v). The samples were eluted (42, 48) and passed through another 0.5-ml DEAE column to remove residual silica chips. The purified preparations were stored dry at −20 °C.
O-Deacylation of L. interrogans Lipid A by Mild Alkaline Hydrolysis
Complete hydrolysis of all ester-linked fatty acids was achieved by resuspending ~0.1 mg of pure lipid A in a 1-ml glass vial equipped with a Teflon-lined cap in 120 μl of CHCl3, MeOH, 0.6 M NaOH (2:3:1, v/v/v) and allowing the hydrolysis to proceed at room temperature for 30 –60 min. The final mild alkaline hydrolysis mixture was converted into an acidic two-phase Bligh-Dyer system by the addition of 220 μl of CHCl3, 210 μl of MeOH, and 206 μl of 0.1 M HCl. The lower phase was dried with a stream of N2, and the sample was stored at −20°C. The deacylated lipid A was further purified using a 0.25-ml DEAE-cellulose column, equilibrated, and eluted as above. The O-deacylated lipid A was eluted with CHCl3, MeOH, 30 mM NH4Ac (2:3:1, v/v/v), was recovered by acidic two-phase Bligh-Dyer partitioning, and was stored dry at −20 °C prior.
To obtain lipid A that is partially O-deacylated, purified lipid A (~0.2 mg) was dissolved in 600 μl of CHCl3, MeOH, 0.6 M NaOH (2:3:1, v/v/v). After incubation for only 5 min at room temperature, the solution was converted into an acidic two-phase Bligh-Dyer mixture by the addition of 300 μl of CHCl3, 200 μl of MeOH, and 350 μl of 0.1 M HCl. After mixing, the lower phase was dried with a stream of N2 and stored at −20 °C.
MALDI-TOF Mass Spectrometry of L. interrogans Lipid A
Spectra were acquired in the negative ion and the positive ion linear modes using a Kratos Analytical (Manchester, UK) MALDI-TOF mass spectrometer, equipped with a 337-nm nitrogen laser, a 20-kV extraction voltage, and time-delayed extraction (42). Each spectrum was the average of 50 shots. The lipid A samples were prepared for MALDI-TOF analysis by depositing 0.3 μl of the sample dissolved in chloroform, methanol (4:1, v/v), followed by 0.3 μl of the matrix, which was a mixture of saturated 6-aza-2-thiothymine in 50% acetonitrile and 10% tribasic ammonium citrate (9:1, v/v). The sample mixtures were allowed to dry at room temperature. Hexa-acylated lipid A 1,4′-bisphosphate from E. coli (Sigma) set at m/z 1797 was used as an external standard for calibration.
Fatty Acid Analysis by Gas Chromatography/Mass Spectrometry
Briefly, ~1 mg of purified L. interrogans lipid A was dissolved in 200 μl of toluene and 400 μl of fresh 1% sulfuric acid in methanol. The sample was then hydrolyzed for 2 h at 100 °C, after which 1 ml of hexane was added. The mixture was mixed for 30 s, followed by centrifugation for ~10 min at room temperature. The upper phase containing the methyl esters was removed and dried down under a stream of nitrogen. Fatty acid methyl esters were analyzed at the University of Minnesota Mass Spectrometry Facility Department of Chemistry using a Finnigan MAT 95 mass spectrometer coupled to a Hewlett-Packard Series II model 5890 gas chromatograph.
NMR Analysis
NMR spectroscopy was carried out at the Duke University NMR Spectroscopy Center (48, 49). The L. interrogans lipid A was dissolved in 0.6 ml of CDCl3, CD3OD, D2O (2:3:1,v/v/v) in a 5-mm NMR tube. Proton and carbon chemical shifts are reported relative to internal tetramethylsilane at 0.00 ppm.
NMR spectra were recorded on Varian Unity 500 or 600 NMR spectrometers, each equipped with a Sun Ultra 10 computer and a 5-mm Varian probe. Two-dimensional NMR experiments (COSY, NOE spectroscopy, total correlation spectroscopy, and HMQC) were performed at 600 MHz (48, 50). Directly detected 1H-decoupled 31P NMR spectra were recorded at 202.37 MHz with a spectral window of 12143.3 Hz digitized into 25,280 data points (digital resolution of 1 Hz/point or ~0.005 ppm/point), a 60° pulse flip angle (8 μs), and a 1.6-s repeat time. 31P chemical shifts were referenced to 85% H3PO4 at 0.000 ppm. Inverse decoupled difference spectra were recorded as 1H-detected 31P-decoupled heteronuclear NMR experiments (49, 50).
RESULTS
Weak Binding of L. interrogans Lipid A to DEAE-cellulose
The lipid A recovered from L. interrogans LPS after hydrolysis at 100 °C in 12.5 mM sodium acetate, pH 4.5, migrated as a discrete band during TLC (Fig. 1). Given its elution from DEAE-cellulose with CHCl3, MeOH, 30 mM aqueous ammonium acetate (2:3:1, v/v/v), this substance is predicted to have a net negative charge of about −1 (42, 45). Typical lipid A 1,4′bisphosphate species from E. coli elute with CHCl3, MeOH, 240 mM ammonium acetate (2:3:1, v/v/v) (46, 50), because they can carry up to four negative charges. The weak binding of the L. interrogans material to DEAE-cellulose resembles what is seen with R. etli lipid A in which both phosphate groups are missing and replaced with carboxylate-containing sugars (42).
Fig. 1. Thin layer chromatography of purified lipid A from L. interrogans serovar Pomona.
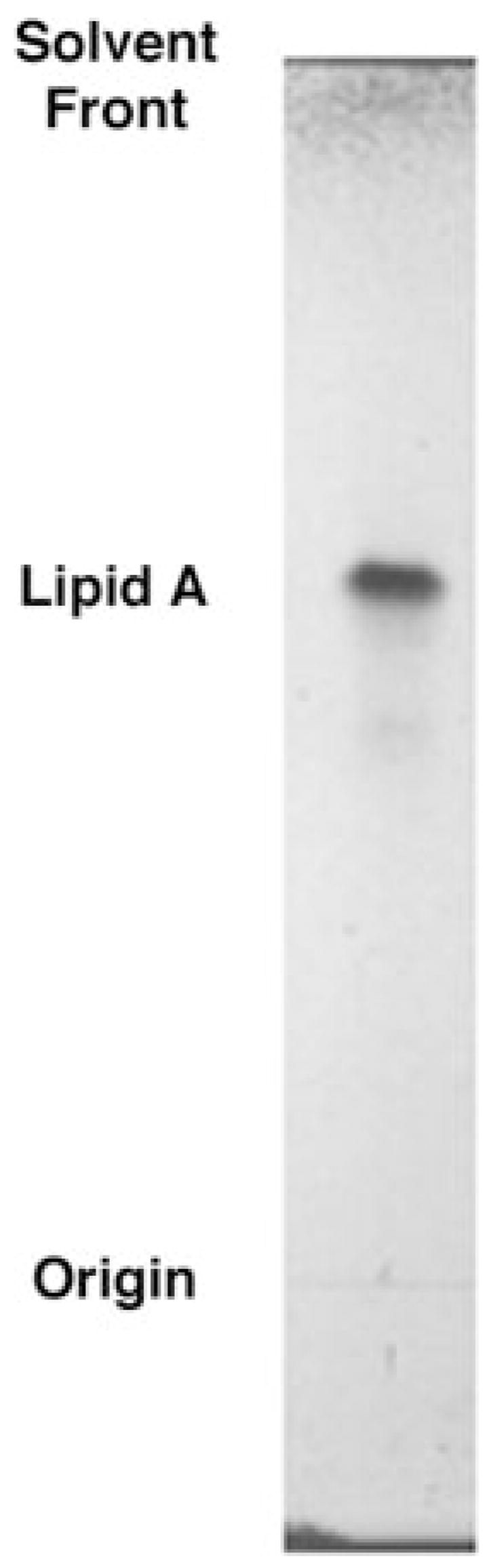
The solvent was CHCl3, pyridine, 88% formic acid/H2O/MeOH (60:35:10:5:2, v/v/v/v/v). The lipid A band was visualized by charring on a hot plate (42).
