Abstract
The “light” signal from the environment sets the circadian clock to regulate multiple physiological processes for optimal rhythmic growth and development. One such process is the control of flowering time by photoperiod perception in plants. In Arabidopsis (Arabidopsis thaliana), the flowering time is determined by the correct interconnection of light input and signal output by the circadian clock. The identification of additional clock proteins will help to better dissect the complex nature of the circadian clock in Arabidopsis. Here, we show LIGHT-REGULATED WD1 (LWD1)/LWD2 as new clock proteins involved in photoperiod control. The lwd1lwd2 double mutant has an early-flowering phenotype, contributed by the significant phase shift of CONSTANS (CO), and, therefore, an increased expression of FLOWERING LOCUS T (FT) before dusk. Under entrainment conditions, the expression phase of oscillator (CIRCADIAN CLOCK ASSOCIATED1 [CCA1], LATE ELONGATED HYPOCOTYL [LHY], TIMING OF CAB EXPRESSION1 [TOC1], and EARLY FLOWERING4 [ELF4]) and output (GIGANTEA, FLAVIN-BINDING, KELCH REPEAT, F-BOX1, CYCLING DOF FACTOR1, CO, and FT) genes in the photoperiod pathway shifts approximately 3 h forward in the lwd1lwd2 double mutant. Both the oscillator (CCA1, LHY, TOC1, and ELF4) and output (COLD, CIRCADIAN RHYTHM, AND RNA BINDING2 and CHLOROPHYLL A/B-BINDING PROTEIN2) genes have a short period length in the lwd1lwd2 double mutant. Our data imply that LWD1/LWD2 proteins function in close proximity to or within the circadian clock for photoperiodic flowering control.
Arabidopsis (Arabidopsis thaliana) flowers early under long days and is thus categorized as a facultative long-day (LD) plant. In the past decade, both genetic and biochemical studies of Arabidopsis have greatly fueled our understanding of photoperiod control in plants (for review, see Imaizumi and Kay, 2006; Kobayashi and Weigel, 2007). The environmental light signals and the internal circadian clock must function in harmony to achieve proper photoperiod control in plants.
Environmental light signals are perceived by plant photoreceptors, including the red/far-red photoreceptors and blue light photoreceptors. This process inputs the light signal to reset the circadian clock for optimal rhythmic growth and development in plants. Recent studies have also revealed a few regulators that are essential for the proper function of the Arabidopsis circadian rhythm (for review, see McClung, 2006; Yakir et al., 2007). Among those, EARLY FLOWERING3 (ELF3), TIME FOR COFFEE, and LIGHT INSENSITIVE PERIOD1 (LIP1) have been implicated as functioning to input the light signal into the core circadian clock (McWatters et al., 2000; Covington et al., 2001; Hall et al., 2003; Ding et al., 2007; Kevei et al., 2007).
The most well-studied Arabidopsis circadian clock is formed by a negative feedback loop composed of the oscillator proteins CIRCADIAN CLOCK ASSOCIATED1 (CCA1), LATE ELONGATED HYPOCOTYL (LHY), and TIMING OF CAB EXPRESSION1 (TOC1; Schaffer et al., 1998; Wang and Tobin, 1998; Strayer et al., 2000; Alabadi et al., 2001). ELF4, GIGANTEA (GI), LUX ARRHYTHMO/PHYTOCLOCK1 (LUX), TOC1 paralogs, and PSEUDO-RESPONSE REGULATOR5 (PPR5), PRR7, and PRR9 represent members of additional feedback loops of the clock (Yamamoto et al., 2003; Farre et al., 2005; Hazen et al., 2005; Kikis et al., 2005; Nakamichi et al., 2005; Onai and Ishiura, 2005; Martin-Tryon et al., 2007; McWatters et al., 2007). More recently, FIONA1 (FIO1) was found to regulate the clock in close association with the central oscillators (Kim et al., 2008). The proper expression of the clock genes is crucial for the function of plant circadian rhythm. In addition to transcriptional control, the protein stability of TOC1 and the activity of CCA1 can be regulated by ZEITLUPE (ZTL) and casein kinase II, respectively (Mas et al., 2003; Daniel et al., 2004). There exists a complex interlocked network within the Arabidopsis circadian clock (McClung, 2006). More effort is still needed to uncover additional clock proteins for a complete understanding of circadian regulation in Arabidopsis.
In the aspect of flowering time control regulated by photoperiod sensing, circadian output from the clock regulates the rhythmic expression of FLAVIN-BINDING, KELCH REPEAT, F-BOX1 (FKF1) and GI (Fowler et al., 1999; Park et al., 1999; Nelson et al., 2000; Imaizumi et al., 2003; Mizoguchi et al., 2005). In a blue light-dependent manner, the complex of FKF1 and GI functions as a positive regulator of CONSTANS (CO) expression via targeted degradation of CYCLING DOF FACTOR1 (CDF1), a repressor of CO (Imaizumi et al., 2005; Sawa et al., 2007). In addition, light also regulates CO protein stability (Valverde et al., 2004), a process regulated by SUPPRESSOR OF PHYA-105 (SPA) proteins (Laubinger et al., 2006) and/or COP1 (for CONSTITUTIVE PHOTOMORPHOGENIC1; Jang et al., 2008; Liu et al., 2008). The timing of CO gene expression and the regulation of CO protein stability together are crucial for Arabidopsis to sustain correct daylength measurement. The accumulation of CO protein by dusk under LD conditions activates the expression of FLOWERING LOCUS T (FT; Suarez-Lopez et al., 2001; Yanovsky and Kay, 2002). FT protein was later proven to be one of the “florigen” molecules generated in photoperiod-induced leaves and translocated to the shoot apex for the stimulation of the vegetative-to-floral transition (Corbesier et al., 2007; Jaeger and Wigge, 2007; Mathieu et al., 2007; Tamaki et al., 2007).
Although a good foundation has been laid for studies of photoperiod sensing in Arabidopsis, missing pieces in this big puzzle of fine-tuning and/or modulating photoperiodism still remain to be discovered. Light appears to be the most effective signal in synchronizing the environmental cue and the internal circadian clock in plants. Previous studies indicated that many of the key regulators in circadian and photoperiod control exhibit light-regulated expression characteristics. These observations prompted us to use a reverse genetics approach to find additional light-regulated genes that contribute to photoperiod regulation. We uncovered an early-flowering Arabidopsis mutant defective in both LIGHT-REGULATED WD1 (LWD1) and LWD2, both of which encode WD (for Trp and Asp)-containing proteins. Here, we show that the increased expression of FT is the likely cause of the early-flowering phenotype in the lwd1lwd2 double mutant. Our data also indicate that LWD1 and LWD2 are new clock proteins. Their presence is essential for the proper expression phase and period length of both the oscillator and output genes known to participate in Arabidopsis photoperiod sensing.
RESULTS
The lwd1lwd2 Double Mutant Has an Early-Flowering Phenotype
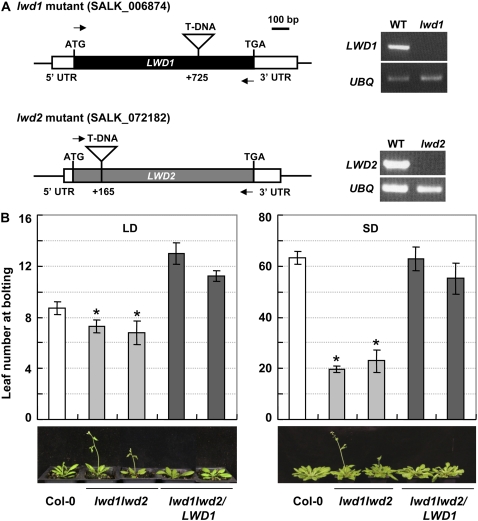
Our previous transcriptome analyses of Arabidopsis seedling photomorphogenesis and dark-treated leaves revealed many light-responsive genes (Lin and Wu, 2004; Chang et al., 2008). Genes encoding unique protein features were selected for further characterization of their possible contribution in light-regulated processes in Arabidopsis, including photoperiod flowering control. Among the genes examined, LWD1 (At1g12910) shares approximately 90% amino acid sequence identity with LWD2 (At3g26640) in Arabidopsis (Supplemental Fig. S1), so the two genes may have overlapping functions. Arabidopsis mutants carrying T-DNA insertions in LWD1 or LWD2 were obtained from the Arabidopsis Biological Resource Center and designated lwd1 (SALK_006874) or lwd2 (SALK_072182; Fig. 1A). LWD1 or LWD2 transcripts were undetectable in homozygous lwd1 or lwd2 single mutants, respectively, on reverse transcription (RT)-PCR analyses with primers spanning the T-DNA insertion sites (Fig. 1A). Phenotype characterization of the lwd1 or lwd2 single mutants did not uncover obvious phenotypic alterations, which suggested that these two proteins have functional redundancy (data not shown).
