Abstract
Both leaf production and leaf expansion are tightly linked to cell expansion and cell division, but the functional relationships between all these variables are not clearly established. To get insight into these relationships, a quantitative genetic analysis was performed in 118 recombinant inbred lines derived from a cross between the Landsberg erecta and Antwerp accessions and was combined with a structural equation modeling approach. Main effects and epistatic interactions at the quantitative trait locus (QTL) level were detected for rosette area, rosette leaf number, leaf 6 area, epidermal cell area and number. A QTL at ERECTA marker (ER) controlled cell expansion and cell division, in interaction with two other QTLs at SNP295 and SNP21 markers. Moreover, both the screening for marker association involved in the variation of the relationships between leaf growth variables and the test of alternative functional models by structural equation modeling revealed that the allelic value at ER controlled epidermal cell area and epidermal cell number in a leaf. These effects are driven both by a whole plant mechanism associated with leaf production and by a single leaf mechanism associated with leaf expansion. The complex effects of the QTL at ER were validated in selected heterogeneous inbred families. The ERECTA gene, which is mutated in the Landsberg erecta parental line, was found to be a putative candidate responsible for these mapped effects by phenotyping mutants of this gene at the cellular level. Together, these results give insight into the complex determination of leaf epidermal cell number and area.
Final leaf area in a plant is an integrated variable depending on many different elementary processes, such as cell production and cell expansion, duration and rate of expansion of each individual leaf, leaf production rate, and duration of the phase of leaf production. As a consequence, leaf growth can be studied through various variables at different organizational levels, such as cellular, individual leaf, and whole plant. As a first step toward a modeling approach of whole plant leaf growth, it is necessary to elucidate how the different leaf growth variables are connected to one another. Until now, the causal or functional links between underlying leaf growth variables have not been clearly identified. Even the coordination of the cellular processes controlling leaf growth is not yet established.
The traditional view is that leaf development is driven by cell cycle-associated processes leading to an accumulation of cells in particular regions of the leaf, thereby driving morphogenesis and determining the size of the leaf (Fleming, 2007). This describes the classical cellular theory of development. Apparently, in agreement with this theory, final individual leaf area has been shown to be tightly correlated to final cell number in the leaf for many species (Francis, 1992; Granier et al., 2000). However, such correlative analyses cannot really be interpreted as a causal or functional link between cell division and leaf expansion. A significant covariation between two variables can be explained by various processes, including one variable acting on the other directly or indirectly (functional link), by the fact that both are driven by the same regulatory pathway, or by random processes.
Evidence that this cellular theory is insufficient in some situations comes from specific manipulation of the cell cycle and cell division during leaf development. Overexpression of CYCLIN-D2 leads to an increase in cell division rate in tobacco (Nicotiana tabacum) leaves, an increase in leaf growth rate but without significant changes in final leaf shape and size (Cockcroft et al., 2000). Similarly, overexpression of CYCLIN-D3 leads to an increase in cell number in Arabidopsis (Arabidopsis thaliana) leaves, but leaf size was not proportionally increased as cell area was reduced (Dewitte et al., 2003). In these studies, the increase in cell number resulting from the manipulation of cell cycle genes was accompanied by a decrease in cell area. Similarly, a decrease in cell number resulting from overexpression of cell cycle inhibitors or from specific reduction in cyclin-dependent kinase activity was accompanied by an increase in cell area in the leaves (Hemerly et al., 1995; De Veylder et al., 2001). Compensatory effects between cell number and cell size in leaves have been reported in many other examples in the literature: for instance, in collections of leaf growth mutants (Tsukaya, 2006; Ferjani et al., 2007) and in leaves of plants altered by different environmental stresses, such as moderate soil water deficit (Aguirrezabal et al., 2006), reduced incident light (Cookson and Granier, 2006), or changes in daylength (Cookson et al., 2007). Complete compensation between cell number and cell size would suggest that the control of organ size takes place at the level of the organ itself. This theory has often been referred to as the organismal view of development (Fleming, 2007). However, because these compensations are partial, Tsukaya proposed a new view of leaf development in which leaf expansion is controlled both at the cellular level and at the organ level with a compensatory system linking the two processes (Tsukaya, 2003, 2006). This compensatory system allows an increase in cell volume to be triggered totally or partially by a decrease in cell number (and vice versa). Regulation of individual leaf development seems even more complex because there is additional evidence that it is also controlled, to some extent, at the whole plant level. For example, the decline in cell area in leaves with increasing leaf position in a plant depends on whole plant control mechanisms related to floral transition as shown by floral bud removal or changes in daylength delaying or accelerating flowering (Ashby, 1948; Cookson et al., 2007).
The research presented here aimed to identify functional links between (1) leaf cellular growth processes themselves (namely, cell division and cell expansion); (2) leaf cellular growth processes and individual leaf expansion; and (3) leaf cellular growth processes and rosette leaf production. As a first step, we phenotyped leaf growth from the cellular level to the whole plant level in a set of recombinant inbred lines (RILs) and we searched for relationships between cellular leaf growth variables and variables at other organizational levels. Then, colocalizations of quantitative trait loci (QTLs) for cellular leaf growth variables and other leaf growth variables were identified and interpreted. Moreover, QTLs for correlations between the leaf growth variables were also detected with a systematic automated analysis of the bivariate correlations. These three steps revealed possible sets of functional links between leaf growth variables, which were tested further with structural equation modeling (Shipley, 2000). Finally, one of the QTLs at ERECTA marker (ER), controlling both epidermal cell number and area, was confirmed in a heterozygous inbred family (HIF). A candidate gene approach was performed, indicating that the ERECTA gene itself could be responsible for these mapped cellular effects.
RESULTS
Variation in Leaf Growth Variables of the Ler × An-1 RIL Population
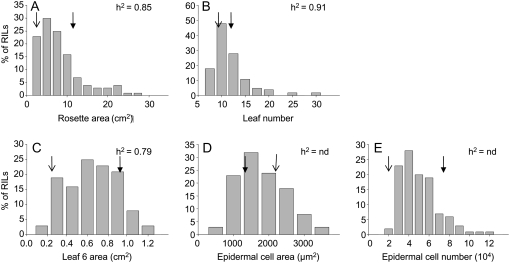
Rosette area, leaf number, leaf 6 area, and epidermal cell number in leaf 6 were significantly lower in Antwerp (An-1) compared with Landsberg erecta (Ler; Table I). In contrast, epidermal cell area in leaf 6 was significantly higher in An-1 compared with Ler. A large phenotypic variation in the population of RILs derived from a cross Ler × An-1 was observed for all variables including those for which parental values hardly differed (Fig. 1; Table I). The broad sense heritabilities ranged from 0.79 to 0.91 for leaf 6 area and leaf number, respectively (Fig. 1).
Table I.