Identical Lipid A Molecules in L. interrogans Serovars Pomona and Icterohaemorrhagiae
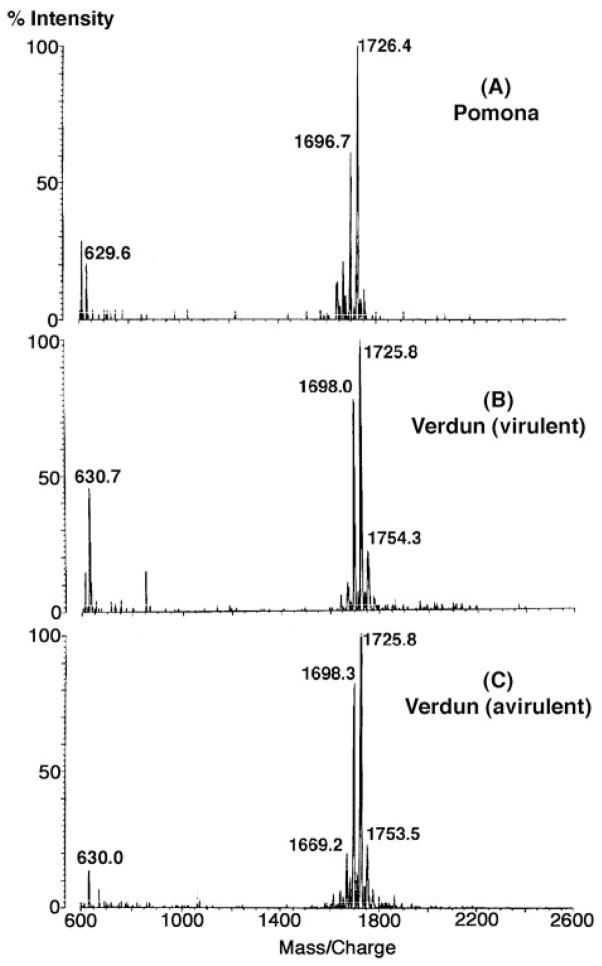
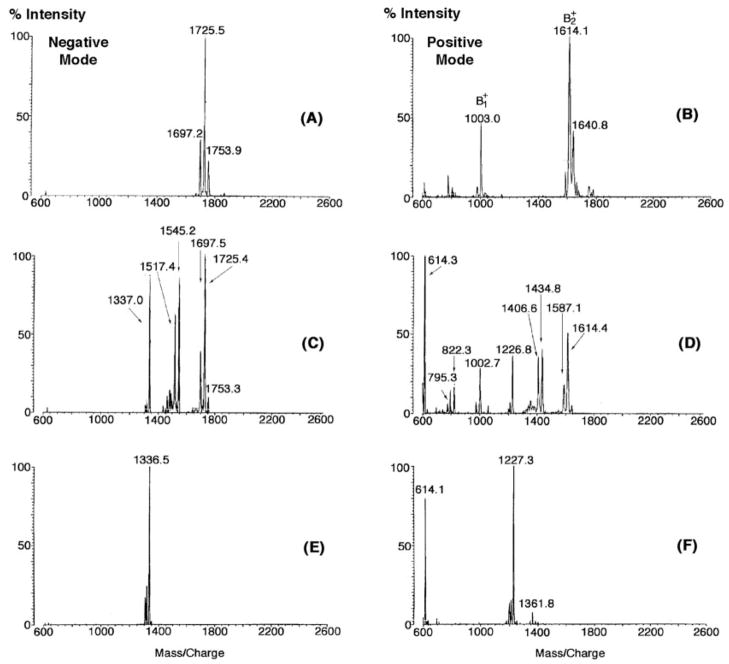
Negative ion mode MALDI-TOF mass spectrometry of the lipid A isolated from L. interrogans serovar Pomona (Fig. 2A) revealed a major peak at m/z 1726.4, likely representing [M − H]− of the major molecular species. Smaller peaks differing by 28 atomic mass units presumably reflect fatty acyl chain length heterogeneity. The lipid A of both the virulent and the avirulent forms L. interrogans strain Verdun (22) yielded virtually identical spectra (Fig. 2, B and C). Because much greater quantities of LPS were available from the Pomona serovar (12, 19), all of the NMR studies were performed with Pomona lipid A.
Fig. 2. Negative ion MALDI-TOF mass spectra of lipid A preparations from different L. interrogans strains.
The major lipid A molecules of these strains appear to be identical.
One-dimensional NMR Spectroscopy L. interrogans Lipid A
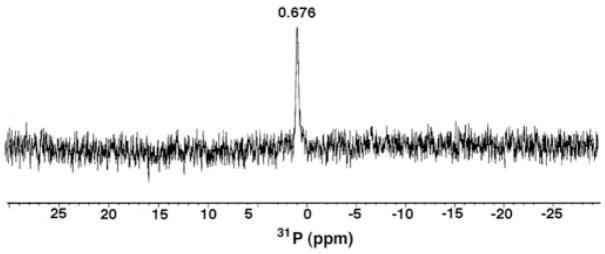
The 31P NMR spectrum of L. interrogans lipid A at 202 MHz revealed a single resonance at 0.676 ppm (Fig. 3), demonstrating that this material contains only one phosphate group. The chemical shift of the phosphorus signal is in the range of what is expected for a monophosphomonoester unit (−0.5 to +2 ppm) under these conditions (50). However, a monophosphodiester group cannot be excluded based solely upon the chemical shift position. The presence of only one phosphate group is consistent with the low affinity of L. interrogans lipid A for DEAE-cellulose.
Fig. 3. 31P NMR spectrum of L. interrogans serovar Pomona lipid A.
The 2-mg lipid A sample was dissolved at 25 °C in 0.6 ml of CDCl3, CD3OD, D2O (2:3:1, v/v/v).
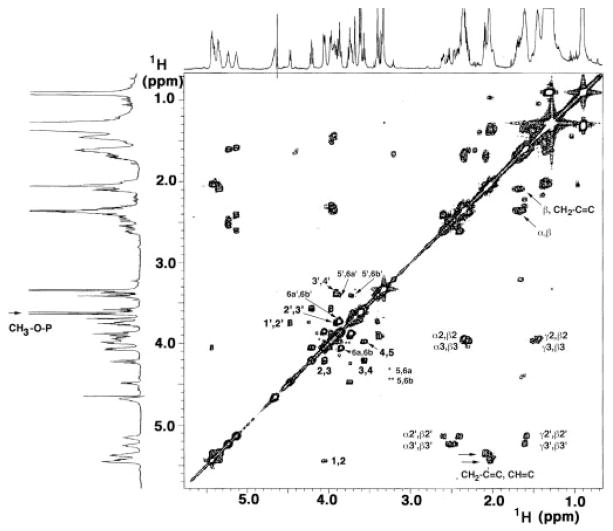
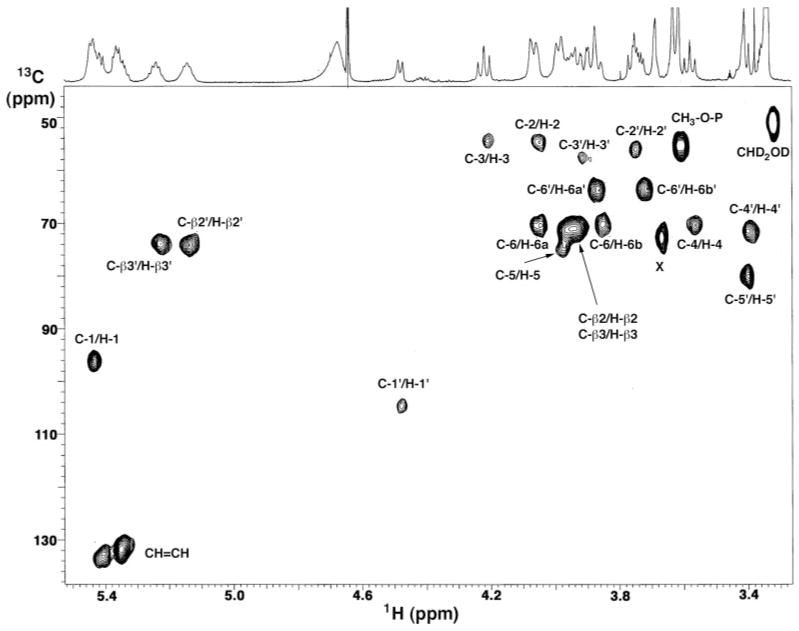
The one-dimensional 600 MHz 1H NMR spectrum of L. interrogans lipid A in CDCl3, CD3OD, D2O (2:3:1, v/v/v) revealed well resolved peaks in the sugar (3.5–5.5 ppm) and acyl chain (0.9 –2.8 ppm) regions (Fig. 4, upper and left margins), similar to E. coli or R. etli lipid A (48–50). However, the L. interrogans spectrum contains an unusually prominent doublet at 3.61 ppm, which integrates to three protons and shows an apparent coupling constant of 10.9 Hz (Fig. 4, left arrow). This kind of signal had not been seen previously in lipid A from diverse sources (47–50).