Figure 1.
LWD1 and LWD2 regulate flowering time in Arabidopsis under both LD and SD conditions. A, Two lines (SALK_006874 [lwd1] and SALK_072182 [lwd2]) carried the T-DNA insertion in the intronless LWD1 and LWD2 genes. The locations of the T-DNA, 5′/3′ untranslated region (UTR), and ATG/TGA are marked on the LWD1/LWD2 gene models. The level of LWD1 (LWD2) transcript in the wild type and lwd1 (lwd2) was assessed by RT-PCR with the primers listed in Supplemental Primer Table S1 (marked with horizontal arrows). RT-PCR was performed with UBQ10-specific primers used as a control for input RNA in the RT reaction. B, Plants of wild-type Arabidopsis (Col-0), lwd1lwd2 double mutant, and two independent complementation lines (lwd1lwd2/LWD1) were grown under LD (16 h of light/8 h of dark) or SD (8 h of light/16 h of dark) conditions. The plants were photographed, and the leaf numbers of each corresponding line were measured at the time of bolting as described in “Materials and Methods.” Asterisks indicate that lwd1lwd2 plants flowered significantly earlier than wild-type plants (Student';s t test; P < 0.0005, n = 10).
The lwd1lwd2 double mutant was next generated by reciprocal crossing of lwd1 and lwd2. Northern-blot analyses confirmed no full-length LWD1 or LWD2 transcripts in the lwd1lwd2 double mutant (Supplemental Fig. S2). The mutant lwd1lwd2 showed an early-flowering phenotype under LD conditions, with significantly fewer leaves than in the wild-type ecotype Columbia-0 (Col-0) prior to bolting (7.05 ± 0.7 versus 8.7 ± 0.48 leaves; Fig. 1B, left). The early-flowering phenotype of lwd1lwd2 was more prominent under short-day (SD) conditions (20.65 ± 3.6 versus 63.3 ± 2.6 leaves; Fig. 1B, right). Under both LD and SD conditions, the early-flowering phenotype in the lwd1lwd2 double mutant could be rescued by introducing a genomic fragment of LWD1 back to the lwd1lwd2 double mutant (lwd1lwd2/LWD1 in Fig. 1B). These results indicate that the lost function of LWD is indeed responsible for the early-flowering phenotype in lwd1lwd2 double mutant plants.
Genes in Photoperiodic Pathways Are Differentially Regulated in the lwd1lwd2 Double Mutant
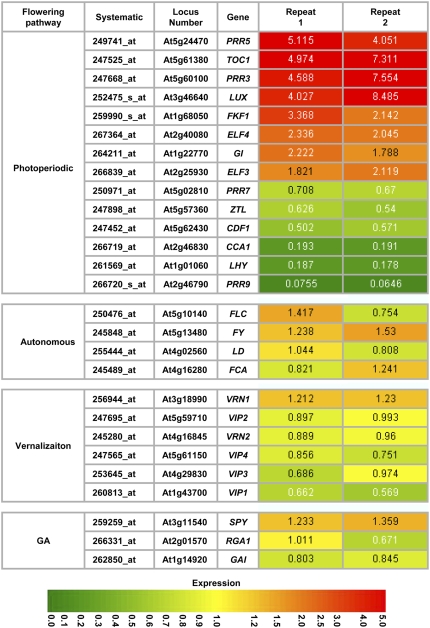
Previous studies indicated that defects in photoperiodic sensing, vernalization, autonomous, or gibberellin pathways could account for the early-flowering phenotype in Arabidopsis (for review, see Blazquez, 2000; Mouradov et al., 2002; Komeda, 2004). The early-flowering phenotype in lwd1lwd2 is likely due to the misregulation of genes in one or more of these pathway(s) by the mutation in LWD1 and LWD2. To clarify this, we performed a transcriptomic comparison between wild-type Arabidopsis and the lwd1lwd2 double mutant with the use of the Affymetrix GeneChip. The expression of genes belonging to the photoperiodic pathways was more severely affected than that of genes in the other three pathways in the lwd1lwd2 double mutant (Fig. 2). This implies that the defect in photoperiod sensing is primarily responsible for the early-flowering phenotype in the lwd1lwd2 double mutant.
Figure 2.
Genes in the photoperiodic pathway are differentially regulated in lwd1lwd2 plants. Transcriptomic comparison was performed to analyze whether genes in the four pathways regulating flowering time are differentially regulated in 31-d-old wild-type and lwd1lwd2 plants. Expression changes between wild-type and lwd1lwd2 plants were color coded with extreme red (for 5-fold up-regulated) and extreme green (for 5-fold down-regulated) in lwd1lwd2. The fold change of gene expression in the lwd1lwd2 double mutant is indicated. CO and FT were not included in this list because they were classified as “absent” in at least one of the four ATH1 hybridization experiments (see “Materials and Methods”). Expression data shown are listed in Supplemental Table S1.
FT Is Highly Expressed in lwd1lwd2 under SD Conditions
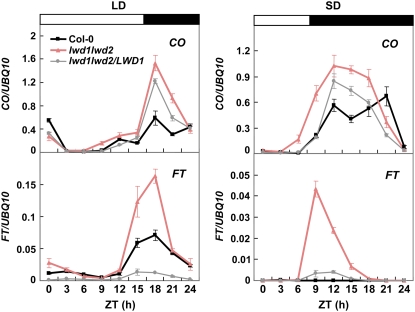
In the photoperiodic pathway, the circadian regulation of CO and FT leads to the correct measurement of daylength information for flowering determination. Previous studies have shown that the higher accumulation of CO transcripts/protein before dusk and/or the increased expression of FT in the photoperiodic pathway lead to an early-flowering phenotype in Arabidopsis (for review, see Imaizumi and Kay, 2006). To relate these observations to the early-flowering phenotype observed in the lwd1lwd2 double mutant, we sought to examine the expression of these two genes in lwd1lwd2. We used real-time quantitative RT-PCR (qRT-PCR) to measure the transcript abundance of both genes during a 24-h period under both LD and SD conditions. An increased expression of CO was observed in lwd1lwd2 under both LD and SD (Fig. 3). The marginal up-regulation of CO resulted in an increased induction of FT in lwd1lwd2 under LD (Fig. 3, left), which was sufficient for lwd1lwd2 to show an early-flowering phenotype under LD (Fig. 1B). Under SD, likely because of an advanced expression phase (see below), the increased expression of CO before dusk led to a higher expression of FT in lwd1lwd2 (Fig. 3, right). The remarkable increase in FT transcript abundance in lwd1lwd2 under SD explains why the lwd1lwd2 double mutant flowers substantially earlier than wild-type plants under SD.
Figure 3.
FT is highly expressed in lwd1lwd2 under SD conditions. Eighteen-day-old wild-type, lwd1lwd2, and lwd1lwd2/LWD1 plants grown under LD or SD conditions were harvested at different ZT times for total RNA isolation. White bars denote the light intervals, and black bars denote darkness. qRT-PCR was used to monitor the expression of CO and FT. Data are means ± SEM from four independent experiments.
