Abstract
Transporters for di- and tripeptides belong to the large and poorly characterized PTR/NRT1 (peptide transporter/nitrate transporter 1) family. A new member of this gene family, AtPTR5, was isolated from Arabidopsis (Arabidopsis thaliana). Expression of AtPTR5 was analyzed and compared with tissue specificity of the closely related AtPTR1 to discern their roles in planta. Both transporters facilitate transport of dipeptides with high affinity and are localized at the plasma membrane. Mutants, double mutants, and overexpressing lines were exposed to several dipeptides, including toxic peptides, to analyze how the modified transporter expression affects pollen germination, growth of pollen tubes, root, and shoot. Analysis of atptr5 mutants and AtPTR5-overexpressing lines showed that AtPTR5 facilitates peptide transport into germinating pollen and possibly into maturating pollen, ovules, and seeds. In contrast, AtPTR1 plays a role in uptake of peptides by roots indicated by reduced nitrogen (N) levels and reduced growth of atptr1 mutants on medium with dipeptides as the sole N source. Furthermore, overexpression of AtPTR5 resulted in enhanced shoot growth and increased N content. The function in peptide uptake was further confirmed with toxic peptides, which inhibited growth. The results show that closely related members of the PTR/NRT1 family have different functions in planta. This study also provides evidence that the use of organic N is not restricted to amino acids, but that dipeptides should be considered as a N source and transport form in plants.
Plants have evolved multiple transport systems for nitrogen (N) to facilitate uptake from soil and internal reallocation to support development, growth, and reproduction (Glass et al., 2002; Rentsch et al., 2007). Although organic N is generally the most prevalent form of N in the soil, it is considered to be a N source for soil microorganisms rather than plants (Kaye and Hart, 1997). However, the view that plants rely entirely on microorganisms for mineralization of organic N has been revised. Plant roots can take up amino acids (Wright, 1962) and the ecological significance of this ability has been recognized (Chapin et al., 1993; Näsholm et al., 1998; Schmidt and Stewart, 1999). Studies with Arabidopsis (Arabidopsis thaliana) mutants atlht1 and ataap1 provided molecular evidence that amino acid transporters facilitate uptake of amino acids into roots (Hirner et al., 2006; Lee et al., 2007; Svennerstam et al., 2007). In addition to amino acids, more complex organic N sources may also be important. There is evidence that plant roots access proteins as a N source without relying on soil microbes or symbiotic fungi (Paungfoo-Lonhienne et al., 2008). Likely mechanisms for protein acquisition include exudation of proteolytic enzymes at the root surface and possibly in the root apoplast and uptake of protein via endocytosis (Paungfoo-Lonhienne et al., 2008). Thus, peptides and proteins may be more important N sources for plants than previously considered. Plants were shown to be able to grow on small peptides, but it was not established whether hydrolysis occurred prior to uptake (Higgins and Payne, 1982).
Rather than being considered a N source for growth, small peptides are viewed in the context of germination and senescence, which require rapid proteolysis and retranslocation of N (Higgins and Payne, 1982). Protein degradation products from the endosperm are transported as di- and oligopeptides into the scutellum of germinating barley (Hordeum vulgare) grains to supply the embryo (Higgins and Payne, 1978). Rates of peptide transport into the scutellum were higher than transport rates of amino acids, which highlights the importance of peptides for supplying sinks (Higgins and Payne, 1978). Peptide transport also occurs in mature broad bean (Vicia faba) leaves (Jamai et al., 1994), which in turn demonstrates that peptide transport is not restricted to specific developmental stages. Rather, mobilization of peptides from proteolysis and transport of peptides throughout the plant appears to be an efficient strategy for rapid reallocation of N.
Data available on concentration of small peptides in soil and plants are limited, mainly hampered by the lack of suitable detection systems. Breakdown of proteins and the rapid conversion of peptides into amino acids are problematic issues when preparing biological samples for determining the concentration of small peptides. Pools of small peptides have been reported, but the size and composition of these pools were determined only in few cases (Higgins and Payne, 1982). The least ambiguous evidence for a significant amount of peptides comes from studies with germinating barley grains, which demonstrated a pool of peptides of two to six amino acids with similar composition to seed storage proteins (Higgins and Payne, 1981, 1982).
Peptide transporters that have been identified in plants belong to three gene families, each recognizing peptides of defined length. Di- and tripeptides are transported by members of the PTR/NRT1 (peptide transporter/nitrate transporter 1) family, transport of peptides of four to five amino acids is mediated by members of the OPT (oligopeptide transporter) family, and transporters for larger peptides, so far only characterized in animals, have been identified among the ATP-binding cassette (ABC) transporters (Rentsch et al., 2007).
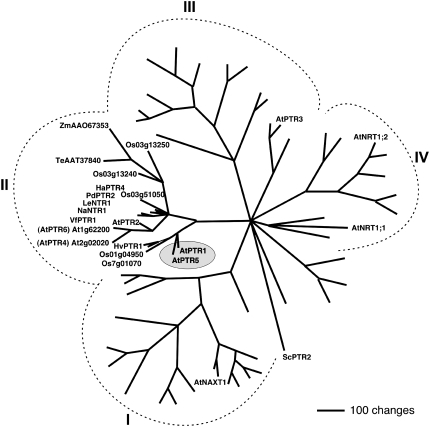
In prokaryotes, fungi, and animals, the PTR/NRT1 gene family has few members, while in plants this gene family is much larger. Arabidopsis has 53 genes in four subfamilies of PTR/NRT1 (Fig. 1; Tsay et al., 2007). For most PTR/NRT1 transporters in plants, substrate selectivity has not been determined and some PTR/NRT1 proteins mediate transport of substrates other than peptides (i.e. uptake or export of nitrate or carboxylates; Jeong et al., 2004; Segonzac et al., 2007; Tsay et al., 2007). Several members of subfamily II (see below) and one member of subgroup III (AtPTR3, At5g46050) are confirmed transporters for di- and tripeptides (Karim et al., 2005, 2007; Rentsch et al., 2007). Species with confirmed peptide transporters in subgroup II include Arabidopsis (AtPTR1, AtPTR2), fava bean (VfPTR1), Hakea actities (HaPTR4), and barley (HvPTR1; Frommer et al., 1994; Rentsch et al., 1995; West et al., 1998; Miranda et al., 2003; Dietrich et al., 2004). Subfamily II also contains several noncharacterized genes from Arabidopsis and other species (Fig. 1).
Figure 1.
Phylogenetic relationship between AtPTR1, AtPTR5, and related proteins. The analysis was performed using the aligned protein sequences of the Arabidopsis PTR/NRT1 gene family, including proteins from other plant species for proteins of subfamily II (PTR1-like proteins; Rentsch et al., 2007; Tsay et al., 2007; VfPTR1, Miranda et al., 2003; NaNTR1, Schulze et al., 1999; HaPTR4, C. Paungfoo-Lonhienne, D. Rentsch, and S. Schmidt, unpublished data; PdPTR2, Campalans et al., 2001; LeNTR1, AF016713; HvPTR1, West et al., 1998). Maximum parsimony analysis was performed using PAUP 4.0b10 with all characters unweighted and gaps scored as missing characters (Swofford, 2003). The complete alignment was based on 1,409 amino acids, 633 characters were parsimony informative. The yeast peptide transporter ScPTR2 was used as the outgroup.
AtPTR1 and AtPTR2 recognize various di- and tripeptides with high affinity (Rentsch et al., 1995; Chiang et al., 2004; Dietrich et al., 2004). Chiang et al. (2004) also established that AtPTR2 transports peptides and protons simultaneously by a random binding mechanism. Evidence for a physiological role of these di-/tripeptide transporters in plants is limited to antisense repression of AtPTR2, which showed that peptide transport is important for flowering and seed growth (Song et al., 1997).
Linking functional characterization of peptide transporters in heterologous expression systems with physiological evidence for peptide transport, we isolated and characterized the Arabidopsis di-/tripeptide transporter AtPTR5. We demonstrate that AtPTR5 mediates uptake of peptides during pollen germination. Further, expression and localization studies suggest a function of AtPTR5 in N transport during ovule and early seed development. Using atptr mutants and AtPTR5-overexpressing lines, we also provide evidence that peptide transporters facilitate uptake of N from the rhizosphere.
