Abstract
Root growth is mainly determined by cell division and subsequent elongation in the root apical area. Components regulating cell division in root meristematic cells are largely unknown. Previous studies have identified rice (Oryza sativa) ROOT ARCHITECTURE ASSOCIATED1 (OsRAA1) as a regulator in root development. Yet, the function of OsRAA1 at the cellular and molecular levels is unclear. Here, we show that OsRAA1-overexpressed transgenic rice showed reduced primary root growth, increased numbers of cells in metaphase, and reduced numbers of cells in anaphase, which suggests that OsRAA1 is responsible for limiting root growth by inhibiting the onset of anaphase. The expression of OsRAA1 in fission yeast also induced metaphase arrest, which is consistent with the fact that OsRAA1 functions through a conserved mechanism of cell cycle regulation. Moreover, a colocalization assay has shown that OsRAA1 is expressed predominantly at spindles during cell division. Yeast two-hybrid and pull-down assays, as well as a bimolecular fluorescence complementation assay, all have revealed that OsRAA1 interacts with a rice homolog of REGULATORY PARTICLE TRIPLE-A ATPASE4, a component that is involved in the ubiquitin pathway. Treating transgenic rice with specific inhibitors of the 26S proteasome blocked the degradation of OsRAA1 and increased the number of cells in metaphase. Mutation of a putative ubiquitination-targeting D-box (RGSLDLISL) in OsRAA1 interrupted the destruction of OsRAA1 in transgenic yeast. These results suggest that ubiquitination and proteasomic proteolysis are involved in OsRAA1 degradation, which is essential for the onset of anaphase, and that OsRAA1 may modulate root development mediated by the ubiquitin-proteasome pathway as a novel regulatory factor of the cell cycle.
The development of multicellular organisms, such as plants, relies on the temporal and spatial control of cell proliferation and growth. The root tip has been widely used to study plant cell proliferation and cell growth due to its relatively simple structure and distinct regions of meristem, elongation, and mature zones. In the mitotic cell cycle program, DNA replication of the S phase is followed by the M phase, a segregation of the duplicated genetic materials into two daughter cells. Preparation gap phases, G1 and G2, precede the S and M phases, respectively.
Several genes are involved in phase transition during the cell cycle. The sequential and transient activation of cyclin-dependent kinases (CDKs) dictates a unidirectional progress through the cell cycle. Although CDKs have been identified to be the key mediators that control basic cell cycle development (Inze and De Veylder, 2006), some other regulatory molecules are also involved. For example, a CDK inhibitor named KRP1, a target of the ubiquitin/proteasome pathway, functions in the G1-S transition in Arabidopsis (Arabidopsis thaliana). KRP1 interacts with the CDKA;1/cyclin D2;1 complex in planta (Ren et al., 2008). Protein kinase CK2 is an evolutionarily conserved Ser/Thr phosphotransferase composed of two distinct subunits, α (catalytic) and β (regulatory). A dominant-negative mutant of CK2 stably expressed in tobacco (Nicotiana tabacum) BY-2 cells interrupted the G1/S and G2 phases (Moreno-Romero et al., 2008).
The ubiquitin-proteasome pathway is known to control root development in Arabidopsis (Smalle et al., 2002; Xie et al., 2002; Ueda et al., 2004). It is also involved in regulation of the phase transition in the plant cell cycle (King et al., 1996; Koepp et al., 1999; Tyers and Jorgensen, 2000). The ubiquitin-proteasome pathway includes several enzymes, such as ubiquitin-activating enzyme (E1), ubiquitin-conjugating enzyme (E2), and ubiquitin ligase (E3). A key process is E3 action specifically recognizing its substrate. AtHUB1 of RING E3 regulates the G2-to-M transition in Arabidopsis (Fleury et al., 2007). Anaphase-promoting complex/cyclosome (APC) of E3 has a conserved role in cell cycle progression during the entire life cycle of plants (Perez-Perez et al., 2008). In the transition from metaphase to anaphase, several APC-mediated key proteins (King et al., 1996; Townsley and Ruderman, 1998) are ubiquitinated on multiple sites and subsequently degraded by an ATP-dependent reaction cascade (Murray, 1995; Cohen-Fix et al., 1996; Funabiki et al., 1996; Juang et al., 1997; Michaelis et al., 1997). The common structural motif of substrates identified by APC is a nine-amino acid sequence [RxxLxx(L/I)xN] termed the destruction box (D-box). In plants, most of the known targets of the APC are cyclins containing the D-box motif (Renaudin et al., 1996; Fang et al., 1998; Hames et al., 2001). The targeted proteins are then recognized and degraded by a macromolecular complex, 26S proteasome, which is composed of the 20S proteolytic core and the 19S regulatory complex (RP; Peters et al., 1994). The 19S RP contains two stable subcomplexes, the lid and the base, which include six different ATPases (REGULATORY PARTICLE TRIPLE-A ATPASE1 [RPT1], RPT2, RPT3, RPT4, RPT5, and RPT6) and three non-ATPase subunits (RPN1, RPN2, and RPN10). Each ATPase, which is proposed to be involved in recognizing the ubiquitinated substrate, is essential for growth in yeast (Rechsteiner et al., 1993; Russell et al., 2001).
RPT4/SUG2 plays essential roles during the entire life of yeast cells. Deletion of the gene causes cell death (Ferrell et al., 2000). RPT4, one of the subunits of 19S RP, recognizes some ubiquitinated substrates, which are involved in regulating spindle-pole-body (SPB) duplication in yeast cells (McDonald and Byers, 1997). As well, RPT4-independent proteolysis is involved in nucleotide excision repair and transcription elongation (Russell et al., 1999; Gonzalez et al., 2002). Although RPT4 has also been identified in Arabidopsis and rice (Oryza sativa), its function in plant cells is entirely unknown (Fu et al., 1999, 2001; Shibahara et al., 2002).
