Abstract
Steady-state labeling experiments with [1-13C]Glc were used to measure multiple metabolic fluxes through the pathways of central metabolism in a heterotrophic cell suspension culture of Arabidopsis (Arabidopsis thaliana). The protocol was based on in silico modeling to establish the optimal labeled precursor, validation of the isotopic and metabolic steady state, extensive nuclear magnetic resonance analysis of the redistribution of label into soluble metabolites, starch, and protein, and a comprehensive set of biomass measurements. Following a simple modification of the cell culture procedure, cells were grown at two oxygen concentrations, and flux maps of central metabolism were constructed on the basis of replicated experiments and rigorous statistical analysis. Increased growth rate at the higher O2 concentration was associated with an increase in fluxes throughout the network, and this was achieved without any significant change in relative fluxes despite differences in the metabolite profile of organic acids, amino acids, and carbohydrates. The balance between biosynthesis and respiration within the tricarboxylic acid cycle was unchanged, with 38% ± 5% of carbon entering used for biosynthesis under standard O2 conditions and 33% ± 2% under elevated O2. These results add to the emerging picture of the stability of the central metabolic network and its capacity to respond to physiological perturbations with the minimum of rearrangement. The lack of correlation between the change in metabolite profile, which implied significant disruption of the metabolic network following the alteration in the oxygen supply, and the unchanging flux distribution highlights a potential difficulty in the interpretation of metabolomic data.
Although the complexity and plasticity of the metabolic network in plants allows them to adapt to fluctuating environmental conditions, the same properties also present a significant obstacle to metabolic engineering (Carrari et al., 2003a; Kruger and Ratcliffe, 2008; Sweetlove et al., 2008). The problem is particularly acute in primary metabolism, where there have been numerous instances of unsuccessful engineering, and reflects the current incomplete understanding of the way in which metabolic networks respond to environmental and genetic perturbations. Fluxes of central carbon metabolism are part of the missing information (Sweetlove et al., 2003), and although they are necessarily related to enzyme abundances, metabolite concentrations, and transcriptional responses (Carrari et al., 2006; Junker et al., 2007), their reliable prediction from the available data remains a non-trivial task (Sweetlove and Fernie, 2005). For this reason, the development and application of techniques for the measurement of flux in plants has become an important area of research (Schwender et al., 2004a; Fernie et al., 2005; Ratcliffe and Shachar-Hill, 2006).
Steady-state metabolic flux analysis (MFA) has the capacity to resolve parallel, cyclic, and reversible fluxes, making it a useful technique for quantifying metabolic fluxes and investigating the factors that control them in plants (Roscher et al., 2000; Ratcliffe and Shachar-Hill, 2006). MFA studies have revealed novel aspects of plant metabolism, as well as providing the first measurements of many fluxes in vivo and independently verifying previous research on plant metabolism (Schwender et al., 2004a). For example, recent work on Brassica napus embryos highlighted the importance of Rubisco in refixation of CO2 for improved conversion of photosynthate to seed storage compounds (Schwender et al., 2004b), while work on sunflower (Helianthus annuus) embryos (Alonso et al., 2007a) provided evidence that pyruvate uptake is not the dominant route for the provision of precursors for plastidic fatty acid synthesis, in agreement with recent work on Arabidopsis (Arabidopsis thaliana; Andre et al., 2007). Recent refinements to MFA theory (Sriram and Shanks, 2004a; Ghosh et al., 2006; Kruger et al., 2007a; Libourel et al., 2007) and the development of specialized software (Wiechert et al., 2001; Sriram et al., 2004) have been accompanied by a rapid increase in the number of species investigated using MFA (Schwender, 2008). However, the approach has yet to be applied to Arabidopsis, and this article describes an MFA study of respiratory and biosynthetic carbon metabolism in a heterotrophic Arabidopsis cell suspension.
A key question in the regulation of central carbon metabolism is how the simultaneous demands of catabolic respiratory metabolism and anabolic biosynthetic metabolism are managed. The tricarboxylic acid (TCA) cycle is central to both processes, generating reducing equivalents for the mitochondrial electron transfer chain, and providing precursors for several biosynthetic pathways (Fernie et al., 2004). Recent studies have helped to demonstrate the degree to which the TCA cycle is connected to other metabolic processes: for example, antisense knockdown of malate dehydrogenase (Nunes-Nesi et al., 2005), and fumarase (Nunes-Nesi et al., 2007), and a mutation within the aconitase gene (Carrari et al., 2003b) all had marked effects on photosynthetic performance in tomato (Solanum lycopersicum), while the effects of oxidative stress on the TCA cycle propagated throughout the metabolic network in Arabidopsis (Baxter et al., 2007). MFA has demonstrated that the TCA cycle operates in different flux modes in different systems. For example, in B. napus embryos, photosynthetic ATP production alleviates the need for any cyclic TCA cycle flux (Schwender et al., 2006), while in other systems, there appears to be more conventional operation of the TCA cycle (Rontein et al., 2002; Alonso et al., 2007b) with both significant respiratory and biosynthetic flux. MFA can also be used to investigate the response of the TCA cycle and its associated pathways to different growth conditions. Recent work revealed that B. napus embryos respond to a switch from an organic to an inorganic nitrogen source by increasing anaplerotic flux through phosphoenolpyruvate carboxylase (PEPC), thereby replacing the carbon removed from the TCA cycle for ammonium assimilation (Junker et al., 2007). In soybean (Glycine max) cotyledons, relative flux through PEPC was reduced by increased growth temperature (Iyer et al., 2008).
Thus, a quantitative picture is emerging of TCA cycle fluxes and the extent to which they vary depending on the need to generate precursors for biosynthesis and reductant for ATP synthesis. However, the factors that may control the rates of biosynthesis and respiration, and the balance between these two competing processes, have not been tested systematically. Accordingly, we varied the concentration of O2 in the medium of an Arabidopsis cell suspension culture with the aim of perturbing the operation of the TCA cycle. The effect of this manipulation on the flux map of central metabolism, and in particular the effect on the balance between respiratory and biosynthetic fluxes, was quantified using steady-state MFA.
RESULTS
Construction and Refinement of a Metabolic Model
The successful application of MFA requires the construction of a model that not only accurately reflects metabolism within the experimental system, but that also can be solved with the quantity and quality of data that are likely to be obtainable. To this end, we constructed an initial model of central carbon metabolism in heterotrophic Arabidopsis cell suspension cultures using a format compatible with the steady-state MFA software 13C-FLUX (Wiechert et al., 2001). The structure of the network was based on information from the literature, principally other MFA studies of heterotrophic metabolism in plants (Schwender et al., 2006; Alonso et al., 2007a; Sriram et al., 2007), and from metabolic databases (Schomburg et al., 2004; Zhang et al., 2005). Reactions primarily associated with photosynthetic metabolism (Calvin cycle, photorespiration) and seed germination (glyoxylate cycle) were not included, because there is no evidence that they occur to any significant extent in dark-grown Arabidopsis cell suspensions. Carbon transitions within the network were derived from the primary literature and standard biochemical textbooks. Accurate representation of carbon transitions around the TCA cycle in the model was confirmed by comparing data from a 13C NMR analysis of the labeling of organic and amino acids in methanolic extracts of heterotrophic cell suspensions fed with [2,3-13C]succinate for 6 or 18 h with the pattern of label distribution predicted by the model (data not shown).
The complexity of the network that can be analyzed is partly determined by the extent to which the redistribution of the 13C-label can be quantified after labeling to isotopic steady state. We used the statistical analysis component of 13C-FLUX, EstimateStat, to predict errors on optimized flux estimates for different network configurations and 13C-labeled precursors and, hence, refined the initial model to the point where it could be solved with the data obtainable from a steady-state labeling experiment. By this method, structurally nonidentifiable fluxes, i.e. those that can take any value without impacting on the observed label distribution (Wiechert et al., 2001), were identified and removed from the network. For example, flux from 2-oxoglutarate to succinate, whether occurring via α-ketoglutarate dehydrogenase or via the γ-aminobutyrate (GABA) shunt, was considered as a single flux, because these parallel pathways produce identical distributions of label in their product, and Suc cycling was excluded from the model for the reasons discussed elsewhere (Kruger et al., 2007a). Simplifications were also introduced where the accuracy of flux estimates was predicted to be poor, providing that such simplifications would not prevent conclusions being drawn about the function of the TCA cycle. The refined network is illustrated in Supplemental Figure S1.
Determination of Optimal 13C-Labeled Precursor
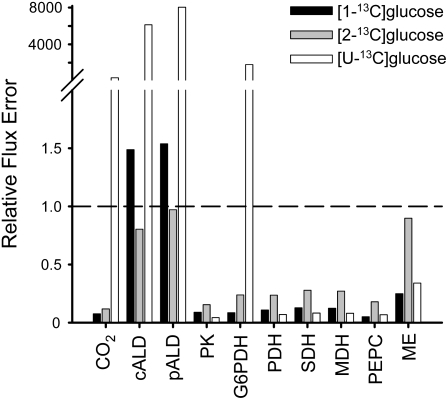
EstimateStat was also used to predict the optimal precursor for estimating TCA cycle fluxes in the refined model. Figure 1 suggests that [1-13C]Glc provides the best estimates of flux through the TCA cycle and elsewhere in the network. [1-13C]Glc and [U-13C6]Glc performed similarly well for TCA cycle fluxes, with predicted errors less than the magnitude of the flux estimates, but this analysis suggested that [1-13C]Glc provided the more accurate estimates of flux elsewhere in the network. [2-13C]Glc appeared to offer no advantage over [1-13C] or [U-13C6]Glc and is significantly more expensive. The analysis was repeated using several different models with more or less explicitly defined subcellular compartmentation of glycolysis and the oxidative pentose phosphate pathway (data not shown). Though the degree of compartmentation greatly affected the predicted relative errors, the qualitative finding that [1-13C]Glc would provide the smallest relative errors for the majority of fluxes remained the same. Post hoc analysis of the final model at the end of the investigation (data not shown) confirmed that labeling with 100% [1-13C]Glc provided the most reliable estimates of flux.
Figure 1.