RESULTS
AtPTR5 Transports Dipeptides When Expressed in Yeast and Xenopus Oocytes
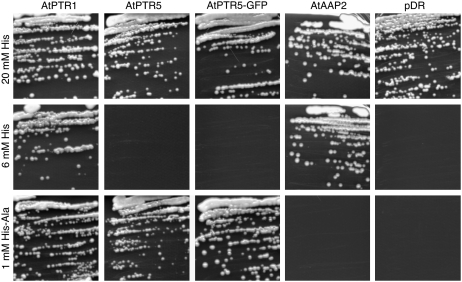
AtPTR5 (At5g01180) belongs to subgroup II of the PTR/NRT1 family and has highest homology to AtPTR1 (74% identity on amino acid level), to barley HvPTR1 (64.7% identity), and two noncharacterized open reading frames (ORFs) from rice (Oryza sativa; Os01g04950, 67% identity and Os7g01070, 62% identity; Fig. 1; Tsay et al., 2007). An AtPTR5-ORF was isolated by reverse transcription (RT)-PCR using total RNA of Arabidopsis flowers. Expression of AtPTR5 in the peptide transport-deficient and His auxotroph yeast (Saccharomyces cerevisiae) strain LR2 showed that, similar to other members of subfamily II, AtPTR5 mediates growth when using selective concentrations of His-Ala (1 mm) as sole source of His (Fig. 2; Frommer et al., 1994; Rentsch et al., 1995; West et al., 1998; Miranda et al., 2003; Dietrich et al., 2004). Interestingly, the Arabidopsis di- and tripeptide transporters AtPTR1 and AtPTR2 also mediated growth on selective concentrations of His (6 mm), although affinity for His and transport rates seem to be rather low (Frommer et al., 1994; Chiang et al., 2004; Dietrich et al., 2004). In contrast, AtPTR5 did not mediate growth on 6 mm His (Fig. 2). As control, vector-transformed cells and LR2 expressing the amino acid permease AtAAP2 that mediates growth on His, but not on dipeptides, is shown (Kwart et al., 1993; Rentsch et al., 1995; Dietrich et al., 2004).
Figure 2.
Complementation of a yeast strain (LR2) deficient in the uptake of di- and tripeptides. Shown is growth of LR2 cells expressing the peptide transporters AtPTR1 (Dietrich et al., 2004), AtPTR5, and AtPTR5-GFP, the amino acid transporter AtAAP2 (Kwart et al., 1993), and the strain transformed with the vector pDR195 on SC medium containing selective concentrations of His (i.e. 6 mm His or 1 mm His-Ala) or nonselective His concentrations (20 mm).
To compare the kinetic properties of AtPTR5 with other peptide transporters, dipeptide-induced currents of neutral, basic, and acidic amino acid-containing dipeptides were analyzed in Xenopus laevis oocytes. At a membrane potential of −140 mV and [H+]out of 3.2 μm (pH 5.5), the addition of 1 mm Ala-Ala, Ala-Lys, and Ala-Asp to the bathing medium induced inward currents with a range of 70 to 740 nA in oocytes expressing AtPTR5. No dipeptide-induced currents were observed in control oocytes. Peptide-induced inward currents were voltage dependent, increasing with hyperpolarizing membrane potential within the range of −140 mV to +20 mV (data not shown). At −140 mV and [H+]out of 3.2 μm, the apparent affinity of AtPTR5 for Ala-Asp (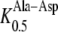 131 ± 1 μm) and Ala-Lys (
131 ± 1 μm) and Ala-Lys (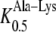 163 ± 7 μm) was similar to the affinity determined for other di-/tripeptide transporters (Chiang et al., 2004; Dietrich et al., 2004).
163 ± 7 μm) was similar to the affinity determined for other di-/tripeptide transporters (Chiang et al., 2004; Dietrich et al., 2004).
AtPTR5 Is Localized at the Plasma Membrane
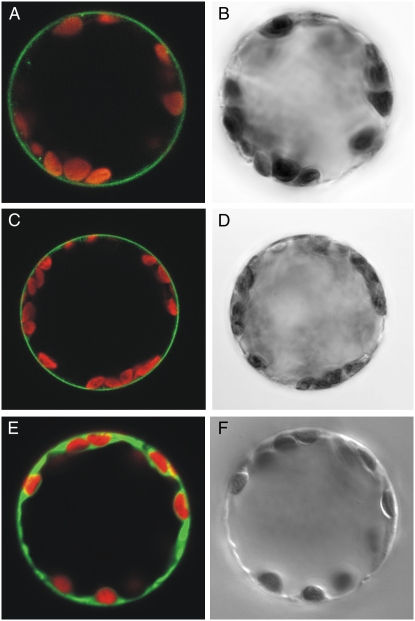
Immunodetection of HvPTR1 on isolated plasma membranes or expression of AtPTR1-GFP and GFP-AtPTR1 fusion proteins have been used to demonstrate the localization at the plasma membrane (Waterworth et al., 2000; Dietrich et al., 2004). Functional complementation of the yeast peptide transport mutant LR2 by AtPTR5 and functional expression of AtPTR5 in X. laevis oocytes indicated that at least part of the AtPTR5 protein is localized at the plasma membrane. To corroborate evidence for localization, N- and C-terminal fusion proteins of AtPTR5 and GFP were transiently expressed in tobacco (Nicotiana tabacum) protoplasts. Fluorescence images showed that AtPTR5-GFP and GFP-AtPTR5 fusion proteins localize at the plasma membrane (Fig. 3, A and C, respectively). Free GFP was localized in the cytosol (Fig. 3E). Fusion to GFP did not disturb peptide transport function because the AtPTR5-GFP fusion protein was functional when expressed in yeast (Fig. 2).
Figure 3.
Localization of AtPTR5/GFP fusion proteins at the plasma membrane of tobacco protoplasts. Confocal laser-scanning microscope images (A, C, E) and corresponding bright-field images (B, D, F) of tobacco protoplasts transiently expressing fusion proteins of GFP with AtPTR5 or GFP. A and B, AtPTR5-GFP. C and D, GFP-AtPTR5. E and F, GFP. Merged images show GFP fluorescence (green) and chlorophyll fluorescence (red). Diameter of protoplasts is approximately 40 μm.
AtPTR5 Is Expressed in Pollen, Ovules, and during Early Seed Development
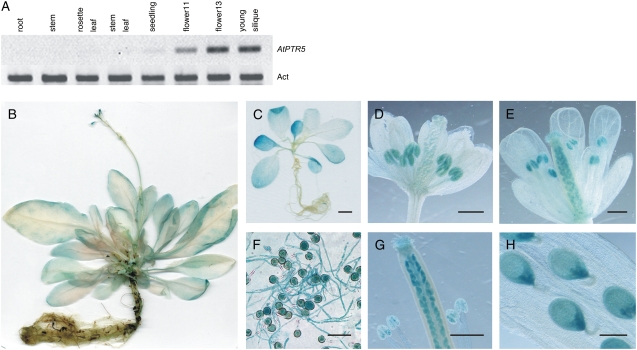
Semiquantitative RT-PCR using RNA from soil-grown plants showed weak AtPTR5 expression in seedlings, flowers, and young siliques (Fig. 4A). This expression pattern was supported by analyses of 37 independent transgenic Arabidopsis lines, expressing the Escherichia coli GUS (uidA) gene under control of a 2,582-bp fragment of the AtPTR5 promoter. All lines displayed highest GUS activity in mature and germinating pollen, ovules, and developing seeds (Fig. 4, D–H). This expression pattern was corroborated by publicly available microarray data (e.g. Genevestigator; Zimmermann et al., 2004). GUS activity decreased after the early torpedo stage of embryo development and was absent in mature and germinating seeds (Fig. 4H; data not shown). One-half of the lines had weak GUS activity in leaves of seedlings, and mature plants of all lines displayed weak GUS activity in leaves and older sections of roots (Fig. 4, B and C).
Figure 4.
Expression analysis of AtPTR5 in Arabidopsis. A, Expression was analyzed by using semiquantitative RT-PCR with the actin mRNA (Act, At3g18780) as a reference. RNA from source leaves, stems, roots, flowers, and siliques of soil-grown plants was analyzed. Similar results were obtained using RNAs from three independent experiments as template. B to H, Expression of the uidA gene under control of the AtPTR5 promoter. AtPTR5-GUS expression in 6-week-old plants (B), 2-week-old seedlings (C), flowers before pollination (stage 12; D), and after pollination (stage 14; E), in vitro-germinated pollen (F), and developing seeds (G and H). No staining could be detected in mature seeds and during seed germination and seedling establishment. Bars = 2,000 μm in C, 25 μm in F, 200 μm in H, and 500 μm in D, E, and G.
AtPTR1 and AtPTR5 Mediate Transport of Peptides in Planta
To assess the role of AtPTR5 and its closest homolog AtPTR1 for uptake of peptides from the growth medium and translocation in plants, we analyzed atptr1 and atptr5 mutants and transgenic Arabidopsis lines overexpressing AtPTR5 under the control of the constitutive 35S promoter.
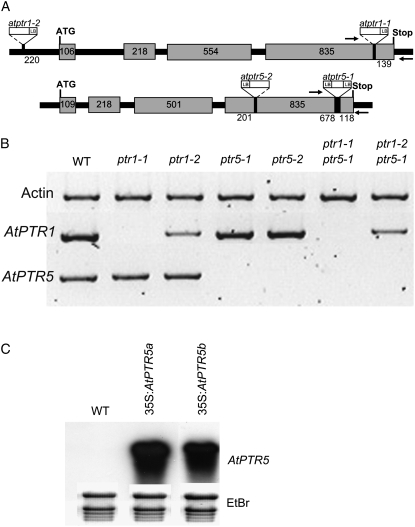
Atptr1-1 carries the T-DNA in the last exon, whereas atptr1-2 has an insertion 220 bp upstream of the start codon, lying in an intron of the 5′-untranslated region (UTR; Fig. 5A). Both atptr5 mutant lines carry an insertion in the last exon. Using total RNA from leaves (AtPTR1) or flowers (AtPTR5) and primers downstream of the insertion or enclosing the T-DNA (Fig. 5A), RT-PCR analysis verified that mRNA of AtPTR1 and AtPTR5 was absent in the atptr1-1 and both atptr5 mutant lines, as well as in the corresponding double mutants. In the atptr1-2 single and atptr1-2 atptr5-1 double mutant, mRNA levels of AtPTR1 were only partially reduced (Fig. 5B). With primers upstream of the T-DNA insertion, AtPTR5 and AtPTR1 transcripts could be amplified in both atptr5 lines and in the atptr1-1 line (data not shown). As the partial mRNA could result in a truncated, but possibly still functional protein, a cDNA was constructed that reflects the genomic situation in the atptr5-1 mutant and consisted of the truncated AtPTR5-ORF and part of the T-DNA. The cDNA was expressed in the yeast strain LR2. This truncated AtPTR5 was not able to restore growth of the yeast mutant, indicating that in the atptr5-1 line AtPTR5-mediated peptide transport does not occur (data not shown).