Previous studies have suggested that rice ROOT ARCHITECTURE ASSOCIATED1 (OsRAA1), a small GTP-binding protein in plants, is involved in rice root development (Ge et al., 2004; Han et al., 2005). OsRAA1 is expressed specifically in the root apical meristem, elongation zone, steles of the branch zone, and young lateral root. Enhanced expression of OsRAA1 results in reduced growth of primary roots, increased numbers of adventitious roots, and a helix primary root (Ge et al., 2004). Transgenic rice lines of AtFPF1, the OsRAA1 homolog from Arabidopsis, showed a similar effect on development, which suggests a conserved function of OsRAA1/AtFPF1 in both rice and Arabidopsis (Xu et al., 2005). Shortened primary roots are a typical phenotype observed in OsRAA1-overexpressed transgenic rice plants. However, the true molecular and cellular mechanisms of OsRAA1 in regulating primary root development have not been unfolded.
In this study, we investigated the mechanisms of OsRAA1 in regulating rice root development using a set of biochemical and cytological approaches. An OsRAA1-interacting protein was obtained by a yeast two-hybrid screen. The in vitro and in vivo interactions between OsRAA1 and this protein were confirmed using bimolecular fluorescence complementation (BiFC), pull-down, and coimmunoprecipitation analyses. Transgenic rice plants, as well as yeast, were used to identify cell cycle regulation. A proteasome inhibition experiment revealed that the 26S proteasome is involved in the cell cycle and OsRAA1 degradation. These results support the hypothesis that the degradation of OsRAA1 is essential for the onset of anaphase in the cell cycle during root development.
RESULTS
OsRAA1 Interaction Protein Screening
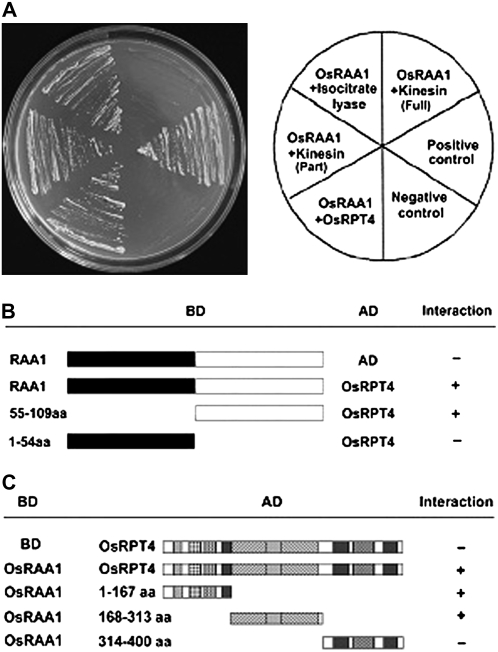
To investigate the biochemical functions of OsRAA1, a yeast two-hybrid approach utilizing OsRAA1 as a bait was conducted to identify the interaction proteins of OsRAA1. Our results indicated that the OsRAA1-binding domain (BD) fusion protein alone could not activate the expression of the reporter gene in the presence of the GAL4-activation domain (AD) vector, which indicates that OsRAA1 does not interact with GAL4-AD and does not activate transcription autonomously (data not shown). A cDNA expression library containing about 3 × 106 transformants was screened. A total of 81 colonies were obtained on a medium lacking His, Trp, and Leu but supplemented with 5 mm 3-amino-1,2,4-triazole. Among them, 32 colonies can activate the lacZ reporter gene. These colonies were able to grow on medium lacking Ade, His, Trp, and Leu. Sequence analysis revealed that they encode 18 independent fragments. Three fragments were further confirmed by transforming OsRAA1-BD back into the AH109 yeast strain, with BD used as a control. Other fragments were recognized as false positives. Homology analysis revealed three putative partner proteins: OsRPT4, chain A of isocitrate lyase, and kinesin-related protein (Fig. 1A). The frequency of OsRPT4 was about 10% in the 32 colonies (data not shown). A fragment of a kinesin-related protein interacted with OsRAA1. Further studies on the interaction mechanism are required to interpret why the full-length kinesin shows an indistinct interaction.
Figure 1.
OsRAA1 interacts with RPT4 in yeast. A, Yeast two-hybrid assay demonstrating OsRAA1 interacting with OsRPT4, partial fragment of kinesin protein, and isocitrate lyase. B, Overview of the OsRAA1 domain interacting with OsRPT4 by two-hybrid assay, with AD used as a control. OsRPT4 strongly bound to the C terminus rather than the N terminus of OsRAA1. C, Derivatives of OsRPT4 interacting with OsRAA1, with BD as a control. OsRAA1 interacted with amino acids 1 to 167 of OsRPT4 at the N terminus, amino acids 168 to 313 containing motifs A and B, but not with amino acids 314 to 400 at the C terminus.
To determine the binding region of OsRAA1 with RPT4, two truncated versions of BD at both the N and C termini of OsRAA1 were constructed. As shown in Figure 1B, RPT4 interacted with the C terminus rather than the N terminus of OsRAA1.
The screened cDNA of OsRPT4 was 1,625 bp with an open reading frame (ORF) of 1,203 bp (Supplemental Fig. S1). The gene encodes a predicted product of 400 amino acid polypeptides of 44.6 kD, pI 7.25. Semiquantitative reverse transcription (RT)-PCR analysis indicated that the gene is expressed in all tissues tested (Supplemental Fig. S2). To determine the region of OsRPT4 that interacts with OsRAA1, we cloned the full-length form and various deleted forms of OsRPT4 into the AD construct. OsRAA1 interacted with the fragments of OsRPT4 containing amino acids 1 to 167 and 168 to 313, which possess the conserved motifs A and B, respectively, but not with the fragment containing amino acids 314 to 400 at the C terminus (Fig. 1C).
OsRAA1 Interacts with OsRPT4 in Vitro and in Vivo
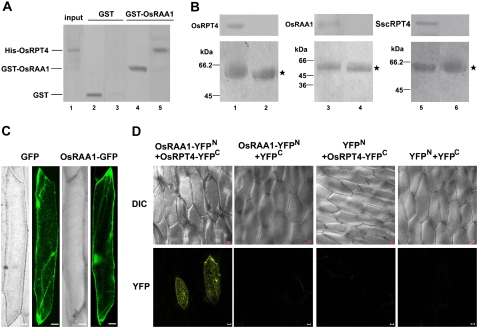
The direct interaction of OsRAA1 and OsRPT4 was further examined by pull-down, immunoprecipitation, and BiFC techniques. After incubating His-RPT4 with glutathione S-transferase (GST)-OsRAA1 or GST alone, the eluted protein complexes from glutathione-Sepharose 4B beads were examined by SDS-PAGE and western-blot analysis against antibodies specific for OsRAA1, RPT4, and GST. His-RPT4 was pulled down with GST-OsRAA1 but not with GST, which indicates that OsRAA1 and OsRPT4 directly bind to each other in vitro (Fig. 2A).