Fig. 4. 1H-1 H COSY of L. interrogans serovar Pomona lipid A at 600 MHz.
The 2-mg lipid A sample was dissolved at 25 °C in 0.6 ml of CDCl3, CD3OD, D2O (2:3:1, v/v/v). The numbering scheme is shown in Fig. 7A. The arrow at the left highlights the doublet near 3.61 ppm that arises from the methylated 1-phosphate. The two arrows at the lower right highlight the strong cross-peaks between the olefinic and vinylic protons of the secondary acyl chains.
NMR Evidence for a Methylated 1-Phosphate Moiety
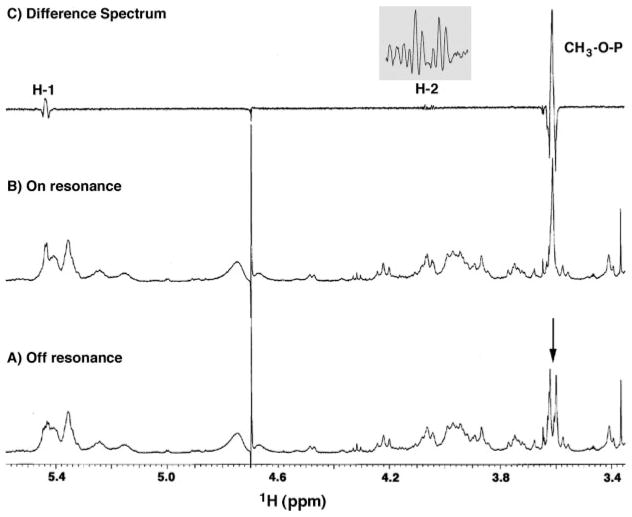
The 3.61-ppm doublet in the 1H NMR spectrum of L. interrogans lipid A does not show any cross-peaks in the 1H-1H COSY analysis (Fig. 4). To determine whether or not this signal arises from a methylated phosphate group, selective inverse decoupling difference spectroscopy was used to detect possible heteronuclear coupling of the phosphorus atom to the 3.61-ppm doublet (49, 50). The difference spectrum (Fig. 5C) of the off and on resonance 31P-decoupled 1H NMR spectra (Figs. 5, A and B) demonstrated the collapse of the 3.61 ppm doublet to a singlet during phosphorus irradiation. In addition, the difference spectrum showed simplification of a proton signal at 5.44 ppm to a doublet and of another proton at 4.05 ppm to a double-doublet (Fig. 5C, inset). The latter chemical shifts and peak shapes are consistent with phosphorus coupling to the H-1 and H-2 atoms, respectively, of a proximal α-linked glucopyranoside unit (49, 50). The heteronuclear coupling data provide strong evidence that L. interrogans lipid A contains an unprecedented mono-phosphodiester unit bridging the C-1 atom of the proximal sugar and a methyl group.
Fig. 5. Selectively 31P-decoupled 1H NMR spectra of L. interrogans serovar Pomona lipid A.
The sample used in Fig. 4 was subjected to 1H NMR analysis with the decoupler either off resonance or on the 31P atom resonance. The doublet at 3.61 ppm (arrow in A) collapses to a singlet (B) when the phosphorus atom is selectively irradiated. The difference spectrum (C) highlights additional coupling of the phosphorus atom to the H-1 and H-2 signals of the proximal sugar. The inset in C shows the H-2 difference spectrum in detail.
Confirmation of a Methyl Group on the 1-Phosphate Moiety by MALDI-TOF Mass Spectrometry
The negative ion MALDI-TOF mass spectra of intact L. interrogans lipid A (Figs. 2 and 6A) show an intense peak with m/z near 1726 atomic mass units, interpreted as the molecular ion [M − H]−. Additional structural information is contained in the positive ion mode spectrum (Fig. 6B). The oxonium ion (Fig. 6B) forms during fragmentation of the disaccharide glycosidic linkage (Fig. 7) (51) and reflects the mass of the distal sugar unit. The ion (Fig. 7) is generated by loss of the substituent attached to the 1-position in the proximal sugar (51). The mass of the group attached to the anomeric carbon is determined by comparing the m/z of the ion to the molecular weight derived from the [M − H]− ion (Fig. 6A). The mass of the proximal unit (without the substituent at the 1-position) is determined from the difference of and .
Fig. 6. MALDI-TOF mass spectrometry of L. interrogans serovar Pomona lipid A subjected to mild NaOH hydrolysis.
The left half of the figure shows the negative ion mass analysis of L. interrogans lipid A before (A), after 5 min (C), or after 30 min of mild NaOH treatment (E). The right half of the figure shows the corresponding positive ion spectra (B, D, and F, respectively). The loss of two O-linked acyl chains from the distal unit with masses corresponding to C12:1 and C14:1 is complete after 30 min.
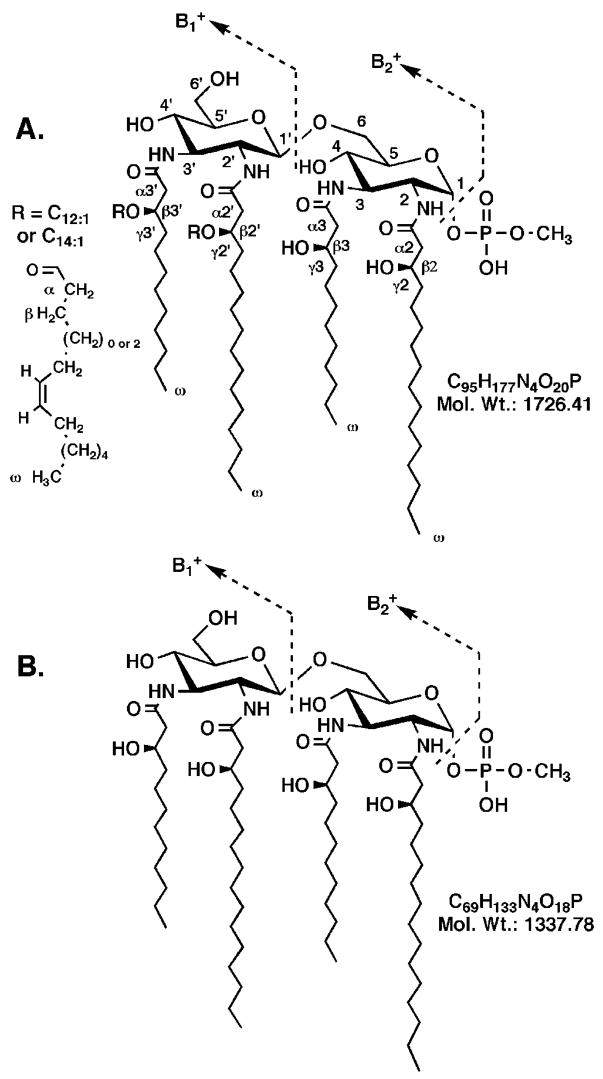
Fig. 7. Proposed structures of intact and O-deacylated L. interrogans serovar Pomona lipid A.
The numbering scheme shown for the intact lipid A in A is used for the NMR assignments. The position of the cis-double bond is inferred based on the mechanism by which unsaturated fatty acids are synthesized in E. coli (58–60). The fully O-deacylated material is shown in B. Fatty acid analysis of L. interrogans lipid A shows equal amounts of 3-hydroxylaurate and 3-hydroxy-palmitate; 3-hydroxylaurate is attached at positions 3 and 3′ (28).