Figure 5.
Characterization of homozygous atptr1, atptr5, and atptr1 atptr5 mutants as well as overexpression lines (35S:AtPTR5). A, Position of T-DNA or transposon insertions in the AtPTR1 or AtPTR5 gene. Gray boxes represent exons. Numbers indicate length in base pairs for each exon or distances between exon borders and insertions. Arrows show position of primers used for RT-PCR in B. ATG, Start codon; Stop, stop codon; LB, left border of the insertion. B, Semiquantitative RT-PCR analysis of expression of AtPTR1, AtPTR5, and actin in wild type, the single mutants atptr1-1, atptr1-2, atptr5-1, and atptr5-2, and the double mutants atptr1-1 atptr5-1 and atptr1-2 atptr5-1 using RNA from flowers (AtPTR5) and leaves (AtPTR1). C, Northern-blot analysis of RNA from mature leaves of wild-type plants (WT) and 35S:AtPTR5-overexpressing lines (35S: AtPTR5a and 35S:AtPTR5b). As a loading control, staining with ethidium bromide (EtBr) is shown.
In addition to the analysis of knockout mutants, 45 independent transgenic Arabidopsis lines overexpressing AtPTR5 under control of the 35S promoter were analyzed for AtPTR5 transcript levels. Two lines (T3 plants) with elevated AtPTR5 mRNA in leaves were selected for further investigation (Fig. 5C). When cultured in soil or on AM medium, no growth phenotype (e.g. shoot growth, root length, flowering time) could be detected between wild type, single and double mutants, or overexpression lines. Furthermore, in all experiments, the corresponding single and double mutants behaved identically. Although the atptr1-2 line showed only a partial reduction in AtPTR1 transcript level in leaves, no difference to atptr1-1 was found.
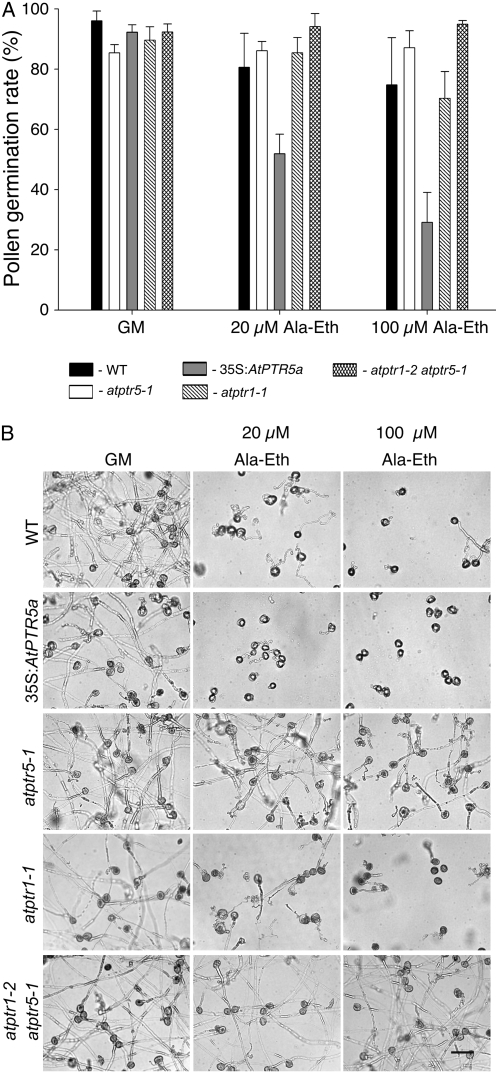
GUS analyses and publicly available microarray data (e.g. Genevestigator; Zimmermann et al., 2004) showed high expression of AtPTR5 in pollen. To assess peptide transport activity during pollen germination, pollen was germinated for 6 and 14 h in the presence of the toxic dipeptide alanyl-ethionine (Ala-Eth) and the effect on germination rate and pollen tube growth was determined. Germination was defined as protrusion of pollen tubes from the pollen grain, whereas elongation of the pollen tube defined growth. On germination medium (GM; Footitt et al., 2007), no differences were observed in germination rate and growth of pollen tubes of wild type and mutants (atptr1-1, atptr1-2, atptr5-1, atptr5-2, atptr1-2 atptr5-1, and 35S:AtPTR5a; Fig. 6, all lines mentioned in the text were tested, but only one of the lines is shown). In the presence of 100 μm Ala-Eth, germination was strongly reduced in the 35S:AtPTR5a line, whereas no difference was observed between wild-type pollen and pollen of the different knockout lines (Fig. 6A). On Ala-Eth-containing medium, growth of the pollen tube was least affected in both atptr5 knockout lines. Reduction of pollen tube elongation was in the order 35S:AtPTR5a > (atptr1-1, atptr1-2, wild type) > (atptr5-1, atptr5-2, atptr1-2 atptr5-1), indicating that AtPTR5 mediates uptake of peptides during pollen germination and tube growth (Fig. 6B).
Figure 6.
In vitro pollen germination of wild type, atptr1-1, atptr5-1, atptr1-2 atptr5-1, and 35S:AtPTR5a. A, Pollen germination rate of wild type, atptr1-1, atptr5-1, atptr1-2 atptr5-1, and 35S:AtPTR5a. Pollen was germinated for 14 h on GM (Footitt et al., 2007) in the absence or presence of 20 or 100 μm of the toxic dipeptide Ala-Eth. Germination was defined as emergence of the pollen tubes. B, Germination and growth of pollen tubes of wild type, atptr1-1, atptr5-1, atptr1-2 atptr5-1, and 35S:AtPTR5a; bar = 25 μm. Germination and pollen tube growth of atptr1-2 and atptr5-2 mutants was comparable to results shown for atptr1-1 and atptr5-1, respectively (data not shown).
Although AtPTR5 expression was detected in the ovule and early stages of seed development, this did not affect mature seeds of atptr5, 35S:AtPTR5a, and wild-type plants as they had a similar N content (data not shown).