Figure 2.
Interaction of OsRAA1 with OsRPT4. A, OsRAA1 interacts with OsRPT4. GST-OsRAA1 fusion protein and GST were expressed and purified from Escherichia coli, adsorbed onto GST-Sepharose 4B, and incubated with His-OsRPT4. After being washed, the protein-bound protein was fractionated by 12% SDS-PAGE and underwent western-blot analysis with antiserum of RAA1 (lane 4), GST antibodies (lane 2), or RPT4 antibodies (lanes 3 and 5). One-tenth of the total input of the E. coli total protein inducing His-RPT4 expression was loaded onto SDS-PAGE gels and detected by RPT4 antibodies (lane 1). B, Western-blot results of in vivo coimmunoprecipitation confirming the interaction between OsRAA1 and OsRPT4. Protein extracts from transgenic rice roots (left and middle) or transgenic yeast cells (right) were immunoprecipitated with 3 μL of anti-OsRAA1 (lanes 1, 3, and 5) or control preimmune serum (lanes 2, 4, and 6). The immunoprecipitated solution then underwent 12% T and 16.5% T, 3% C SDS-PAGE, then was incubated with anti-RPT4 (1:400; lanes 1, 2, 5, and 6) and anti-OsRAA1 (1:400; lanes 3 and 4; top images). The bottom images show Coomassie Brilliant Blue staining of loading controls. Molecular mass markers are indicated at left. Stars indicate immunoglobulin heavy chains. C, Localization of OsRAA1-GFP fusion protein in onion cells. D, BiFC view of OsRAA1 and OsRPT4 interaction in Chinese cabbage epidermal cells on transient expression assay. Corresponding differential interference contrast images are shown at top. Bars = 20 μm.
Coimmunoprecipitation was performed to further verify the interaction between OsRAA1 and OsRPT4 in planta. Our results indicated that RPT4 can be coimmunoprecipitated with OsRAA1 using antibodies against OsRAA1 (Fig. 2B). Similarly, in transgenic yeast cells, SscRPT4 could be coprecipitated with OsRAA1 (Fig. 2B).
Transiently transformed onion (Allium cepa) epidermal layer containing the fusion protein OsRAA1-GFP was used to monitor the localization of OsRAA1. Confocal analysis using this transformed tissue indicated that the fluorescence signals are evenly distributed in the cell, similar to the pattern of GFP alone (Fig. 2C). In addition, the BiFC technique was also used to demonstrate the interaction between OsRAA1 and OsRPT4. The pUCSPYNE-OsRAA1 vector contains a fusion of OsRAA1 at the N terminus of yellow fluorescent protein (YFP; OsRAA1-YFPN), and the pUCSPYCE-OsRPT4 vector contains a fusion of OsRPT4 at the C terminus of YFP (OsRPT4-YFPC). The two recombinant DNA constructs were introduced into Chinese cabbage (Brassica campestris pekinensis) epidermal cells by particle bombardment. Strong fluorescence signals were observed with the OsRAA1-YFPN and OsRPT4-YFPC combinations but were not observed when coexpressed with empty vector (Fig. 2D). The interaction of OsRAA1 and OsRPT4 showed cytoplasmic and nuclear localization, which was consistent with the OsRAA1-GFP fusion protein localization in onion epidermal cells (Fig. 2C).
Aberrant Mitosis Occurs in OsRAA1 Transgenic Rice and Transgenic Yeast Cells
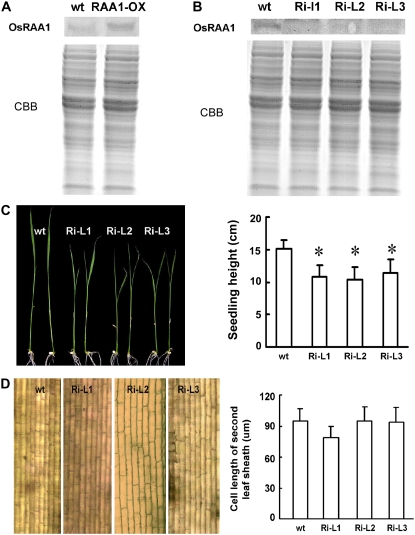
Transgenic rice overexpressing OsRAA1 with increased expression at the protein level (Fig. 3A) clearly showed suppressed growth of the primary roots in comparison with those of wild-type plants (Ge et al., 2004). Knockdown plants were generated using RNA interference (RNAi) technology. Western-blot analysis confirmed that these lines are true RNAi knockdown transgenic lines (Fig. 3B). These lines showed a reduced seedling height phenotype (Fig. 3C), which was opposite the increased height phenotype seen in OsRAA1-overexpressing transgenic plants (Ge et al., 2004). It appears that reducing the expression of OsRAA1 does not affect primary root development (Fig. 3C). Interestingly, in knockdown lines, the cell length of the leaf sheath with reduced height was the same as that of the wild type (Fig. 3D), which suggests that the reduced height of seedlings in the knockdown lines was caused by reduced cell numbers due to abnormal cell division. Since yeast two-hybrid results indicated that OsRAA1 interacts with OsRPT4, which is a known cell cycle regulator, we investigated the cell cycle in transgenic roots to unveil the mechanism of OsRAA1 regulation in root growth.
Figure 3.
Expression of OsRAA1 and root morphological responses in transgenic lines. A, Identification of OsRAA1 expression in transgenic line (OsRAA1-OX) and wild-type (wt) plants by western-blot analysis. Coomassie Brilliant Blue (CBB) staining of a loading control is shown at bottom. B, Identification of OsRAA1 expression in RNAi lines (Ri-L) and wild-type (wt) plants by western-blot analysis. CBB staining of a loading control is shown at bottom. C, Phenotypes (left) and statistics (right) of RNAi transgenic seedling height aboveground. Significant differences between wild-type and RNAi lines were determined by two-sample t test (* P < 0.01). D, The unchanged cell length of RNAi transgenic rice lines (left), and the cell length with no significant differences (right).