Mean values of the five leaf growth variables measured in the parental lines Ler and An-1 (n = 8)
*, **, and ***, Significant difference between the two accessions with P values <0.05, <0.01, and <0.001, respectively.
| Leaf Growth Variables (Abbreviations, Units) | Mean for Ler | Mean for An-1 |
|---|---|---|
| Rosette leaf area (RA, cm2) | 11.6 | 2.05*** |
| Rosette leaf number (LN, leaf) | 12 | 9.18* |
| Leaf 6 area (AL6, cm2) | 0.86 | 0.22*** |
| Epidermal cell area (CA, μm2) | 1,354 | 2,293*** |
| Epidermal cell number (CN, cells) | 75,645 | 18,329*** |
Figure 1.
Frequency distribution of five leaf growth variables in the Ler × An-1 population. Growth variables are rosette area (A); leaf number (B); leaf 6 area (C); epidermal cell area in leaf 6 (D); and epidermal cell number in leaf 6 (E). Broad sense heritabilities are given in the top right corner of each image when they could be calculated (h2) and are denoted by “nd” otherwise (n = 4). Mean values of each variable for the two parental lines are shown by open and closed arrows on corresponding images for An-1 and Ler, respectively (n = 8).
Genotypic Correlations among Leaf Growth Variables and Colocalization of QTLs in the Ler × An-1 RIL Population
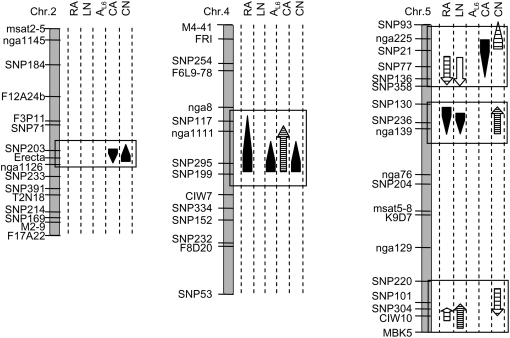
Epidermal cell number in leaf 6 was not significantly correlated to rosette area, but was negatively correlated to leaf number, indicating that, in this population, plants having a higher number of leaves have a lower number of cells in leaf 6 (Table II). The absence of an overall correlation between rosette area and epidermal cell number can be explained by colocalization of QTLs with similar or opposite allelic effects. A QTL for rosette area, on chromosome IV around SNP295 marker, collocated with a QTL for epidermal cell number both with the same allelic effect (Fig. 2; Table III), whereas two other QTL clusters were detected on chromosome V and have opposite allelic effects for rosette area and epidermal cell number (Fig. 2; Table III). The negative correlations found between epidermal cell number in leaf 6 and leaf production variables (Table II) could also be explained by colocalizations of QTLs with opposite allelic effects on chromosome V (Fig. 2; Table III).
Table II.
Pearson's correlations between leaf growth variables in the Ler × An-1 RIL population as calculated with the mean value of the variables of 118 F11 lines (n = 4)
*, **, and ***, Significant correlation with P values <0.05, <0.01, and <0.001, respectively. RA, Rosette area; LN, leaf number; AL6, leaf 6 area; CA, epidermal cell area in leaf 6; CN, epidermal cell number in leaf 6.
| RA | LN | AL6 | CA | CN | |
|---|---|---|---|---|---|
| cm2 | leaves | cm2 | μm2 | cells | |
| RA (cm2) | 1 | 0.87** | 0.23* | 0.43*** | −0.06 |
| LN (leaves) | 1 | 0.16 | 0.13 | −0.29** | |
| AL6 (cm2) | 1 | 0.58*** | 0.57*** | ||
| CA (μm2) | 1 | −0.05 | |||
| CN (cells) | 1 |
Figure 2.
The Ler × An-1 linkage map showing QTLs for the five leaf growth variables: rosette area (RA), leaf number (LN), leaf 6 area (AL6), epidermal cell area in leaf 6 (CA), and epidermal cell number in leaf 6 (CN). Only groups of colocalization of QTLs with at least one of the cellular leaf growth variables are shown. For the complete set of detected QTLs, see Table III. QTLs are represented by arrows and the lengths of the arrows indicate the 2-LOD support intervals. The direction of arrowheads indicates the sign of the additive effect: Arrows pointing upward indicate that Ler alleles have a positive effect. The shape of the arrow indicates the nature of the QTL: main effects (headed arrows); in epistatic interactions (nonheaded arrows). The grayscale of the arrows indicates the percentage of phenotypic variance explained by the QTL, respectively, 0% to 10%, 10% to 25%, 25% to 50%, and 50% to 100% from the whitest to the darkest.
Table III.
Characteristics of the detected QTLs for leaf growth variables in the Ler × An-1 population
RA, Rosette area (cm2); LN, leaf number; AL6, leaf 6 area (cm2); CA, epidermal cell area in leaf 6 (μm2); CN, epidermal cell number in leaf 6.
| Traits | r2Ga | r2Pb | Chromosome Markerc | Position (cM) | F | P Valued | r2Pe | 2af |
|---|---|---|---|---|---|---|---|---|
| RA | 80.1 | 68.3 | 4-FRI | 3.0 | 10.16 | 0.002 | 10.2 | 2.25 |
| 4-SNP295 | 34.4 | 11.39 | 0.001 | 11.6 | 2.46 | |||
| 5-SNP77 | 13.3 | 9.35 | 0.004 | 9.2 | −2.45 | |||
| 5-SNP304 | 79.7 | 18.55 | <0.001 | 19.8 | 2.94 | |||
| 3*5-nga172*SNP236 | 0.0*28.4 | 25.70 | <0.001 | 47.0 | – | |||
| LN | 80.8 | 73.5 | 5-SNP77 | 13.3 | 8.15 | <0.001 | 8.7 | −1.32 |
| 5-CIW10 | 81.3 | 34.25 | <0.001 | 28.5 | 2.40 | |||
| 2*4-SNP71*M4-41 | 25.7*0.0 | 7.66 | <0.001 | 21.1 | – | |||
| 3*5-nga172*SNP236 | 3.7*28.4 | 39.02 | <0.001 | 57.6 | – | |||
| AL6 | 74.4 | 58.8 | 5-SNP204 | 45.3 | 7.08 | 0.002 | 7.5 | 0.11 |
| 1*3-SNP132*SNP225 | 15.0*40.6 | 4.74 | 0.001 | 14.0 | – | |||
| 2*4-F12A24b*SNP295 | 17.8*34.4 | 31.56 | <0.001 | 52.1 | – | |||
| CA | – | 67.5 | 4-SNP295 | 34.4 | 31.71 | <0.001 | 29.2 | 623 |
| 1*4-SNP107*nga8 | 7.9*20.1 | 6.83 | <0.001 | 21.0 | – | |||
| 2*5-Erecta*SNP21 | 34.8*9.1 | 37.97 | <0.001 | 59.7 | – | |||
| CN | – | 69.0 | 5-SNP236 | 28.4 | 21.57 | <0.001 | 25.5 | 14,219 |
| 5-SNP101 | 77.9 | 17.42 | <0.001 | 21.7 | 12,609 | |||
| 2*4-Erecta*SNP295 | 34.8*34.4 | 28.09 | <0.001 | 57.2 | – | |||
| 3*5-CF7M19*nga225 | 56.1*4.8 | 7.85 | <0.001 | 27.2 | – |
Percentage of genotypic variance explained by the QTL model.