The effect of labeled precursor on predicted relative flux errors (flux error/flux). Simulations used the network illustrated in Supplemental Figure S1. A relative error of less than one (indicated by the dashed line) indicates that the predicted flux error is less than the magnitude of the flux itself. Abbreviations: CO2 (CO2 output), cALD (cytosolic aldolase), pALD (plastidic aldolase), PK (pyruvate kinase), G6PDH (Glc-6-P dehydrogenase), PDH (pyruvate dehydrogenase), SDH (succinate dehydrogenase), MDH (malate dehydrogenase), PEPC (PEP carboxylase), ME (mitochondrial NAD-ME).
Elevated O2 Conditions Perturb Cell Suspension Culture Metabolism
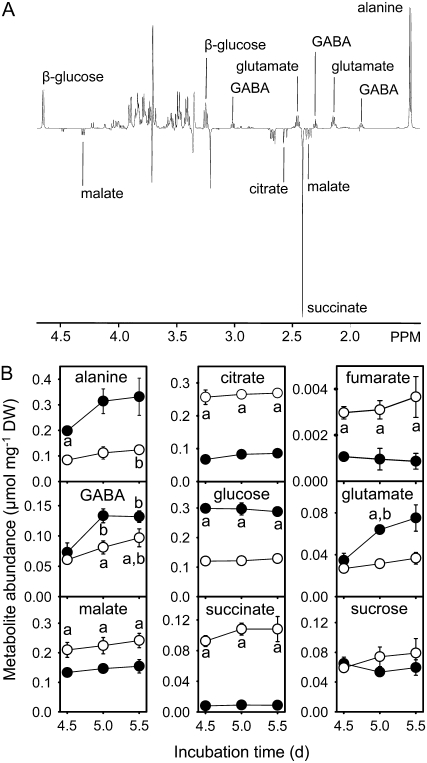
As oxygen is required to support respiration, the availability of oxygen to the cell suspension cultures might be expected to influence overall rates of metabolism and/or the partitioning of carbon entering the TCA cycle between respiration and biosynthesis. To test this hypothesis, we established a system for culturing cells at elevated O2 concentration by replacing the aluminum foil used to seal the flasks with Miracloth. After 5 d of growth, there was consistently more oxygen dissolved in the medium of cultures covered with Miracloth (elevated O2) than those covered with foil (standard O2); in a representative experiment, the oxygen concentration of the culture medium was 161.5 ± 12.5 μm for elevated O2 cells and 76.0 ± 2.5 μm for standard O2 cells, both of which are lower than the 270 μm expected for air-saturated water at 21°C (Truesdale and Downing, 1954). This difference was sufficient to cause significant increases in the abundance of amino acids (Glu, GABA, and Ala) and sugars (Glc) and decreases in the abundance of organic acids (malate, succinate, citrate, and fumarate) under elevated O2 conditions (Fig. 2). In addition, the rates of biomass accumulation and Glc consumption were both greater under elevated O2 conditions; Glc consumption increased from 214 ± 45 mg d−1 per flask under standard conditions to 335 ± 26 mg d−1 per flask under elevated conditions, while biomass accumulation increased from 174 ± 17 mg d−1 per flask to 250 ± 19 mg d−1 per flask. However, the relative biomass composition of the cell suspensions after 5 d of growth (percentage of biomass that consisted of starch, cell wall, protein, lipids) was the same under both conditions (Table I). No significant differences could be detected in the composition of the growth medium using 1H NMR (data not shown). These results suggest that a rearrangement of the metabolic network leads to increased biosynthetic fluxes and a switch from accumulation of organic acids to accumulation of amino acids in response to elevated O2. Such a rearrangement could be in the vicinity of the TCA cycle, given its involvement in both respiratory and biosynthetic processes, and this hypothesis was tested using MFA.
Figure 2.
A, Representative 1H NMR difference spectrum for elevated minus standard O2 conditions. NMR spectra of soluble metabolite extracts of cells grown for 5 d under standard O2 conditions were subtracted from the corresponding spectra for cells grown under elevated O2 conditions. Metabolites giving positive signals in this difference spectrum are more abundant under elevated O2 conditions. B, The abundance of soluble metabolites in Arabidopsis cell suspension cells after 4.5 to 5.5 d incubation under standard (○) or elevated (•) O2 conditions. Errors are ±1 sd (n = 2 or 3). “a” indicates statistically significant differences (Student's t test, P < 0.05) in metabolite abundances between the two conditions for that time point. “b” indicates statistically significant differences (Student's t test, P < 0.05) in metabolite abundance under a particular condition between the indicated time point and 4.5 d growth. No significant differences in abundance were detected between 5 and 5.5 d growth.
Table I.
Biomass composition of Arabidopsis cell cultures under standard and elevated levels of oxygen
All measurements were made on cell suspension cultures grown at the same time as those labeled to isotopic steady state with [1-13C]Glc. Each value is the mean ± sd of two or three biological replicates as described in “Materials and Methods.” The data for soluble metabolites are based on the measurements in Figure 2.
| Biomass Component | Mass of Biomass Component
|
|
|---|---|---|
| Standard O2 | Elevated O2 | |
| % dry wt | ||
| Cell wall | 32.03 ± 1.01 | 35.53 ± 3.27 |
| Starch | 2.06 ± 0.87 | 2.86 ± 0.63 |
| Lipids | 9.64 ± 0.54 | 10.94 ± 0.83 |
| Protein | 19.05 ± 1.00 | 19.12 ± 1.07 |
| Soluble metabolites | 16.27 ± 0.69 | 15.90 ± 0.61 |
Validation of Isotopic and Metabolic Steady State
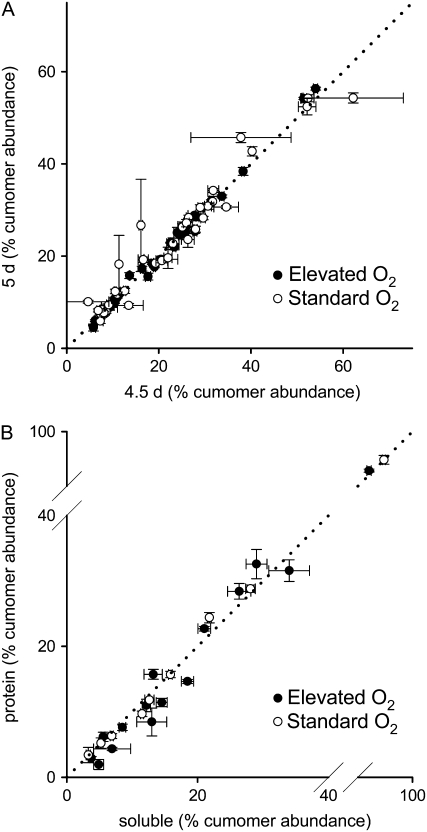
Steady-state flux analysis requires isotopomer abundances to be measured when the system is at isotopic and metabolic steady state, i.e. when metabolic fluxes are constant and when the distribution of label throughout the network has stabilized (Ratcliffe and Shachar-Hill, 2006). To confirm that isotopic steady state was reached after 5 d of growth under both elevated and standard O2 conditions, cells were fed with [U-13C6]Glc (6% of total Glc supplied) for 4.5 or 5 d and then the distribution of 13C within soluble metabolites was analyzed using one-dimensional (1D) 13C NMR. Cumomer abundances as a percentage of total labeling of a metabolite were calculated and compared between 4.5 and 5 d. There was no significant variation in abundance for the majority of measurements, and although several measurements showed significant variation, these reflected changes in abundance of <2.5%, strongly suggesting that these metabolites had reached isotopic steady state after 5 d (Fig. 3A). Comparison of percentage cumomer abundances between soluble and protein-derived amino acids after labeling with [1-13C]Glc also revealed few significant differences (Fig. 3B). Such differences as were present reflected changes in abundance of <3.9%, suggesting that turnover of protein is sufficiently rapid for it to be labeled to isotopic steady state (Roscher et al., 2000). Thus, it was concluded that the cells were at isotopic steady state after 5 d of growth.
Figure 3.
A, Demonstration of isotopic steady state of soluble metabolites after 5 d growth for cells grown at standard and elevated O2 with [U-13C6]Glc (6% of total Glc supplied) as label source. Errors are ±1 se (n = 3 or 4). B, Demonstration of isotopic steady state of protein amino acids after 5 d growth for cells grown at standard and elevated O2 with [1-13C]Glc as label source. Errors are ±1 sd (n = 2 or 3). In both cases, cumomer abundances are expressed as a percentage of the total label within a metabolite. Dotted lines indicate where cumomer abundances at isotopic steady state should lie.
The existence of an isotopic steady state suggests that any changes in metabolic fluxes over time must be relatively slow (Roscher et al., 2000). Stronger evidence for a metabolic steady state in our system is provided by the fact that the abundance of most soluble metabolites did not change from 4.5 to 5.5 d (Fig. 2). In particular, there were no significant changes in abundance between 5 and 5.5 d of growth, suggesting that the cells reached metabolic steady state after 5 d. In addition, the rate of Glc consumption and biomass increase was constant from 4.5 to 5.5 d (data not shown).
Label Measurements and Estimation of Instrument Precision
Cell cultures were labeled to isotopic steady state by growth on 100% [1-13C]Glc for 5 d under elevated O2 and standard O2 conditions. Quantitative 1D 13C NMR spectroscopy was used to obtain the label measurements necessary for estimation of metabolic fluxes from soluble metabolites, protein amino acids, and Glc digested from starch. The contributions to each assigned peak were analyzed by line-fitting (Fig. 4, inset), and the resulting label measurements were combined appropriately to give relative cumomer abundances. This procedure yielded, from three biological replicates, a total of 389 relative cumomer abundance measurements for the standard O2 condition and 429 measurements for the elevated O2 condition. All measurements for a single biological replicate came from the same batch of cells. The complete measurement dataset is given in Supplemental Table S1.
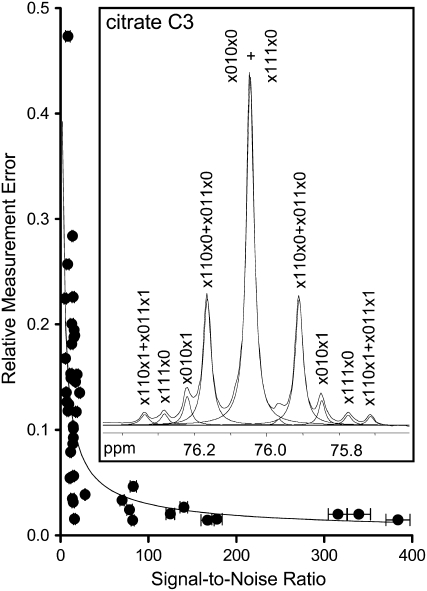
Figure 4.