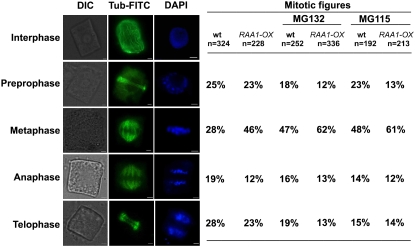
Immunofluorescence analysis of microtubules during mitosis showed no abnormal phenotypes in transgenic rice compared with those of the wild type. However, OsRAA1-overexpressing rice plants showed 46% of cells localized in metaphase, whereas wild-type rice plants showed only about 28%. In addition, the proportion of cells in anaphase was lower in transgenic rice than in the wild type (12% versus 19%; Fig. 4). The proportion of cells in preprophase was comparable between transgenic and wild-type rice, but that in telophase was lower in transgenic rice (Fig. 4). Thus, the mitosis process was blocked to a certain extent in the transition from metaphase to anaphase in transgenic rice.
Figure 4.
Effects of MG132 and MG115 on the mitotic index in immunofluorescence. Transgenic (OsRAA1-OX) and wild-type (wt) root tip cells were double stained with β-tubulin antibody, anti-mouse IgG fluorescein isothiocyanate (FITC) conjugate shown in green, and DAPI shown in blue in the presence of 100 μm MG132 or MG115. Mitotic phase is indicated at left. DIC, Differential interference contrast. Bars = 2 μm.
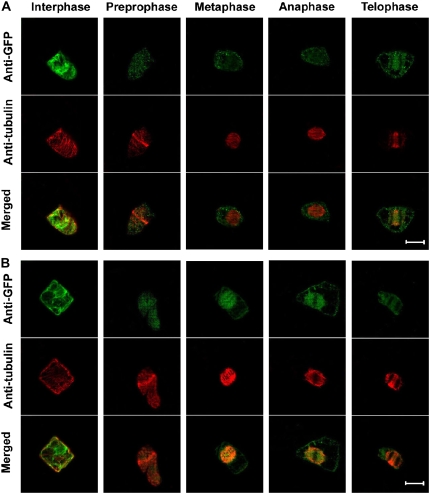
To understand the localization of OsRAA1 during cell mitosis, we stably transformed a construct harboring the OsRAA1-GFP fusion gene, as well as GFP alone, into tobacco BY-2 cells. Immunoassay of a cell line with GFP alone showed immunofluorescence in the whole cell during the cell cycle (Fig. 5A). In contrast, in the OsRAA1-GFP transgenic line, OsRAA1-GFP was enriched at spindles, including tubulins, from metaphase to anaphase during cell division (Fig. 5B), which suggests that OsRAA1 may function at the transition from metaphase to anaphase during cell division.
Figure 5.
Localization of OsRAA1 during cell mitosis. Immunolocalization of OsRAA1 and tubulin in BY-2 cells stably expressing OsRAA1-GFP or GFP. Confocal images (GFP in green and β-tubulin in red) were collected from various cell phases of the cell cycle. A, Localization of GFP and tubulin as a control. Merged images are shown at bottom. B, Localization of OsRAA1-GFP and tubulin. Merged images are shown at bottom. Bars = 20 μm.
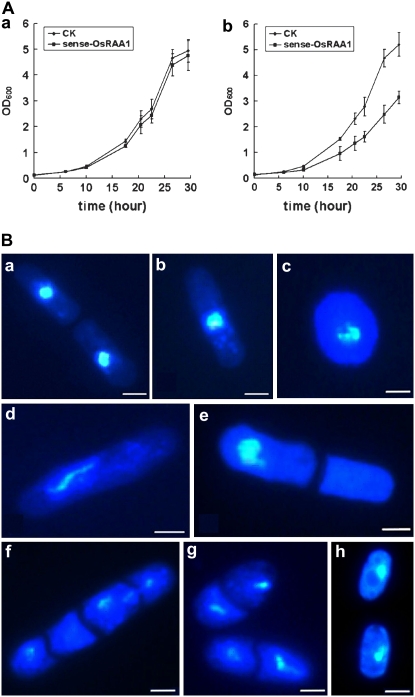
To confirm the function of OsRAA1 in the cell cycle, we studied the cell system of the fission yeast Schizosaccharomyces pombe. Amino acid sequence alignment showed high identity (71%) between OsRPT4 and S. pombe RPT4 and 99% identity between the conserved motifs A and B (Supplemental Fig. S1). A BLAST search revealed no homolog of OsRAA1 in S. pombe. The constructs of the OsRAA1 overexpression vector, as well as the control vector pESPM, were transformed into yeast cells of the Leu− strain SPQ-01, and the transgenic yeast was identified subsequently by RT-PCR (Supplemental Fig. S3). The NMT1 promoter inducible by thiamine removal controls the production of OsRAA1. The immunoprecipitation results showed that OsRAA1 interacted with yeast RPT4 (Fig. 2B). Cells transformed with OsRAA1 showed reduced growth when thiamine was absent (Fig. 6Ab), although they grew normally on medium supplemented with thiamine (Fig. 6Aa). Strains with OsRAA1 overexpressed, however, exhibited abnormal cells (Fig. 6B, c–g), whereas all cells transformed with empty vector as a control segregated normally (Fig. 6B, a and b). The growth of more than half of cells (52%) in transgenic strains was blocked at metaphase (Fig. 6Bd). These results further confirmed that overexpression of OsRAA1 causes metaphase arrest and reduced cell growth.
Figure 6.