LWD1/LWD2 Set the Correct Expression Phase of Circadian Clock-Regulated Genes
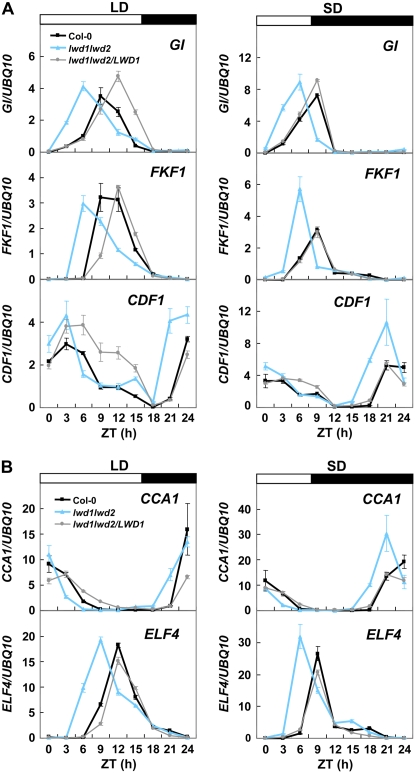
Previous reports indicated that CDF1 is a negative regulator of CO gene expression. This negative regulation could be derepressed by the targeted degradation of CDF1 by a protein complex composed of FKF1 and GI (Imaizumi et al., 2005; Sawa et al., 2007). Similar to CO and FT, GI, FKF1, and CDF1 are all circadian clock-regulated genes. Since CO showed a clear advanced expression phase in the lwd1lwd2 double mutant grown under SD (Fig. 3), we next sought to examine whether this is caused by the alteration of circadian expression of GI, FKF1, and CDF1. For this purpose, the expression kinetics of these genes was measured in wild-type, lwd1lwd2 double mutant, and lwd1lwd2/LWD1 complementation plants. As shown in Figure 4A, the expression level of CDF1 was higher in lwd1lwd2 double mutant than in wild-type plants under both LD and SD, whereas FKF1 showed increased expression in lwd1lwd2 only under SD. An approximately 3-h advanced expression phase was observed for all three genes known to regulate CO expression in the lwd1lwd2 double mutant under both LD and SD conditions (Fig. 4A). Thus, the mutation in LWD1 and LWD2 results in advanced expression of GI and advanced and increased expression of FKF1 and CDF1. This change of GI, FKF1, and CDF1 expression pattern subsequently leads to an advanced expression phase (approximately 3 h) and, eventually, the accumulation of CO and FT before dusk in the lwd1lwd2 double mutant (Fig. 3).
Figure 4.
LWDs regulate the expression phase of circadian clock-regulated genes under both LD and SD conditions. Eighteen-day-old wild-type, lwd1lwd2, and lwd1lwd2/LWD1 plants grown under LD or SD conditions were harvested at different ZT times for total RNA isolation. White bars denote the light intervals, and black bars denote darkness. qRT-PCR was used to monitor the expression of GI, FKF1, and CDF1 (A) or CCA1 and ELF4 (B). Data are means ± SEM from four independent experiments.
The next question is whether the abnormal function of central oscillators is responsible for the advanced expression phase of these circadian clock-regulated genes in the lwd1lwd2 double mutant. To answer this, we tested the expression patterns of CCA1 (morning gene) and ELF4 (evening gene) under both LD and SD conditions. As shown in Figure 4B, a 3-h advanced expression phase was seen for both genes. Two additional oscillator genes, LHY and TOC1, also possessed an advanced expression phase (data not shown).
Functional complementation of lwd1lwd2 by LWD1 could restore the expression phase of all genes tested to that seen in wild-type Arabidopsis (Fig. 4, A and B), which indicates that the loss of LWD1/LWD2 is responsible for maintaining the expression phase of both oscillator and output genes in Arabidopsis.
These expression data suggest that LWD1/LWD2 are needed to control the proper expression phase of the central oscillator genes. Mutation in LWD1/LWD2 will advance the expression of the oscillator genes during a 24-h period under both LD and SD conditions. Consequently, the output genes we tested, FKF1, GI, and CDF1, are expressed approximately 3 h earlier under both conditions. Increased transcript levels of CO and FT in the lwd1lwd2 double mutant were observed under both LD and SD, whereas the advanced expression phase of these two genes was only seen under SD.
LWD1/LWD2 Regulate the Period Length and Amplitude of Oscillator and Output Genes
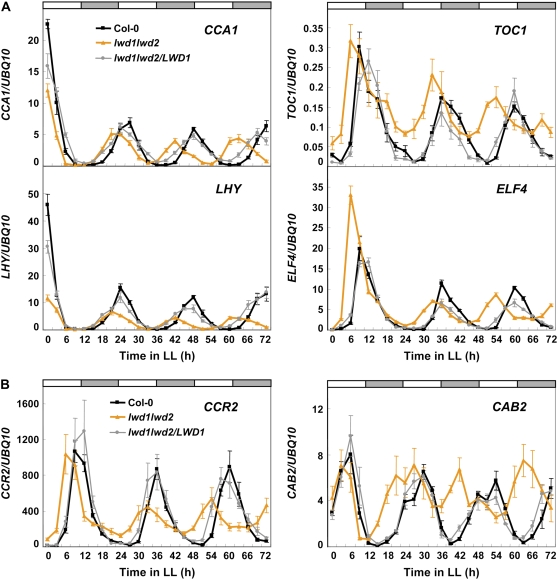
Since the advanced expression phase could be coupled with the shortened period length for circadian clock-regulated genes in Arabidopsis (Portoles and Mas, 2007), we next examined the period length of four oscillator genes, CCA1, LHY, TOC1, and ELF4, which have an advanced expression phase in the lwd1lwd2 double mutant (Fig. 4B; data not shown). For this study, wild-type, lwd1lwd2 double mutant, and lwd1lwd2/LWD1 complementation plants were entrained under 12 h of light and 12 h of dark for 18 d and then released to continuous light (LL). Samples were collected every 3 h for a total of 72 h for RNA extraction and qRT-PCR analyses. Although these oscillator genes still exhibited a rhythmic expression pattern under LL, the period length of each gene was shorter (approximately 21 h) in the lwd1lwd2 double mutant than in the wild type under LL (Fig. 5A). On examining the period length of two output genes, COLD, CIRCADIAN RHYTHM, AND RNA BINDING2 (CCR2) and CHLOROPHYLL A/B-BINDING PROTEIN2 (CAB2; Millar and Kay, 1991; Carpenter et al., 1994), we found their period length also shortened (Fig. 5B). Of note, the circadian amplitude of LHY and CCR2 was significantly reduced in the lwd1lwd2 double mutant under LL.
Figure 5.
LWDs regulate the period length and amplitude of circadian clock-regulated genes in LL. Eighteen-day-old wild-type, lwd1lwd2, and lwd1lwd2/LWD1 plants grown under 12 h of light and 12 h of dark were transferred to LL (time 0). Samples were harvested at 3-h intervals for a total of 72 h. qRT-PCR was used to monitor the expression of CCA1, LHY, TOC1, and ELF4 (A) or CCR2 and CAB2 (B). Expression is relative to that of UBQ10. Data are means ± SEM from four independent experiments.
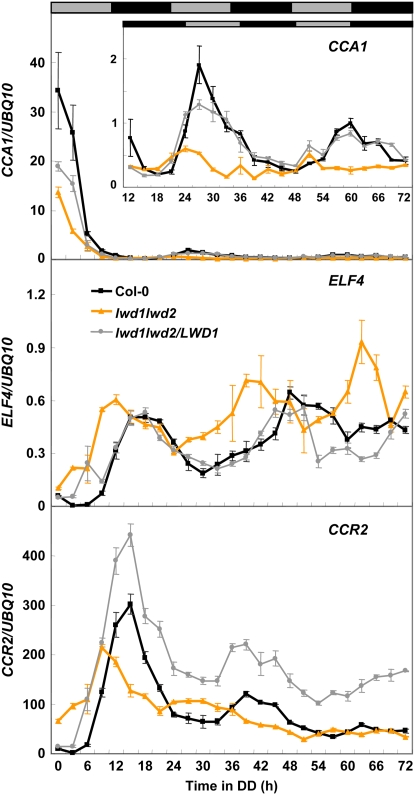
To test if the short period phenotype of the lwd1lwd2 double mutant is dependent on the light condition, we further analyzed the expression of CCA1, ELF4, and CCR2 in the wild-type, lwd1lwd2 double mutant, and lwd1lwd2/LWD1 complementation plants entrained under 12 h of light and 12 h of darkness for 18 d and then released to continuous dark (DD). As shown in Figure 6, ELF4 and CCR2 still maintained a rhythmic expression pattern with a shorter (approximately 3 h) period length in the lwd1lwd2 double mutant than in the wild type under DD. The expression amplitude for CCA1 was reduced under DD (Fig. 6).