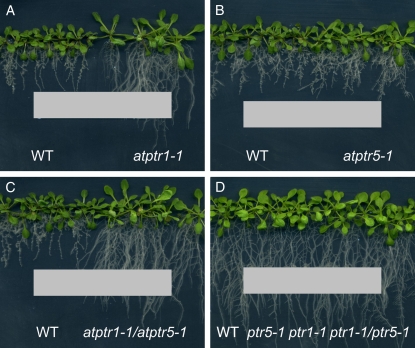
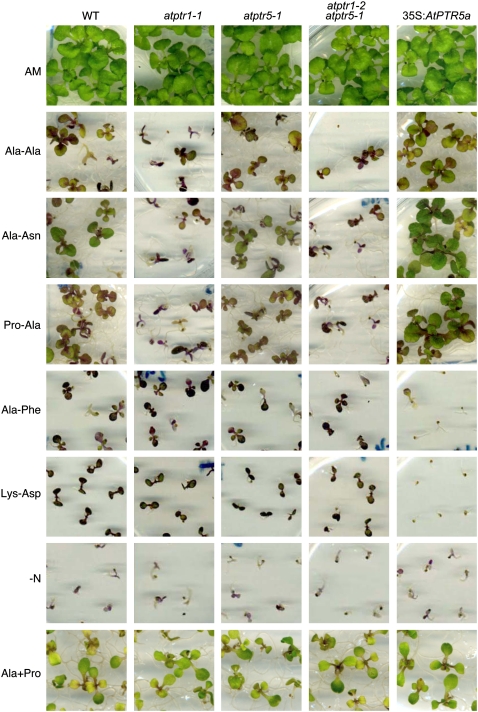
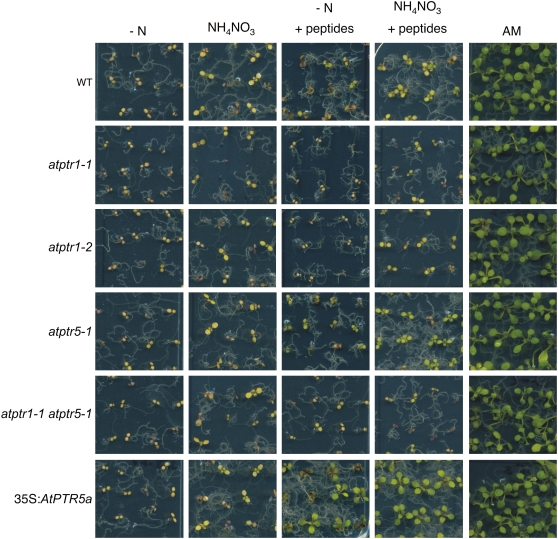
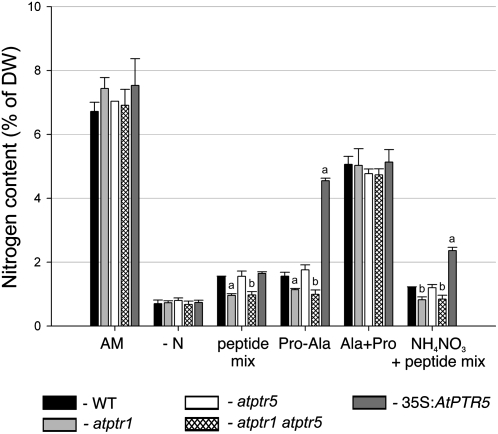
In contrast to AtPTR5, AtPTR1 expression was also detected in roots (Dietrich et al., 2004). In agreement with these findings, in the presence of the toxic dipeptide Ala-Eth, root growth of both atptr1 lines and the atptr1 atptr5 double mutants was less affected than wild type or the atptr5 single mutants (Fig. 7, A–C). On AM medium, no differences in root growth could be observed among the lines (Fig. 7D). To investigate whether AtPTR1 and/or AtPTR5 is required for uptake of small peptides from the medium and whether these peptides can be used as the sole N source, wild-type, single and double mutants (atptr1-1, atptr1-2, atptr5-1, atptr5-2, atptr1-1 atptr5-1, atptr1-2 atptr5-1), as well as both 35S:AtPTR5 lines were germinated on minimal medium (−N) containing 20 mm of a dipeptide as sole source of N (Fig. 8). After 2 weeks on AM control medium, no differences in shoot growth were observed between wild-type and mutant lines. On medium with any of the dipeptides as sole N source, all seedlings produced more anthocyanins and shoot growth was reduced, while thickness of shoots seemingly increased. On Ala-Ala, Ala-Asn, and Pro-Ala no difference in growth could be observed between wild type and both atptr5 mutants, whereas atptr1 single and atptr1 atptr5 double mutants were more strongly reduced in shoot growth. No difference between the atptr1 mutants, wild type, and the atptr5 mutants was observed on Ala-Phe or Lys-Asp (Fig. 8). In contrast, growth of both overexpressing lines (35S:AtPTR5) was improved on Ala-Ala, Pro-Ala, and Ala-Asn and was strongly reduced on Ala-Phe and Lys-Asp (Fig. 8). As in soil, peptides are expected to be present as a mixture rather than as a single dipeptide, seedlings were also cultivated on a mixture of dipeptides (2.5 mm) in the presence or absence of low concentrations (0.1 mm) of ammonium nitrate (Fig. 9). This mixture contained five different dipeptides (0.5 mm each), consisting of amino acids that were described to be nontoxic at low concentrations (Voll et al., 2004; Lee et al., 2007). All lines grew equally well on control AM medium, growth was reduced on low ammonium nitrate, and poor on medium lacking N. In contrast, on medium containing dipeptides (with and without ammonium nitrate), overexpressing lines produced more leaves and were less chlorotic than wild type or knockout mutants, and wild type and atptr5 mutants performed slightly better than the atptr1 and atptr1 atptr5 mutants. These visual phenotypic changes were confirmed by analysis of biomass. Compared to wild type, shoot dry weight of overexpressing lines was significantly increased up to 3-fold on Ala-Ala, Pro-Ala, and the peptide mixture, or decreased up to nearly 4-fold on Ala-Phe (Table I). In contrast, shoot dry weight was reduced by approximately 50% in atptr1 and atptr1 atptr5 seedlings on Ala-Ala and Pro-Ala. Shoot biomass of atptr5 lines, on the other hand, was similar to wild type. Analysis of N content of shoots (% of dry weight) corroborates the evidence that changes in seedling growth and shoot dry weight are due to differences in N uptake (Fig. 10). N content of atptr1 and atptr1 atptr5 seedlings cultivated on dipeptides was significantly reduced, whereas the N content of atptr5 lines was similar to wild type. Analysis of 35S:AtPTR5 lines revealed an increase in shoot N content of up to 3-fold when grown on Pro-Ala as sole N source (Fig. 10). Shoot N content was also elevated when seedlings were cultivated on dipeptides in the presence of low concentrations of ammonium nitrate.
Figure 7.
Root growth of Arabidopsis wild-type (WT), atptr1-1, atptr5-1, and atptr1-1 atptr5-1 seedlings in the presence of the toxic dipeptide Ala-Eth. Arabidopsis plants were grown for 7 d on AM medium and subsequently for 10 d on AM medium containing a filter strip soaked with 1.1 μmol Ala-Eth (A–C) or with water (D). Data shown are representative for the independent single and double mutants.
Figure 8.
Growth of Arabidopsis wild-type (WT), atptr1-1, atptr5-1, atptr1-2 atptr5-1, and 35S:AtPTR5a seedlings on medium containing peptides as sole N source. Seeds were germinated and plants cultivated for 2 weeks either on AM medium or on one-half-strength Murashige and Skoog medium lacking NH4NO3, and KNO3 replaced by KCl (−N) in the presence or absence of 20 mm of a dipeptide as a sole N source. Alternatively, seedlings were cultivated on −N medium supplemented with 10 mm Pro and 10 mm Ala (+Ala+Pro). Data shown are representative for the independent mutants and overexpressing lines, respectively.
Figure 9.
Growth of wild-type (WT), atptr1-1, atptr1-2, atptr5-1, atptr1-1 atptr5-1, and 35S:AtPTR5a seedlings on medium containing a mixture of peptides. Seeds were germinated and cultivated for 2 weeks either on AM medium or on one-half-strength Murashige and Skoog medium lacking NH4NO3 and KNO3 replaced by KCl (−N) or on −N medium containing 0.1 mm NH4NO3 (NH4NO3) in the presence or absence of 2.5 mm of a peptide mixture (0.5 mm each of Ala-Ala, Pro-Ala, Ala-Asp, Arg-Glu, and Ala-Gly). Data shown are representative for the independent mutants and overexpressing lines, respectively.
Table I.
Shoot dry weight (DW) of 2-week-old Arabidopsis seedlings grown on medium with different N sources
Medium and experimental setup are as described in the Figure 8 and 9 legends. Data were obtained from three independent experiments. Values are the means ± se per plant. Different letters indicate statistically significant differences compared to wild type (Student's t test). *, Whole seedling.
| Growth Medium | Shoot DW/Seedling
|
||||||||
|---|---|---|---|---|---|---|---|---|---|
| Wild Type | atptr1-1 | atptr1-2 | atptr5-1 | atptr5-2 | atptr1-1 atptr5-1 | atptr1-2 atptr5-1 | 35S:PTR5a | 35S:PTR5b | |
| μg | |||||||||
| AM | 555 ± 34 | 539 ± 71 | 569 ± 14 | 641 ± 43 | 493 ± 28 | 539 ± 17 | 509 ± 6 | 574 ± 17 | 645 ± 14 |
| −N | 55 ± 3 | 53 ± 1 | 49 ± 1 | 52 ± 2 | 44 ± 1 | 44 ± 3 | 55 ± 2 | 49 ± 1 | 45 ± 1 |
| NH4NO3 | 79 ± 7 | 67 ± 7 | 73 ± 5 | 65 ± 3 | 63 ± 13 | 75 ± 5 | 66 ± 13 | 79 ± 9 | 79 ± 16 |
| −N + peptide mix | 149 ± 8 | 86 ± 4a | 92 ± 2a | 139 ± 6 | 136 ± 3 | 74 ± 2a | 78 ± 5a | 228 ± 12a | 231 ± 15a |
| NH4NO3 + peptide mix | 169 ± 22 | 103 ± 7a | 103 ± 10b | 151 ± 12 | 135 ± 10 | 101 ± 6a | 101 ± 7b | 267 ± 19b | 259 ± 16b |
| −N + Ala-Ala | 245 ± 17 | 109 ± 8a | 129 ± 5a | 194 ± 2 | 208 ± 13 | 115 ± 4a | 107 ± 1a | 561 ± 27a | 518 ± 19a |
| −N + Pro-Ala | 186 ± 8 | 100 ± 9a | 108 ± 2a | 152 ± 14 | 155 ± 16 | 79 ± 2a | 95 ± 5a | 498 ± 40a | 447 ± 39a |
| −N + Ala-Phe | 147 ± 6 | 125 ± 8 | 151 ± 4 | 128 ± 1 | 109 ± 14 | 148 ± 4 | 137 ± 3 | 40 ± 3*b | 39 ± 0.1*b |
| −N + Ala + Pro | 309 ± 17 | 352 ± 19 | 326 ± 21 | 347 ± 3 | 317 ± 17 | 361 ± 23 | 281 ± 67 | 378 ± 26 | 361 ± 14 |
P < 0.005.