The dependence of relative error of peak areas on peak SNR in 1D 13C NMR spectra. From these data, power regression was used to define an empirical relationship between relative error (RE) and SNR: RE = 0.62(SNR)−0.66. RE was calculated as the sd of three measurements made on the same sample divided by the average peak area. Measurements were made on a sample of [U-13C6]Glc with a low SNR and on a mixture of Ala, citrate, Glu, Asp, and malate at higher SNR. Errors are ±1 sd (n = 3). Inset, Line-fitting of a representative peak from a 1D 13C NMR spectrum. Line-fitted peaks are indicated with dashed lines. Peak annotations indicate the cumomer abundance defined by the area of the line-fitted peak; 1 corresponds to the presence of 13C at positions 1 to 6 in citrate (standard numbering), 0 corresponds to the presence of 12C, and x corresponds to either 13C or 12C.
To optimize flux estimates, 13C-FLUX minimizes the variance-weighted sum of squared differences between the experimental labeling data and simulated labeling data generated by 13C-FLUX using the flux estimates. The algorithm is therefore guided to an optimal flux solution by the size of the errors assigned to individual label measurements, and thus the accurate assignment of errors is likely to be important for the reproducible determination of the true flux solution. To accommodate this in the analysis, an empirical relationship was established between relative peak error and peak signal to noise ratio (SNR; Fig. 4). This formula was used to assign error estimates to individual 13C NMR labeling measurements and hence to relative cumomer abundances.
Exploration of Network Structure and Active Reactions
The 13C-FLUX implementation of the sequential quadratic programming algorithm Donlp2 (Peter Spellucci, Technische Universität Darmstadt, Germany) was used to fit the free net and exchange fluxes to the measured isotopomer data. Fluxes were fitted to all three biological replicates for a single treatment simultaneously to give a single solution that should represent the average flux state of the three replicates. To constrain the flux solution within known bounds, we used the rates of Glc consumption and biomass accumulation, and the biomass composition (Table I) to calculate input and output fluxes for cell suspensions grown under elevated and standard O2 (Sriram et al., 2006; Alonso et al., 2007a; Supplemental Table S2). Fluxes derived from biomass measurements were constrained to the mean measured value and were not allowed to vary during the fitting procedure (Schwender et al., 2006). Allowing these output fluxes to vary (Alonso et al., 2007a) led to unacceptably large discrepancies between the fitted biomass fluxes and the measured values. Free net and exchange fluxes were chosen as recommended elsewhere (Wiechert et al., 2001), and exchange fluxes were constrained to zero for steps considered to be thermodynamically irreversible. At this stage, modifications to the network were incorporated to reflect the available isotopomer and biomass data and to test whether alterations to the network structure could improve the fit of the data as measured by the sum of squared, weighted differences between the real and simulated data.
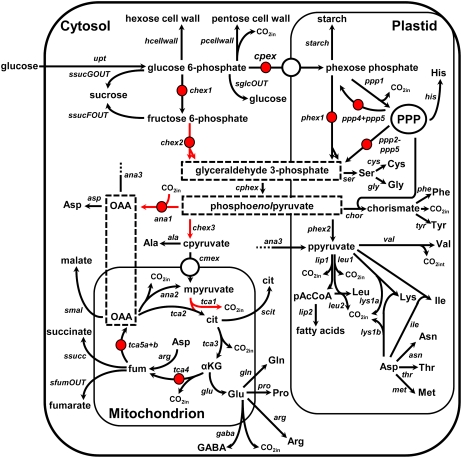
The presence of phosphoenolpyruvate carboxykinase (PEPCK) in the cell cultures is supported by proteomic analysis (L. Miguet, unpublished data; Baerenfaller et al., 2008), and the possibility of a significant flux through PEPCK was investigated by allowing a reversible flux through the PEPC step (ana1; Fig. 5). This exchange flux was consistently assigned a small but significant value during fitting of the elevated O2 dataset, suggesting that the PEPCK reaction occurs in Arabidopsis cells. A similar flux has been detected in developing sunflower embryos (Alonso et al., 2007a).
Figure 5.
Metabolic model used for the determination of intracellular fluxes. Free net fluxes are indicated with red arrows, while free exchange fluxes are indicated with red circles. All other exchange fluxes were constrained to zero during parameter fitting. The direction of arrows indicates the direction of positive flux as defined in the model. Flux names are given in italics. Abbreviations: fum (fumarate), OAA (oxaloacetate), cit (citrate), αKG (α-ketoglutarate). Standard abbreviations of amino acid names are used. Dashed boxes indicated where the subcellular localization of a metabolite or reaction cannot be inferred from the data or from the literature. The letters “p”, “c,” and “m” preceding metabolite names indicate separate pools of that metabolite in the plastid, cytosol, and mitochondrion, respectively. PPP indicates the pentose phosphate pathway. Output of CO2 from the system is included in the model but not illustrated here.
The labeling data from amino acids synthesized from cytosolic (Ala; see Miyashita et al., 2007) and plastidic (Val, Ile, and Leu) pyruvate indicated that pyruvate in these compartments is not at isotopic equilibrium. This was modeled by introducing separate pools of pyruvate in the plastid and cytosol (ppyruvate and cpyruvate; Fig. 5). However, the introduction of an irreversible uptake of pyruvate into the plastid did not improve the fit of the data, suggesting that the labeling data contained little information on the uptake of pyruvate. A similar result was obtained with sunflower embryos (Alonso et al., 2007a), and a recent investigation, in which decreased plastidic pyruvate kinase produced severe decreases in lipid accumulation in Arabidopsis, is consistent with the hypothesis that uptake from the cytosol is not the main route by which the plastidic pyruvate pool is maintained (Andre et al., 2007).
Recent data also suggest that cytosolic and plastidic isoforms of NADP-malic enzyme (ME) are expressed constitutively in heterotrophic tissues of Arabidopsis (Gerrard Wheeler et al., 2008). While addition of a plastidic NADP-ME (ana3 oxaloacetate → ppyruvate; Fig. 5) to the model did not improve the fit of the data, analysis using EstimateStat suggested that this flux could be determined with good precision from our labeling data and it was therefore included in the final model. Addition of a cytosolic isoform of NADP-ME (oxaloacetate → cpyruvate; Fig. 5) did not improve the fit above that obtained with PEPCK and the plastidic NADP-ME, and, in addition, its presence greatly increased the flux errors predicted by EstimateStat. For this reason, cytosolic NADP-ME was not included in the final model. However, in trials where cytosolic NADP-ME was included, the fitting process consistently assigned it a small but significant flux, and the flux through PEPCK decreased to zero. This suggests that what we describe as PEPCK flux (the reversibility of PEPC) could equally be described as flux through cytosolic ME.
Model Validation and Statistical Analysis
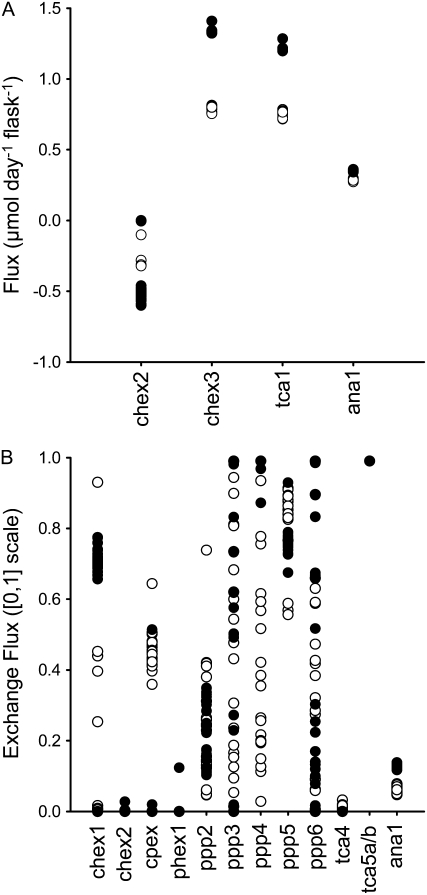
The final network structure is shown in Figure 5, and in 13C-FLUX format, in Supplemental Table S3. The fitting procedure was initiated 150 times for both datasets with random initial flux estimates, and a feasible flux solution was found in over 85% of fits under both conditions. Some free fluxes converged to similar values in each run of the fitting algorithm, and for these fluxes the distribution of solutions was unimodal (Fig. 6). These fluxes included all the net fluxes, apart from flux through cytosolic aldolase under elevated O2 conditions, and all the exchange fluxes associated with the TCA cycle. In contrast, some of the remaining free fluxes did not converge to similar values in each run, with the solutions appearing to adopt a random distribution, suggesting that there is little information in the labeling data to constrain the fluxes to a particular value. The flux solution giving the lowest sum of squared weighted differences for each dataset was taken forward for further analysis and is referred to as the optimal flux solution.
Figure 6.
Distribution of flux solutions during replicate fitting for the standard (○) and elevated (•) O2 datasets. A, Net fluxes. B, Exchange [0,1] fluxes. The best (lowest sum of squared residuals) 25 solutions for each dataset are shown, with each point representing a single solution. Exchange fluxes (Xch [0,1]) are expressed on a normalized scale following hyperbolic transformation, defined by Xch [0,1] = JXch/(1 + JXch), where JXch is the true exchange flux. Net (J), exchange, forward (Jf), and reverse (Jr) fluxes are related by J = Jf − Jr and JXch = min (Jf, Jr) (Wiechert et al., 2001).