Phenotypic characterization of overproduced OsRAA1 yeast strains. A, Overexpressed OsRAA1 suppresses the growth rate of the yeast cells. SPQ-01 yeast cells were grown overnight at 30°C, and 0.2 × 106 cells mL−1 were inoculated into 50 mL of Edinburgh minimal medium and grown in a medium containing thiamine (a) or not (b). Growth was monitored by optical density at 600 nm (OD600). B, Chromosome segregation defects in yeast cells. Transgenic cells were grown with vigorous agitation at 30°C to log phase in the presence of thiamine. After being washed, cells were put in liquid culture with no thiamine for 24 h. A vector-transformed cell that just completed separation (a) and one in interphase (b) are shown as controls. c to g show aberrant transgenic cells, and their percentage location is 5%, 52%, 4%, 3%, and 7%, respectively. For every panel, more than 500 cells were examined. The transgenic line with mutant OsRAA1 showed a normal shape (h). Bars = 10 μm.
Degradation of OsRAA1 Depends on the Proteasome
The interaction of OsRAA1 with OsRPT4 suggests that OsRAA1 is likely to be involved in regulating protein degradation. To further investigate the possible involvement of the 26S proteasome in OsRAA1-mediated root development, we examined the effects of the specific 26S proteasome inhibitors MG132 (Z-Leu-Leu-Leu-al) and MG115 (Z-leu-leu-Norvalinal) on mitosis of both wild-type and OsRAA1-overexpressing transgenic rice plants. In the presence of MG132, mitosis of the wild type showed metaphase arrest, with 47% of cells localized in metaphase compared with 28% of wild-type cells in the absence of MG132 (Fig. 4). Similarly, overexpressed OsRAA1 transgenic plants with MG132 possessed substantially more metaphase cells (62%) than those without MG132 (46%). As a control, the solvent dimethyl sulfoxide (DMSO) alone produced no effect on mitotic progression (data not shown). Thus, the activity of the 26S proteasome is likely involved in separating sister chromatids in OsRAA1-overexpressing rice; we confirmed this concept with MG115 treatment, showing 48% of metaphase cells in the wild type and 61% of such cells in transgenic rice (Fig. 4).
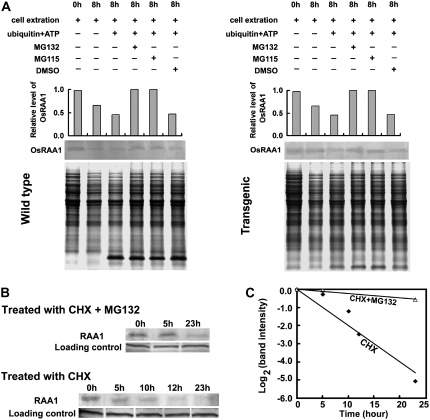
To study the effect of proteasome inhibitors on OsRAA1 stability in vitro, we incubated protein extracts with MG132 or MG115 for 8 h and used the antiserum of OsRAA1 for the detection of the protein. After incubation, the amount of OsRAA1 in the wild type was clearly decreased and severely disrupted with the addition of His-ubiquitin and ATP (Fig. 7A). OsRAA1 level was increased with the addition of MG132 or MG115. DMSO treatment had no effect on protein level (Fig. 7A). These results suggested that the protein level of OsRAA1 was regulated by the 26S proteasome. Similarly, in OsRAA1-overexpressing rice, OsRAA1 level was slightly reduced after incubation, and its presence was significantly disrupted by the addition of ubiquitin and ATP (Fig. 7A). OsRAA1 level in vivo was gradually reduced within 23 h of treatment with cycloheximide (CHX), an inhibitor of protein synthesis (Fig. 7, B and C). The calculated half-life was about 5 h. The half-life of OsRAA1 was extended when seedlings were treated with both MG115 and CHX (Fig. 7, B and C). With MG132 or MG115 treatment, the disruption of OsRAA1 expression was abolished, which was analogous to such treatment in the wild type. Similarly, MG132 and MG115 treatment suppressed the degradation reaction in OsRAA1-overexpressing transgenic yeast cells (Fig. 8A).
Figure 7.
Effects of the inhibitors on OsRAA1 degradation mediated by the 26S proteasome, and half-life of OsRAA1. The mixture was separated on 16.5% T, 3% C SDS-PAGE and underwent western-blot analysis with antiserum of OsRAA1. A, Effect of the inhibitor on OsRAA1 degradation was monitored with the antibody. OsRAA1 of wild-type and transgenic rice and detected with the addition of 10 μg of ubiquitin in the presence of 100 μm MG132 or MG115 with 25°C incubation for 8 h. The columns show relative levels of OsRAA1 detected by the antibody. DMSO was used as a control. Coomassie Brilliant Blue staining of a loading control is shown at bottom. B, OsRAA1 half-life in rice seedlings. Protein synthesis was inhibited by treatment with CHX (100 μm) for the indicated times. At the same time, seedlings were treated with MG132 (100 μm). The OsRAA1 was monitored at the indicated times. Data represent results from more than three experiments. C, Quantitation of the OsRAA1 destabilization shown in B.
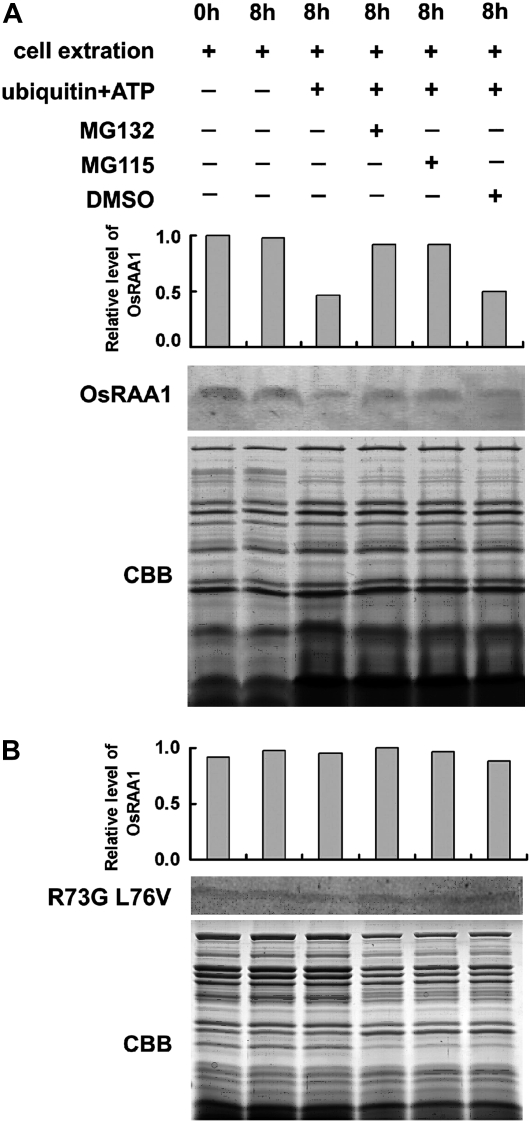
Figure 8.