P < 0.05.
Figure 10.
N content of wild-type (WT), atptr1, atptr5, atptr1 atptr5, and 35S:AtPTR5 seedlings on medium containing different N sources. Seeds were germinated and cultivated for 2 weeks on different media as described in the Figure 8 and 9 legends. Data were obtained from three independent experiments. Values are the means of N ±sd (% of dry weight [DW]). Different letters indicate statistically significant differences compared to wild type (Student's t test). a, P < 0.005; b, P < 0.05.
We examined whether amino acids resulting from extracellular cleavage of the dipeptides might lead to the differences observed. HPLC analyses of medium containing Ala-Ala or Ala-Pro showed that no amino acids were generated after 2 weeks in the absence or presence of plants (data not shown). Furthermore, on medium with an equimolar mixture of the amino acids Ala and Pro, no differences in growth, biomass, and shoot N levels were observed between mutants, overexpressing lines, and wild type, while differences were seen for Pro-Ala (Figs. 8 and 10; Table I). This demonstrates that the observed growth differences on Pro-Ala were not due to extracellular cleavage of the dipeptide and subsequent uptake of amino acids, but originate from the uptake of the dipeptides; together, the data indicate that peptides are efficiently taken up and metabolized as a source of N.
DISCUSSION
AtPTR5 Mediates High-Affinity Transport of Dipeptides
AtPTR5 had similar apparent affinities for Ala-Lys (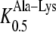 163 μm) and Ala-Asp (
163 μm) and Ala-Asp (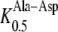 131 μm) as its closest homolog AtPTR1 (
131 μm) as its closest homolog AtPTR1 (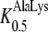 112 μm,
112 μm, 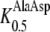 416 μm; Dietrich et al., 2004), but a much higher affinity for Ala-Asp when compared to AtPTR2 (
416 μm; Dietrich et al., 2004), but a much higher affinity for Ala-Asp when compared to AtPTR2 (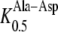 1832 μm; Chiang et al., 2004). While the latter differences in K0.5 values for Ala-Asp might be due to the higher membrane potential used for the electrophysiological measurements with AtPTR2, they most probably reflect true differences because transport of dipeptides by AtPTR2 seems to be largely independent of the membrane potential (Chiang et al., 2004). Nevertheless, K0.5 values of AtPTR2 for other dipeptides composed of neutral and basic amino acids were similar to those determined for AtPTR5 and AtPTR1, indicating that affinities for peptides are generally conserved among the PTR transporters (Chiang et al., 2004; Dietrich et al., 2004).
1832 μm; Chiang et al., 2004). While the latter differences in K0.5 values for Ala-Asp might be due to the higher membrane potential used for the electrophysiological measurements with AtPTR2, they most probably reflect true differences because transport of dipeptides by AtPTR2 seems to be largely independent of the membrane potential (Chiang et al., 2004). Nevertheless, K0.5 values of AtPTR2 for other dipeptides composed of neutral and basic amino acids were similar to those determined for AtPTR5 and AtPTR1, indicating that affinities for peptides are generally conserved among the PTR transporters (Chiang et al., 2004; Dietrich et al., 2004).
AtPTR5 Is Localized at the Plasma Membrane and Facilitates Transport of Peptides into Reproductive Organs
AtPTR5 expression was detected in developing and germinating pollen grains as well as in ovules and during early stages of seed development (Fig. 4). Further, GFP fusion proteins showed that AtPTR5 is localized at the plasma membrane (Fig. 3). Together, these results indicate a role of AtPTR5 in uptake of di- and tripeptides into cells of reproductive organs. While AtPTR1 is also targeted to the plasma membrane, proteome analyses suggest that AtPTR2 is localized at the tonoplast, which points to a role in intracellular rather than intercellular transport (Carter et al., 2004; Dietrich et al., 2004; Shimaoka et al., 2004; Dunkley et al., 2006). The more distantly related AtPTR3 in subgroup III mediates transport in a peptide transport-deficient yeast strain, but affinities or intracellular localization have not been studied (Karim et al., 2005, 2007). Additionally, AtPTR3 seems to be involved in defense reactions during abiotic and biotic stresses rather than in transport of organic N during plant growth (Karim et al., 2007).
Partitioning of N to reproductive structures is a key determinant for development of viable pollen and seeds and thus for yield and successful reproduction (Pate, 1980; Gifford et al., 1984). During development and germination, pollen remains symplastically isolated from the surrounding tissue and depends entirely on nutrients supplied via the phloem from source tissues and their uptake from the apoplast. Studies of the Arabidopsis pollen transcriptome and localization analyzes identified several amino acid transporters, mainly of the LHT family, Pro transporter AtProT1, several members of the OPT family, but also AtPTR5 among the genes expressed specifically or preferentially in pollen, supporting the importance of N supply for pollen production (Fig. 4; Honys and Twell, 2004; Lee and Tegeder, 2004; Pina et al., 2005; Bock et al., 2006). AtPTR5 is expressed both in early and late stages with highest expression at the trinuclear stage of pollen development (Honys and Twell, 2004; Bock et al., 2006). The pollen germination assay showed that AtPTR5-mediated peptide uptake activity is present during early stages of germination (Fig. 6A). In addition, growth of the pollen tubes was negatively affected by Ala-Eth, indicating that AtPTR5-mediated peptide transport also occurs during pollen tube growth (Fig. 6B). However, the latter effect could also be explained by uptake of Ala-Eth during germination that subsequently affects growth of pollen tubes. Therefore, protein localization studies are needed to further determine whether AtPTR5 contributes to N transport during pollen germination and pollen tube growth.
AtPTR5-GUS activity was also detected in ovules and during early stages of seed development, suggesting a role of AtPTR5 in delivery of organic N for establishing the next generation (Fig. 4). Seed loading with N occurs mainly by organic N forms that are delivered via the phloem (Pate et al., 1977; Pate, 1980). Several amino acid transport systems are involved during this phase. Ataap8 mutants showed that AtAAP8 is important for early seed development, while expression of AtAAP1 and AtAAP2 points to a complementary role during seed development (Hirner et al., 1998; Schmidt et al., 2007). Further, the oligopeptide transporter atopt3 mutant showed arrested seed development; however, as recently demonstrated, AtOPT3 seems to be involved in iron transport rather than transfer of organic N (Stacey et al., 2002, 2008). With respect to dipeptide transport, arrested seed development has been described for atptr2 antisense lines (Song et al., 1997). However, this effect could not be observed in atptr2 T-DNA insertion lines, indicating that down-regulation of other PTRs in the antisense lines might have contributed to this phenotype (D. Dietrich and D. Rentsch, unpublished data). Similar to the amino acid transporters AtAAP1 and AtAAP2, the dipeptide transporters AtPTR1 and AtPTR5 were expressed in the silique vasculature and developing seeds, with their expression preceding the protein-filling phase, suggesting a role in di- and tripeptide supply to and uptake into the seeds, respectively (Fig. 4; Hirner et al., 1998; Dietrich et al., 2004). Nonetheless, mature seeds of atptr5 and wild-type plants did not differ in their total N content, indicating that AtPTR5 mediated di- and tripeptide transport does not represent a limiting step for final N content in seeds and/or that missing AtPTR5 activity is being compensated by other peptide or amino acid transporters.
Peptide Transporters Mediate Uptake of Peptides into Roots
Most soils contain a high content of organic N compounds that are, however, hardly accessible for degradation because they are protected by minerals and humus (Leinweber and Schulten, 1998). Proteolysis is generally considered as the rate-limiting step in protein degradation and released organic N is used by microorganisms and plants as N sources (Cunningham and Wetzel, 1989; Asmar et al., 1994). Paungfoo-Lonhienne et al. (2008) showed that plant roots can use larger proteins as N sources and that the proteins are hydrolyzed at the root surface and potentially in the root apoplast. While in these studies the sizes of the peptide fragments generated were not determined, import of peptides into root cells could potentially be mediated by transporters of the PTR, OPT, and/or ABC transporter families.
Here, we present evidence for uptake of dipeptides into roots by members of the PTR family. We demonstrate that atptr1 mutants exhibit reduced growth and lower N content when cultivated on Ala-Ala, Pro-Ala, or a mixture of dipeptides, supporting a function of AtPTR1 in peptide acquisition (Figs. 8–10; Table I). Conversely, when overexpressing a peptide transporter (35S:AtPTR5), plant growth was enhanced and N content increased (Fig. 8–10; Table I). Because AtPTR5 is not expressed in roots, as expected, atptr5 mutants behaved like wild-type plants. Under the conditions tested, AtPTR5 did not seem to be up-regulated in the absence of AtPTR1 because the double mutants behaved like the single atptr1 mutants. Whereas certain dipeptides, including Ala-Ala, Ala-Asn, and Pro-Ala promoted better growth of AtPTR5-overexpressing lines, Ala-Phe and Lys-Asp inhibited growth, indicating that their increased uptake caused a toxic effect either of the peptides or more likely by the amino acids generated (Fig. 8; Table I). Imbalance of the amino acid pools disturbs homeostasis, which can be lethal for the plant, a fact that was used to identify mutants in amino acid metabolism and transport (Voll et al., 2004; Lee et al., 2007). The predicted peptide transport activity 35S:AtPTR5 > wild type > atptr1 mutants correlated with the observed effect, indicating the dependence of the phenotypes on peptide transporter function. Because AtPTR1 did not mediate transport of the amino acids Ala, Lys, Asp, or Asn, and because His was only transported with low affinity (Dietrich et al., 2004), uptake of amino acids generated by extracellular cleavage and subsequent transport by a peptide transporter seemed unlikely, even more so because no amino acids could be detected in the growth medium. This notion was corroborated by the finding that growth of atptr1 mutants and 35S:AtPTR5 lines did not reveal differences to wild-type plants when cultured on amino acids (Figs. 8 and 10; Table I). It remains to be investigated how much AtPTR1 contributes to N acquisition from the organic N pool of soil as opposed to acquisition of peptides retrieved from senescing root cells. Taken together, our results provide evidence that AtPTR1 has a crucial role in uptake of peptides into roots and that increased expression of peptide transporters can enhance uptake of N and plant growth.