Figure 6.
LWDs regulate the period length and amplitude of circadian clock-regulated genes in DD. Eighteen-day-old wild-type, lwd1lwd2, and lwd1lwd2/LWD1 plants grown under 12 h of light and 12 h of dark were transferred to DD (time 0). Samples were harvested at 3-h intervals for a total of 72 h. qRT-PCR was used to monitor the expression of CCA1, ELF4, and CCR2. Expression is relative to that of UBQ10. Data are means ± SEM from four independent experiments.
Taken together, our data indicate that functional LWD1/LWD2 are required for maintaining the period length and amplitude of both oscillator and output genes in Arabidopsis. Because the period length of these genes was shortened to approximately 3 h under both LL and DD in the lwd1lwd2 double mutant, LWD1/LWD2 are more likely to function close to the central oscillator.
LWD1 Has a Diurnal Expression Pattern
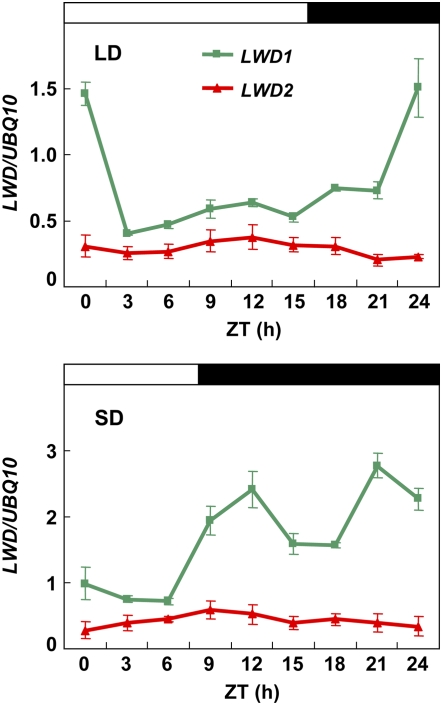
The alteration in both phase and period length of circadian-regulated genes in the lwd1lwd2 double mutant prompted us to examine whether the expression of LWD1 and LWD2 was under the control of the circadian clock in wild-type Arabidopsis. LWD1 exhibited a recognizable diurnal expression pattern under LD and SD conditions (Fig. 7). LWD2, however, has a constant expression level under both LD and SD conditions. The functional redundancy of LWD1 and LWD2 prompted us to examine the transcript abundance of both genes. In light of the high sequence homology between the LWD1 and LWD2 coding regions, the LWD1- and LWD2-specific primers used here were located at the more divergent 3′ untranslated region and thus could be used to unambiguously differentiate the expression patterns of these two genes. qRT-PCR showed that the peak expression of LWD1 was approximately 2-fold higher in plants grown under SD than LD conditions (Supplemental Fig. S4). The steady-state transcript level of LWD1 is apparently much higher than that of LWD2, especially before dawn, approximately in the range of 40-fold (ZT24 [for zeitgeber time in hours]) and 100-fold (ZT21) higher under LD and SD conditions, respectively.
Figure 7.
Quantitative expression analyses of LWD1 and LWD2 in Arabidopsis. Eighteen-day-old wild-type Arabidopsis plants grown under LD or SD conditions were harvested at different ZT times for total RNA isolation. qRT-PCR was used to monitor the expression of LWD1 and LWD2. Expression is relative to that of UBQ10. Data are means ± SEM from four independent experiments.
DISCUSSION
LWD1 and LWD2 Are New Players in Arabidopsis Photoperiod Sensing
We have adopted a reverse genetics approach to characterize two previously uncharacterized light-regulated genes, LWD1 and LWD2. Our data support the notion that LWD1 and LWD2 function in Arabidopsis photoperiod sensing. That the early-flowering phenotype could only be observed in the lwd1lwd2 double mutant indicates that LWD1 and LWD2 work redundantly in this respect (Fig. 1B). The successful complementation of this phenotype with just LWD1 further supports this notion. This also explained why these genes were not uncovered in previous genetic screening for Arabidopsis mutants with aberrant flowering time.
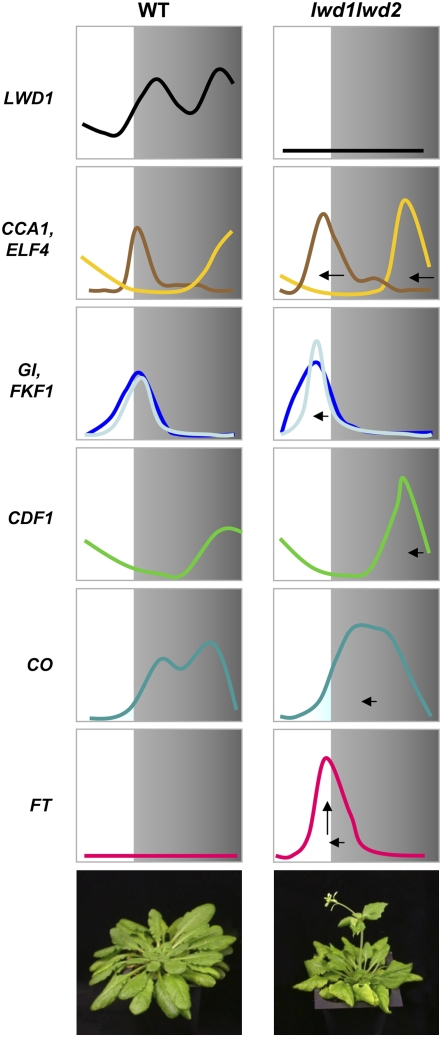
Transcriptome analysis revealed the impact of the mutation in LWD1 and LWD2 on the photoperiodic pathway (Fig. 2). A detailed comparison of the expression profiles for photoperiodic genes provided an explanation for the early-flowering phenotype in the lwd1lwd2 double mutant. As summarized in Figure 8, in wild-type Arabidopsis (left), LWD1 expresses in a diurnal pattern and regulates the expression of oscillator genes by a molecular mechanism yet to be identified. Under SD, the circadian clock-regulated FKF1 and GI do not reach their expression peak until dusk approaches. Under this circumstance, CDF1 protein is still present at a sufficient level to repress the expression of CO before dusk. Thus, FT could only express at a low level that is insufficient to induce flowering. On the contrary, Arabidopsis defective in both LWD1 and LWD2 (lwd1lwd2; Fig. 8, right) possesses perturbed circadian regulation with an advanced expression phase (left-pointing arrow) for the oscillator and output genes examined. The advanced expression of CO results in a higher CO transcript level before dusk in lwd1lwd2 plants (light-blue area). As a result, FT is highly expressed in lwd1lwd2 (up-pointing arrow), which leads to the early-flowering phenotype. Although not as dramatic, a similar scenario could explain the subtle but significant early-flowering phenotype of lwd1lwd2 under LD (Supplemental Fig. S5). While the aberrant clock function provides the simplest explanation for the increased expression of FT in the lwd1lwd2 double mutant, a slim possibility still exists that LWD1/LWD2 may directly regulate the expression of FT independent of their impact on circadian oscillators and output genes upstream of FT.
Figure 8.
An illustration showing how LWDs regulate the temporal expression pattern of oscillator and output genes in photoperiod sensing under SD conditions. The expression kinetics of LWD1, oscillator genes (CCA1 and ELF4), and output genes (GI, FKF1, CDF1, CO, and FT) under SD conditions show their expression phase and amplitude in wild-type and lwd1lwd2 plants. Yellow line, CCA1; brown line, ELF4; blue line, GI; light-blue line, FKF1. The light-blue area highlights the accumulation of CO transcripts. The left-pointing and up-pointing arrows refer to phase shift and higher expression level, respectively.