Fig. 6B shows prominent peaks at m/z 1003.0 and 1614.1, which are interpreted as the and ions, respectively, of L. interrogans lipid A (Fig. 7). Under the ionization conditions employed, the molecular ion [M − H]− (m/z = 1727.4) is not detected (Fig. 6B). The ion at m/z 1614.1 is 112.4 atomic mass units less than the molecular weight deduced from the negative mode spectrum in Fig. 6A (1726.5). If L. interrogans lipid A contained an unsubstituted phosphate group at the 1-position, as is present in E. coli lipid A, the ion should be 97.0 atomic mass units smaller than the molecular weight predicted from the negative mode spectrum (52). Given the experimental error of the MALDI-TOF measurements, the discrepancy of 15.4 atomic mass units (112.4 -97.0) is consistent with the presence of a methyl substituent on the 1-phosphate group of L. interrogans lipid A, as deduced from the above NMR experiments (Fig. 5).
The difference in mass between the and ions is 611.1 atomic mass units (Fig. 6B), suggesting the presence of two acyl chains on the proximal unit of L. interrogans lipid A. Given the absolute selectivity of L. interrogans LpxA for 3-hydroxylauroyl-ACP and UDP-GlcNAc3N (28), the GlcN3N 3-position must be acylated with 3-hydroxylaurate. To account for the 611.1 atomic mass units difference in the and ions (Fig. 6B), the acyl chain at the GlcN3N 2-position could be 3-hydroxypalmitate (Fig. 7). In fact, 3-hydroxylaurate and 3-hydroxypalmitate were predominant components in the fatty acid analysis (data not shown). The size of the ion at m/z 1003.0 atomic mass units suggests that four acyl chains are attached to the distal unit (Fig. 7).
Mild Alkaline Hydrolysis of L. interrogans Lipid A
Exposure of lipid A or lipid A precursors to aqueous triethylamine at 37 °C releases unsubstituted O-linked 3-hydroxyacyl chains (42, 53). Mild triethylamine does not remove O-linked acyloxyacyl moieties or O-linked normal fatty acids under standard conditions. Exposure of L. interrogans lipid A to aqueous triethylamine at 37 °C did not alter its molecular weight, as judged by mass spectrometry (not shown), demonstrating the absence of unsubstituted, O-linked 3-hydroxyacyl chains.
Treatment of L. interrogans lipid A with 0.1 M NaOH for 30 min shifted [M − H]− from m/z 1725.5 to 1336.5 atomic mass units (Fig. 6, A and C), suggesting the release of two ester-linked acyl chains with the masses of C12:1 and C14:1 (Fig. 7A). A 5-min exposure to 0.1 M NaOH yielded partially O-deacylated intermediates, as judged by the appearance of peaks at m/z 1545.2 and 1517.4 atomic mass units (Fig. 6B), consistent with the presence of ester-linked C12:1 and C14:1 moieties.
The MALDI-TOF analyses of the partially and completely hydrolyzed L. interrogans lipid A samples in the positive mode (Fig. 6, D and F, respectively) are in accord with the negative mode data (Fig. 6, C and E). Importantly, the ion is shifted from m/z 1003.0 atomic mass units to m/z 614.1 atomic mass units after complete hydrolysis (Fig. 6, B versus F), demonstrating conclusively that both ester-linked acyl chains must be located on the distal unit of L. interrogans lipid A. The difference in mass between the and ions following complete hydrolysis is 613.2 atomic mass units (Fig. 6F), which is the essentially same as observed for the untreated material (Fig. 6B) and in good agreement with the expected value of 612.9 for the structure shown in Fig. 7.
The molecular weight of the dilute NaOH treated L. interrogans lipid A is 1337.5 (Fig. 6E). Given that the corresponding ion is observed at m/z 1227.3 atomic mass units (Fig. 6F), the molecular mass of the substituent present at the 1-position of this substance is 110.2 (i.e. 1337.5 -1227.3). This result in good agreement with what is expected for the loss of a methylated phosphate residue (111.0 atomic mass units) (Fig. 7).
Two-dimensional 1H NMR Analysis of L. interrogans Lipid A
All of the chemical shifts and coupling constants for L. interrogans lipid A are summarized in Table I, using the proposed structure and numbering scheme shown in Fig. 7A. Many of the protons assigned in the 1H-1H COSY of L. interrogans lipid A (Fig. 4) appear at similar shifts as their E. coli counterparts (49, 50). For instance, the H-1 anomeric proton of the proximal sugar at 5.44 ppm and the H-1′ anomeric proton of the distal sugar at 4.47 ppm are easily recognized (Fig. 4) and serve as convenient entry points for the evaluation of the sugar region connectivity (3.5–4.5 ppm). The COSY (Fig. 4) and total correlation spectroscopy (not shown) data permit the sequential identification of H-2 through H-6a and H-6b for each hexose ring (Fig. 4 and Table I). The small J1,2 coupling (3.2 Hz) and the large J2,3, J3,4 and J4,5 couplings (9 to 11 Hz) suggest that the proximal pyranose ring is in the α-anomeric configuration with axially disposed H-2, H-3, H-4, and H-5 protons (Fig. 7). The large J1′,2′ coupling (7.1 Hz) shows that the distal sugar is in the β-configuration (Fig. 7). NOE spectroscopy analysis (not shown) demonstrates the following NOE dipolar interactions: 1) from the resolved H-1′ to H-3′ and to H-5′ within the distal pyranose unit and to the H-6a and H-6b of the proximal sugar; 2) from H-1 to H-2 and from H-2 to H-4 in the proximal sugar; and 3) from H-5 to H-3 and to H-6a and H-6b in the proximal sugar. The NOE from H-1′ to H-6a and H-6b is diagnostic for the β-1′,6 linkage (49, 50). The multiple 1,3 diaxial and single axial-equatorial (H-1 to H-2) intramolecular NOEs confirm that both sugar rings adopt the chair conformations with a β-linkage between the proximal α-glucopyranose and the distal β-glucopyranose rings.
Table I. 1H, 13C, and 31P NMR chemical shifts of L. interrogans lipid A.
1H and 13C chemical shifts at 25 °C in CDCl3:CD3OD:D2O (2:3:1, v/v/v) relative to internal tetramethylsilane derived from the two-dimensional NMR experiments (Figs. 4 and 8). The 31P chemical shift was relative to 85% H3PO4. The numbering scheme is shown in Fig. 7A.
| Position | δC | δH,[(mult), J(Hz)] | δP |
|---|---|---|---|
| Proximal GlcN3N | |||
| 1 | 95.3 | 5.44 [dd, J1,2 = 3.2]a | +0.676 |
| 2 | 53.52 | 4.05 [ddd, J,2,3 = 11.23]a | |
| 3 | 53.46 | 4.21 [dd, J3,4 = 9.8] | |
| 4 | 69.00 | 3.57 [dd, J4,5 = 8.9] | |
| 5 | 73.63 | 3.98 [m] | |
| 6a | 69.29 | 4.04 [m] | |
| 6b | 3.85 [m] | ||
| Distal GlcN3N | |||
| 1′ | 101.9 | 4.47 3dd, J13,23 = 7.1] | |
| 2′ | 55.08 | 3.75 [m] | |
| 3′ | 56.71 | 3.90 [m] | |
| 4′ | 70.31 | 3.39 [m] | |
| 5′ | 79.05 | 3.40 [m] | |
| 6′a | 62.66 | 3.87 [m] | |
| 6′b | 3.73 [m] | ||
| 3-Hydroxyacyl | |||
| α2 | 45.0 | ~2.35, 2.30 | |
| β2 | 69.8 | ~3.93 [m] | |
| γ2 | 38.5 | ~1.44 [m] | |
| α3 | 42.3 | ~2.40 | |
| β3 | 70.0 | ~3.95 [m] | |
| γ3 | 38.5 | ~1.44 [m] | |
| α2′ | 42.4 | ~2.58, 2.45 | |
| β2′ | 73.3 | ~5.13 [m] | |
| γ2′ | ~1.63 [m] | ||
| α3′ | 42.3 | ~2.52 | |
| β3′ | 73.5 | ~5.22 [m] | |
| γ3′ | ~1.63 [m] | ||
| Other | |||
| α CH2 | 35.3 | ~2.34 [m] | |
| β CH2 | ~1.61 [m] | ||
| (CH2) | 30.4 | ~ 1.28 [m] | |
| ω CH3 | 15.2 | 0.891 [t] | |
| CH=CH | 131.24 | 5.35 | |
| CH=CH | 132.5 | 5.40 | |
| CH2-C=C | 28.06 | 2.08 | |
| CH2-C=C | 28.57 | 2.03 | |
| CH3-OP | 54.34 | 3.61 [d, JH,P = 10.9] | |
Measured from the resolved H-2 double-doublet in the 31P-decoupled 1H NMR difference spectrum (Fig. 5C).