CONCLUSION
Our study shows that AtPTR transporters have different roles within plants. AtPTR5 supplies germinating pollen with peptides and most likely functions in supplying peptides to maturating pollen, developing ovules, and seeds. In contrast, AtPTR1 mediates uptake of dipeptides into root cells. Overexpression of AtPTR5 resulted in enhanced growth; thus, the ability to transport peptides across the plasma membrane appears to be generic and regulated primarily via tissue-specific expression. Further, our study provides novel evidence that the use of organic forms of N is not restricted to amino acids, but that di- and tripeptides should be considered as a potentially important N source as well.
MATERIALS AND METHODS
Plant Material and Growth Conditions
Seeds of the Arabidopsis (Arabidopsis thaliana ecotype Columbia [Col-0]) mutant lines atptr1-1, atptr1-2, and atptr5-1 were obtained from the SALK collection (Alonso et al., 2003; stock numbers Salk_131550, Salk_130803, and Salk_008661 respectively). The atptr5-2 mutant line was obtained from the Wisconsin DsLox transposon-tagged lines (Woody et al., 2007, line WiscDsLox342H06). The locations of the T-DNA insertion sites were verified by sequencing PCR products amplified by using T-DNA left-border primers (for SALK lines, 5′-GCGTGGACCGCTTGCTGCAACT-3′; for Wisconsin line, 5′-AACGTCCGCAATGTGTTATTAAGTTGTC-3′) and gene-specific primers (for atptr1 lines, 5′-AACACACCAACAACAACTAGACCACATAC-3′; for atptr5 lines, 5′-ATATCAGCTTCGGTTCCTGGTCTAACAC-3′ and 5′-ATCAGAATTCGTGATTGCAGCCATC-3′). Lines were back-crossed twice with the wild type (Col-0) and selfed to isolate homozygous lines. Double knockout lines were isolated by crossing homozygous single atptr1 mutants with the atptr5-1 line; homozygous double mutants were identified in the BC2 progeny. The homozygous genotype of the insertion lines was confirmed by PCR on genomic DNA. All knockout lines were analyzed by RT-PCR for the absence of mRNA of AtPTR1 (leaf tissue), and AtPTR5 (flower tissue) or both transcripts as described below.
Arabidopsis ecotype Col-0 and mutant lines were grown in soil in a growth chamber at 22°C/18°C, 65% humidity, and 16 h of light (240 μmol m−2 s−1). Plants were transformed by Agrobacterium tumefaciens (GV3101 pMP90) mediated gene transfer (Koncz and Schell, 1986) using the floral-dip method (Clough and Bent, 1998) as described earlier (Grallath et al., 2005).
For axenic culture, sterilized Arabidopsis seeds were vernalized in 0.1% agarose at 4°C in the dark for 2 d. Seeds were germinated and cultivated on either of the following media: (1) AM medium (2.16 g L−1 Murashige and Skoog salts [Duchefa]); (2) one-half-strength Murashige and Skoog medium (Murashige and Skoog, 1962) lacking NH4NO3 and KNO3 replaced by KCl (−N; replacing KNO3 by KCl did not have any effect on plant growth [e.g. biomass]); (3) −N medium containing 20 mm of a dipeptide as a sole nitrogen source; (4) −N medium containing 2.5 mm of an equimolar peptide mixture (Ala-Ala, Pro-Ala, Ala-Asp, Arg-Glu, and Ala-Gly); (5) −N containing 0.1 mm NH4NO3 and the peptide mixture. As a control, plants were cultivated on −N medium containing a mixture of the amino acids Ala and Pro (10 mm each). All media were adjusted to pH 5.8, completed with 1% Suc, and solidified with Oxoid agar (Oxoid). Seventy-five milliliters of medium was used per 12 × 12-cm plate and six lines, approximately 42 seeds each, were cultivated per plate. Alternatively, four lines and approximately 11 seeds per line were cultivated on 10-mL medium (plate diameter 5 cm). Plants were cultivated for 2 weeks in a growth chamber under the above-mentioned conditions, except that the light intensity was 140 μmol m−2 s−1.
For root growth experiments, seedlings were grown (vertically) on AM medium for 7 d. Sterile filter paper stripes soaked with water as a control or with a solution containing 1.1 μmol Ala-Eth were placed in front of the growing roots. The bottom of the plates was covered with aluminum foil and the plates were cultivated for another 10 d.
In Vitro Pollen Germination
For each genotype, pollen from three flowers was cultured in suspended drops in either GM as described by Footitt et al. (2007) or in GM containing 20 or 100 μm Ala-Eth (JPT Peptide Technologies). Germination was scored by microscopic examination and the germination rate was calculated from three independent experiments. Germinated pollen was visualized with a Zeiss Axioskop 2 microscope equipped with an Axiocam camera (Zeiss).
DNA Work
Standard methods (PCR, cloning procedure, transformation of bacteria, plasmid preparation, and DNA cleavage with restriction enzymes) were performed as described by Sambrook et al. (1989) and Ausubel et al. (1994).
AtPTR5-cDNA was isolated by RT-PCR using primer 5′-TCACACTTAAACTTGTCGAAACCAAAC-3′, primer 5′-TCAAAGCGCATGCCCGGTCGT-3′, and RNA extracted from flowers of Arabidopsis ecotype Col-0 as template. The AtPTR5-cDNA was cloned in the SmaI site of pDR196 (Rentsch et al., 1995) and verified by sequencing.
For translational fusions with GFP, the ORF of the AtPTR5 cDNA was amplified by PCR and cloned in pUC18-spGFP6 and pUC18-GFP5Tsp (M. Suter-Grotemeyer and D. Rentsch, unpublished data). AtPTR5-GFP fusion, 5′-CTAGCTAGCATGGAAGATGACAAGGATATATACAC-3′, 5′-AATAGATCTTCAAGCGCATGCCCGGTCGTTTTC-3′ (ORF cloned into SpeI/BglII site). GFP-AtPTR5 fusion, 5′-CTAGCTAGCATGCGAAGATGACAAGGATATATACAC-3′, 5′-AATGTCGACTCAAAGCGCATGCCCGGTCG-3′ (ORF cloned into NheI/SalI site). Sequence identity of all PCR-amplified fragments was verified by sequencing. For expression of the fusion proteins in yeast, the fragments were transferred to pDR-spGFP6 and pDR-GFP5-sp (M. Suter-Grotemeyer and D. Rentsch, unpublished data).
The AtPTR5-promoter region (2,582 bp upstream of the start codon) was amplified by PCR using genomic DNA of Arabidopsis ecotype Col-0 and primers 5′-ACCTACAAGGGACAATTGATTTTGG-3′ and 5′-TGTTGTTTGGTTTCGACAAGTTTAAGT-3′. The PCR product was cloned into the blunted SpeI site of pBluescript SK− (Stratagene) and sequenced. For plant transformation, the promoter was excised with BamHI and NotI and transferred into the SmaI site of the binary vector pCB308 (Xiang et al., 1999). For overexpression in Arabidopsis, AtPTR5 was used to circumvent cosuppression of AtPTR1 in roots. The AtPTR5-cDNA was excised from pDR-AtPTR5 with BamHI and SalI and cloned into corresponding sites of the binary vector pBinAR (Röber et al., 1996) downstream of the cauliflower mosaic virus 35S promoter (Gatz et al., 1992).
RNA Work
To quantify expression, total RNA was extracted using a method based on phenol-SDS extraction (Ausubel et al., 1994) including an additional DNase I treatment. RT was performed using the Moloney murine leukemia virus reverse transcriptase (USB) according to the manufacturer's instructions with oligo(dT) primers and 1.5 μg of total RNA as template, followed by PCR amplification of the 3′-cDNA region using cDNA-specific primers (for AtPTR1, 5′-AACACACCAACAACAACTAGACCACATAC-3′ and 5′-GTCAGGCTTGATTATGTC-3′; for AtPTR5, 5′-TGAGGAGTCTCTGCTCGG-3′ and 5′-ATCAGAATTCGTGATTGCAGCCATC-3′). For amplification of the region upstream of the insertion, the following primers were used: 5′-TTGTGCACACAGATAACTTAAAGTTT-3′ and 5′-TTGCCAGCGGGATAATGAACT-3′ (for AtPTR1), and 5′-GCTTCGGTTCCTGGTCTAACACCAAC-3′ and 5′-CAAACGAGAACAGAGGAAGCAATCA-3′ (for AtPTR5). The Arabidopsis actin gene AtACT2 was used as a reference (5′-ATTCAGATGCCCAGAAGTCTTGTT-3′ and 5′-GAAACATTTTCTGTGAACGATTCCT-3′).