Effects of mutation in the D-box motif of OsRAA1 on its degradation by the 26S proteasome. The yeast protein extracts were separated on 16.5% T, 3% C SDS-PAGE and underwent western-blot analysis with the antibody to OsRAA1. OsRAA1 (A) and D-box mutant OsRAA1 R73G L76V (B) were detected with the addition of 10 μg of ubiquitin in the presence of 100 μm MG132 or MG115 with 25°C incubation for 8 h. The columns show relative levels of OsRAA1 detected by the antibody. DMSO was used as a control for the inhibitor treatment. Coomassie Brilliant Blue (CBB) staining of a loading control is shown at bottom.
The specific E3, APC, recognizes the key proteins in the transition from metaphase to anaphase, which are subsequently ubiquitinated on multiple sites and are degraded by the 26S proteasome (Murray, 1995; Cohen-Fix et al., 1996; Funabiki et al., 1996; King et al., 1996; Juang et al., 1997; Michaelis et al., 1997; Townsley and Ruderman, 1998). The common motif of the APC substrate is the D-box, a nine-amino acid sequence [RxxLxx(L/I)xN]. OsRAA1 possesses the D-box motif RGSLDLISL at amino acids 73 to 81, with R and L conserved and essential for its function. Therefore, we transformed yeast cells with mutations of Arg-73 to Gly-73 and Leu-76 to Val-76 in the D-box. With the mutated D-box, the level of OsRAA1 did not differ between nonubiquitinated controls and cells treated with inhibitors (Fig. 8B). Similarly, transgenic yeast cells with the mutated OsRAA1 did not differ in phenotype from those with the empty vector (Fig. 6Bh). The mutated OsRAA1 was not degraded by yeast 26S proteasome. Thus, these results suggest that the disruption of OsRAA1 depends on the ubiquitin-proteasome pathway.
DISCUSSION
OsRAA1 Regulates Primary Root Development by Modulating Mitosis
In Arabidopsis, several genes controlling division and differentiation in root growth and development have been identified through mutation genetics. The Arabidopsis root mutants scarecrow (scr) and short root (shr) cause reduced primary root length by degenerated cell division (Scheres et al., 1995; Di Laurenzio et al., 1996). SCR encoding a putative transcription factor is required for asymmetric cell division in roots and shoots. SHR is upstream of SCR and is required for the initiation of middle cortex formation (Paquette and Benfey, 2005). In contrast, root development of rice is being characterized with studies on a growing number of mutants involving unknown genes that affect root formation, such as rm1 and rm2 (Ichii and Ishikawa, 1997) and rrl1 and rrl2 (Inukai et al., 2001). In rice, genes such as OsCKI1 and OsGNA1 were found to be involved in root development, but the regulatory mechanism is still unknown (Liu et al., 2003; Jiang et al., 2005). Since previous studies identified OsRAA1 as regulating root development, we investigated its function by biochemical and cytological approaches. We found that OsRAA1-overexpressing rice plants showed reduced primary root growth but increased numbers of cells in metaphase and reduced numbers of cells in anaphase. To investigate the aberrant mitosis, yeast two-hybrid and pull-down assays, as well as a BiFC assay, revealed that OsRAA1 interacts with a rice homolog of RPT4, a component of the ubiquitin pathway. Treating transgenic rice with specific inhibitors of the 6S proteasome blocked the degradation of OsRAA1 and increased the number of cells in metaphase. The 26S proteasome and ubiquitination may be involved in the degradation of OsRAA1, which is essential for the onset of anaphase and root growth.
The expression pattern of OsRAA1 is always in rapidly growing cells, such as primordia, meristem, and the division zone of the root apex. Overexpression of OsRAA1 results in sterile florets and longer leaves at the last stage of plant development, which might be caused by both the extension of cells and increased cell numbers (Ge et al., 2004). Our RNAi knockdown transgenic rice showed a normal phenotype in root development. In contrast, the height of the knockdown plants, comparable to that in overexpressed transgenic rice plants (Ge et al., 2004), was suppressed, which may have resulted from reduced cell division (Fig. 3). The phenotypes of the knockdown transgenic rice plants in root development may be explained by OsRAA1 being involved in a mechanism with functional redundancy of genes induced by auxin in root development and action in the differentiation of primordia in leaf development (Ge et al., 2004). Our data support the notion that OsRAA1 is involved in the regulation of cell division.
OsRAA1 Interacts with OsRPT4 and Functions in Sister Chromatid Separation in Rice Root Cells
Yeast two-hybrid analysis, together with pull-down assay, showed that OsRPT4 is a partner of OsRAA1, with the N terminus of OsRAA1 and the C terminus of RPT4 free from the interaction. Immunoprecipitation in rice, as well as BiFC data, revealed OsRAA1 interacting with OsRPT4 in vivo. The interaction was in the cytoplasm and nucleus, which is consistent with the localization of OsRAA1 alone (Fig. 2C). RPT4, a member of a very conserved AAA ATPase family, is a component of the 19S proteasome cap (Russell et al., 1996; Fu et al., 1999, 2001). RPT4 plays important roles in the yeast cell cycle and is involved in SPB duplication, which is perhaps related to the abnormal degradation of some proteins required for SPB duplication (McDonald and Byers, 1997).
The exact function of RPT4 and its mechanism have not been reported in plants, although some other 19S complex subunits have been studied. RPT2a is essential for maintaining meristems in Arabidopsis (Ueda et al., 2004). halted root, with a mutant RPT2a gene, caused abnormal cell organization in root and shoot meristems, which suggests that proteasome-dependent programmed proteolysis is required to maintain meristem integrity (Ueda et al., 2004). The 26S proteasome pathway was assumed to play an essential role in maintaining meristem integrity in Arabidopsis (Ueda et al., 2004). Disruption of RPN12 by a T-DNA insertion caused reduced root elongation and altered growth response to exogenous applied cytokinins in Arabidopsis (Smalle et al., 2002). RPN12 modulates the stability of some factors responsible for cytokinin regulation (Smalle et al., 2002), but the substrates recognized by RPN12 and RPT2 are unknown. Our studies suggest that OsRAA1 interacting with RPT4 is involved in root growth and development.