LWD1 and LWD2 Are New Clock Components
Previous reports showed that mutation of some Arabidopsis genes results in the alteration of period length for circadian clock-regulated genes in Arabidopsis. Both lengthened and shortened period lengths have been observed in Arabidopsis carrying mutations in the circadian clock-regulated genes. For example, mutation in photoreceptors (PHYTOCHROME A [PHYA], PHYB, CRYPTOCHROME1, and ZTL1), PPR7, and FIO1 resulted in a longer period length in Arabidopsis (Somers et al., 1998, 2000; Michael et al., 2003; Kim et al., 2008). In contrast, a shorter period length was observed previously in Arabidopsis mutants defective in CCA1, LHY, TOC1, PRR3, PRR5, GI, LIP1, and SENSITIVITY TO RED LIGHT REDUCED1 (Millar et al., 1995; Green and Tobin, 1999; Park et al., 1999; Mizoguchi et al., 2002; Michael et al., 2003; Staiger et al., 2003; Kevei et al., 2007; Martin-Tryon et al., 2007). Our results show that mutation in LWD1/LWD2 affects the period length of the oscillator genes CCA1, LHY, TOC1, and ELF4 (Fig. 5). Short period length was also observed for the output genes CCR2 and CAB2 in the lwd1lwd2 double mutant (Fig. 5). Since CCR2 and CAB2 are not directly involved in regulation of the flowering process, this indicates that LWD1 and LWD2 have broader influences on clock functions than just regulating flowering time in Arabidopsis. A shorter period length for circadian clock-regulated genes was observed in the lwd1lwd2 double mutant under both LL and DD (Figs. 5 and 6), indicating that LWD1 and LWD2 are important for maintenance of the period length of circadian clock-regulated genes regardless of light inputs (Figs. 5 and 6). This suggests that LWD1 and LWD2 more likely function in close proximity to or within the clock rather than in the light input pathway. In conclusion, LWD1/LWD2 act as new clock components that play a crucial role in the photoperiodic pathway for flowering time control in Arabidopsis as well as regulate the proper rhythmic expression of genes, CCR2 and CAB2, for the other physiological processes.
We speculate that LWD1 functions in keeping the correct expression phase and amplitude of the morning genes CCA1 and LHY because of the following observations. First, the expression of LWD1 precedes that of the morning genes and peaks before dawn (Fig. 7). Second, the expression amplitude was significantly diminished only for the morning genes CCA1 and LHY under DD and LL, respectively (Figs. 5 and 6). Third, the expression pattern and transcript level of LWD1 remained indistinguishable among wild-type, cca1 mutant, and transgenic plants overexpressing CCA1 (J.F. Wu and S.H. Wu, unpublished data), which indicates that the expression of LWD1 does not depend on CCA1. LWD1 might function to delay the expression of the morning genes until dawn approaches. Apparently, the expression of morning genes guarded by LWD1 must occur at the correct phase to reach a desirable amplitude under free-running conditions. That LWD1/LWD2 function to delay the expression phase of the morning genes CCA1 and LHY until dawn in wild-type Arabidopsis is of great interest. Whether LWD1/LWD2 act in the delay mechanism between TOC1 and CCA1/LHY is also worth testing.
Molecular Characteristics of LWDs
A total of 237 WD-containing proteins are annotated in the Arabidopsis genome (van Nocker and Ludwig, 2003). To date, only a few members of the Arabidopsis WD protein superfamily have been characterized. Supplemental Figure S1 shows that LWD1 and LWD2 have five WD repeats. It has been proposed that the WD repeats form a propeller structure and serve as a protein-protein interaction platform (Smith et al., 1999).
Several WD proteins were found to contribute to circadian control. For example, FWD1 (F-box/WD-40 repeat-containing protein 1) was reported to modulate circadian rhythm in Neurospora by regulating the degradation of the clock protein FREQUENCY (He et al., 2003; He and Liu, 2005). In Arabidopsis, SPA1 and COP1 modulate the abundance of the circadian protein CO to regulate flowering time (Laubinger et al., 2006; Jang et al., 2008; Liu et al., 2008). Here, we report to our knowledge the first WD proteins, LWD1 and LWD2, that function as clock proteins and regulate Arabidopsis photoperiodic flowering.
One common feature of these WD proteins is that, in addition to WD repeats, extension or additional protein domains are present in these proteins (e.g. a protein kinase domain for SPA1, a RING finger for COP1, and an F-box domain for FWD1). However, no known protein domains in addition to the WD repeats could be recognized in LWD1 and LWD2. The best studied case for WD proteins comprising only WD repeats is TRANSPARENT TESTA GLABRA1 (TTG1), which is the closest homolog of LWD1/LWD2 in Arabidopsis (BLASTP P value of <10−110). LWD1, LWD2, and TTG1 form a distinct group of WD-repeat proteins in Arabidopsis, as described previously (van Nocker and Ludwig, 2003). Pair-wise sequence identity comparison and alignment of LWD1, LWD2, and TTG1 are shown in Supplemental Figure S6. TTG1 functions in regulating flavonoid biosynthesis and epidermal cell fate determination through interaction with key bHLH and MYB transcriptional regulators (Broun, 2005; Gonzalez et al., 2008; Zhao et al., 2008). Of interest will be testing whether LWD1/LWD2 directly interact with specific proteins, such as the bHLH and MYB transcriptional regulators in the photoperiodic pathway, to achieve their functions as Arabidopsis clock proteins. Also, one of the TTG1 mutant alleles, ttg1-9, possesses a S282F mutation (Walker et al., 1999). This amino acid is equivalent to Thr-285 of LWD1 and Ser-285 of LWD2, as highlighted in Supplemental Figure S6. Whether this amino acid contributes to the structural integrity of functions of LWD1/LWD2 could be further investigated.
LWD Orthologous Proteins Are Present in Multiple Organisms
At a cutoff of 50% amino acid identity, orthologous proteins of LWD1/LWD2 could be found in a wide spectrum of organisms (HomoloGene:55930 in the National Center for Biotechnology Information database; http://www.ncbi.nlm.nih.gov/sites/entrez?db=homologene). These organisms include Oryza sativa (Os02g0524600; NP_001046989.1), Chlamydomonas reinhardtii (CHLREDRAFT_130509; XP_001695930.1), Homo sapiens (WDR68; NP_005819.3), Mus musculus (Wdr68; NP_082222.1), Xenopus laevis (MGC82392; NP_001086790), Danio rerio (wdr68; NP_956363.1), and Drosophila melanogaster (CG14614; NP_608461.1). A multiple sequence alignment of these orthologous proteins is shown in Supplemental Figure S7. The prevalence of LWD1/LWD2 orthologs in a wide spectrum of organisms implies a general involvement of these proteins in growth and/or developmental processes. Reports of biological functions for most of these proteins remain limited. Thus, the further characterization of Arabidopsis LWD1/LWD2 is expected to provide hints for the functional elucidation of these orthologous proteins. Interestingly, a recent report described the high resemblance of the transcriptional feedback loops in circadian clocks of Chlamydomonas and Arabidopsis (Matsuo et al., 2008). It will be worthwhile to test whether CHLREDRAFT_130509, the LWD1/LWD2 orthologous protein, also functions in regulating the circadian clock in Chlamydomonas.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
Two T-DNA insertion lines (Alonso et al., 2003), SALK_006874 (lwd1) and SALK_072182 (lwd2), were obtained from the Arabidopsis Biological Resource Center. lwd1 and lwd2 were crossed to generate the lwd1lwd2 double mutant used in this study. More than six independent lwd1lwd2/LWD1 lines were constructed by introducing a 1.4-kb (−263 to +1,175) genomic fragment of LWD1 into lwd1lwd2 by floral dipping (Clough and Bent, 1998). The LWD1 expression in these complementation lines was confirmed by northern-blot analysis (data not shown). Two representative lwd1lwd2/LWD1 lines, 36-6-3-4 and 32-2-1-2, are shown in Figure 1B. lwd1lwd2/LWD1 line 36-6-3-4 was used for the qRT-PCR experiments shown in Figures 3 to 6. Seeds of Arabidopsis (Arabidopsis thaliana) Col-0, mutant, and transgenic plants were germinated directly in soil and placed at 4°C for 4 d to synchronize the germination. For photoperiod treatment, the plants were grown under LD (16 h of light/8 h of dark) or SD (8 h of light/16 h of dark) at a fluence rate of 80 to 120 μmol m−2 s−1.