Northern-blot analyses were performed as described in Ausubel et al. (1994) according to the formaldehyde/formamide protocol using 15 μg of total RNA isolated from leaves and the cDNA of AtPTR5 as a probe.
Determination of Peptide Concentration and Total N Content
Concentration of peptides and amino acids in the medium was determined by HPLC at ARS, University of Bern (Bidlingmeyer et al., 1984). Total N content of shoots was determined at the Washington State University Analytical Core laboratory; shoot material (1.5 mg) was combusted in a Costech ECS 4010 elemental analyzer, the resulting N2 separated by gas chromatography and then analyzed using a Micromass Isoprime isotope ratio mass spectrometer (IRMS).
Yeast Growth, Transformation, and Selection
Yeast strain LR2 (MATα hip1-614 his4-401 can1 ino1 ura3-52 ptr2Δ∷hisG; Rentsch et al., 1995) was transformed according to Dohmen et al. (1991), and transformants were selected on SC medium supplemented with 20 mm of His. To test for peptide transport activity, transformants were selected on SC medium containing 1 mm His-Ala as sole source of the required amino acid His (Rentsch et al., 1995).
Transient Expression in Protoplasts
Transient expression of GFP fusion proteins in tobacco (Nicotiana tabacum) protoplasts was performed as described (Dietrich et al., 2004). Samples were examined with a SP2 AOBS confocal microscope (Leica Microsystems). Filter settings were 500 to 520 nm for GFP and 628 to 768 nm for chlorophyll epifluorescence detection.
Staining for GUS Activity
Histochemical localization of GUS activity was performed as described previously (Dietrich et al., 2004). Pictures were taken with a Leica MZFLIII imaging system (Leica Microsystems) or a Zeiss Axioskop 2 microscope equipped with an Axiocam camera (Zeiss).
Expression in Xenopus Oocytes
Expression of AtPTR5 in Xenopus oocytes was performed as described in Meyer et al. (2006). Substrate-induced currents were measured 2 to 4 d after injection. The voltage protocol was as described in Dietrich et al. (2004), with the following modification: test potential (Vm) between +20 to −140 mV. Each voltage pulse was applied for 150 ms and the currents were filtered at 100 Hz.
Acknowledgments
We wish to thank Susanne Schmidt (University of Queensland) and Hanjo Hellmann (Washington State University, Pullman) for helpful comments on the manuscript. We are grateful to Liliane Sticher (University of Fribourg) for help with taking images, Hans Weber (IPK Gatersleben) for determining total N in seeds, Christopher Ball and Rebecca Alder (University of Bern) for taking care of the plants, and Erwin Sigel (University of Bern) for providing oocytes of X. laevis. We wish to thank Urs Kämpfer and Johann Schaller (University of Bern) for determining concentrations of peptides and amino acids in the medium and Silke Lehmann, Anna Meier, Sara Hirsbrunner, and Mirco Hecht for help with harvesting Arabidopsis seedlings.
This work was supported by the Swiss National Foundation (grant no. 3100A0–107507).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Doris Rentsch (doris.rentsch@ips.unibe.ch).
Open Access articles can be viewed online without a subscription.
References
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen HM, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, et al (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301 653–657 [DOI] [PubMed] [Google Scholar]
- Asmar F, Eiland F, Nielsen NE (1994) Effect of extracellular-enzyme activities on solubilization rate of soil organic nitrogen. Biol Fertil Soils 17 32–38 [Google Scholar]
- Ausubel F, Brent R, Kingston R, Moore D, Seidman J, Smith J, Struhl K (1994) Current Protocols in Molecular Biology. Wiley, New York
- Bidlingmeyer BA, Cohen SA, Tarvin TL (1984) Rapid analysis of amino acids using pre-column derivatization. J Chromatogr 336 93–104 [DOI] [PubMed] [Google Scholar]
- Bock KW, Honys D, Ward JM, Padmanaban S, Nawrocki EP, Hirschi KD, Twell D, Sze H (2006) Integrating membrane transport with male gametophyte development and function through transcriptomics. Plant Physiol 140 1151–1168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campalans A, Pages M, Messeguer R (2001) Identification of differentially expressed genes by the cDNA-AFLP technique during dehydration of almond (Prunus amygdalus). Tree Physiol 21 633–643 [DOI] [PubMed] [Google Scholar]
- Carter C, Pan SQ, Jan ZH, Avila EL, Girke T, Raikhel NV (2004) The vegetative vacuole proteome of Arabidopsis thaliana reveals predicted and unexpected proteins. Plant Cell 16 3285–3303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapin FS, Moilanen L, Kielland K (1993) Preferential use of organic nitrogen for growth by a nonmycorrhizal arctic sedge. Nature 361 150–153 [Google Scholar]
- Chiang CS, Stacey G, Tsay YF (2004) Mechanisms and functional properties of two peptide transporters, AtPTR2 and fPTR2. J Biol Chem 279 30150–30157 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16 735–743 [DOI] [PubMed] [Google Scholar]
- Cunningham HW, Wetzel RG (1989) Kinetic-analysis of protein-degradation by a fresh-water wetland sediment community. Appl Environ Microbiol 55 1963–1967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietrich D, Hammes U, Thor K, Suter-Grotemeyer M, Fluckiger R, Slusarenko AJ, Ward JM, Rentsch D (2004) AtPTR1, a plasma membrane peptide transporter expressed during seed germination and in vascular tissue of Arabidopsis. Plant J 40 488–499 [DOI] [PubMed] [Google Scholar]
- Dohmen RJ, Strasser AW, Honer CB, Hollenberg CP (1991) An efficient transformation procedure enabling long-term storage of competent cells of various yeast genera. Yeast 7 691–692 [DOI] [PubMed] [Google Scholar]
- Dunkley TPJ, Hester S, Shadforth IP, Runions J, Weimar T, Hanton SL, Griffin JL, Bessant C, Brandizzi F, Hawes C, et al (2006) Mapping the Arabidopsis organelle proteome. Proc Natl Acad Sci USA 103 6518–6523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Footitt S, Dietrich D, Fait A, Fernie AR, Holdsworth MJ, Baker A, Theodoulou FL (2007) The COMATOSE ATP-binding cassette transporter is required for full fertility in Arabidopsis. Plant Physiol 144 1467–1480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frommer WB, Hummel S, Rentsch D (1994) Cloning of an Arabidopsis histidine transporting protein related to nitrate and peptide transporters. FEBS Lett 347 185–189 [DOI] [PubMed] [Google Scholar]
- Gatz C, Frohberg C, Wendenburg R (1992) Stringent repression and homogeneous de-repression by tetracycline of a modified Camv 35s promoter in intact transgenic tobacco pants. Plant J 2 397–404 [DOI] [PubMed] [Google Scholar]
- Gifford RM, Thorne JH, Hitz WD, Giaquinta RT (1984) Crop productivity and photoassimilate partitioning. Science 225 801–808 [DOI] [PubMed] [Google Scholar]
- Glass ADM, Britto DT, Kaiser BN, Kinghorn JR, Kronzucker HJ, Kumar A, Okamoto M, Rawat S, Siddiqi MY, Unkles SE, Vidmar JJ (2002) The regulation of nitrate and ammonium transport systems in plants. J Exp Bot 53 855–864 [DOI] [PubMed] [Google Scholar]
- Grallath S, Weimar T, Meyer A, Gumy C, Suter-Grotemeyer M, Neuhaus JM, Rentsch D (2005) The AtProT family. Compatible solute transporters with similar substrate specificity but differential expression patterns. Plant Physiol 137 117–126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins CF, Payne JW (1978) Peptide transport by germinating barley embryos: uptake of physiological di- and oligopeptides. Planta 138 211–215 [DOI] [PubMed] [Google Scholar]
- Higgins CF, Payne JW (1981) The peptide pools of germinating barley grains: relation to hydrolysis and transport of storage proteins. Plant Physiol 67 785–792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins CF, Payne JW (1982) Plant peptides. In D Boulter, B Parthier, eds, Nucleic Acids and Proteins in Plants, Encyclopedia of Plant Physiology, Vol 14A. Springer, Berlin, pp 438–458
- Hirner A, Ladwig F, Stransky H, Okumoto S, Keinath M, Harms A, Frommer WB, Koch W (2006) Arabidopsis LHT1 is a high-affinity transporter for cellular amino acid uptake in both root epidermis and leaf mesophyll. Plant Cell 18 1931–1946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirner B, Fischer WN, Rentsch D, Kwart M, Frommer WB (1998) Developmental control of H+/amino acid permease gene expression during seed development of Arabidopsis. Plant J 14 535–544 [DOI] [PubMed] [Google Scholar]