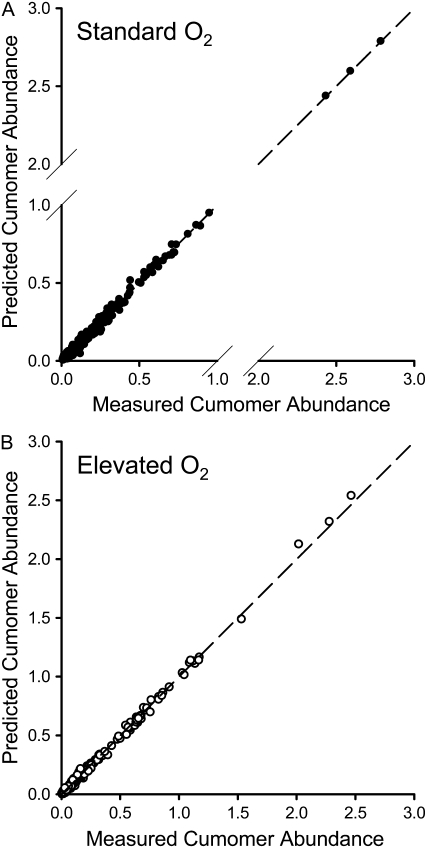
Figure 7 shows that there was good agreement between the observed labeling data for the standard O2 and elevated O2 conditions and the labeling data predicted from the optimal flux solutions. Moreover, all measurements contributing more than 1% of the total sum of squared weighted differences could be removed from the fit (24 measurements contributing 40% of the residuum for standard O2 conditions and 21 measurements contributing 29% of the residuum for elevated O2 conditions) without altering the optimum flux solution, demonstrating that these poorly fitting measurements were not important in constraining the fit. It can be concluded that the optimal flux solutions provide adequate descriptions of the labeling data.
Figure 7.
Comparison of experimental and simulated labeling data for the optimal flux solutions under standard O2 (A) and elevated O2 (B) conditions. The input labeling data (measured cumomer abundance) is compared to the value predicted by 13C-FLUX for the optimum flux solution (predicted cumomer abundance) under standard and elevated O2 conditions.
The errors for the optimal fluxes were derived using EstimateStat and are summarized in Table II. To ensure that biological error present within the replicate label measurements and biomass fluxes was translated into errors in the flux estimates, the labeling data, excluding the poorly fitting data described above, were reduced to a single set of measurements (see “Materials and Methods”). It was then possible to use EstimateStat in combination with replicate fitting experiments (Fig. 6) to define a list of fluxes that were statistically well determined (Wiechert et al., 2001).
Table II.
Metabolic fluxes in Arabidopsis cell cultures under standard and elevated levels of oxygen
Fluxes were determined by fitting labeling and biomass data to the model shown in Figure 5. Net fluxes are the optimum estimate ± sd as determined by EstimateStat. A negative value indicates that the direction of net flux is opposite to that assumed in the model (Fig. 5). Exchange fluxes (JXch) are given as the optimum estimate, with lower and upper asymmetric bounds corresponding to the optimum normalized [0,1] exchange flux ±1 sd (see legend to Fig. 6). Where the bounds of the normalized exchange flux lie outside the [0,1] range, an exchange flux of 0 (≤0) or infinity (∞,≥1) is given. Where no exchange flux is given, the value was constrained to zero in the fit. See Supplemental Table S4 for the complete flux solution. DH, Dehydrogenase.
| Flux Name | Description | Standard O2
|
Elevated O2
|
||
|---|---|---|---|---|---|
| Net Flux | Exchange Flux | Net Flux | Exchange Flux | ||
| mmol d−1 per flask | mmol d−1 per flask | ||||
| upt | Glc uptake | 1.322 ± 0.071 | 2.066 ± 0.077 | ||
| CO2 | CO2 output | 2.772 ± 0.349 | 4.498 ± 0.291 | ||
| Hexose/triose metabolism | |||||
| chex1 | Cytosolic phosphoglucoseisomerase | 0.010 ± 0.425 | 0.000 (0.000, ∞) | −0.481 ± 0.396 | 2.196 (1.770, 2.778) |
| chex2 | Cytosolic aldolase | −0.003 ± 0.425 | 0.000 (0.000, 0.429) | −0.494 ± 0.396 | 0.000 (0.000, 0.864) |
| chex3 | Cytosolic pyruvate kinase | 0.805 ± 0.070 | 1.330 ± 0.083 | ||
| cpex | Hexose phosphate uptake by plastid | 0.921 ± 0.448 | 0.755 (0.212, 2.184) | 1.893 ± 0.407 | 0.000 (0.000, 0.223) |
| phex1 | Plastidic aldolase | 0.752 ± 0.424 | 0.000 (0.000, 2.206) | 1.630 ± 0.401 | 0.000 (0.000, 2.389) |
| phex2 | Plastidic pyruvate kinase | 0.422 ± 0.050 | 0.622 ± 0.050 | ||
| cphex | Glyceraldehyde-3-P DH | 1.570 ± 0.084 | 2.380 ± 0.110 | ||
| ppp1 | Oxidative pentose phosphate pathway | 0.402 ± 0.200 | 0.599 ± 0.161 | ||
| Biomass accumulation | |||||
| hcellwall | Cell wall (hexose component) | 0.293 ± 0.031 | 0.466 ± 0.053 | ||
| pcellwall | Cell wall (pentose component) | 0.063 ± 0.007 | 0.101 ± 0.012 | ||
| starchOUT | Starch | 0.022 ± 0.010 | 0.044 ± 0.010 | ||
| lipidOUT | Acetyl-CoA in phospholipid | 0.275 ± 0.031 | 0.446 ± 0.047 | ||
| glycerolOUT | Glycerol in phospholipid | 0.003 ± 0.000 | 0.004 ± 0.000 | ||
| TCA cycle | |||||
| cmex | Pyruvate uptake by mitochondrion | 0.754 ± 0.069 | 1.205 ± 0.082 | ||
| tca1 | Mitochondrial pyruvate DH | 0.773 ± 0.068 | 1.205 ± 0.081 | ||
| tca2 | Citrate synthase | 0.773 ± 0.068 | 1.205 ± 0.081 | ||
| tca3 | Aconitase and isocitrate DH | 0.727 ± 0.067 | 1.185 ± 0.081 | ||
| tca4 | α-Ketoglutarate DH and succinate DH | 0.649 ± 0.066 | 0.003 (0.000, 0.019) | 1.050 ± 0.079 | 0.000 (0.000, 0.031) |
| tca5a+b | Fumarase and malate DH | 0.646 ± 0.066 | 99.000 (19.075, ∞) | 1.074 ± 0.080 | 99.000 (20.365, ∞) |
| Anaplerotic fluxes | |||||
| ana1 | PEPC/PEPCK | 0.286 ± 0.043 | 0.068 (0.033, 0.105) | 0.349 ± 0.048 | 0.149 (0.131, 0.167) |
| ana2 | Mitochondrial ME | 0.019 ± 0.014 | 0.000 ± 0.048 | ||
| ana3 | Plastidic ME | 0.000 ± 0.035 | 0.035 ± 0.007 | ||
| Amino acid and organic acid synthesis | |||||
| ala | Ala synthesis | 0.051 ± 0.007 | 0.125 ± 0.015 | ||
| asp | Asp synthesis | 0.102 ± 0.006 | 0.145 ± 0.011 | ||
| glu | Glu synthesis | 0.078 ± 0.005 | 0.134 ± 0.006 | ||
| scit | Citrate accumulation | 0.046 ± 0.005 | 0.021 ± 0.002 | ||
| smal | Malate accumulation | 0.039 ± 0.006 | 0.037 ± 0.004 | ||
| ssucc | Succinate accumulation | 0.019 ± 0.002 | 0.002 ± 0.000 | ||
| sfumOUT | Fumarate accumulation | 0.001 ± 0.000 | 0.000 ± 0.000 | ||
Flux Maps of Central Carbon Metabolism under Standard and Elevated Oxygen Conditions
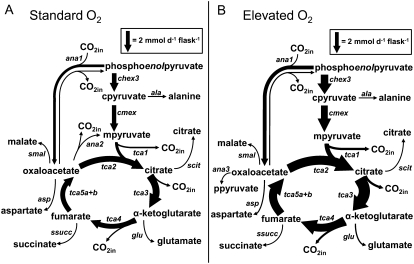
Table II contains net and exchange fluxes together with errors calculated using EstimateStat for the optimal flux solutions under standard and elevated O2 conditions. Figure 8 shows the TCA cycle and its associated biosynthetic fluxes in detail. From Figure 8 and the data in Table II, it is clear that elevated O2 brings about an increase in flux through the TCA cycle and through the biosynthetic pathways associated with it. However, the proportion of carbon entering the TCA cycle that is used for biosynthesis is unaffected by the increased O2 concentration, with 38% ± 5% of carbon used for biosynthesis under standard O2 conditions and 33% ± 2% under elevated O2 conditions. Moreover, if the net fluxes throughout the network are expressed relative to the rate of Glc uptake, it is apparent that elevated O2 did not bring about a major rearrangement of the metabolic network, either at the level of the TCA cycle or at the level of the whole network (Fig. 9).
Figure 8.
The effect of altered O2 concentration on metabolic fluxes. A, Standard O2; B, Elevated O2. The width of arrows is proportional to flux of carbon through a particular step calculated from the flux estimates in Table II and the number of carbon atoms in the metabolites involved in each reaction. Reactions that carry no flux under the specified condition are not included. Forward and reverse fluxes for ana1 (PEPCK) are illustrated. Exchange fluxes for tca4 and tca5a + 5b were also determined (Table II) but are omitted for clarity. The fraction of carbon entering the TCA cycle used for biosynthesis was calculated as: (4ana3 + 4asp + 5glu + 6scit + 4smal + 4ssucc + 4sfumOUT)/(3cmex + 4ana1).
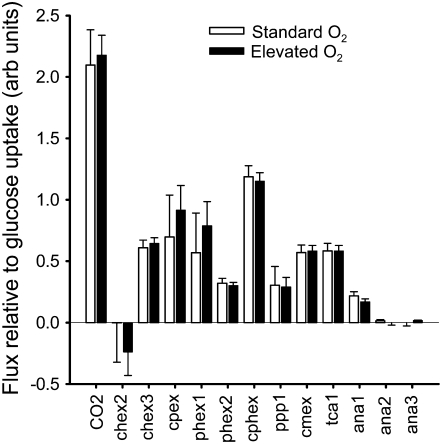
Figure 9.