Determination of Flowering Time
The number of rosette leaves equal to or greater than 2 mm long was recorded for each plant when the primary florescence reached 1 cm above the rosette leaves. This phenotype observation was repeated at least three times. Four to 10 plants for each genotype were planted for scoring for each biological replicate.
Constructs
Sequences for all primers used in this study are listed in Supplemental Primer Table S1. pCAMBIA1390 (CSIRO, Australia) was used to generate lwd1lwd2/LWD1 complementation lines. A 1.4-kb (−263 to +1,175) genomic fragment of LWD1 was amplified with the primers pLWD1-PstI-S and LWD1-SmaI-2-AS and subcloned into pCAMBIA1390. All constructs used in this study were confirmed by sequencing.
RNA Isolation
Total RNA was isolated as described previously (Chang et al., 1993) with minor modifications. Plant tissues were frozen and ground in liquid nitrogen and extracted by vortexing with 8 volumes of extraction buffer (2% hexadecyltrimethylammonium bromide, 2% polyvinylpyrrolidone K 30, 100 mm Tris-HCl, pH 8.0, 25 mm EDTA, 2.0 m NaCl, 0.5 g L−1 spermidine, and 2% 2-mercaptoethanol) prewarmed at 65°C. The homogenate was then extracted twice with an equal volume of chloroform:isoamyl alcohol (24:1) by vortexing and centrifugation for 15 min at 12,000g. One-quarter volume of 10 m LiCl was then added to the aqueous phase for selective precipitation of RNA molecules. After overnight incubation at 4°C, the RNA pellet was harvested by centrifugation at 12,000g for 30 min, washed with 75% ethanol, and dissolved in 20 μL of RNase-free water.
Affymetrix ATH1 Genome Array Hybridization and Data Analyses
ATH1 Genome Array hybridization involved use of the Arabidopsis ATH1 Genome Array (Affymetrix). Plants (31-d-old wild-type and lwd1lwd2 double mutant plants) were grown under 12 h of light and 12 h of dark and harvested at ZT5 to ZT9. Ten micrograms of total RNA was used for cDNA synthesis, labeled by in vitro transcription, followed by fragmentation according to the manufacturer's suggestion (GeneChip Expression Analysis Technical Manual, Rev. 5; Affymetrix). Eleven-microgram labeled samples were hybridized to the ATH1 Genome Array at 45°C for 16.5 h. Washing and staining involved Fluidic Station-450, and the ATH1 Genome Array was scanned with use of the Affymetrix GeneChip Scanner 7G. The results were quantified and analyzed by use of MicroArray Suite 5.0 software (Affymetrix).
Gene expression data for Affymetrix ATH1 were analyzed as described previously (Lin and Wu, 2004). The average intensity of all probe sets of each chip was scaled to 500 so that the hybridization intensity of all chips was equivalent. “Set measurements less than 0.01 to 0.01,” “Per Chip: Normalize to 50th percentile,” and “Per Gene: Normalize to control mean” were applied for data normalization when Affymetrix data files were imported into GeneSpring 7.2 (Agilent) for further analyses. Genes marked as “present” in all chips analyzed were used for further data analysis shown in Figure 2 and Supplemental Figure S3. Raw data associated with Figure 2 are shown in Supplemental Table S1. The data sets have been deposited in the National Center for Biotechnology Information's Gene Expression Omnibus (Edgar et al., 2002) and are accessible through GEO Series accession number GSE 11762.
qRT-PCR
Total RNA was isolated as described above and quantified by use of a NanoDrop ND-1000 spectrophotometer (NanoDrop Technologies). cDNA was synthesized from 2 μg of DNase-treated total RNA with the use of SuperScript II reverse transcriptase (Invitrogen) and poly(T) primer. All primers were designed by Primer Express (Applied Biosystems). An amount of 50 μL of real-time PCR contained the following: primers, 5 μL of cDNA (equivalent to approximately 0.25 ng of mRNA), and 25 μL of SYBR Green PCR Master Mix (Applied Biosystems). The names of the primer pairs used for each gene are UBQ10-ABI-1, UBQ10-ABI-2, LWD1-1242-ABI-S, LWD1-1293-ABI-AS, LWD2-1098-ABI-S, LWD2-1231-ABI-AS, CCA1-1695-ABI-S, CCA1-1768-ABI-AS, LHY-1991-ABI-S, LHY-2067-ABI-AS, TOC1-725-ABI-S, TOC1-803-ABI-AS, ELF4-185-ABI-S, ELF4-260-ABI-AS, GI-3513-ABI-S, GI-3563-ABI-AS, FKF1-1583-ABI-S, FKF1-1652-ABI-AS, CDF1-678-ABI-S, CDF1-732-ABI-AS, CO-811-ABI-S, CO-861-ABI-AS, FT-336-ABI-S, FT-388-ABI-AS, CCR2-593-ABI-S, CCR2-679-ABI-AS, CAB2-950-ABI-S, and CAB2-1099-ABI-AS. Sequences and ratios of the primers (5 μm each) were determined experimentally as suggested by the manufacturer and listed in Supplemental Primer Table S1. Real-time PCR involved use of the ABI Prism 7000 sequence detection system (Applied Biosystems) with programs recommended by the manufacturer (2 min at 50°C, 10 min at 95°C, and 40 cycles of 95°C for 15 s and 60°C for 1 min). The comparative threshold cycle (CT) method was used to determine the relative amount of gene expression, with the expression of UBQ10 used as an internal control. For clarity, mean values of 2−ΔCT (ΔCT = CT, gene of interest − CT, UBQ10) calculated from four independent experiments were multiplied by 100 for the results plotted in Figures 3 to 7.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Amino acid sequence alignment of LWD1 and LWD2.
Supplemental Figure S2. Northern-blot analyses of LWD1 and LWD2 in lwd1lwd2 double mutant plants.
Supplemental Figure S3. Differential gene expression is highly correlated between the two biological replicates of ATH1 hybridization.
Supplemental Figure S4. Absolute quantitation of LWD1 and LWD2 transcripts in Arabidopsis.
Supplemental Figure S5. A model showing how LWDs regulate the temporal expression pattern of oscillator and output genes in photoperiod sensing under LD conditions.
Supplemental Figure S6. Sequence comparison of Arabidopsis LWD1, LWD2, and TTG1.
Supplemental Figure S7. Amino acid sequence alignment of LWD1 orthologous proteins.
Supplemental Table S1. ATH1 expression data for genes in four different pathways regulating flowering time.
Supplemental Primer Table S1.
Supplemental Materials and Methods S1.
Supplementary Material
Acknowledgments
We express our gratitude to the anonymous reviewers for their very constructive suggestions for the revision of the manuscript. We thank Kuo-Chen Yeh and Long-Chi Kevin Wang for helpful discussions. We also thank Mei-Jane Fang for technical assistance. Affymetrix GeneChip assays were performed by the Affymetrix Gene Expression Service Laboratory (http://ipmb.sinica.edu.tw/affy/) supported by Academia Sinica.
This work was supported by Academia Sinica (grant no. AS91IB1PP to S.-H.W.).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Shu-Hsing Wu (shuwu@gate.sinica.edu.tw).