Percentage of phenotypic variance explained by the QTL model.
Marker determined in cofactor selection (see “Materials and Methods”).
Significance of the term of the QTL model (see “Materials and Methods”).
Percentage of phenotypic variance explained by terms of the QTL model.
Mean effect of the replacement of both An-1 alleles by Ler alleles at the QTL.
Epidermal cell area in leaf 6 was positively correlated to rosette area and leaf 6 area, but not to leaf number (Table II). This is explained by a QTL for rosette area, which coincided with QTLs for leaf 6 area and for epidermal cell area in leaf 6 in the middle of chromosome 4 at SNP295 (Fig. 2; Table III). The Ler alleles increased the values of all the variables at this marker.
A QTL for cell area centered on ER accounted for 34.9% of the phenotypic variance (Fig. 2) and colocalized with a QTL for epidermal cell number, which accounted for 8.4% of the phenotypic variance (Fig. 2). At this chromosomal position, only these QTLs were detected with opposite allelic effects. In addition, a cluster of QTLs at SNP295 included QTLs for epidermal cell area and number accounting for 14.2% and 11.3%, respectively, of the phenotypic variance. At this position, QTLs for both variables had the same allelic effect (Fig. 2). This cluster included also a QTL for leaf 6 area, accounting for 36.2% of the phenotypic variance (Fig. 2).
Interactions among QTLs for Cellular Leaf Growth Variables
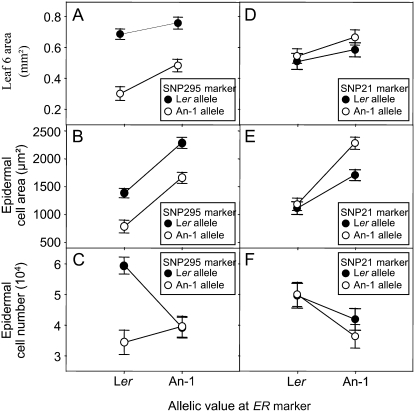
The interaction between the QTLs at markers SNP295 and ER was revealed by using EPISTAT (Table III). ANOVAs were performed using SPSS to further analyze this interaction (see “Materials and Methods”), which is presented in Figure 3, A to C. Epidermal cell area in leaf 6 was systematically higher in lines carrying the An-1 allele at the ER marker whatever the allele at SNP295 (Fig. 3B). Leaf 6 area and epidermal cell area in leaf 6 were both systematically higher when lines contained the Ler allele at SNP295 independently of the allelic value at ER (Fig. 3, A and B), indicating additive effects of both loci for these traits. However, for epidermal cell number in leaf 6, both QTLs showed strong interaction as epidermal cell number was increased by a factor of 2 only when lines had Ler alleles both at ER and at SNP295 (Fig. 3C) compared to the other three combinations of alleles at these markers.
Figure 3.
Allelic values for leaf 6 area, epidermal cell area, and epidermal cell number in leaf 6 at ER and SNP295 markers (A–C, respectively) and at ER and SNP21 markers (D–F, respectively) identified in epistatic interaction. Vertical bars are ses.
The phenotypic effect explained by the QTL at ER for epidermal cell area was increased when this QTL was considered in interaction with the QTL at SNP21 (Fig. 3E; Table III). The An-1 allele at SNP21 caused a slight, but significant, increase in epidermal cell area in leaf 6 without changing significantly leaf 6 area and epidermal cell number (Fig. 3, D–F). However, this same allele caused a significant increase in epidermal cell area with a decrease in epidermal cell number when plants had the An-1 allele at ER (Fig. 3, D–F). The Ler allele at ER therefore reduced the positive effect of the An-1 allele at SNP21 on epidermal cell area (Fig. 3E).
Genetic Control of Correlations between Leaf Growth Variables
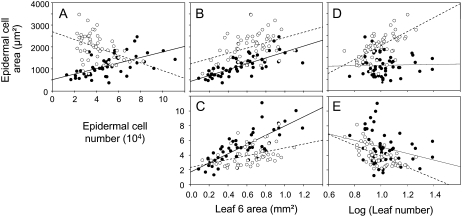
We tested whether the relationships between leaf growth variables were affected by allelic segregation at specific loci. For this, a script was developed in R (R Development Core Team, 2007) to automatically test to which extent the slopes of the relationships between the leaf growth variables were affected by considering separately the two alleles at each marker. On all 810 possible pairs of relationships (10 correlations at 81 markers), 41 had significantly different slopes when separately considering the alleles at each marker (data not shown). In general, among all the markers tested, the alleles at the ER marker had a more drastic effect than the others (higher P value) on a larger set of couples of variables (see Supplemental Fig. S1). Overall, the highest effect detected was one at the ER locus for the slope of the linear regressions between epidermal cell area and epidermal cell number in leaf 6 (see Supplemental Fig. S1C). A more detailed analysis of the effects on this correlation of the two alleles at ER position revealed that the absence of correlation between epidermal cell number and cell area when the whole population was analyzed (Table II) masked two strong correlations in opposite directions (Fig. 4A). Indeed, epidermal cell number was positively correlated to epidermal cell area for lines with the Ler allele at the ER marker (Fig. 4A; r2 = 0.58; P < 0.001) and negatively otherwise (Fig. 4A; r2 = 0.41; P = 0.002). In addition, alleles at ER modified the slopes of the relationships between epidermal cell area in leaf 6 and leaf number (Fig. 4D). A positive correlation between leaf number and epidermal cell area in leaf 6 was observed only for lines with the An-1 allele at ER (Fig. 4D; r2 = 0.62; P < 0.001), reflecting that epidermal cell area in leaf 6 was increased when leaf production was increased. In contrast, in lines carrying the Ler allele at ER, cell area in leaf 6 was totally independent of the number of leaves produced by the plants (Fig. 4D; r2 = 0.03; P = 0.85). Alleles at ER also significantly affected the slopes of the relationships between epidermal cell number in leaf 6 and area of the sixth leaf (Fig. 4C; P = 0.001), but neither the slopes of relationships between epidermal cell area and leaf 6 area (Fig. 4B; P = 0.71) nor those between epidermal cell number in leaf 6 and the number of leaves (Fig. 4E; P = 0.264).
Figure 4.