The effect of elevated O2 on metabolic fluxes relative to the rate of Glc uptake. Fluxes corresponding to the optimal solution are divided by the rate of Glc uptake, thereby permitting changes in organization of the metabolic network to be distinguished. Errors are ±1 sd as determined using EstimateStat. Flux names correspond to those given in Figure 5 and Table II.
DISCUSSION
This study aimed to quantify multiple fluxes within the central carbon metabolism of Arabidopsis and to investigate the effect of an altered O2 concentration on the relationship between respiratory and biosynthetic fluxes around the TCA cycle. The flux maps reported here are the first, to our knowledge, to be obtained using steady-state MFA for Arabidopsis, and they show that while an elevated O2 concentration in a cell culture can increase fluxes throughout the metabolic network and alter the abundance of soluble metabolites, these changes do not require either a major reorganization of the network or a change in the balance between respiratory and biosynthetic flux.
Reliable Quantification of Metabolic Fluxes
Flux quantification was carried out using data obtained by 1D 13C NMR from three biological replicates of a [1-13C]Glc labeling experiment, leading to a set of statistically well-determined flux estimates that appears to represent the global optimum flux solution. The MFA protocol incorporated several refinements aimed at improving the precision and reliability of the flux estimates.
First, to increase the likelihood of being able to accurately quantify TCA cycle fluxes, we began our investigation by predicting the optimal 13C-labeled precursor to use for isotopic steady-state labeling experiments. While the approach used here considered fewer parameters than recent work in this area (Ghosh et al., 2006; Libourel et al., 2007), it nevertheless indicated that the use of [1-13C]Glc would permit the accurate quantification of TCA cycle fluxes and associated biosynthetic and anapleurotic fluxes. To extend this relatively simple approach beyond the most commonly available isotopomers of Glc, it will be necessary to develop methods for predicting how the precise mixture of 13C-labeled precursors influences the labeling measurements that can be made.
Second, to quantify the biosynthetic capacity of the TCA cycle more completely than hitherto, measurements of organic and amino acids derived from the TCA cycle were incorporated into the model (Fig. 2, analysis of protein hydrolysate). While the list of quantified biomass components deriving from the TCA cycle is not exhaustive, accounting for these extra biosynthetic demands in the model should provide a more accurate picture of how the TCA cycle functions in respiration and biosynthesis. In contrast to previous work in this field, all of the biomass and labeling measurements were made on cell cultures initiated from the same stock cultures and grown concurrently. This should ensure that the labeling data and biomass data are consistent with each other, which may not otherwise be the case, particularly for soluble metabolites that show significant batch-to-batch variability.
Finally, to avoid the need to calculate averages of relative cumomer abundances, which are nonlinear functions of metabolic fluxes, fluxes were fitted simultaneously to three separate biological replicates. This process was assisted by assigning specific errors to individual measurements on the basis of SNR (Fig. 4). Biological error was applied to the estimates of flux measurement precision (Table II) by assessing the relative error present in replicate cumomer abundance measurements. Repeated fitting of the model combined with statistical analysis then indicated fluxes that were both statistically well determined and which took similar values in each run. Fulfillment of both of these criteria is important, as statistically well-defined fluxes may not always have a single optimal value. In our investigation, this is the case for the net flux chex2 under elevated O2 conditions (Fig. 6), and, hence, biological conclusions made on the basis of consideration of a single flux solution, or indeed a subset of flux solutions, risk being erroneous.
The relative simplicity of the protocol developed in this study suggests that it may be possible to determine metabolic fluxes in heterotrophic tissue cultures of Arabidopsis reasonably easily, potentially allowing a wide range of genotypes and environmental conditions to be analyzed. The large datasets collected in this study will allow us, by exploiting the ability of 13C-FLUX to determine the sensitivity of individual label measurements to changes in fluxes (see Schwender et al., 2006), to identify the minimum dataset needed to acquire accurate and robust estimates of metabolic fluxes in Arabidopsis. Because the acquisition and analysis of large numbers of 1D 13C NMR measurements from multiple sources (soluble metabolites, starch, and protein amino acids) is currently a time-consuming process, this refinement should reduce the experimental effort required for future investigations. To apply our approach to different genotypes, it will, however, be necessary adapt the method for systems other than cell suspension cultures, which require considerable time and effort to establish.
The TCA Cycle in Arabidopsis
While the flux distribution centered on the TCA cycle in heterotrophic Arabidopsis cell suspensions under standard O2 conditions is qualitatively similar to flux distributions in other plant species (Rontein et al., 2002; Alonso et al., 2007a, 2007b; Sriram et al., 2007), there are nonetheless quantitative differences that may reflect the precise function of the TCA cycle in different systems. Entry of carbon into the TCA cycle via both pyruvate dehydrogenase and PEPC has been observed in all MFA studies of plants. In the Arabidopsis cells, the flux through PEPC was 38% of pyruvate uptake by the mitochondrion, identical to the value obtained for soybean embryos (38%; Sriram et al., 2004b) and similar to values obtained for tomato cell suspension cultures (27% in pre-stationary phase; Rontein et al., 2002) and Catharanthus roseus hairy root cultures (23%; Sriram et al. 2007). While these values for PEPC broadly agree with those measured in other tissues, the flux through NAD-ME as a percentage of pyruvate uptake was the lowest yet measured in plants (2.5%), contrasting with the previous lowest value found in tomato cell suspensions at pre-stationary phase (6%) and values as high as 66% found in B. napus embryos (Schwender et al., 2006). This result suggests that a dominant role of this enzyme in pyruvate production, as proposed by others (Tronconi et al., 2008), may be a specific feature of leaves in which malate that accumulates during the light is metabolized in the subsequent dark period. Because our cells show no sign of hypoxia under standard O2 conditions, the lack of NAD-ME flux is consistent with the proposed role for NAD-ME under low O2 (Roberts et al., 1992; Edwards et al., 1998).
In Arabidopsis under standard O2 conditions, 38% of carbon entering the TCA cycle was used for biosynthetic processes, including the synthesis of protein and accumulation of amino acids and organic acids (Fig. 8; Table II). Little to no carbon was withdrawn from the TCA cycle for plastidic fatty acid synthesis, consistent with the view that the main route of carbon supply for fatty acid synthesis is via plastidic pyruvate kinase and not via plastidic ME or uptake of pyruvate from the cytosol (Schwender et al., 2006; Alonso et al., 2007a; Andre et al., 2007). Withdrawal of TCA cycle carbon for biosynthesis has also been observed in several MFA analyses, and the proportion of carbon removed presumably reflects the balance between the demand for biosynthetic precursors and the ATP required to convert those biosynthetic precursors into end products and maintain other cell functions. For example, in B. napus embryos (Schwender et al., 2006), 70% of carbon entering the TCA cycle was used for biosynthetic processes. Here, photosynthetic production of ATP reduced the need for the TCA cycle to generate reductant for the mitochondrial electron transport chain, and most of the carbon entering the TCA cycle was removed for the elongation of fatty acids in the cytosol. On the other hand, in sunflower embryos (Alonso et al., 2007a), only 6% of carbon entering the TCA cycle was used for biosynthesis, confirming the importance of the TCA cycle in meeting cellular energy demand in this system.
The Metabolic Response to Increased O2 Concentration
The increased rates of biomass accumulation and Glc consumption in the Arabidopsis cell suspension culture at elevated O2 concentrations were associated with increases in net fluxes throughout the metabolic network (Fig. 8; Table II). These higher fluxes corresponded to higher rates of ATP synthesis; calculations based on the optimal flux solutions, assuming that 2.5 molecules of ATP are produced for each NADH and 1.5 molecules produced for each FADH2 (Brand, 1994), suggest that the rate of ATP production increased by around 50%, from 15 mmol d−1 per flask to 23 mmol d−1 per flask, under elevated O2 conditions. Increased ATP levels and rates of protein synthesis have also been detected in response to increased O2 levels in cell suspensions of peanut (Arachis hypogaea; Verma and Marcus, 1974). While an increase in O2 concentration would increase ATP production if the oxygen availability under standard conditions limited the activity of cytochrome oxidase, this seems unlikely given that the O2 levels in the cell suspension culture were well above the Km of cytochrome oxidase (0.13 μm in soybean root mitochondria; Millar et al., 1994). Thus, there would have to be a large O2 concentration gradient between the medium and the mitochondrion for this explanation to be plausible. Moreover, measurements of soluble metabolites gave no indication that the cells were hypoxic under standard conditions. For example, the abundance of Ala and GABA was significantly lower under standard conditions than under elevated O2 conditions (Fig. 2), whereas these metabolites typically accumulate during hypoxia (Miyashita and Good, 2008). It is possible that the effect on metabolic fluxes is related to the observations that have been made on growing potato (Solanum tuberosum) tubers (Geigenberger et al., 2000), where glycolytic flux increased toward the outside of the tuber where the O2 concentration was higher. This effect was not limited to glycolysis, as rates of starch synthesis, lipid synthesis, and protein synthesis all increased at higher O2 concentrations (van Dongen et al., 2004, and refs. therein). On this basis, it has been proposed that variation in O2 concentration may affect metabolism independently of any effect on cytochrome oxidase activity, perhaps as an adaptation that adjusts metabolism to the availability of O2 and prevents respiration driving a tissue into anoxia when oxygen supply is limited (Geigenberger, 2003).
While manipulation of O2 concentration brought about marked changes in absolute fluxes, expressing flux relative to the rate of Glc uptake showed that there was no major rearrangement of the metabolic network despite the associated changes in soluble metabolite levels (Fig. 2). In particular, MFA indicates that ratios of internal fluxes in the TCA cycle and elsewhere remained almost constant (Fig. 9), a result in keeping with the unchanged biomass proportions (Table I). Thus, the changes in levels of organic acids, amino acids, and sugars can be produced without major changes in relative fluxes within central metabolism. The fluxes that lead to the accumulation of soluble metabolites in this system are very small compared to fluxes though the core of carbon metabolism (Table II), so only small net changes in flux are required to produce large changes in the relative abundance of the soluble metabolites. For example, the 72% decrease in citrate accumulation relative to Glc uptake that occurred at elevated O2 could be produced with a change in citrate synthase flux of only 4%. Similarly, subtle changes in relative fluxes around oxaloacetate may be responsible for some of the changes in soluble metabolite abundances (Fig. 2). For example, under elevated O2 conditions, there was a detectable (22%) decrease in relative flux through PEPC (Figs. 8 and 9), while flux through PEPCK increased (Table II), possibly contributing to the decreased abundance of organic acids. However, in general, it appears that the differences in rates of accumulation of soluble metabolites arose from rearrangements of the central metabolic network that are smaller than can currently be detected using MFA.