The online version of this article contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Alabadi D, Oyama T, Yanovsky MJ, Harmon FG, Mas P, Kay SA (2001) Reciprocal regulation between TOC1 and LHY/CCA1 within the Arabidopsis circadian clock. Science 293 880–883 [DOI] [PubMed] [Google Scholar]
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, et al (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301 653–657 [DOI] [PubMed] [Google Scholar]
- Blazquez M (2000) Flower development pathways. J Cell Sci 113 3547–3548 [DOI] [PubMed] [Google Scholar]
- Broun P (2005) Transcriptional control of flavonoid biosynthesis: a complex network of conserved regulators involved in multiple aspects of differentiation in Arabidopsis. Curr Opin Plant Biol 8 272–279 [DOI] [PubMed] [Google Scholar]
- Carpenter CD, Kreps JA, Simon AE (1994) Genes encoding glycine-rich Arabidopsis thaliana proteins with RNA-binding motifs are influenced by cold treatment and an endogenous circadian rhythm. Plant Physiol 104 1015–1025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang CS, Li YH, Chen LT, Chen WC, Hsieh WP, Shin J, Jane WN, Chou SJ, Choi G, Hu JM, et al (2008) LZF1, a HY5-regulated transcriptional factor, functions in Arabidopsis de-etiolation. Plant J 54 205–219 [DOI] [PubMed] [Google Scholar]
- Chang S, Puryear J, Cainey J (1993) A simple and efficient method for isolating RNA from pine trees. Plant Mol Biol Rep 11 113–116 [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16 735–743 [DOI] [PubMed] [Google Scholar]
- Corbesier L, Vincent C, Jang S, Fornara F, Fan Q, Searle I, Giakountis A, Farrona S, Gissot L, Turnbull C, et al (2007) FT protein movement contributes to long-distance signaling in floral induction of Arabidopsis. Science 316 1030–1033 [DOI] [PubMed] [Google Scholar]
- Covington MF, Panda S, Liu XL, Strayer CA, Wagner DR, Kay SA (2001) ELF3 modulates resetting of the circadian clock in Arabidopsis. Plant Cell 13 1305–1315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniel X, Sugano S, Tobin EM (2004) CK2 phosphorylation of CCA1 is necessary for its circadian oscillator function in Arabidopsis. Proc Natl Acad Sci USA 101 3292–3297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Z, Millar AJ, Davis AM, Davis SJ (2007) TIME FOR COFFEE encodes a nuclear regulator in the Arabidopsis thaliana circadian clock. Plant Cell 19 1522–1536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R, Domrachev M, Lash AE (2002) Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res 30 207–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farre EM, Harmer SL, Harmon FG, Yanovsky MJ, Kay SA (2005) Overlapping and distinct roles of PRR7 and PRR9 in the Arabidopsis circadian clock. Curr Biol 15 47–54 [DOI] [PubMed] [Google Scholar]
- Fowler S, Lee K, Onouchi H, Samach A, Richardson K, Morris B, Coupland G, Putterill J (1999) GIGANTEA: a circadian clock-controlled gene that regulates photoperiodic flowering in Arabidopsis and encodes a protein with several possible membrane-spanning domains. EMBO J 18 4679–4688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez A, Zhao M, Leavitt JM, Lloyd AM (2008) Regulation of the anthocyanin biosynthetic pathway by the TTG1/bHLH/Myb transcriptional complex in Arabidopsis seedlings. Plant J 53 814–827 [DOI] [PubMed] [Google Scholar]
- Green RM, Tobin EM (1999) Loss of the circadian clock-associated protein 1 in Arabidopsis results in altered clock-regulated gene expression. Proc Natl Acad Sci USA 96 4176–4179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall A, Bastow RM, Davis SJ, Hanano S, McWatters HG, Hibberd V, Doyle MR, Sung S, Halliday KJ, Amasino RM, et al (2003) The TIME FOR COFFEE gene maintains the amplitude and timing of Arabidopsis circadian clocks. Plant Cell 15 2719–2729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazen SP, Schultz TF, Pruneda-Paz JL, Borevitz JO, Ecker JR, Kay SA (2005) LUX ARRHYTHMO encodes a Myb domain protein essential for circadian rhythms. Proc Natl Acad Sci USA 102 10387–10392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Q, Cheng P, Yang Y, He Q, Yu H, Liu Y (2003) FWD1-mediated degradation of FREQUENCY in Neurospora establishes a conserved mechanism for circadian clock regulation. EMBO J 22 4421–4430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Q, Liu Y (2005) Degradation of the Neurospora circadian clock protein FREQUENCY through the ubiquitin-proteasome pathway. Biochem Soc Trans 33 953–956 [DOI] [PubMed] [Google Scholar]
- Imaizumi T, Kay SA (2006) Photoperiodic control of flowering: not only by coincidence. Trends Plant Sci 11 550–558 [DOI] [PubMed] [Google Scholar]
- Imaizumi T, Schultz TF, Harmon FG, Ho LA, Kay SA (2005) FKF1 F-box protein mediates cyclic degradation of a repressor of CONSTANS in Arabidopsis. Science 309 293–297 [DOI] [PubMed] [Google Scholar]
- Imaizumi T, Tran HG, Swartz TE, Briggs WR, Kay SA (2003) FKF1 is essential for photoperiodic-specific light signalling in Arabidopsis. Nature 426 302–306 [DOI] [PubMed] [Google Scholar]
- Jaeger KE, Wigge PA (2007) FT protein acts as a long-range signal in Arabidopsis. Curr Biol 17 1050–1054 [DOI] [PubMed] [Google Scholar]
- Jang S, Marchal V, Panigrahi KC, Wenkel S, Soppe W, Deng XW, Valverde F, Coupland G (2008) Arabidopsis COP1 shapes the temporal pattern of CO accumulation conferring a photoperiodic flowering response. EMBO J 27 1277–1288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kevei E, Gyula P, Feher B, Toth R, Viczian A, Kircher S, Rea D, Dorjgotov D, Schafer E, Millar AJ, et al (2007) Arabidopsis thaliana circadian clock is regulated by the small GTPase LIP1. Curr Biol 17 1456–1464 [DOI] [PubMed] [Google Scholar]
- Kikis EA, Khanna R, Quail PH (2005) ELF4 is a phytochrome-regulated component of a negative-feedback loop involving the central oscillator components CCA1 and LHY. Plant J 44 300–313 [DOI] [PubMed] [Google Scholar]
- Kim J, Kim Y, Yeom M, Kim JH, Nam HG (2008) FIONA1 is essential for regulating period length in the Arabidopsis circadian clock. Plant Cell 20 307–319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi Y, Weigel D (2007) Move on up, it's time for change: mobile signals controlling photoperiod-dependent flowering. Genes Dev 21 2371–2384 [DOI] [PubMed] [Google Scholar]
- Komeda Y (2004) Genetic regulation of time to flower in Arabidopsis thaliana. Annu Rev Plant Biol 55 521–535 [DOI] [PubMed] [Google Scholar]
- Laubinger S, Marchal V, Le Gourrierec J, Wenkel S, Adrian J, Jang S, Kulajta C, Braun H, Coupland G, Hoecker U (2006) Arabidopsis SPA proteins regulate photoperiodic flowering and interact with the floral inducer CONSTANS to regulate its stability. Development 133 3213–3222 [DOI] [PubMed] [Google Scholar]
- Lin JF, Wu SH (2004) Molecular events in senescing Arabidopsis leaves. Plant J 39 612–628 [DOI] [PubMed] [Google Scholar]
- Liu LJ, Zhang YC, Li QH, Sang Y, Mao J, Lian HL, Wang L, Yang HQ (2008) COP1-mediated ubiquitination of CONSTANS is implicated in cryptochrome regulation of flowering in Arabidopsis. Plant Cell 20 292–306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin-Tryon EL, Kreps JA, Harmer SL (2007) GIGANTEA acts in blue light signaling and has biochemically separable roles in circadian clock and flowering time regulation. Plant Physiol 143 473–486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mas P, Kim WY, Somers DE, Kay SA (2003) Targeted degradation of TOC1 by ZTL modulates circadian function in Arabidopsis thaliana. Nature 426 567–570 [DOI] [PubMed] [Google Scholar]