Relationships between epidermal cell area and epidermal cell number in leaf 6 (A); epidermal cell area in leaf 6 and leaf 6 area (B); epidermal cell number in leaf 6 and leaf 6 area (C); epidermal cell area in leaf 6 and leaf number (D); and epidermal cell number in leaf 6 and leaf number in two subpopulations from the Ler × An-1 population (E): for all RILs with the Ler allele at ER (n = 58; black circles) and for RILs with the An-1 allele at ER (n = 57; white circles).
Structural Equation Models
We further used structural equation modeling to investigate the functional relationships among leaf growth variables. The purpose of structural equation modeling is to quantify the relative contributions of correlated causal sources of variance once a certain network of interrelated variables with biological significance has been accepted (Shipley, 2000). We first constructed an initial causal model linking our five growth-related variables based on the results obtained combining the colocalizations between QTLs and the bivariate correlations (Supplemental Fig. S2). In this initial model, rosette area was determined both by leaf number and leaf 6 area. Leaf 6 area was determined by epidermal cell area and epidermal cell number and, in addition, a link was established between leaf number and epidermal cell number in leaf 6 to take into account the tradeoff explained before (Supplemental Fig. S2). Although this model seemed meaningful biologically and all the specified relationships were statistically significant, it had to be rejected by the analysis when tested against our dataset (P = 0.014; Supplemental Fig. S2A). An additional free correlation between epidermal cell area and cell number was added in a second model to reflect the tradeoff suggested by a QTL near ER in our analysis that is also often described in the literature (Cookson et al., 2007; Ferjani et al., 2007). The resulting model was also rejected (Supplemental Fig. S2B). Subsequent models were tested, but none of them provided an acceptable fit to our dataset in the whole population.
The analysis of the interactions between QTLs controlling epidermal cell area and/or cell number was subsequently combined with the detection of QTLs modifying correlations between cellular leaf growth variables and others. This allowed us to set up new hypotheses for the functional links between leaf growth variables: (1) Epidermal cell area is positively determined by the number of leaves (locus at ER) and also by the expansion of leaf 6 itself (locus at SNP295); (2) epidermal cell number is negatively determined by the number of leaves produced by the rosette (locus at CIW10 and SNP77) and, to some extent, positively by the expansion of leaf 6 itself (interaction between a locus at ER and a locus at SNP295); (3) both epidermal cell area and cell number contribute to leaf 6 area; (4) both the individual leaf area reflected by leaf 6 and the number of leaves contribute to rosette area; and (5) epidermal cell area in leaf 6 contributes to rosette area.
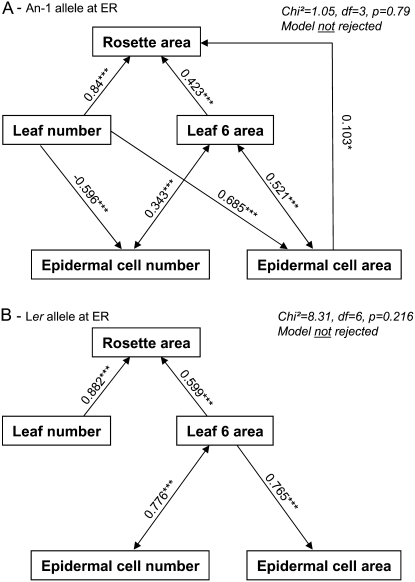
These hypotheses were integrated into a new structural equation model. The model provided a very good fit to the data (P = 0.79; root mean square error approximation [RMSEA] <0.05; comparative fit index [CFI] = 0.99) in the subpopulation with an An-1 allele at ER (Fig. 5A). All the path coefficients were significantly different from zero. Additionally, a substantial proportion of variance of the response variables, namely, epidermal cell area, epidermal cell number, and rosette area, was explained by the model (r2 = 0.43, 0.38, and 0.96, respectively).
Figure 5.
Two path diagrams tested in each subpopulation from the Ler × An-1 population: for all RILs with the An-1 allele at ER (A) and for RILs with the Ler allele at ER (B). Arrows represent linear functional relations between leaf growth variables. Single-headed arrows represent causal relationships and double-headed arrows represent free correlations. Standardized path coefficients are indicated on each arrow with level of significance (***, P < 0.001; *, P < 0.05). Both models were tested against our data and results are given in the top right corner.
This model was rejected in the subpopulation with a Ler allele at ER (P < 0.001). However, genetic results described above permitted the establishment of different hypotheses in this case and a new model was constructed for this second subpopulation. We have shown that Ler allele at ER (1) abolished the relationship between leaf number and epidermal cell area; (2) abolished the relationship between epidermal cell area and rosette area; (3) affected the relationship between leaf number and epidermal cell number in leaf 6 in interaction with SNP295; and (4) affected the relationship between epidermal cell area in leaf 6 and leaf 6 area.
This second model (Fig. 5B) with deletion of four arrows compared to the first one was not rejected in the subpopulation carrying a Ler allele at ER (P = 0.22) and provided a good fit to the data (RMSEA = 0.09; CFI = 0.99). All the path coefficients were significantly different from zero and a substantial proportion of variance of the response variables, namely, epidermal cell area and rosette area, was explained by the model (r2 = 0.59 and 0.92, respectively). This model was strongly rejected in the subpopulation carrying an An-1 allele at ER (P < 0.001).
Confirmation of the QTLs at ER and Test of a Candidate Gene
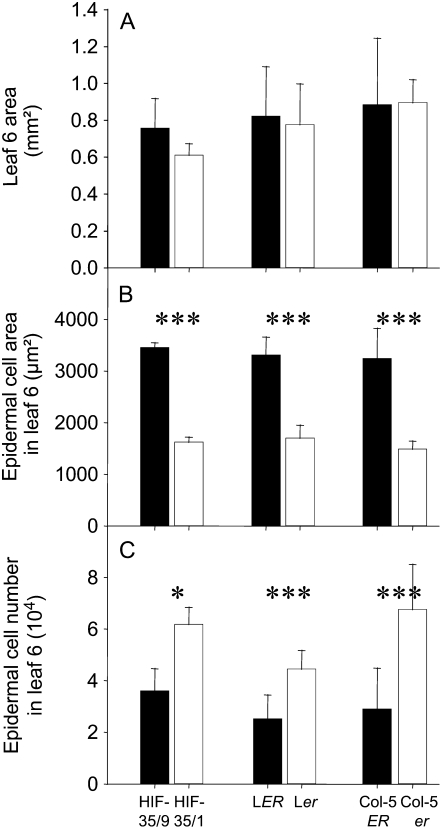
Measurements of final epidermal cell area and number in HIF derived from the RIL 35 (heterozygous line at ER) confirmed the effect of the QTLs clustered at ER marker (Fig. 6). Epidermal cell area was twice as large in HIF-35/9 carrying the An-1 allele at this marker compared with HIF-35/1, which remained heterozygous at this marker (Fig. 6B). In addition, epidermal cell number was twice as low in HIF-35/9 as in HIF-35/1 (Fig. 6C). As a consequence, final leaf area did not differ significantly between the two lines (Fig. 6A).