Cytoskeletal motors, such as the kinesin family, are involved in spindle morphogenesis at the transition from metaphase to anaphase in mitosis in yeast, animals, and plants (Lee and Liu, 2004). The kinesins convert the energy from ATP hydrolysis into movement along microtubles (Ambrose et al., 2005). The Arabidopsis kinesin ATK5 localizes to microtubules in interphase cells and uses pulse-end tracking to reach spindle midzones (Ambrose et al., 2005). OsRAA1 also interacts with a protein that shares homology with a kinesin-related protein (Fig. 1), although the mechanism of their interaction is unknown. OsRAA1 was localized in spindles during cell division (Fig. 5). Therefore, the interaction of OsRAA1 with both RPT4 and kinesin was involved in sister chromatid separation in mitosis.
The Degradation of OsRAA1 by Proteasome Is Important for the Onset of Anaphase
The different RPT4 subunit in the 19S RP might serve to recognize a distinct substrate, and a working model has been proposed (Rechsteiner et al., 1993). The alignment of amino acids between these ATPases showed their C terminus more conserved than the N terminus, which possesses the “helix shuffle” interacting with target proteins for disruption (Rechsteiner et al., 1993). In the process, ATP hydrolysis is required for supplying energy to lead the substrate to the 20S core. The protein sequence of yeast RPT4 shares the helix shuffle motif composed of Leu zippers or α-helical coiled coils (McDonald and Byers, 1997). Our data support OsRAA1 interacting with the N terminus and the middle region containing the conserved motifs A and B rather than the C terminus of OsRPT4 (Fig. 1B). OsRPT4 might serve to specifically degrade OsRAA1.
Other results of RPT4 in yeast suggest a role for RPT4 in degrading OsRAA1. Yeast cell division in the temperature-sensitive mutant rpt4td ceased when cells were transferred at a restriction temperature, because the SPB cannot be duplicated in the mutant cells, although the cytoplasmic microtubule showed the normal phenotype (McDonald and Byers, 1997). Yeast RPT4 contributes to targeting the degradation of some unknown nuclear proteins, which are required for SPB duplication (McDonald et al., 2002). In yeast, many other RPT subunits in 19S RP play distinct roles in cell cycle regulation, such as rpt6 and cim5/rpt1 mutants causing cells to arrest in G2/M. NIN1/RPN12, which acts as a subunit in 19S RP but does not belong to the ATPase family, is responsible for the degradation of CDK inhibitor (Kominami et al., 1995). RPT6 is involved in the degradation of a specific substrate in mammalian cells (Wang et al., 1996). The distinct 19S regulatory particles are subunits of the multiple 26S proteasome complexes, which regulate the degradation of different proteins (McDonald and Byers, 1997). A different ATPase showing different activity in Manduca sexta development also supports this concept (Dawson et al., 1995). In plants, previous reports showed that another subunit, RPN10, composed of RP base subcomplexes with RPT4 as well as other seven subunits, functions in the degradation of specific proteins, especially the abscisic acid pathway regulator ABI5 in Arabidopsis (Smalle et al., 2003). Our results suggest that OsRPT4 might be required for the destruction of OsRAA1 induced by auxin (Ge et al., 2004), which modulates the transition from metaphase to anaphase in rice.
The destruction of key proteins by the ubiquitin/26S proteasome pathway in the transition from metaphase to anaphase is an important event in the cell cycle. The ubiquitin pathway is conserved in eukaryotic organisms and is involved in modulating the cell cycle (Hochstrasser, 1995). Degradation of cyclin B is associated with the cell cycle (Evans et al., 1983). So far, reports have described the selective disruption of many proteins such as CDK inhibitor and those participating in separating sister chromatids and the spindle complex, which is required for the onset of anaphase (Peter and Herskowitz, 1994; Murray, 1995; King et al., 1996; Hoyt, 1997).
The cell mitosis phenotype of root tips in our transgenic plants was sensitive to the specific proteasome inhibitors MG132 and MG115. Mitosis in the wild type was blocked to a certain extent in metaphase on incubation with the inhibitors (Fig. 4), which is consistent with the results of similarly treated tobacco BY-2 cells (Genschik et al., 1998). The changes in mitosis in transgenic plants on treatment were similar to those of the wild type, and the arrested metaphase in the transgenic plant cells was enhanced by treatment with proteasome inhibitors (Fig. 4). OsRAA1 degradation was efficiently suppressed by either MG132 or MG115. As well, the half-life was delayed on treatment with the inhibitor (Fig. 7). These results suggest that proteasome-mediated degradation of RAA1 is involved in transition of metaphase to anaphase in the cell cycle.
The substrate degraded by proteasome was conjugated with ubiquitin by E1, E2, and E3 enzymes in the proteolytic process. APC acts as a specific E3 enzyme for the degradation of proteins regulating mitosis (King et al., 1996; Townsley and Ruderman, 1998). Many key proteins are destroyed at the end of metaphase and in anaphase, such as the anaphase inhibitor Pds1p (Saccharomyces cerevisiae) and Cut2p (S. pombe; Cohen-Fix et al., 1996; Funabiki et al., 1996) and mitosis cyclins (Murray, 1995), and Ase1p is associated with the mitotic spindle (Juang et al., 1997). Arabidopsis HOBBIT, a homolog of the CDC27 subunit of APC, may couple cell division to differentiation by regulating the cell cycle progress in meristems (Blilou et al., 2002).