- Mathieu J, Warthmann N, Kuttner F, Schmid M (2007) Export of FT protein from phloem companion cells is sufficient for floral induction in Arabidopsis. Curr Biol 17 1055–1060 [DOI] [PubMed] [Google Scholar]
- Matsuo T, Okamoto K, Onai K, Niwa Y, Shimogawara K, Ishiura M (2008) A systematic forward genetic analysis identified components of the Chlamydomonas circadian system. Genes Dev 22 918–930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClung CR (2006) Plant circadian rhythms. Plant Cell 18 792–803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McWatters HG, Bastow RM, Hall A, Millar AJ (2000) The ELF3 zeitnehmer regulates light signalling to the circadian clock. Nature 408 716–720 [DOI] [PubMed] [Google Scholar]
- McWatters HG, Kolmos E, Hall A, Doyle MR, Amasino RM, Gyula P, Nagy F, Millar AJ, Davis SJ (2007) ELF4 is required for oscillatory properties of the circadian clock. Plant Physiol 144 391–401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michael TP, Salome PA, Yu HJ, Spencer TR, Sharp EL, McPeek MA, Alonso JM, Ecker JR, McClung CR (2003) Enhanced fitness conferred by naturally occurring variation in the circadian clock. Science 302 1049–1053 [DOI] [PubMed] [Google Scholar]
- Millar AJ, Carre IA, Strayer CA, Chua NH, Kay SA (1995) Circadian clock mutants in Arabidopsis identified by luciferase imaging. Science 267 1161–1163 [DOI] [PubMed] [Google Scholar]
- Millar AJ, Kay SA (1991) Circadian control of cab gene transcription and mRNA accumulation in Arabidopsis. Plant Cell 3 541–550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizoguchi T, Wheatley K, Hanzawa Y, Wright L, Mizoguchi M, Song HR, Carre IA, Coupland G (2002) LHY and CCA1 are partially redundant genes required to maintain circadian rhythms in Arabidopsis. Dev Cell 2 629–641 [DOI] [PubMed] [Google Scholar]
- Mizoguchi T, Wright L, Fujiwara S, Cremer F, Lee K, Onouchi H, Mouradov A, Fowler S, Kamada H, Putterill J, et al (2005) Distinct roles of GIGANTEA in promoting flowering and regulating circadian rhythms in Arabidopsis. Plant Cell 17 2255–2270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouradov A, Cremer F, Coupland G (2002) Control of flowering time: interacting pathways as a basis for diversity. Plant Cell (Suppl) 14 S111–S130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamichi N, Kita M, Ito S, Yamashino T, Mizuno T (2005) PSEUDO-RESPONSE REGULATORS, PRR9, PRR7 and PRR5, together play essential roles close to the circadian clock of Arabidopsis thaliana. Plant Cell Physiol 46 686–698 [DOI] [PubMed] [Google Scholar]
- Nelson DC, Lasswell J, Rogg LE, Cohen MA, Bartel B (2000) FKF1, a clock-controlled gene that regulates the transition to flowering in Arabidopsis. Cell 101 331–340 [DOI] [PubMed] [Google Scholar]
- Onai K, Ishiura M (2005) PHYTOCLOCK 1 encoding a novel GARP protein essential for the Arabidopsis circadian clock. Genes Cells 10 963–972 [DOI] [PubMed] [Google Scholar]
- Park DH, Somers DE, Kim YS, Choy YH, Lim HK, Soh MS, Kim HJ, Kay SA, Nam HG (1999) Control of circadian rhythms and photoperiodic flowering by the Arabidopsis GIGANTEA gene. Science 285 1579–1582 [DOI] [PubMed] [Google Scholar]
- Portoles S, Mas P (2007) Altered oscillator function affects clock resonance and is responsible for the reduced day-length sensitivity of CKB4 overexpressing plants. Plant J 51 966–977 [DOI] [PubMed] [Google Scholar]
- Sawa M, Nusinow DA, Kay SA, Imaizumi T (2007) FKF1 and GIGANTEA complex formation is required for day-length measurement in Arabidopsis. Science 318 261–265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaffer R, Ramsay N, Samach A, Corden S, Putterill J, Carre IA, Coupland G (1998) The late elongated hypocotyl mutation of Arabidopsis disrupts circadian rhythms and the photoperiodic control of flowering. Cell 93 1219–1229 [DOI] [PubMed] [Google Scholar]
- Smith TF, Gaitatzes C, Saxena K, Neer EJ (1999) The WD repeat: a common architecture for diverse functions. Trends Biochem Sci 24 181–185 [DOI] [PubMed] [Google Scholar]
- Somers DE, Devlin PF, Kay SA (1998) Phytochromes and cryptochromes in the entrainment of the Arabidopsis circadian clock. Science 282 1488–1490 [DOI] [PubMed] [Google Scholar]
- Somers DE, Schultz TF, Milnamow M, Kay SA (2000) ZEITLUPE encodes a novel clock-associated PAS protein from Arabidopsis. Cell 101 319–329 [DOI] [PubMed] [Google Scholar]
- Staiger D, Allenbach L, Salathia N, Fiechter V, Davis SJ, Millar AJ, Chory J, Fankhauser C (2003) The Arabidopsis SRR1 gene mediates phyB signaling and is required for normal circadian clock function. Genes Dev 17 256–268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strayer C, Oyama T, Schultz TF, Raman R, Somers DE, Mas P, Panda S, Kreps JA, Kay SA (2000) Cloning of the Arabidopsis clock gene TOC1, an autoregulatory response regulator homolog. Science 289 768–771 [DOI] [PubMed] [Google Scholar]
- Suarez-Lopez P, Wheatley K, Robson F, Onouchi H, Valverde F, Coupland G (2001) CONSTANS mediates between the circadian clock and the control of flowering in Arabidopsis. Nature 410 1116–1120 [DOI] [PubMed] [Google Scholar]
- Tamaki S, Matsuo S, Wong HL, Yokoi S, Shimamoto K (2007) Hd3a protein is a mobile flowering signal in rice. Science 316 1033–1036 [DOI] [PubMed] [Google Scholar]
- Valverde F, Mouradov A, Soppe W, Ravenscroft D, Samach A, Coupland G (2004) Photoreceptor regulation of CONSTANS protein in photoperiodic flowering. Science 303 1003–1006 [DOI] [PubMed] [Google Scholar]
- van Nocker S, Ludwig P (2003) The WD-repeat protein superfamily in Arabidopsis: conservation and divergence in structure and function. BMC Genomics 4 50–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker AR, Davison PA, Bolognesi-Winfield AC, James CM, Srinivasan N, Blundell TL, Esch JJ, Marks MD, Gray JC (1999) The TRANSPARENT TESTA GLABRA1 locus, which regulates trichome differentiation and anthocyanin biosynthesis in Arabidopsis, encodes a WD40 repeat protein. Plant Cell 11 1337–1350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang ZY, Tobin EM (1998) Constitutive expression of the CIRCADIAN CLOCK ASSOCIATED 1 (CCA1) gene disrupts circadian rhythms and suppresses its own expression. Cell 93 1207–1217 [DOI] [PubMed] [Google Scholar]
- Yakir E, Hilman D, Harir Y, Green RM (2007) Regulation of output from the plant circadian clock. FEBS J 274 335–345 [DOI] [PubMed] [Google Scholar]
- Yamamoto Y, Sato E, Shimizu T, Nakamich N, Sato S, Kato T, Tabata S, Nagatani A, Yamashino T, Mizuno T (2003) Comparative genetic studies on the APRR5 and APRR7 genes belonging to the APRR1/TOC1 quintet implicated in circadian rhythm, control of flowering time, and early photomorphogenesis. Plant Cell Physiol 44 1119–1130 [DOI] [PubMed] [Google Scholar]
- Yanovsky MJ, Kay SA (2002) Molecular basis of seasonal time measurement in Arabidopsis. Nature 419 308–312 [DOI] [PubMed] [Google Scholar]
- Zhao M, Morohashi K, Hatlestad G, Grotewold E, Lloyd A (2008) The TTG1-bHLH-MYB complex controls trichome cell fate and patterning through direct targeting of regulatory loci. Development 135 1991–1999 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.