Figure 6.
Leaf 6 area (A), epidermal cell area in leaf 6 (B), and epidermal cell number in leaf 6 (C) measured in six genotypes: HIF-35/9 (n = 5) and HIF-35/1 (n = 5), LER (n = 10) and Ler (n = 10), Col-5ER (n = 10) and Col-5er (n = 10). Vertical bars are sds. *, **, and ***, Significant difference between the two lines with P values <0.05, <0.01, and <0.001, respectively.
Considering the traits analyzed, these QTLs could be attributable to the erecta mutation that is carried by the Ler line and segregates in the mapping population. Single gene mutants confirmed that this candidate gene could be responsible of the mapped cellular effects by measuring epidermal cell area and epidermal cell number in leaves of two erecta mutants with two different genetic backgrounds. The two mutants showed significantly reduced epidermal cell area and increased epidermal cell number without changes in final leaf 6 area (Fig. 6, A–C), revealing complete compensation between the two growth variables measured at the cellular level.
DISCUSSION
Both Epidermal Cell Number and Cell Area Are Controlled by Whole Plant Processes Associated with Leaf Production
Short-day conditions, increasing the number of leaves on the rosette, cause a decrease in epidermal cell number in each individual leaf (Cookson et al., 2007). Consistently, epidermal cell number in leaf 6 of the RILs analyzed here was negatively correlated to rosette leaf number. Such correlations could be interpreted as a tradeoff between meristematic activity associated with individual leaf expansion and with leaf production. This tradeoff has already been suggested by Ter Steege et al. (2005), who found similar tendencies in leaves of Aegilops also using a QTL analysis and is also consistent with older work, suggesting that cell division in a leaf depends on the number of leaves growing together (Wilson, 1966). In our study, these negative correlations were illustrated at the genetic level by two clusters of QTLs. In this context, these QTLs can be interpreted as being involved in leaf production through the duration of the vegetative phase, which has a tradeoff effect on epidermal cell number in leaf 6.
Both epidermal cell number and area vary in leaves depending on the rank of their emergence. A decline in cell area with increasing leaf position is commonly observed in plants, whereas epidermal cell number is generally increased as reported in Ipomoea (Ashby, 1948), sunflower (Helianthus annuus; Granier and Tardieu, 1998), and Arabidopsis (Cookson et al., 2007). Epidermal cell area distributions between different individual leaves have been discussed in terms of whole plant control mechanisms with a role for floral development (Ashby, 1948). Consistently, it has recently been shown that the control of epidermal cell area in a leaf depended on flowering. Inducing flowering initiation with long-daylength treatments or delaying flowering by removing floral buds caused, respectively, a decrease and an increase in cell area in a given leaf (Cookson et al., 2007). In this context, a positive relationship between epidermal cell area in leaf 6 and the number of rosette leaves was expected in the population of RILs analyzed here. However, this correlation was not found when the whole population was analyzed. By using an analysis of the correlations on subgroups of genotypes with different alleles at each locus, the variation in the slopes of the relationships between these traits was shown to depend on the allele at ER. Indeed, the absence of correlation within the whole population was due to the Ler allele at ER. On the other hand, the correlation was positive and highly significant. This result suggested that the Ler allele at ER completely abolished the relationship between the number of leaves and the extent of cell expansion in a leaf. The absence of this relationship in plants carrying the Ler allele at ER was confirmed by the structural equation models.
Relationships between Leaf Area, Epidermal Cell Number, and Cell Area
Many studies have shown that the final area for a leaf at a given rank on the plant was more related to its final epidermal cell number than to its final epidermal cell area (Dale, 1992; Granier et al., 2000; Cookson et al., 2005). In addition, the variability in leaf area along the stem of the same plant is also more related to differences in cell number than cell size (Cookson et al., 2007). In our study, two strong positive correlations were found, one between epidermal cell number in leaf 6 and final leaf 6 area, and the other between epidermal cell area in leaf 6 and final leaf 6 area. Colocalization of QTLs involved in the variation of these two correlations was found on chromosome IV, around SNP295. This locus could be interpreted as a QTL controlling leaf expansion at the leaf level itself and, depending on other loci, it caused either an increase in cell area or in cell number or both. The An-1 allele at this marker reduced by approximately a factor of 2.3 final leaf 6 area, which is sufficient to explain the large difference in size between the two parental accessions.
As described in the introduction of this article, it is often shown that there is a balance between both cell area and cell number in plants (Ter Steege et al., 2005; Ferjani et al., 2007). In the first general analysis, we found that it was not the case in the whole population studied here: Both variables were positively correlated with final leaf 6 area, but there was no correlation between them. Despite the absence of negative correlation, QTLs for cell number and cell area coincided at ER, with opposite allelic effects. This locus could be interpreted as a QTL for compensation between cell area and epidermal cell number with the possibility that ERECTA itself controls the balance between the two processes. Accordingly, it was shown here, in two different genetic backgrounds (Columbia [Col-5] and Ler), that ERECTA promotes cell expansion and limits cell division. This is also consistent with our results on the HIFs and with QTL mapping, which detected two QTLs with opposite additive effects at ER, one for epidermal cell area and the other for epidermal cell number. However, it is clearly shown here that the increase in cell number is not only due to the Ler allele at ER itself, but to an interaction with the Ler allele at SNP295. If leaf expansion is limited, as is the case when plants had an An-1 allele at SNP295, the Ler (mutant erecta) allele at ER could not promote cell division. This result indicates that, at least in some cases, leaf expansion itself is a necessary driving force for epidermal cell division in the leaf and is in agreement with partial control of organ growth at the scale of the organ itself (Green, 1976). This result was integrated in the two structural equation models retained for the two subpopulations at ER and formalized by the presence of a double arrow between leaf 6 area to epidermal cell number in leaf 6. Our interpretation of this result was reinforced by the structural equation models because it was a necessary condition to avoid model rejection.
Role for ER in the Determination of Epidermal Cell Area and Cell Number in Leaves
Arabidopsis Ler is one of the most popular laboratory strains that have been widely used as a wild-type background for collections of mutants (Berná et al., 1999) and as a parental line for populations of RILs and near isogenic lines extensively used in quantitative genetics (El-Lithy et al., 2006). It harbors the erecta mutation and therefore has a characteristic visible phenotype that has been described for years in the literature as conferring a compact inflorescence, blunt fruits, short petiole, and modified organ shape (Redei, 1962). This typical phenotype is not observed in the An-1 accession, suggesting that An-1 has a functional ERECTA gene.