The low field position of H-1 at 5.44 ppm (Fig. 4) is characteristic of an anomeric proton of a sugar 1-phosphate, similar to H-1 in E. coli lipid A, which resonates at 5.46 ppm (49, 50). However, H-4′ of L. interrogans lipid A (3.40 ppm) resonates at significantly higher field than the H-4′ of E. coli lipid A (4.17 ppm), suggesting that the 4′-position of L. interrogans lipid A is not phosphorylated. The ion detected by mass spectrometry in the positive ion mode (Fig. 6B) likewise suggests the absence of a phosphate group at position 4′. The 31P NMR data (Fig. 3) conclusively demonstrate only a single phosphate group in L. interrogans lipid A with heteronuclear coupling between the phosphorus atom and H-1 of the proximal sugar (Fig. 5).
H-3 and H-3′ of L. interrogans lipid A resonate near 4.2 ppm and 3.9 ppm, respectively (Fig. 4 and Table I), significantly upfield of the H-3 and H-3′ signals in E. coli lipid A at 5.25 and 5.18 ppm (49, 50). Both the 3- and 3′-positions of E. coli lipid A are substituted with ester-linked acyl chains, and therefore H-3 and H-3′ of E. coli lipid A are shifted considerably down-field relative to typical nonesterified sugar oxymethines groups (49, 50). L. interrogans lipid A does not contain esterified sugar oxymethine groups at positions 3 and 3′.
13C NMR Evidence for a GlcN3N Disaccharide in L. interrogans Lipid A
In lipid A disaccharides consisting of two glucosamine units (49, 50), two cross-peaks (originating from C-2 and C-2′) are observed in the 52–57-ppm region of the HMQC spectrum. As shown in Fig. 8, the HMQC spectrum of L. interrogans lipid A reveals four sugar resonances between 52 and 58 ppm. Two of these cross-peaks are attributed to C-2 (53.5 ppm) and C-2′ (55.1 ppm), because they correlate to H-2 at 4.05 ppm and H-2′ at 3.75 ppm, respectively. The cross-peaks at 53.5 ppm and 56.7 ppm correlate with H-3 (4.21 ppm) and H-3′ (3.90 ppm), demonstrating unequivocally that both C-3 and C-3′ are substituted with nitrogen atoms in L. interrogans lipid A. The presence of aminomethine compared with oxymethine groups accounts for the large differences in the chemical shifts observed for H-3 and H-3′ of L. interrogans lipid A versus E. coli lipid A. Moreover, the C-2 and C-3 shifts of L. interrogans lipid A agree with those reported for the α form of 2,3-diamino-2,3-dideoxyglucose in a 4:1 benzene-dimethyl sulfoxide mixture (54), whereas the C-2′ and C-3′ shifts are close to those of the β form of 2,3-diamino-2,3-dideoxyglucose (54).
Fig. 8. Partial HMQC spectrum of L. interrogans serovar Pomona lipid A at 600 MHz.
The 2-mg lipid A sample was dissolved at 25 °C in 0.6 ml of CDCl3, CD3OD, D2O (2:3:1, v/v/v). The cross-peaks are labeled according to the scheme shown in Fig. 7A.
The HMQC spectrum (Fig. 8) confirms two anomeric protons. H-1 of the proximal sugar at 5.44 ppm correlates with C-1 at 96 ppm, whereas H-1′ of the distal sugar at 4.48 ppm connects to C-1′ at 105 ppm. These C-1 and C-1′ chemical shifts are characteristic of the α- and β-anomeric configurations, respectively (54), and are consistent with the 1H NMR data (Fig. 4). The prominent three-proton doublet of L. interrogans lipid A at 3.61 ppm correlates to a carbon signal at 54.34 ppm, close to that of the CD2HOD signal from the methanol solvent and in accord with the proposal that the methyl doublet arises from a methylated phosphate group (Fig. 7). The striking cross-peaks within the olefinic carbon region near 132 ppm (Fig. 8) correlate with proton signals at 5.35 and 5.40 ppm (Fig. 4), diagnostic for the presence of unsaturated acyl chains.
1H NMR Analysis of the Acyloxyacyl Residues and Mono-unsaturated Acyl Chains in L. interrogans Lipid A
The R-3-hydroxyacyl chains that are the hallmark of all lipid A molecules are readily detected in L. interrogans lipid A by 1H NMR (Fig. 4). The β-oxymethine protons of these acyl chains (Fig. 7A) resonate between 3.7 and 4.2 ppm when the β-OH group is not substituted, but they are shifted to about 5.2 ppm when a secondary acyl chain is present (48–50, 55–57) (Fig. 4). The four α/β and four γ/β cross-peaks (Fig. 4) confirm that there are four β-hydroxyacyl chains in L. interrogans lipid A. Two of the four α/β cross-peaks overlap near 2.4 and 3.95 ppm (α2,β2 and α3,β3). Two of the four γ/β cross-peaks are detected near 1.5 and 3.95 ppm (γ2,β2 and γ3,β3). These signals are characteristic of α- and γ-methylene protons adjacent to β-oxymethines of unsubstituted β-hydroxyacyl chains (48–50). The two remaining sets of α/β and γ/β cross-peaks (Fig. 4) are detected near 2.4 –2.6 and 5.2 ppm and near 1.6 and 5.2 ppm, respectively. The downfield shift of the β2′ and β3′ protons versus the β2 and β3 protons (Fig. 4) confirms the presence of two acyloxyacyl moieties in L. interrogans lipid A (Fig. 7A).
Prominent cross-peaks are also observed near 5.38 and 2.1 ppm and near 5.42 and 2.05 ppm (Fig. 4). These signals arise from the spin coupling of olefinic protons to adjacent vinylic methylenes in the secondary acyl chains (Fig. 7A). The COSY analysis is therefore consistent with both the HMQC and the mass spectrometry in demonstrating the presence of unsaturated secondary acyl chains in L. interrogans lipid A. The exact location and stereochemistry of the double bonds remains to be determined. However, if L. interrogans generates fatty acid cis-double bonds by the same anaerobic pathway as E. coli (58–60), one would expect the double bonds of both the C12:1 and the C14:1 chains to be located at position ω-7 (Fig. 7A). The COSY analysis supports this idea, because it shows a cross-peak between at least one vinylic methylene and one β-methylene group (Fig. 4), as expected for the proposed structure of the C12:1 chain (Fig. 7A). The total correlation spectroscopy data (not shown) confirm the connectivity of olefinic protons to a subset of α- and β-methylenes within the secondary acyl chains, as well as showing the expected strong connectivity to vinylic and aliphatic methylenes.
DISCUSSION
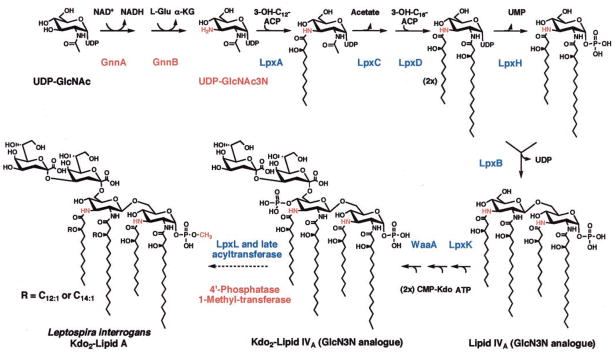
The studies presented above document for the first time the existence of a lipid A molecule in L. interrogans with the proposed structure shown in Fig. 7A. The identification of this substance is consistent with the fact that the L. interrogans genome encodes a complete set of Lpx proteins (13), which catalyze the biosynthesis of the lipid A anchor of LPS in virtually all other Gram-negative bacteria (Fig. 9) (2). L. interrogans is the first spirochete shown to possess the lpx genes, in contrast to T. pallidum, T. denticola, and B. burgdorferi, which do not make lipid A (3, 6, 7). The absence of lipid A and LPS in the latter organisms (3–5) may explain their restricted ability to grow outside of their mammalian hosts. It would be of great interest to inactivate the lpxA gene (2, 61–63) in L. interrogans to determine whether or not this organism is viable in the absence of its LPS. However, at present no mutagenesis system is available for pathogenic Leptospira spp. Lipid A is essential for growth in all Gram-negative bacteria examined to date (2, 64) with the exception of Neisseria meningitidis strains containing a polysialic acid capsule (65, 66). In the latter, the lpxA gene can be deleted with the consequence that the bacteria grow slowly and now require their polysialic acid capsule for viability (66).