- Honys D, Twell D (2004) Transcriptome analysis of haploid male gametophyte development in Arabidopsis. Genome Biol 5 R85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamai A, Chollet JF, Delrot S (1994) Proton-peptide co-transport in broad bean leaf tissues. Plant Physiol 106 1023–1031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong JY, Suh S, Guan CH, Tsay YF, Moran N, Oh CJ, An CS, Demchenko KN, Pawlowski K, Lee Y (2004) A nodule-specific dicarboxylate transporter from alder is a member of the peptide transporter family. Plant Physiol 134 969–978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karim S, Holmstrom KO, Mandal A, Dahl P, Hohmann S, Brader G, Palva ET, Pirhonen M (2007) AtPTR3, a wound-induced peptide transporter needed for defence against virulent bacterial pathogens in Arabidopsis. Planta 225 1431–1445 [DOI] [PubMed] [Google Scholar]
- Karim S, Lundh D, Holmstrom KO, Mandal A, Pirhonen M (2005) Structural and functional characterization of AtPTR3, a stress-induced peptide transporter of Arabidopsis. J Mol Model 11 226–236 [DOI] [PubMed] [Google Scholar]
- Kaye JP, Hart SC (1997) Competition for nitrogen between plants and soil microorganisms. Trends Ecol Evol 12 139–143 [DOI] [PubMed] [Google Scholar]
- Koncz C, Schell J (1986) The promoter of Tl-DNA gene 5 controls the tissue-specific expression of chimeric genes carried by a novel type of Agrobacterium binary vector. Mol Gen Genet 204 383–396 [Google Scholar]
- Kwart M, Hirner B, Hummel S, Frommer WB (1993) Differential expression of two related amino acid transporters with differing substrate specificity in Arabidopsis thaliana. Plant J 4 993–1002 [DOI] [PubMed] [Google Scholar]
- Lee YH, Tegeder M (2004) Selective expression of a novel high-affinity transport system for acidic and neutral amino acids in the tapetum cells of Arabidopsis flowers. Plant J 40 60–74 [DOI] [PubMed] [Google Scholar]
- Lee YH, Foster J, Chen J, Voll LM, Weber APM, Tegeder M (2007) AAP1 transports uncharged amino acids into roots of Arabidopsis. Plant J 50 305–319 [DOI] [PubMed] [Google Scholar]
- Leinweber P, Schulten HR (1998) Nonhydrolyzable organic nitrogen in soil size separates from long-term agricultural experiments. Soil Sci Soc Am J 62 383–393 [Google Scholar]
- Meyer A, Eskandari S, Grallath S, Rentsch D (2006) AtGAT1, a high affinity transporter for gamma-aminobutyric acid in Arabidopsis thaliana. J Biol Chem 281 7197–7204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miranda M, Borisjuk L, Tewes A, Dietrich D, Rentsch D, Weber H, Wobus U (2003) Peptide and amino acid transporters are differentially regulated during seed development and germination in faba bean. Plant Physiol 132 1950–1960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murashige T, Skoog F (1962) A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiol Plant 15 473–497 [Google Scholar]
- Näsholm T, Ekblad A, Nordin A, Giesler R, Hogberg M, Hogberg P (1998) Boreal forest plants take up organic nitrogen. Nature 392 914–916 [Google Scholar]
- Pate JS (1980) Transport and partitioning of nitrogenous solutes. Annu Rev Plant Physiol Plant Mol Biol 31 313–340 [DOI] [PubMed] [Google Scholar]
- Pate JS, Sharkey PJ, Atkins CA (1977) Nutrition of a developing legume fruit: functional economy in terms of carbon, nitrogen, water. Plant Physiol 59 506–510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paungfoo-Lonhienne C, Lonhienne TGA, Rentsch D, Robinson N, Christie M, Webb RI, Gamage HK, Carroll BJ, Schenk PM, Schmidt S (2008) Plants can use protein as a nitrogen source without assistance from other organisms. Proc Natl Acad Sci USA 105 4524–4529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pina C, Pinto F, Feijo JA, Becker JD (2005) Gene family analysis of the Arabidopsis pollen transcriptome reveals biological implications for cell growth, division control, and gene expression regulation. Plant Physiol 138 744–756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rentsch D, Laloi M, Rouhara I, Schmelzer E, Delrot S, Frommer WB (1995) NTR1 encodes a high affinity oligopeptide transporter in Arabidopsis. FEBS Lett 370 264–268 [DOI] [PubMed] [Google Scholar]
- Rentsch D, Schmidt S, Tegeder M (2007) Transporters for uptake and allocation of organic nitrogen compounds in plants. FEBS Lett 581 2281–2289 [DOI] [PubMed] [Google Scholar]
- Röber M, Geider K, Müller Röber B, Willmitzer L (1996) Synthesis of fructans in tubers of transgenic starch-deficient potato plants does not result in an increased allocation of carbohydrates. Planta 199 528–536 [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T (1989) Molecular Cloning: A Laboratory Manual, Ed 2. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY
- Schmidt R, Stransky H, Koch W (2007) The amino acid permease AAP8 is important for early seed development in Arabidopsis thaliana. Planta 226 805–813 [DOI] [PubMed] [Google Scholar]
- Schmidt S, Stewart GR (1999) Glycine metabolism by plant roots and its occurrence in Australian plant communities. Aust J Plant Physiol 26 253–264 [Google Scholar]
- Schulze W, Frommer WB, Ward JM (1999) Transporters for ammonium, amino acids and peptides are expressed in pitchers of the carnivorous plant Nepenthes. Plant J 17 637–646 [DOI] [PubMed] [Google Scholar]
- Segonzac C, Boyer JC, Ipotesi E, Szponarski W, Tillard P, Touraine B, Sommerer N, Rossignol M, Gibrat R (2007) Nitrate efflux at the root plasma membrane: Identification of an Arabidopsis excretion transporter. Plant Cell 19 3760–3777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimaoka T, Ohnishi M, Sazuka T, Mitsuhashi N, Hara-Nishimura I, Shimazaki KI, Maeshima M, Yokota A, Tomizawa KI, Mimura T (2004) Isolation of intact vacuoles and proteomic analysis of tonoplast from suspension-cultured cells of Arabidopsis thaliana. Plant Cell Physiol 45 672–683 [DOI] [PubMed] [Google Scholar]
- Song W, Koh S, Czako M, Marton L, Drenkard E, Becker JM, Stacey G (1997) Antisense expression of the peptide transport gene AtPTR2-B delays flowering and arrests seed development in transgenic Arabidopsis plants. Plant Physiol 114 927–935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stacey MG, Koh S, Becker J, Stacey G (2002) AtOPT3, a member of the oligopeptide transporter family, is essential for embryo development in Arabidopsis. Plant Cell 14 2799–2811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stacey MG, Patel A, McClain WE, Mathieu M, Remley M, Rogers EE, Gassmann W, Blevins DG, Stacey G (2008) The Arabidopsis AtOPT3 protein functions in metal homeostasis and movement of iron to developing seeds. Plant Physiol 146 589–601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svennerstam H, Ganeteg U, Bellini C, Nasholm T (2007) Comprehensive screening of Arabidopsis mutants suggests the lysine histidine transporter 1 to be involved in plant uptake of amino acids. Plant Physiol 143 1853–1860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swofford DL (2003) PAUP*. Phylogenetic Analysis Using Parsimony and Other Methods, Version 4.0b10. Sinauer Associates, Sunderland, MA
- Tsay YF, Chiu CC, Tsai CB, Ho CH, Hsu PK (2007) Nitrate transporters and peptide transporters. FEBS Lett 581 2290–2300 [DOI] [PubMed] [Google Scholar]
- Voll LM, Allaire EE, Fiene G, Weber APM (2004) The Arabidopsis phenylalanine insensitive growth mutant exhibits a deregulated amino acid metabolism. Plant Physiol 136 3058–3069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterworth WM, West CE, Bray CM (2000) The barley scutellar peptide transporter: biochemical characterization and localization to the plasma membrane. J Exp Bot 51 1201–1209 [PubMed] [Google Scholar]
- West CE, Waterworth WM, Stephens SM, Smith CP, Bray CM (1998) Cloning and functional characterisation of a peptide transporter expressed in the scutellum of barley grain during the early stages of germination. Plant J 15 221–229 [DOI] [PubMed] [Google Scholar]
- Woody ST, Austin-Phillips S, Amasino RM, Krysan PJ (2007) The WiscDsLox T-DNA collection: an Arabidopsis community resource generated by using an improved high-throughput T-DNA sequencing pipeline. J Plant Res 120 157–165 [DOI] [PubMed] [Google Scholar]
- Wright DE (1962) Amino acid uptake by plant roots. Arch Biochem Biophys 97 174–180 [DOI] [PubMed] [Google Scholar]
- Xiang C, Han P, Lutziger I, Wang K, Oliver DJ (1999) A mini binary vector series for plant transformation. Plant Mol Biol 40 711–717 [DOI] [PubMed] [Google Scholar]
- Zimmermann P, Hirsch-Hoffmann M, Hennig L, Gruissem W (2004) GENEVESTIGATOR. Arabidopsis microarray database and analysis toolbox. Plant Physiol 136 2621–2632 [DOI] [PMC free article] [PubMed] [Google Scholar]