Overall, the O2 concentration did not exert significant control over the balance between respiration and biosynthesis under these conditions, even though it had a significant influence on the growth of the cells. Heterotrophic Arabidopsis cultures may therefore respond to changes in O2 concentration by altering rates of respiration, biosynthesis, and ultimately growth proportionally, such that the demand for ATP to support biosynthesis is balanced by the rate of respiratory processes that generate ATP.
Network Stability
Stability of relative fluxes in carbon metabolism has become a recurring theme in plant MFA studies. While certain environmental and genetic perturbations have been shown to alter the flux distribution in the central metabolic network (Spielbauer et al., 2006; Junker et al., 2007; Iyer et al., 2008), it is also possible for the fluxes to vary in absolute magnitude without any change in their relative values. This is certainly the case in the Arabidopsis cell study reported here, and a similar result was obtained in a less statistically rigorous flux analysis of a tomato cell suspension, where the relative fluxes through glycolysis, the TCA cycle, and the oxidative pentose phosphate pathway remained similar, even though the net fluxes decreased over the culture period (Rontein et al., 2002). Similarly analysis of wild-type, hybrid, and starch-deficient maize (Zea mays) lines (Spielbauer et al., 2006) revealed few differences in relative fluxes despite the great variation in accumulation of starch and seed weight across the genotypes. A similar trend has also been observed in microorganisms (Fischer and Sauer, 2003, 2005) and yeast (Saccharomyces cerevisiae; Blank et al., 2005). For example, the systematic flux ratio analysis of 137 Bacillus subtilis gene deletion mutants revealed that although there was more than 30% variation in growth rate in 90% of the mutants examined, relative fluxes varied by only 3% to 8% (Fischer and Sauer, 2005). Thus, the emerging picture is one in which central metabolism is well adapted to the demands that can be put on it, and it seems likely that only environmental or genetic perturbations that have a major impact on overall biomass composition (Iyer et al., 2008) or uptake fluxes (Junker et al., 2007) are likely to affect metabolism to a degree that an effect on relative fluxes in central carbon metabolism can be detected using MFA.
The stability of the central metabolic network also complicates the interpretation of metabolite profiling data. The levels of most metabolites represent only a very small fraction of the total biomass, making it unlikely that changes in level will reflect significant changes in flux within the central network. Thus, the changes in metabolite abundance caused by altering the availability of oxygen (Fig. 2) do not lead to easily identifiable perturbations in the flux map (Fig. 8). Further improvements in the accuracy and precision of MFA may alleviate this problem, but it will also be important to complement MFA with the continued development of sophisticated models of plant metabolism (Sweetlove et al., 2008). Ultimately, this can be expected to establish the relative merits of composition and flux as the basis for defining metabolic phenotypes (Ratcliffe and Shachar-Hill, 2005).
CONCLUSION
A comprehensive description of the fluxes through the TCA cycle and associated pathways in an Arabidopsis cell suspension has been obtained using a robust steady-state stable isotope-labeling protocol. Increasing the concentration of dissolved oxygen increased fluxes throughout the network and caused changes in the soluble metabolite profile, while at the same time having no effect on the proportion of carbon entering the TCA cycle that was used for biosynthesis and no significant impact on the relative flux distribution in the central network. Ultimately, while the mechanism by which the cells respond to increased oxygen has yet to be established, the study demonstrates the utility of MFA as a tool for probing the impact of an environmental perturbation on the operation of the central metabolic network.
MATERIALS AND METHODS
Experimental System
Cell suspensions of Arabidopsis (Arabidopsis thaliana) ecotype Landsberg erecta (May and Leaver, 1993) were maintained in 250-mL Erlenmeyer flasks under a 16-h-light, 8-h-dark cycle at 21°C on an orbital shaker. Every 7 d, 15 mL cell suspension was subcultured into 90 mL fresh Murashige and Skoog medium supplemented with 3% (w/v) Glc, 0.5 mg/L naphthylacetic acid, and 0.05 mg/L kinetin. Heterotrophic cell suspensions were produced by subculturing 15 mL of a 7-d-old, light-grown cell suspension into 90 mL fresh medium and incubating in the dark at 21°C on an orbital shaker. The concentration of oxygen in the growth medium was altered by controlling the rate of diffusion of air into the flask air space via the material used to seal the cultures. Standard O2 cultures were sealed with a double layer of aluminum foil, while elevated O2 cultures were sealed with four layers of Miracloth (Merck Chemicals) secured with Micropore (3M) surgical tape.
Oxygen Electrode Measurements
Measurements of dissolved oxygen concentration were made using a Clark type oxygen electrode at 21°C. Arabidopsis cell suspension (1 mL) was transferred to the electrode chamber, and oxygen consumption was monitored until the trace became linear. The linear portion was extrapolated to time zero to determine the oxygen concentration at the point of sample addition.
Isotopic Steady-State Labeling
Cells were labeled to isotopic steady state by subculturing light-grown cell suspension into medium where a proportion of the unlabeled Glc was replaced with 13C-labeled Glc (Cambridge Isotope Laboratories and Sigma-Aldrich). This procedure has been shown to have no discernible effect on the flux distribution through the Arabidopsis metabolic network (Kruger et al., 2007b). Biological variation was assessed by subculturing from three separate light-grown stocks that had been maintained independently for several weeks. Cultures were incubated on an orbital shaker in the dark at 21°C for 4.5, 5, or 5.5 d, as appropriate. Cells were harvested by vacuum filtration through a single paper filter, washed with 210 mL Glc-free growth medium, weighed, and immediately frozen in liquid N2. Tissue was stored at −80°C prior to analysis.
Soluble metabolites were extracted from frozen tissue labeled to isotopic steady state using perchloric acid (Kruger et al., 2007b). Following the final freeze-drying step, samples were redissolved in 10% 2H2O, with 10 mm EDTA, 25 mm 1,4-dioxane, and 10 mm KH2PO4/K2HPO4, pH 7.5, for NMR spectroscopy. Starch was extracted from the insoluble residue remaining from perchloric acid extractions. The residue was washed and autoclaved in 100 mm sodium acetate, pH 4.8, for 2 h. Gelatinized starch was then enzymatically digested overnight at 37°C with 30 units of α-amylase (Roche) and 5 units of amyloglucosidase (Roche). The supernatant, containing Glc released from starch, was freeze dried and redissolved in 10% 2H2O with 25 mm 1,4-dioxane for 13C NMR analysis.
Protein was extracted by repeated washing of ground, lyophilized tissue with phosphate buffered saline (130 mm NaCl, 100 mm Na2HPO4/NaH2PO4, pH 7.0). Prior to hydrolysis, protein was precipitated using 12% TCA, washed with ice-cold acetone, and resuspended in 6 m HCl. Hydrolysis was carried out in Pierce hydrolysis tubes; samples were degassed and flushed with N2 three times, then heated at 95°C for 24 h under vacuum. Samples were freeze dried to remove HCl and redissolved in 10% 2H2O with 25 mm 1,4-dioxane, pH 7.5, for 13C NMR analysis.
Biomass Analysis
Growth rate of cell suspensions was determined from fresh weight recorded during harvest and converted to change in dry weight by assuming that fresh cells contained 95% water by weight. This value was supported by experiments in which cell mass was determined before and after freeze drying. Measurements of the abundance of protein, amino acids, cell wall, starch, lipids, and soluble metabolites were made using either tissue labeled to isotopic steady state with [1-13C]Glc or tissue grown concurrently from the same stock cultures.
Protein extracted with phosphate buffered saline was quantified using the Bradford assay. The amino acid content of labeled protein hydrolysates was determined by HPLC (Bruckner et al., 1995) after 1D 13C NMR analysis; amino acids were derivatized with O-phthaldialdehyde, separated using a reverse phase C18 column, and quantified by fluorescence using standard curves.
Starch was quantified by autoclaving duplicate samples of ground, unlabeled, lyophilized tissue for 1 h in 25 mm sodium acetate, pH 4.8. Fifteen units of α-amylase (Roche) and 5 units of amyloglucosidase (Sigma-Aldrich) were added to one of the samples, and both samples were placed at 37°C overnight. Glc in the supernatants from both samples was quantified using a spectrophotometric assay (Sweetlove et al., 1996), and the abundance of starch was calculated from the difference in the amount of Glc in the enzyme treated and untreated samples. The same spectrophotometric assay was used to quantify Glc in the growth medium.
Cell wall was extracted by repeated washing of a known mass of unlabeled ground lyophilized tissue with a mixture of phenol, acetic acid, and water in the ratio 2:1:2 (Sriram et al., 2006). Insoluble material remaining was washed with distilled water to remove residual phenol, freeze dried, and weighed. The mass was corrected for the presence of contaminating starch using the starch quantification protocol above.
Lipids were extracted from a known mass of ground, labeled, lyophilized tissue using hexane and isopropanol according to an established protocol (Hara and Radin, 1978; Mhaske et al., 2005). Solvent was removed by gentle heating and lipids quantified by weight.
Soluble metabolites were extracted with methanol as described elsewhere (Le Gall et al., 2003). A known mass of ground, lyophilized tissue was shaken with 70% methanol, 30% buffer (1 mm sodium 3-trimethylsilylpropionate 2H4 [TSP], 1 mm EDTA, 50 mm KH2PO4, 50 mm KH2PO4) for 30 min. Solvent was removed under vacuum and samples were redissolved in 800 μL 2H2O for 1H NMR analysis. Quantification was based on the addition of a known quantity of TSP to representative samples.