The ERECTA gene was cloned (Torii et al., 1996) and encodes a Leu-rich repeat receptor-like Ser/Thr kinase. In addition to their modified plant architecture, the leaves of erecta mutants have been extensively characterized and show altered stomatal patterning and differentiation (Shpak et al., 2003, 2004, 2005). Several QTLs, mapping close to ER, have been identified in Arabidopsis in different populations of RILs for the area of juvenile leaves (Perez-Perez et al., 2002), the pedicel and floral organ lengths (Juenger et al., 2000), and the petiole length (Swarup et al., 1999; Perez-Perez et al., 2002). It was also recently shown by a QTL approach that the high stomatal density (high number of stomata/mm2) in erecta mutants caused an decrease in their water use efficiency (Masle et al., 2005).
Our QTL analysis shows that QTLs at ER act in regulatory pathways of cell expansion and cell division, by interaction with at least two other genes or groups of genes around marker SNP295 and SNP21 on chromosomes IV and V, respectively. The interaction involving SNP295 and ER for epidermal cell number is noteworthy because its effect on epidermal cell number results from a unique combination of alleles. The analyses performed here with partial correlations at ER present evidence that this QTL drastically alters relationships between leaf growth variables at all organizational levels. For example, an An-1 allele at ER gives a negative correlation between the cell number and cell size, whereas a Ler allele at ER gives a positive correlation. In addition, an An-1 allele at ER confers a strong positive correlation between epidermal cell area and rosette leaf number, which vanishes when plants have the allele from Ler at ER.
Even if a causal connection between ERECTA and the QTLs detected in its region cannot been firmly established in our study, results presented here with the erecta mutants (both in Col and Ler backgrounds) indicate a role of ERECTA on epidermal cell expansion in the leaf. Further analyses will be needed to determine whether ERECTA itself is responsible of the polymorphism between Ler and An-1 in the interval of the QTL at ER and then regulates the timing of cell expansion and cell division in a leaf, integrating both signals at the leaf level and signals at the plant level.
CONCLUSION
Combination of quantitative genetics and statistical modeling approaches allowed us to show that both epidermal cell area and number depend on growth at the leaf level and at the plant level via leaf production. This finding is particularly important because many attempts to increase leaf size by modifying cell division or expansion have failed. Our results indicate that these two variables are, to some extent, retrocontrolled by whole leaf and whole plant processes, therefore limiting their impact on leaf growth itself with maybe a role for the ERECTA gene in the second control. In a more general way, our data show that functional models relating leaf growth variables, as formalized here by path models and tested using structural equation modeling, can strongly depend on the genetic makeup of the plant materials that are tested. The complete genetic analysis performed in this study revealed that crucial relationships between variables combined in a model can differ significantly, depending on the allelic variation at a specific locus. Correlative analyses in unstructured populations of a species would not detect such genetic differences in the correlation structure and this would result in incorrect or poor-fitting models.
MATERIALS AND METHODS
Plant Material
For the QTL mapping, 120 RILs were previously generated from a cross between Ler and An-1 (El-Lithy et al., 2006). Simple sequence length polymorphic markers have been added on all the RILs to increase the density of markers on the genetic map and a new genetic map was generated using JoinMap4. Two (RIL-103 and RIL-114) of 120 RILs were rejected from the analysis because of suspicious genotyping data. All the remaining RILs were grown together in four replicates with the two parental lines grown in eight replicates.
An additional experiment was performed to confirm the QTLs mapped at ER and analyze the effect of ERECTA on the measured variables. Eleven lines derived from the progeny of RIL-35, heterozygous only in the region with the ERECTA gene, were selected for this experiment. Five replicates of each line were grown and the visible phenotype due to the mutation of ERECTA was noted for each plant (observing the compact typical inflorescence). Two lines were identified as having the same inflorescence phenotype for each of the five replicates: HIF-35/1 had a Ler inflorescence phenotype and HIF-35/9 an An-1 inflorescence phenotype. These two lines were genotyped in the ER region (markers Msat2-17, 10.7 Mb and nga1126, 11.7 Mb; data not shown) and HIF-35/1 and HIF-35/9 were heterozygous and homozygous for the An-1 allele at the ER marker, respectively. Leaf and cellular variables were measured on these two lines.
Moreover, two different genetic backgrounds (Ler and Col-5 ER) and their erecta mutant (Ler and Col-5er) have also been phenotyped during the same experiment in 10 replicates (Godiard et al., 2003).
For each experiment, seeds were stored at 4°C and were imbibed with water 30 min before sowing. They were sown in cylindrical pots (9-cm height and 4.5-cm diameter) filled with a mixture (1:1 [v/v]) of a loamy soil and organic compost.
Growth Conditions
The two experiments were performed in a growth chamber equipped with the automated phenotyping platform PHENOPSIS (Granier et al., 2006).
Micrometeorological Conditions
Air temperature and air humidity were measured with an HMP35A Vaisala (Oy) and were homogeneous in the growth chamber with means of 21.4°C and 75%, respectively. Light in the growth chamber was on during 12 h of the day and was provided by a bank of cool-white fluorescent tubes and HQi lamps. Incident light measured at the level of the plants with a light sensor over the waveband of 400 to 700 nm (LI-190SB; LICOR) was of 284 μmol m−2 s−1. During the experiments, all micrometeorological data were measured every 30 s, averaged, and stored every 600 s in a data logger (LTD-CR10 wiring panel; Campbell Scientific).
Control of Soil Water Content
Soil water content was determined before planting to estimate the amount of dry soil and water in each pot. Subsequent changes in pot weight were due to changes in soil water status. This allowed the calculation and automatic daily adjustment of soil water content to 0.40 g water g−1 dry soil with a modified one-tenth-strength Hoagland solution (Hoagland and Arnon, 1950) from sowing until the end of the experiments.
Leaf Growth Variable Measurements
PHENOPSIS took digital pictures of all individual pots on a daily basis during the experiments. On these pictures, stages of leaf development were scored for each individual plant three times a week as described in Boyes et al. (2001). At the end of the experiments, when plants reached stage 6.00 (first flower open; Boyes et al., 2001), the rosettes were cut, each individual leaf was detached, the lamina was separated from the petiole, and each lamina was forced to be flat and stuck with double-sided adhesive on a sheet of paper in order of their emergence on the rosette. The sheet of paper was then scanned for further measurements. In addition, a transparent negative film of the adaxial epidermis of leaf 6 was obtained after evaporation of a varnish spread on the upper surface of the leaf.
Leaf Area
Rosette area (cm2) was determined as the sum of the individual leaf blade area measured on the scans with image analysis software (Bioscan-Optimas, version 4.10). Leaf 6 area (cm2) was also measured on these scans with the same method.
Leaf Production
Leaf number (leaves) was estimated at stage 6.00 by counting the number of leaves formed after the two cotyledons.
Cellular Development
Films of epidermal imprints of the sixth leaf were placed under a microscope (Leitz DM RB; Leica) coupled to an image analyzer. Epidermal cell area (μm2) was estimated by measuring 25 epidermal cell areas at four different zones on each leaf, near the base, near the tip, and one on each side of the leaf with image analysis software (Bioscan-Optimas, version 4.10). Mean epidermal cell area is the mean of these 100 cells. Epidermal cell number was estimated from epidermal cell density by counting the number of epidermal cells in three different zones on each leaf.