Fig. 9. Proposed biosynthetic pathway for L. interrogans lipid A.
Highly significant orthologs of all of the indicated enzymes (2) are encoded in the L. interrogans serovar Lai genome (13). The methyl transferase responsible for the formation of the unusual 1-methylphosphate group of L. interrogans lipid A utilizes S-adenosyl-methionine as the donor (85).
The NMR studies shown in Figs. 4 and 8, in conjunction with the mass spectrometry of native and NaOH treated lipid A (Fig. 6), demonstrate unequivocally that L. interrogans lipid A consists of a hexa-acylated β-1′,6-linked disaccharide in which the usual glucosamine residues are replaced with the more stable GlcN3N analog (28, 29, 67). The selectivity of L. interrogans LpxA for UDP-GlcNAc3N and 3-hydroxylauroylacyl carrier protein, documented in the preceding manuscript (28), independently supports this structural assignment. The two secondary acyl chains of L. interrogans lipid A appear to be unsaturated (Figs. 4, 6, 7A, and 8), which is unusual but not without precedent (68–70). Additional structural studies will be required to determine the exact location of the double bonds in the secondary acyl chains.
The 4′-position of L. interrogans lipid A is not phosphorylated. The latter finding indicates that a 4′ phosphatase must be present in this organism, as in R. etli and R. leguminosarum in which the 4′-phosphate group is also missing (42, 48, 71, 72). Removal of the 4′-phosphate group, when it occurs, appears to be a late step in lipid A biosynthesis, given that the 4′-phosphate residue is actually necessary for the attachment of the Kdo sugars (71, 73, 74). All bacteria with a 4′-phosphatase, including L. interrogans, retain the 4′-kinase encoded by lpxK (13, 75) (Fig. 9). In preliminary studies, 4′-phosphatase activity was observed using washed L. interrogans membranes (not shown) with the hexa-acylated substrate [4′-32P]Kdo2-lipid A from E. coli (76). No dephosphorylation of the tetra-acylated precursors [4′-32P]lipid IVA or [4′-32P]Kdo2-lipid IVA (77) was detected.
The most unique aspect of L. interrogans lipid A is the finding that its 1-phosphate group is methylated (Figs. 4–6). This structural feature is without precedent in lipid A biochemistry (2, 33). In fact, the enzymatic methylation of phosphate groups appears to be very rare in all of biology. To our knowledge, Kates et al. (31) have reported the only other example of a methylated lipid phosphate residue, found in the halophile Halobacterium salinarium, which synthesizes a methylated phosphatidylglycerophosphate analog. The enzymatic and genetic mechanisms for this type of lipid phosphate group methylation have not been explored. Similarly, the origin of the unusual methylated γ-phosphate cap found at the 5′ end of the 7SK, B2, and U6 small RNAs in eucaryotic cells (32) has not been studied at the level of enzymology.
In contrast to the relatively uncommon biological methylation of phosphate residues, enzymes catalyzing the N-methylation of phosphatidylethanolamine (78, 79) or the O-methylation of membrane proteins on selected carboxylate residues (80) are widely distributed. Reversible methylation of the methyl accepting chemotaxis proteins is essential for the proper response of E. coli to chemical signals (80, 81). C-terminal methylation of Ras proteins in higher eucaryotic cells (82, 83) or a-factor mating pheromone of yeast (84) is essential for membrane association and signaling. In these well documented examples of membrane lipid and membrane protein methylation, S-adenosyl-methionine serves as the methyl donor. We have recently found that membranes of L. interrogans catalyze the S-adenosyl-methionine-dependent methylation of Kdo2-lipid A (85).
The characterization of the structure of L. interrogans lipid A sets the stage for the analysis of its biosynthesis and bioactivity. An initial survey of the lipid A described above demonstrates that it is inactive in the limulus lysate assay and against human THP-1 cells,2 indicating that it is not contaminated with a classical endotoxin, such as E. coli lipid A. However, when assayed with mouse RAW 264.7 cells, L. interrogans lipid A induces tumor necrosis factor with about one-tenth the potency of E. coli lipid A.2 We are currently evaluating L. interrogans lipid A with macrophages derived from various mouse TLR knockout strains. It may be that the robust TLR2 activating activity seen with intact L. interrogans LPS (22) requires more than just the lipid A moiety. Isolation of LPS from L. interrogans mutants blocked in defined steps of O-antigen and/or core biosynthesis (12) might address this question. In addition, chemically synthesized versions of L. interrogans lipid A need to be prepared to validate our proposed structure and to determine whether or not the activity seen with L. interrogans lipid A is real or is due to other biologically active impurities.
Footnotes
This work was supported by National Institutes of Health Grants GM-51310 and GM-51796 (to C. R. H. R.) and GM-54882 (to R. J. C.), by Grant PTR94 from the Institut Pasteur (to C. W.), and by a grant from the National Health and Medical Research Council of Australia (to B. A.). The Duke NMR Center is partially supported by National Institutes of Health Grant NCI P30-CA-14236. NMR instrumentation in the Duke NMR Center was funded by the National Science Foundation, the National Institutes of Health, the North Carolina Biotechnology Center, and Duke University.
The abbreviations used are: LPS, lipopolysaccharide; Kdo, 2-keto-3-deoxy-D-manno-octulosonic acid; UDP-GlcNAc3N, UDP 2-acet-amido-3-amino-2,3-dideoxy-α-D-glucopyranose; GlcN3N, 2,3-diamino-2,3-dideoxy-D-glucopyranose; COSY, correlation spectroscopy; HMQC, heteronuclear multiple-quantum coherence; NOE, nuclear Overhauser effect; MALDI-TOF, matrix-assisted laser desorption ionization time-of-flight.