NMR Spectroscopy
Spectra were recorded on a Varian Unity Inova 600 spectrometer (Varian). 1D 13C NMR spectra were recorded at 150.9 MHz using either a 10-mm broadband or a 10-mm 13C/31P switchable probe, and in all experiments Waltz16 decoupling was applied during the detection period to decouple 1H signals. Spectra were referenced to 1,4-dioxane at 67.3 ppm. Where absolute quantification of labeling was not required, a recycle delay of 6 s was used, and the NOE was induced during the relaxation delay to increase SNR values. For quantitative data, the recycle delay was extended to 19 s and the nuclear Overhauser effect was not induced. Acquisition times of the order of 60 h (10,240 scans) were required to obtain suitable spectra from labeled samples for accurate line-fitting. Spectra were acquired in blocks of 1,024 scans that were manually summed following inspection to confirm that there was no significant degradation during acquisition. 1D 1H NMR spectra were recorded at 600 MHz using a 5-mm HCN triple resonance probe and the standard Varian pulse program. Presaturation was applied during the relaxation delay to suppress the water signal, and spectra were referenced to TSP at 0 ppm. Two-dimensional 1H/13C gHMBC and gHSQC spectra were recorded at 600 MHz (1H) and 150.9 MHz (13C) using a 5-mm HCN triple resonance probe and standard Varian pulse programs. WURST-40 or Garp1 decoupling was applied during the detection period to remove 13C-1H coupling.
All spectra were processed and analyzed using NUTS (Acorn NMR). 1D 13C spectra were processed using a line-broadening of 2.5 Hz for spectra requiring line-fitting or 1 Hz for spectra requiring integration. 1D 1H spectra were processed using a line-broadening of 1 Hz. 1H and 13C assignments were based on literature values, comparison with pure standards, and the results of two-dimensional NMR experiments. Spectral deconvolution (line-fitting) of 1D 13C spectra was carried out using the line-fitting subroutine in NUTS. During line-fitting, resonance frequency, signal intensity, line-width, and fraction Lorentzian lineshape were varied to minimize the difference between the real and simulated spectra.
The SNR values for the 1D 13C NMR signals from soluble extracts of cells labeled to isotopic steady state with [1-13C]Glc varied over more than two orders of magnitude between different metabolites and different experimental conditions. Variation in extraction efficiency and sample fresh weights also contributed to considerable variation in signal intensity for the same metabolites between biological replicates. Because instrumental precision depends on SNR, it would have been incorrect to assign the same relative error to every label measurement during optimization of fluxes. We therefore recorded replicate quantitative 1D 13C NMR spectra of standard samples of organic and amino acids (25 mm Ala, 50 mm citrate, 5 mm Glu, 0.5 mm Asp, and 1 mm malate) and [U-13C6]Glc. Signal intensities were measured by line-fitting, and the relative error of the same signal over three replicate spectra was correlated with the corresponding SNR (Fig. 4). An empirical relationship between SNR and relative peak error was determined (Fig. 4), and this formula was used to assign error estimates to 13C NMR labeling measurements.
Metabolic Modeling
Metabolic modeling was carried out using 13C-FLUX (version 20050329; Wiechert et al., 2001, and refs. therein). The EstimateStat component of 13C-FLUX was used to refine the metabolic network. To do this, initial flux estimates were taken from the literature (Rontein et al., 2002), and lists of 1D 13C NMR measurements were taken from steady-state experiments carried out previously in our lab (P. Lelay, unpublished data). The CumoNet component of 13C-FLUX was then used to predict a set of cumomer and isotopomer abundances consistent with the initial flux estimates, and these abundances were combined appropriately to produce pseudo 1D 13C NMR datasets in which the label measurements perfectly reflected the underlying fluxes. An error of 5% was assumed for each measurement within this dataset. EstimateStat was then used to determine the errors associated with the flux estimates. The same approach was used to predict the optimum labeled precursor for minimization of predicted flux estimate errors. Here, the refined network was used in conjunction with sets of 1D 13C NMR measurements derived from experiments specific to the fed precursor.
The estimated flux errors produced by EstimateStat are sensitive to measurement configuration and relative measurement errors but do not depend on absolute label measurements (Wiechert et al., 2001). To derive flux error estimates that incorporate biological error, we determined the relative error in cumomer abundances between the different biological replicates. To do this, we first normalized the labeling data from the three biological replicates using the group scaling factors estimated during the fitting process (Wiechert et al., 2001). The average and sd of replicate normalized cumomer abundances were then calculated, and these measurements used with EstimateStat to estimate flux errors. To combine the errors in fluxes derived from the biomass data with errors associated with the labeling data, the biomass-derived fluxes were set as free fluxes for this part of the analysis, and the biomass-derived fluxes and their sds were defined in the Flux Measurements section of the input file.
Statistical Analysis
All indications of statistical significance are based on a Student's t test with P < 0.05 unless otherwise indicated.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Diagram of model used for optimal precursor prediction.
Supplemental Table S1. Complete measurement configuration.
Supplemental Table S2. Calculation of biomass derived fluxes.
Supplemental Table S3. Complete metabolic model (13C-FLUX network definition) used for fitting.
Supplemental Table S4. Complete optimal flux solutions for Standard and Elevated oxygenation conditions.
Supplementary Material
Acknowledgments
We thank Dr. W. Wiechert (Department of Simulation, University of Siegen, Germany) for permission to use 13C-FLUX.
This work was supported by the Biotechnology and Biological Sciences Research Council, United Kingdom.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: R. George Ratcliffe (george.ratcliffe@plants.ox.ac.uk).
The online version of this article contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Alonso AP, Goffman FD, Ohlrogge JB, Shachar-Hill Y (2007. a) Carbon conversion efficiency and central metabolic fluxes in developing sunflower (Helianthus annuus L.) embryos. Plant J 52 296–308 [DOI] [PubMed] [Google Scholar]
- Alonso AP, Raymond P, Hernould M, Rondeau-Mouro C, de Graaf A, Chourey P, Lahaye M, Shachar-Hill Y, Rolin D, Dieuaide-Noubhani M (2007. b) A metabolic flux analysis to study the role of sucrose synthase in the regulation of the carbon partitioning in central metabolism in maize root tips. Metab Eng 9 419–432 [DOI] [PubMed] [Google Scholar]
- Andre C, Froehlich JE, Moll MR, Benning C (2007) A heteromeric plastidic pyruvate kinase complex involved in seed oil biosynthesis in Arabidopsis. Plant Cell 19 2006–2022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baerenfaller K, Grossmann J, Grobei MA, Hull R, Hirsch-Hoffmann M, Yalovsky S, Zimmermann P, Grossniklaus U, Gruissem W, Baginsky S (2008) Genome-scale proteomics reveals Arabidopsis thaliana gene models and proteome dynamics. Science 320 938–941 [DOI] [PubMed] [Google Scholar]
- Baxter CJ, Redestig H, Schauer N, Repsilber D, Patil KR, Nielsen J, Selbig J, Liu J, Fernie AR, Sweetlove LJ (2007) The metabolic response of heterotrophic Arabidopsis cells to oxidative stress. Plant Physiol 143 312–325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blank LM, Kuepfer L, Sauer U (2005) Large-scale 13C-flux analysis reveals mechanistic principles of metabolic network robustness to null mutations in yeast. Genome Biol 6 R49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brand MD (1994) The stoichiometry of proton pumping and ATP synthesis in mitochondria. The Biochemist 16 20–24 [Google Scholar]
- Bruckner H, Langer M, Lupke M, Westhauser T, Godel H (1995) Liquid chromatographic determination of amino acid enantiomers by derivatization with o-phthaldialdehyde and chiral thiols. Applications with reference to food science. Chromatogr A 697 229–245 [Google Scholar]
- Carrari F, Baxter C, Usadel B, Urbanczyk-Wochniak E, Zanor MI, Nunes-Nesi A, Nikiforova V, Centero D, Ratzka A, Pauly M, et al (2006) Integrated analysis of metabolite and transcript levels reveals the metabolic shifts that underlie tomato fruit development and highlight regulatory aspects of metabolic network behavior. Plant Physiol 142 1380–1396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrari F, Nunes-Nesi A, Gibon Y, Lytovchenko A, Loureiro ME, Fernie AR (2003. b) Reduced expression of aconitase results in an enhanced rate of photosynthesis and marked shifts in carbon partitioning in illuminated leaves of wild species tomato. Plant Physiol 133 1322–1335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrari F, Urbanczyk-Wochniak E, Willmitzer L, Fernie AR (2003. a) Engineering central metabolism in crop species: learning the system. Metab Eng 5 191–200 [DOI] [PubMed] [Google Scholar]
- Edwards S, Nguyen BT, Do B, Roberts JKM (1998) Contribution of malic enzyme, pyruvate kinase, phosphoenolpyruvate carboxylase, and the krebs cycle to respiration and biosynthesis and to intracellular pH regulation during hypoxia in maize root tips observed by nuclear magnetic resonance imaging and gas chromatography-mass spectrometry. Plant Physiol 116 1073–1081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernie AR, Carrari F, Sweetlove LJ (2004) Respiratory metabolism: glycolysis, the TCA cycle and mitochondrial electron transport. Curr Opin Plant Biol 7 254–261 [DOI] [PubMed] [Google Scholar]
- Fernie AR, Geigenberger P, Stitt M (2005) Flux an important, but neglected, component of functional genomics. Curr Opin Plant Biol 8 174–182 [DOI] [PubMed] [Google Scholar]