Statistical Analyses
All statistical analyses were done using the computer package SPSS 11.0.1 for Windows (SPSS) and R software (R Development Core Team, 2007). Statistical differences between parental lines were tested by ANOVA with R. The skewness of the distributions was quantified to estimate their normality. To reduce the positive skewness of the distribution for rosette area and leaf number (Fig. 1), square root, log, and reciprocal transformations were tested. A natural logarithmic (base e) transformation of the data approximated a normal distribution. The detection of QTL has been done with both nontransformed and transformed data and the two methods did not modify the QTL detection (data not shown).
Correlations between leaf growth variables were tested using mean value of each leaf growth variable for each RIL. Both the Pearson and Spearman correlation coefficients were computed and their significance was tested. These two tests of correlations gave similar results. In a second step, a script was developed on R to detect loci affecting the slopes of the correlation between two variables when the correlation was considered as linear. The effect of each marker on the correlation between each couple of variables was determined by analysis of covariance with generalized linear model (GLM). The marker was considered to affect the slope of the linear regression between two variables only when the P value given by GLM was below a threshold of 0.001. This analysis was done both on nontransformed and transformed variables without any changes in the interpretation of the results.
Structural equation modeling is a generalized method for the analysis of covariance relationships and is used to evaluate the fit of data to a priori causal hypotheses about the functioning of a system (Shipley, 2000; Pugesek et al., 2003). These multivariate hypotheses are represented as graphical path models. Structural equation modeling then allows the assessment of the degree of fit between the observed and expected covariance structures, which is expressed as a goodness-of-fit χ2 statistic. Here, the aim was to impose a theoretical structure relating the direct and indirect relationships between leaf growth traits taking into account the results of QTL analyses and bivariate correlations. Transformed variables were used for rosette area and leaf number, to achieve linearity of the bivariate relationships and normal distribution of the residuals. This also helped to satisfy the assumption of univariate as well as multivariate normality. Structural equation models were tested in R using the structural equation modeling package (Fox, 2006), which uses the standard maximum likelihood estimator. A significant goodness-of-fit χ2 statistic indicates that the model does not fit the data. Once a model has not been rejected and considered biologically plausible, parameter estimates can be used to study direct, as well as indirect, effects of the variables. In particular, standardized path coefficients quantify the strength of a relationship, whereas the effects of the other variables are held constant. Parameter estimates are tested for significance using z statistics. Two other indices, RMSEA and CFI, were also used to assess the closeness of fit. Good models have a RMSEA <0.05 and a CFI >0.95.
QTL Mapping
QTLs were first identified using single interval mapping with the software package MapQTL 5 (Van Ooijen, 2004) software for the mapping of QTL in experimental populations (Kyazma B.V.). Cofactors were then selected using the automatic cofactor selection chromosome per chromosome. The selected cofactors were used in the composite interval mapping. The cofactors for which no QTL were detected (LOD under a 95%, LOD threshold <2.4 estimated by performing permutation tests implemented in MapQTL5 using at least 1,000 permutations of the original dataset) were removed successively. Detection and test of epistatic interactions between loci were performed using the software Epistat (Chase et al., 1997). Both epistatic interaction and QTL in main effects were statistically tested using the GLM of the statistical package of SPSS 11.0.1 for Windows (SPSS). QTL models were composed of all statistically significant (P value <0.05) main and interaction effects. The estimated additive genetic effect, the percentage of variance explained by each QTL, and the total variance explained by all the QTL of the QTL models were obtained using the statistical package of SPSS. Heritability (broad sense) was estimated as the proportion of variance explained by between-line differences based on measurements of four plants per genotype. Broad sense heritability could not be estimated for the epidermal cell area and epidermal cell number in leaf 6 because these two variables were estimated only in one repetition.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Identification of markers at which the alleles affect the slope of the correlations between leaf growth variables.
Supplemental Figure S2. Two path diagrams tested in the whole population of RILs from the Ler × An-1 cross.
Supplementary Material
Acknowledgments
We thank Jean-Jacques Thioux and Crispulo Balsera for help with plant growth measurements, the R community for software development, and Sarah Cookson for English corrections.
This work was supported by GENOPLANTE (grant no. GPLA–06014G to S.T.), an European Integrated Project in the 6th Framework Program (AGRON-OMICS grant no. LSHG–CT–2006–037704 to J.F.), as well as the PROCOPE and ARABRAS (ERAPG–003–03) program (financial support and exchange visits between the Montpellier and Cologne groups).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Christine Granier (granier@supagro.inra.fr).
The online version of this article contains Web-only data.
References
- Aguirrezabal L, Bouchier-Combaud S, Radziejwoski A, Dauzat M, Cookson SJ, Granier C (2006) Plasticity to soil water deficit in Arabidopsis thaliana: dissection of leaf development into underlying growth dynamic and cellular variables reveals invisible phenotypes. Plant Cell Environ 29 2216–2227 [DOI] [PubMed] [Google Scholar]
- Ashby E (1948) Studies in the morphogenesis of leaves. 2. The area, cell size and cell number of leaves of Ipomoea in relation to their position on the shoot. New Phytol 47 177–195 [Google Scholar]
- Berná G, Robles P, Micol JL (1999) A mutational analysis of leaf morphogenesis in Arabidopsis thaliana. Genetics 152 729–742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyes DC, Zayed MA, Ascenzi R, McCaskill AJ, Hoffman NE, Davis KR, Görlach J (2001) Growth stage–based phenotypic analysis of Arabidopsis a model for high throughput functional genomics in plants. Plant Cell 13 1499–1510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chase K, Adler FR, Lark KG (1997) Epistat: a computer program for identifying and testing interactions between pairs of quantitative trait loci. Theor Appl Genet 94 724–730 [Google Scholar]
- Cockcroft CE, Den Boer BGW, Healy JMS, Murray JAH (2000) Cyclin D control of growth rate in plants. Nature 405 575–579 [DOI] [PubMed] [Google Scholar]