References
- 1.Brade H, Opal SM, Vogel SN, Morrison DC, editors. Endotoxin in Health and Disease. Marcel Dekker, Inc.; New York: 1999. [Google Scholar]
- 2.Raetz CRH, Whitfield C. Annu Rev Biochem. 2002;71:635–700. doi: 10.1146/annurev.biochem.71.110601.135414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Norris SJ, Cox DL, Weinstock GM. J Mol Microbiol Biotechnol. 2001;3:37–62. [PubMed] [Google Scholar]
- 4.Schultz CP, Wolf V, Lange R, Mertens E, Wecke J, Naumann D, Zähringer U. J Biol Chem. 1998;273:15661–15666. doi: 10.1074/jbc.273.25.15661. [DOI] [PubMed] [Google Scholar]
- 5.Takayama K, Rothenberg RJ, Barbour AG. Infect Immun. 1987;55:2311–2313. doi: 10.1128/iai.55.9.2311-2313.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fraser CM, Casjens S, Huang WM, Sutton GG, Clayton R, Lathigra R, White O, Ketchum KA, Dodson R, Hickey EK, Gwinn M, Dougherty B, Tomb JF, Fleischmann RD, Richardson D, Peterson J, Kerlavage AR, Quackenbush J, Salzberg S, Hanson M, van Vugt R, Palmer N, Adams MD, Gocayne J, Weidman J, Ulterback T, Watthey L, McDonald L, Artiach P, Bowman C, Garland S, Fujii C, Cotton MD, Horst K, Roberts K, Hatch B, Smith HO, Venter JC. Nature. 1997;390:580–586. doi: 10.1038/37551. [DOI] [PubMed] [Google Scholar]
- 7.Fraser CM, Norris SJ, Weinstock GM, White O, Sutton GG, Dodson R, Gwinn M, Hickey EK, Clayton R, Ketchum KA, Sodergren E, Hardham JM, McLeod MP, Salzberg S, Peterson J, Khalak H, Richardson D, Howell JK, Chidambaram M, Utterback T, McDonald L, Artiach P, Bowman C, Cotton MD, Fujii C, Garland S, Hatch B, Horst K, Roberts K, Sandusky M, Weidman J, Smith HO, Venter JC. Science. 1998;281:375–388. doi: 10.1126/science.281.5375.375. [DOI] [PubMed] [Google Scholar]
- 8.Radolf JD, Robinson EJ, Bourell KW, Akins DR, Porcella SF, Weigel LM, Jones JD, Norgard MV. Infect Immun. 1995;63:4244–4252. doi: 10.1128/iai.63.11.4244-4252.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Beermann C, Lochnit G, Geyer R, Groscurth P, Filgueira L. Biochem Biophys Res Commun. 2000;267:897–905. doi: 10.1006/bbrc.1999.2057. [DOI] [PubMed] [Google Scholar]
- 10.Akins DR, Bourell KW, Caimano MJ, Norgard MV, Radolf JD. J Clin Invest. 1998;101:2240–2250. doi: 10.1172/JCI2325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Porcella SF, Schwan TG. J Clin Invest. 2001;107:651–656. doi: 10.1172/JCI12484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bulach DM, Kalambaheti T, de la Pena-Moctezuma A, Adler B. J Mol Microbiol Biotechnol. 2000;2:375–380. [PubMed] [Google Scholar]
- 13.Ren SX, Fu G, Jiang XG, Zeng R, Miao YG, Xu H, Zhang YX, Xiong H, Lu G, Lu LF, Jiang HQ, Jia J, Tu YF, Jiang JX, Gu WY, Zhang YQ, Cai Z, Sheng HH, Yin HF, Zhang Y, Zhu GF, Wan M, Huang HL, Qian Z, Wang SY, Ma W, Yao ZJ, Shen Y, Qiang BQ, Xia QC, Guo XK, Danchin A, Saint Girons I, Somerville RL, Wen YM, Shi MH, Chen Z, Xu JG, Zhao GP. Nature. 2003;422:888–893. doi: 10.1038/nature01597. [DOI] [PubMed] [Google Scholar]
- 14.Levett PN. Clin Microbiol Rev. 2001;14:296–326. doi: 10.1128/CMR.14.2.296-326.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Farr RW. Clin Infect Dis. 1995;21:1–6. doi: 10.1093/clinids/21.1.1. [DOI] [PubMed] [Google Scholar]
- 16.Vinh T, Adler B, Faine S. J Gen Microbiol. 1986;132:103–109. doi: 10.1099/00221287-132-1-103. [DOI] [PubMed] [Google Scholar]
- 17.Shimizu T, Matsusaka E, Nagakura N, Takayanagi K, Masuzawa T, Iwamoto Y, Morita T, Mifuchi I, Yanagihara Y. Microbiol Immunol. 1987;31:717–725. doi: 10.1111/j.1348-0421.1987.tb03133.x. [DOI] [PubMed] [Google Scholar]
- 18.Vinh TU, Shi MH, Adler B, Faine S. J Gen Microbiol. 1989;135:2663–2673. doi: 10.1099/00221287-135-10-2663. [DOI] [PubMed] [Google Scholar]
- 19.Adler B, Ballard SA, Miller SJ, Faine S. FEMS Microbiol Immunol. 1989;1:213–218. doi: 10.1111/j.1574-6968.1989.tb02385.x. [DOI] [PubMed] [Google Scholar]
- 20.Midwinter A, Vinh T, Faine S, Adler B. Infect Immun. 1994;62:5477–5482. doi: 10.1128/iai.62.12.5477-5482.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Adler B, Faine S. In: The Prokaryotes. Dworkin M, editor. Springer-Verlag; New York: 2003. [Google Scholar]
- 22.Werts C, Tapping RI, Mathison JC, Chuang TH, Kravchenko V, Saint Girons I, Haake DA, Godowski PJ, Hayashi F, Ozinsky A, Underhill DM, Kirschning CJ, Wagner H, Aderem A, Tobias PS, Ulevitch RJ. Nat Immunol. 2001;2:346–352. doi: 10.1038/86354. [DOI] [PubMed] [Google Scholar]
- 23.Poltorak A, He X, Smirnova I, Liu MY, Huffel CV, Du X, Birdwell D, Alejos E, Silva M, Galanos C, Freudenberg M, Ricciardi-Castagnoli P, Layton B, Beutler B. Science. 1998;282:2085–2088. doi: 10.1126/science.282.5396.2085. [DOI] [PubMed] [Google Scholar]
- 24.Lien E, Ingalls RR. Crit Care Med. 2002;30:S1–11. [PubMed] [Google Scholar]
- 25.Janeway CA, Jr, Medzhitov R. Annu Rev Immunol. 2002;20:197–216. doi: 10.1146/annurev.immunol.20.083001.084359. [DOI] [PubMed] [Google Scholar]
- 26.Takeda K, Akira S. Cell Microbiol. 2003;5:143–153. doi: 10.1046/j.1462-5822.2003.00264.x. [DOI] [PubMed] [Google Scholar]
- 27.de la Pena-Moctezuma A, Bulach DM, Adler B. FEMS Immunol Med Microbiol. 2001;31:73–81. doi: 10.1111/j.1574-695X.2001.tb01589.x. [DOI] [PubMed] [Google Scholar]
- 28.Sweet CR, Williams AH, Karbarz MJ, Werts C, Kalb SR, Cotter RJ, Raetz CRH. J Biol Chem. 2004;279:25411–25419. doi: 10.1074/jbc.M400597200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sweet CR, Ribeiro AA, Raetz CRH. J Biol Chem. 2004;279:25400–25410. doi: 10.1074/jbc.M400596200. [DOI] [PubMed] [Google Scholar]
- 30.Singh R, Reddy R. Proc Natl Acad Sci U S A. 1989;86:8280–8283. doi: 10.1073/pnas.86.21.8280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kates M, Moldoveanu N, Stewart LC. Biochim Biophys Acta. 1993;1169:46–53. doi: 10.1016/0005-2760(93)90080-s. [DOI] [PubMed] [Google Scholar]
- 32.Shumyatsky G, Shimba S, Reddy R. Gene Expr. 1994;4:29–41. [PMC free article] [PubMed] [Google Scholar]
- 33.Zähringer U, Lindner B, Rietschel ET. In: Endotoxin in Health and Disease. Brade H, Opal SM, Vogel SN, Morrison DC, editors. Marcel Dekker, Inc.; New York: 1999. pp. 93–114. [Google Scholar]
- 34.Que NLS, Ramirez S, Werts C, Ribeiro AA, Bulach DM, Cotter RJ, Raetz CRH. J Endotoxin Res. 2002;8:165. [Google Scholar]
- 35.Johnson RC, Harris VG, Walby J, Henry RA, Auran NE. Appl Microbiol. 1973;26:118–119. doi: 10.1128/am.26.1.118-119.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ellinghausen HC, McCulloch WG. Am J Vet Res. 1965;26:45–51. [PubMed] [Google Scholar]