- Fischer E, Sauer U (2003) Metabolic flux profiling of Escherichia coli mutants in central carbon metabolism using GC-MS. Eur J Biochem 270 880–891 [DOI] [PubMed] [Google Scholar]
- Fischer E, Sauer U (2005) Large-scale in vivo flux analysis shows rigidity and suboptimal performance of Bacillus subtilis metabolism. Nat Genet 37 636–640 [DOI] [PubMed] [Google Scholar]
- Geigenberger P (2003) Response of plant metabolism to too little oxygen. Curr Opin Plant Biol 6 247–256 [DOI] [PubMed] [Google Scholar]
- Geigenberger P, Fernie AR, Gibon Y, Christ M, Stitt M (2000) Metabolic activity decreases as an adaptive response to low internal oxygen in growing potato tubers. Biol Chem 381 723–740 [DOI] [PubMed] [Google Scholar]
- Gerrard Wheeler MC, Arias CL, Tronconi MA, Maurino VG, Andreo CS, Drincovich MF (2008) Arabidopsis thaliana NADP-malic enzyme isoforms: high degree of identity but clearly distinct properties. Plant Mol Biol 67 231–242 [DOI] [PubMed] [Google Scholar]
- Ghosh S, Grossmann IE, Ataai MM, Domach MM (2006) A three-level problem-centric strategy for selecting NMR precursor labeling and analytes. Metab Eng 8 491–507 [DOI] [PubMed] [Google Scholar]
- Hara A, Radin NS (1978) Lipid extraction of tissues with a low-toxicity solvent. Anal Biochem 90 420–426 [DOI] [PubMed] [Google Scholar]
- Iyer VV, Sriram G, Fulton DB, Zhou R, Westgate ME, Shanks JV (2008) Metabolic flux maps comparing the effect of temperature on protein and oil biosynthesis in developing soybean cotyledons. Plant Cell Environ 31 506–517 [DOI] [PubMed] [Google Scholar]
- Junker BH, Lonien J, Heady LE, Rogers A, Schwender J (2007) Parallel determination of enzyme activities and in vivo fluxes in Brassica napus embryos grown on organic or inorganic nitrogen source. Phytochemistry 68 2232–2242 [DOI] [PubMed] [Google Scholar]
- Kruger NJ, Huddleston JE, Le Lay P, Brown ND, Ratcliffe RG (2007. b) Network flux analysis: impact of 13C-substrates on metabolism in Arabidopsis thaliana cell suspension cultures. Phytochemistry 68 2176–2188 [DOI] [PubMed] [Google Scholar]
- Kruger NJ, Le Lay P, Ratcliffe RG (2007. a) Vacuolar compartmentation complicates the steady-state analysis of glucose metabolism and forces reappraisal of sucrose cycling in plants. Phytochemistry 68 2189–2196 [DOI] [PubMed] [Google Scholar]
- Kruger NJ, Ratcliffe RG (2008) Metabolic organization in plants: a challenge for the metabolic engineer. In HJ Bohnert, H Nguyen, NG Lewis, eds, Bioengineering and Molecular Biology of Plant Pathways, Vol 1. Elsevier, Amsterdam, pp 1–27
- Le Gall G, Colquhoun IJ, Davis AL, Collins GJ, Verhoeyen ME (2003) Metabolite profiling of tomato (Lycopersicon esculentum) using 1H NMR spectroscopy as a tool to detect potential unintended effects following a genetic modification. J Agric Food Chem 51 2447–2456 [DOI] [PubMed] [Google Scholar]
- Libourel IGL, Gehan JP, Shachar-Hill Y (2007) Design of substrate label for steady state flux measurements in plant systems using the metabolic network of Brassica napus embryos. Phytochemistry 68 2211–2221 [DOI] [PubMed] [Google Scholar]
- May MJ, Leaver CJ (1993) Oxidative stimulation of glutathione synthesis in Arabidopsis thaliana suspension cultures. Plant Physiol 103 621–627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mhaske V, Beldjilali K, Ohlrogge J, Pollard M (2005) Isolation and characterization of an Arabidopsis thaliana knockout line for phospholipid: diacylglycerol transacylase gene (At5g13640). Plant Physiol Biochem 43 413–417 [DOI] [PubMed] [Google Scholar]
- Millar AH, Bergersen FJ, Day DA (1994) Oxygen affinity of terminal oxidases in soybean mitochondria. Plant Physiol Biochem 32 847–852 [Google Scholar]
- Miyashita Y, Dolferus R, Ismond KP, Good AG (2007) Alanine aminotransferase catalyses the breakdown of alanine after hypoxia in Arabidopsis thaliana. Plant J 49 1108–1121 [DOI] [PubMed] [Google Scholar]
- Miyashita Y, Good AG (2008) Contribution of the GABA shunt to hypoxia-induced alanine accumulation in roots of Arabidopsis thaliana. Plant Cell Physiol 49 92–102 [DOI] [PubMed] [Google Scholar]
- Nunes-Nesi A, Carrari F, Gibon Y, Sulpice R, Lytovchenko A, Fisahn J, Graham J, Ratcliffe RG, Sweetlove LJ, Fernie AR (2007) Deficiency of mitochondrial fumarase activity in tomato plants impairs photosynthesis via an effect on stomatal function. Plant J 50 1093–1106 [DOI] [PubMed] [Google Scholar]
- Nunes-Nesi A, Carrari F, Lytovchenko A, Smith AMO, Loureiro ME, Ratcliffe RG, Sweetlove LJ, Fernie AR (2005) Enhanced photosynthetic performance and growth as a consequence of decreasing mitochondrial malate dehydrogenase activity in transgenic tomato plants. Plant Physiol 137 611–622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratcliffe RG, Shachar-Hill Y (2005) Revealing metabolic phenotypes in plants: inputs from NMR analysis. Biol Rev Camb Philos Soc 80 27–43 [DOI] [PubMed] [Google Scholar]
- Ratcliffe RG, Shachar-Hill Y (2006) Measuring multiple fluxes through plant metabolic networks. Plant J 45 490–511 [DOI] [PubMed] [Google Scholar]
- Roberts JKM, Hooks MA, Miaullis AP, Edwards S, Webster C (1992) Contribution of malate and amino acid metabolism to cytoplasmic pH regulation in hypoxic maize root tips studied using nuclear magnetic resonance spectroscopy. Plant Physiol 98 480–487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rontein D, Dieuaide-Noubhani M, Dufourc EJ, Raymond P, Rolin D (2002) The metabolic architecture of plant cells. Stability of central metabolism and flexibility of anabolic pathways during the growth cycle of tomato cells. J Biol Chem 277 43948–43960 [DOI] [PubMed] [Google Scholar]
- Roscher A, Kruger NJ, Ratcliffe RG (2000) Strategies for metabolic flux analysis in plants using isotope labelling. J Biotechnol 77 81–102 [DOI] [PubMed] [Google Scholar]
- Schomburg I, Chang A, Ebeling C, Gremse M, Heldt C, Huhn G, Schomburg D (2004) BRENDA, the enzyme database: updates and major new developments. Nucleic Acids Res 32 D431–433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwender J (2008) Metabolic flux analysis as a tool in metabolic engineering of plants. Curr Opin Biotechnol 19 131–137 [DOI] [PubMed] [Google Scholar]
- Schwender J, Goffman F, Ohlrogge JB, Shachar-Hill Y (2004. b) Rubisco without the Calvin cycle improves the carbon efficiency of developing green seeds. Nature 432 779–782 [DOI] [PubMed] [Google Scholar]
- Schwender J, Ohlrogge J, Shachar-Hill Y (2004. a) Understanding flux in plant metabolic networks. Curr Opin Plant Biol 7 309–317 [DOI] [PubMed] [Google Scholar]
- Schwender J, Shachar-Hill Y, Ohlrogge JB (2006) Mitochondrial metabolism in developing embryos of Brassica napus. J Biol Chem 281 34040–34047 [DOI] [PubMed] [Google Scholar]
- Spielbauer G, Margl L, Hannah LC, Romisch W, Ettenhuber C, Bacher A, Gierl A, Eisenreich W, Genschel U (2006) Robustness of central carbohydrate metabolism in developing maize kernels. Phytochemistry 67 1460–1475 [DOI] [PubMed] [Google Scholar]
- Sriram G, Fulton DB, Iyer VV, Peterson JM, Zhou R, Westgate ME, Spalding MH, Shanks JV (2004) Quantification of compartmented metabolic fluxes in developing soybean embryos by employing biosynthetically directed fractional 13C labeling, two-dimensional [13C, 1H] nuclear magnetic resonance, and comprehensive isotopomer balancing. Plant Physiol 136 3043–3057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sriram G, Fulton DB, Shanks JV (2007) Flux quantification in central carbon metabolism of Catharanthus roseus hairy roots by 13C labeling and comprehensive bondomer balancing. Phytochemistry 68 2243–2257 [DOI] [PubMed] [Google Scholar]
- Sriram G, Gonzalez-Rivera O, Shanks JV (2006) Determination of biomass composition of Catharanthus roseus hairy roots for metabolic flux analysis. Biotechnol Prog 22 1659–1663 [DOI] [PubMed] [Google Scholar]
- Sriram G, Shanks JV (2004) Improvements in metabolic flux analysis using carbon bond labeling experiments: bondomer balancing and Boolean function mapping. Metab Eng 6 116–132 [DOI] [PubMed] [Google Scholar]
- Sweetlove LJ, Burrell MM, ap Rees T (1996) Starch metabolism in tubers of transgenic potato (Solanum tuberosum) with increased ADPglucose pyrophosphorylase. Biochem J 320 493–498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweetlove LJ, Fell D, Fernie AR (2008) Getting to grips with the plant metabolic network. Biochem J 409 27–41 [DOI] [PubMed] [Google Scholar]
- Sweetlove LJ, Fernie AR (2005) Regulation of metabolic networks: understanding metabolic complexity in the systems biology era. New Phytol 168 9–24 [DOI] [PubMed] [Google Scholar]
- Sweetlove LJ, Last RL, Fernie AR (2003) Predictive metabolic engineering: a goal for systems biology. Plant Physiol 132 420–425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tronconi MA, Fahnenstich H, Gerrard Wheeler MC, Andreo CS, Flugge UI, Drincovich MF, Maurino VG (2008) Arabidopsis NAD-malic enzyme functions as a homodimer and heterodimer and has a major impact on nocturnal metabolism. Plant Physiol 146 1540–1552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truesdale GA, Downing AL (1954) Solubility of oxygen in water. Nature 173 1236 [Google Scholar]
- van Dongen JT, Roeb GW, Dautzenberg M, Froehlich A, Vigeolas H, Minchin PEH, Geigenberger P (2004) Phloem import and storage metabolism are highly coordinated by the low oxygen concentrations within developing wheat seeds. Plant Physiol 135 1809–1821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma DPS, Marcus A (1974) Oxygen availability as a control factor in the density-dependent regulation of protein synthesis in cell culture. J Cell Sci 14 331–337 [DOI] [PubMed] [Google Scholar]
- Wiechert W, Mollney M, Petersen S, de Graaf AA (2001) A universal framework for 13C metabolic flux analysis. Metab Eng 3 265–283 [DOI] [PubMed] [Google Scholar]
- Zhang P, Foerster H, Tissier CP, Mueller L, Paley S, Karp PD, Rhee SY (2005) MetaCyc and AraCyc. Metabolic pathway databases for plant research. Plant Physiol 138 27–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.