- Cookson SJ, Chenu K, Granier C (2007) Day-length affects the dynamics of leaf expansion and cellular development in Arabidopsis thaliana partially through floral transition timing. Ann Bot (Lond) 99 703–711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cookson SJ, Granier C (2006) A dynamic analyses of the shaded-induced plasticity in Arabidopsis thaliana rosette leaf development reveals new components of the shade-adaptative response. Ann Bot (Lond) 97 443–452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cookson SJ, Van Lijsebettens M, Granier C (2005) Correlation between leaf growth variables suggest intrinsic and early controls of leaf size in Arabidopsis thaliana. Plant Cell Environ 28 1355–1366 [Google Scholar]
- Dale JE (1992) How do leaves grow? Advances in cell and molecular biology are unravelling some of the mysteries of leaf development. Bioscience 42 423–432 [Google Scholar]
- De Veylder L, Beeckman T, Beemster GTS, Krols L, Terras F, Landrieu I, Van Der Schueren E, Maes S, Naudts M, Inzé D (2001) Functional analysis of cyclin-dependent kinase inhibitors of Arabidopsis. Plant Cell 13 1653–1667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewitte W, Riou-Khamlichi C, Scofield S, Healy JMS, Jacqmard A, Kilby NJ, Murray JAH (2003) Altered cell cycle distribution, hyperplasia, and inhibited differentiation in Arabidopsis caused by the D-type cyclin CYCD3. Plant Cell 15 79–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Lithy ME, Bentsink L, Hanhart CJ, Ruys GJ, Rovito D, Broekhof JLM, van der Poel HJA, van Eijk MTJ, Vreugdenhil D, Koornneef M (2006) New Arabidopsis recombinant inbred line populations genotyped using SNPWave and their use for mapping flowering-time quantitative trait loci. Genetics 172 1867–1876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferjani A, Horiguchi G, Yano S, Tsukaya H (2007) Analysis of leaf development in fugu mutants of Arabidopsis reveals three compensation modes that modulate cell expansion in determinate organs. Plant Physiol 144 988–999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleming AJ (2007) Cell cycle control during leaf development. In D Inzé, ed, Cell Cycle Control and Plant Development. Annual Plant Reviews. Blackwell Publishing, Oxford, pp 203–226
- Fox J (2006) Structural equation modeling with the sem package in R. Structural Equation Modeling 13: 465–486
- Francis D (1992) The cell cycle in plant development. New Phytol 122 1–20 [DOI] [PubMed] [Google Scholar]
- Godiard L, Sauviac L, Torii KU, Grenon O, Mangin B, Grimsley NH, Marco Y (2003) ERECTA, an LRR receptor-like kinase protein controlling development pleiotropically affects resistance to bacterial wilt. Plant J 36 353–365 [DOI] [PubMed] [Google Scholar]
- Granier C, Aguirrezabal L, Chenu K, Cookson SJ, Dauzat M, Hamard P, Thioux JJ, Rolland G, Bouchier-Combaud S, Lebaudy A, et al (2006) PHENOPSIS, an automated platform for reproducible phenotyping of plant responses to soil water deficit in Arabidopsis thaliana permitted the identification of an accession with low sensitivity to soil water deficit. New Phytol 169 623–635 [DOI] [PubMed] [Google Scholar]
- Granier C, Tardieu F (1998) Spatial and temporal analyses of expansion and cell cycle in sunflower leaves. A common pattern of development for all zones of a leaf and different leaves of a plant. Plant Physiol 116 991–1001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granier C, Turc O, Tardieu F (2000) Co-ordination of cell division and tissue expansion in sunflower, tobacco and pea leaves. Dependence or independence of both processes? J Plant Growth Regul 19 45–54 [DOI] [PubMed] [Google Scholar]
- Green PB (1976) Growth and cell pattern formation on an axis: critique of concepts, terminology, and mode of study. Bot Gaz 137 187–202 [Google Scholar]
- Hemerly A, de Almeida Engler J, Bergounioux C, Van Montagu M, Engler G, Inzé D, Ferreira P (1995) Dominant negative mutants of the Cdc2 kinase uncouple cell division from iterative plant development. EMBO J 14 3925–3936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoagland DR, Arnon DI (1950) The water culture method for growing plants without soil. Californian Agricultural Experimental Station Circular 347 1–32 [Google Scholar]
- Juenger T, Purugganan M, Mackay TFC (2000) Quantitative trait loci for floral morphology in Arabidopsis thaliana. Genetics 156 1379–1392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masle J, Gilmore SR, Farquhar GD (2005) The ERECTA gene regulates plant transpiration efficiency in Arabidopsis. Nature 436 866–870 [DOI] [PubMed] [Google Scholar]
- Perez-Perez JM, Serrano-Cartagena J, Micol JL (2002) Genetic analysis of natural variations in the architecture of Arabidopsis thaliana vegetative leaves. Genetics 162 893–915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pugesek BH, Tomer A, von Eye A (2003) Structural Equation Modelling: Applications in Ecological and Evolutionary Biology. Cambridge University Press, Cambridge, UK
- R Development Core Team (2007) R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna
- Redei GP (1962) Single locus heterosis. Z Vererbungsl 93 164–170 [Google Scholar]
- Shipley B (2000) Cause and Correlation in Biology: A User's Guide to Path Analysis, Structural Equations, and Causal Inference. Cambridge University Press, Cambridge, UK
- Shpak ED, Berthiaume CT, Hill EJ, Torii KU (2004) Synergistic interaction of three ERECTA-family receptor-like kinases controls Arabidopsis organ growth and flower development by promoting cell proliferation. Development 131 1491–1501 [DOI] [PubMed] [Google Scholar]
- Shpak ED, Lakeman MB, Torii KU (2003) Dominant-negative receptor uncovers redundancy in the Arabidopsis ERECTA leucine-rich repeat receptor-like kinase signalling pathway that regulates organ shape. Plant Cell 15 1095–1110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shpak ED, McAbee JM, Pillitteri LJ, Torii KU (2005) Stomatal patterning and differentiation by synergistic interactions of receptor kinases. Science 309 290–293 [DOI] [PubMed] [Google Scholar]
- Swarup K, Alonso-Blanco C, Lynn JR, Michaels SD, Amasino RM, Koornneef M, Millar AJ (1999) Natural allelic variation identifies new genes in the Arabidopsis circadian system. Plant J 20 67–77 [DOI] [PubMed] [Google Scholar]
- Ter Steege MW, den Ouden FM, Lambers H, Stam P, Peeters AJM (2005) Genetic and physiological architecture of early vigor in Aegilops tauschii, the D-genome donor of hexaploid wheat. A quantitative trait loci analysis. Plant Physiol 139 1078–1094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torii KU, Mitsukawa N, Oosumi T, Matsuura Y, Yokoyama R, Whittier RF, Komeda Y (1996) The Arabidopsis ERECTA gene encodes a putative receptor protein kinase with extracellular leucine-rich repeats. Plant Cell 8 735–746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukaya H (2003) Organ shape and size: a lesson from studies of leaf morphogenesis. Curr Opin Plant Biol 6 57–62 [DOI] [PubMed] [Google Scholar]
- Tsukaya H (2006) Mechanism of leaf-shape determination. Annu Rev Plant Biol 57 477–496 [DOI] [PubMed] [Google Scholar]
- Van Ooijen JW (2004) MapQTL 5, Software for the Mapping of Quantitative Trait Loci in Experimental Populations. Kyazma B.V., Wageningen, The Netherlands
- Wilson GL (1966) Studies on the expansion of the leaf surface. V-cell division and expansion in a developing leaf as influenced by light and upper leaves. J Exp Bot 17 440–451 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.