- 37.Johnson RC, Harris VG. J Bacteriol. 1967;94:27–31. doi: 10.1128/jb.94.1.27-31.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Westphal O, Jann K. Methods Carbohydr Chem. 1965;5:83–91. [Google Scholar]
- 39.Manthey CL, Vogel SL. J Endotoxin Res. 1994;1:84–91. [Google Scholar]
- 40.Hirschfeld M, Ma Y, Weis JH, Vogel SN, Weis JJ. J Immunol. 2000;165:618–622. doi: 10.4049/jimmunol.165.2.618. [DOI] [PubMed] [Google Scholar]
- 41.Tapping RI, Akashi S, Miyake K, Godowski P, Tobias PS. J Immunol. 2000;165:5780–5787. doi: 10.4049/jimmunol.165.10.5780. [DOI] [PubMed] [Google Scholar]
- 42.Que NLS, Lin S, Cotter RJ, Raetz CRH. J Biol Chem. 2000;275:28006–28016. doi: 10.1074/jbc.M004008200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bligh EG, Dyer JJ. Can J Biochem Physiol. 1959;37:911–917. doi: 10.1139/o59-099. [DOI] [PubMed] [Google Scholar]
- 44.Raetz CRH, Kennedy EP. J Biol Chem. 1973;248:1098–1105. [PubMed] [Google Scholar]
- 45.Raetz CRH, Purcell S, Meyer MV, Qureshi N, Takayama K. J Biol Chem. 1985;260:16080–16088. [PubMed] [Google Scholar]
- 46.Zhou Z, Lin S, Cotter RJ, Raetz CRH. J Biol Chem. 1999;274:18503–18514. doi: 10.1074/jbc.274.26.18503. [DOI] [PubMed] [Google Scholar]
- 47.Zhou Z, Ribeiro AA, Lin S, Cotter RJ, Miller SI, Raetz CRH. J Biol Chem. 2001;276:43111–43121. doi: 10.1074/jbc.M106960200. [DOI] [PubMed] [Google Scholar]
- 48.Que NLS, Ribeiro AA, Raetz CRH. J Biol Chem. 2000;275:28017–28027. doi: 10.1074/jbc.M004009200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ribeiro AA, Zhou Z, Raetz CRH. Magn Res Chem. 1999;37:620–630. [Google Scholar]
- 50.Zhou Z, Ribeiro AA, Raetz CRH. J Biol Chem. 2000;275:13542–13551. doi: 10.1074/jbc.275.18.13542. [DOI] [PubMed] [Google Scholar]
- 51.Costello CE, Vath JE. Methods Enzymol. 1990;193:738–768. doi: 10.1016/0076-6879(90)93448-t. [DOI] [PubMed] [Google Scholar]
- 52.Sweet CR, Preston A, Toland E, Ramirez SM, Cotter RJ, Maskell DJ, Raetz CRH. J Biol Chem. 2002;277:18281–18290. doi: 10.1074/jbc.M201057200. [DOI] [PubMed] [Google Scholar]
- 53.Basu SS, White KA, Que NL, Raetz CRH. J Biol Chem. 1999;274:11150–11158. doi: 10.1074/jbc.274.16.11150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Agrawal PK. Phytochemistry. 1992;31:3307–3330. doi: 10.1016/0031-9422(92)83678-r. [DOI] [PubMed] [Google Scholar]
- 55.Takayama K, Qureshi N, Mascagni P, Nashed MA, Anderson L, Raetz CRH. J Biol Chem. 1983;258:7379–7385. [PubMed] [Google Scholar]
- 56.Strain SM, Armitage IM, Anderson L, Takayama K, Qureshi N, Raetz CRH. J Biol Chem. 1985;260:16089–16098. [PubMed] [Google Scholar]
- 57.Brozek KA, Bulawa CE, Raetz CRH. J Biol Chem. 1987;262:5170–5179. [PubMed] [Google Scholar]
- 58.Kass LR, Bloch K. Proc Natl Acad Sci U S A. 1967;58:1168–1173. doi: 10.1073/pnas.58.3.1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rando RR, Bloch K. J Biol Chem. 1968;243:5627–5634. [PubMed] [Google Scholar]
- 60.Magnuson K, Jackowski S, Rock CO, Cronan JE., Jr Microbiol Rev. 1993;57:522–542. doi: 10.1128/mr.57.3.522-542.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Galloway SM, Raetz CRH. J Biol Chem. 1990;265:6394–6402. [PubMed] [Google Scholar]
- 62.Wyckoff TJ, Raetz CRH. J Biol Chem. 1999;274:27047–27055. doi: 10.1074/jbc.274.38.27047. [DOI] [PubMed] [Google Scholar]
- 63.Raetz CRH, Roderick SL. Science. 1995;270:997–1000. doi: 10.1126/science.270.5238.997. [DOI] [PubMed] [Google Scholar]
- 64.Onishi HR, Pelak BA, Gerckens LS, Silver LL, Kahan FM, Chen MH, Patchett AA, Galloway SM, Hyland SA, Anderson MS, Raetz CRH. Science. 1996;274:980–982. doi: 10.1126/science.274.5289.980. [DOI] [PubMed] [Google Scholar]
- 65.Steeghs L, den Hartog R, den Boer A, Zomer B, Roholl P, van der Ley P. Nature. 1998;392:449–450. doi: 10.1038/33046. [DOI] [PubMed] [Google Scholar]
- 66.Steeghs L, de Cock H, Evers E, Zomer B, Tommassen J, van der Ley P. EMBO J. 2001;20:6937–6945. doi: 10.1093/emboj/20.24.6937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Weckesser J, Mayer H. FEMS Microbiol Rev. 1988;4:143–153. doi: 10.1111/j.1574-6968.1988.tb02740.x. [DOI] [PubMed] [Google Scholar]
- 68.Carty SM, Sreekumar KR, Raetz CRH. J Biol Chem. 1999;274:9677–9685. doi: 10.1074/jbc.274.14.9677. [DOI] [PubMed] [Google Scholar]
- 69.Vorachek-Warren MK, Carty SM, Lin S, Cotter RJ, Raetz CRH. J Biol Chem. 2002;277:14186–14193. doi: 10.1074/jbc.M200408200. [DOI] [PubMed] [Google Scholar]
- 70.Vorachek-Warren MK, Ramirez S, Cotter RJ, Raetz CRH. J Biol Chem. 2002;277:14194–14205. doi: 10.1074/jbc.M200409200. [DOI] [PubMed] [Google Scholar]
- 71.Price NJP, Jeyaretnam B, Carlson RW, Kadrmas JL, Raetz CRH, Brozek KA. Proc Natl Acad Sci U S A. 1995;92:7352–7356. doi: 10.1073/pnas.92.16.7352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Brozek KA, Kadrmas JL, Raetz CRH. J Biol Chem. 1996;271:32112–32118. [PubMed] [Google Scholar]
- 73.Clementz T, Raetz CRH. J Biol Chem. 1991;266:9687–9696. [PubMed] [Google Scholar]
- 74.Belunis CJ, Raetz CRH. J Biol Chem. 1992;267:9988–9997. [PubMed] [Google Scholar]
- 75.Garrett TA, Kadrmas JL, Raetz CRH. J Biol Chem. 1997;272:21855–21864. doi: 10.1074/jbc.272.35.21855. [DOI] [PubMed] [Google Scholar]
- 76.Kanipes MI, Lin S, Cotter RJ, Raetz CRH. J Biol Chem. 2001;276:1156–1163. doi: 10.1074/jbc.M009019200. [DOI] [PubMed] [Google Scholar]
- 77.Brozek KA, Hosaka K, Robertson AD, Raetz CRH. J Biol Chem. 1989;264:6956–6966. [PubMed] [Google Scholar]
- 78.Cui Z, Vance JE, Chen MH, Voelker DR, Vance DE. J Biol Chem. 1993;268:16655–16663. [PubMed] [Google Scholar]
- 79.Arondel V, Benning C, Somerville CR. J Biol Chem. 1993;268:16002–16008. [PubMed] [Google Scholar]
- 80.Toews ML, Goy MF, Springer MS, Adler J. Proc Natl Acad Sci U S A. 1979;76:5544–5548. doi: 10.1073/pnas.76.11.5544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Armitage JP. Adv Microb Physiol. 1999;41:229–289. doi: 10.1016/s0065-2911(08)60168-x. [DOI] [PubMed] [Google Scholar]
- 82.Dai Q, Choy E, Chiu V, Romano J, Slivka SR, Steitz SA, Michaelis S, Philips MR. J Biol Chem. 1998;273:15030–15034. doi: 10.1074/jbc.273.24.15030. [DOI] [PubMed] [Google Scholar]
- 83.Bergo MO, Leung GK, Ambroziak P, Otto JC, Casey PJ, Young SG. J Biol Chem. 2000;275:17605–17610. doi: 10.1074/jbc.C000079200. [DOI] [PubMed] [Google Scholar]
- 84.Marr RS, Blair LC, Thorner J. J Biol Chem. 1990;265:20057–20060. [PubMed] [Google Scholar]
- 85.Boon Hinckley M, Werts C, Raetz CRH. FASEB J. 2004 in press. [Google Scholar]