The D-box RxxLxx(L/I)xN, as the common motif, is conserved in APC substrates (Plesse et al., 1998; Renaudin et al., 1998). OsRAA1 possesses the D-box motif RGSLDLISL at amino acids 73 to 81. Some members of the OsRAA family also share RGCLDLISL with WhRoP1 (BE428690) from wheat (Triticum aestivum) and BaSpP2 (BE196402) from barley (Hordeum vulgare). The R and L in the motif are consistent in all of these proteins. The degradation of cyclin B1 was abolished when the motif was mutated as GxxVxx(L/I)xN in tobacco BY-2 cells, results that are analogous to those in vertebrates (Brandeis and Hunt, 1996; Genschik et al., 1998; Zur and Brandeis, 2002). Similarly, mutation at the D-box of OsRAA1 resulted in the suppression of proteasome-dependent programmed proteolysis (Fig. 8). Our results suggest that the D-box is required for the ubiquitin-mediated degradation of OsRAA1, which further suggests that OsRAA1 might be important for the transition from metaphase to anaphase of cell division.
Taken together, our results show that OsRAA1 modulates mitosis in rice, and this mitotic instability is required for the onset of anaphase, which is similar to the effect of the destruction of cyclins. In transgenic rice with overexpressed OsRAA1, the increased accumulation of OsRAA1 exceeded the regulation ability of the 26S proteasome and caused metaphase arrest and reduced growth of primary roots. In rice, OsRAA1 induced by auxin (Ge et al., 2004) interacts with RPT4, a regulatory subunit of the 26S proteasome, and functions in mitosis. RPT4 may regulate another gene's transcription by binding to the promoter (Gonzalez et al., 2002). So, in nature, its negative regulatory function in root mitosis may be balanced with other positive regulators to ensure a high fidelity in rate and quality control of anaphase in mitosis.
In this study, we have shown that OsRAA1 functions in the transition from metaphase to anaphase during cell division, thus having an important role in anaphase onset during mitosis in plants. The disruption of OsRAA1 mediated by the ubiquitin pathway was required for the onset of anaphase in mitosis during root development. OsRAA1 was degraded by the 26S proteasome through interaction with OsRPT4, and APC perhaps serves to recognize OsRAA1 as E3.
MATERIALS AND METHODS
Plant Materials and Yeast Strains
Rice (Oryza sativa japonica ‘Zhonghua 10’) and overexpressed OsRAA1 transgenic rice lines were described previously (Ge et al., 2004). A tobacco (Nicotiana tabacum ‘BY-2’) cell line (Nagata et al., 1992) was used for immunolocalization analysis. Yeast (Saccharomyces cerevisiae) strain AH109 was used for yeast two-hybrid screening. Yeast (Schizosaccharomyces pombe) strain SPQ-01 was used in phenotype analysis.
Two-Hybrid cDNA Library Construction and Screening
Yeast two-hybrid screening for proteins interacting with OsRAA1 involved the use of the Matchmaker system (Clontech). The ORF of OsRAA1 was amplified by PCR with the primers 5′-ATCGAATTCATGTCAGGGGTTTGGGTGTTCAAG-3′ and 5′-TATCTGCAGGTCAATTTAGGCGTCGACGACGCG-3′ and the restriction sites EcoRI and PstI and then constructed in frame into the BD cloning vector pGBKT7 to act as bait. pGBKT7-OsRAA1 was transformed into AH109 cells by the lithium acetate-mediated method. Total RNA of rice seedling roots was isolated by use of the Qiagen RNeasy plant mini kit, and the cDNA library was constructed and screened according to the manufacturer's instructions (Clontech). The yeast colonies were put on plates in medium-stringency medium in the absence of Trp, Leu, and His with 5 mm 3-aminotriazol and were tested on high-stringency medium SD/−Ade−His−Leu−Trp. Colonies showing a positive signal were subsequently examined by activating the lacZ reporter gene.
Protein Deletion Mutation and Yeast Plasmid Construction
The bait of OsRAA1 with the N or C terminus was generated by PCR and cloned into pGBKT7. The deletion of RPT4 was generated by PCR, and the fragments encoding amino acids 1 to 167, 168 to 313, and 314 to 400 were amplified and cloned into pGADT7.
Pull-Down Assay
GST and GST-OsRAA1 fusion protein were purified as described previously (Han et al., 2005). The beads binding GST or GST-OsRAA1 were incubated with His-RPT4 overnight at 4°C. The eluted solution was boiled with SDS sample buffer after being washed several times with 1× phosphate-buffered saline (PBS) and subjected to 12% SDS-PAGE. Western analysis of the resolved proteins involved use of antiserum of OsRAA1, anti-GST monoclonal antibodies (Pharmacia), and anti-RPT4. The antiserum of OsRAA1 was raised in a rabbit, with a synthetic 15-residue peptide (YYEDPSLFQFHKRGS) used as an antigen, as described previously (Han et al., 2005).
Coimmunoprecipitation and Tris-Tricine Gel Electrophoresis
Immunoprecipitation involved the use of the IP-50 kit (Sigma-Aldrich). Approximately 100 μg of protein extracts from roots was incubated for 8 h with constant rotation at 4°C with 3 μL of the antiserum of OsRAA1 or preimmune serum. At the end of the incubation, 50 μL of protein G-Sepharose was added to immunoprecipitate the proteins for another 8-h incubation at 4°C; the precipitate was washed five times with a solution of 0.5 m NaCl and 50 mm Tris-HCl (pH 8.0), and the proteins were loaded onto polyacrylamide gels of 12% T, 3% C and 16.5% T, 3% C. T denotes the total concentration of both monomers (acrylamide and bisacrylamide), and C denotes the concentration of the cross-linker relative to the total concentration T. After gel electrophoresis, proteins were assayed with the use of anti-RPT4 antibody or antiserum of OsRAA1.
Tricine-SDS-PAGE gel electrophoresis was performed as described (Schagger and von Jagow, 1987). Briefly, the stacking and separating gels were 4% T, 3% C and 16.5% T, 3% C, respectively. The anode buffer was 0.2 m Tris-HCl buffer (pH 8.9), and the cathode buffer was Tris-Tricine buffer (pH 8.25). Electrophoresis was performed at 40 V constant for approximately 6 h at room temperature.
Transient Subcellular Localization
The PCR product of the OsRAA1 gene was ligated with XbaI and KpnI double-digested pGFP221 to create pGFP-OsRAA1, in which the cDNA region covering an ORF of OsRAA1 was fused to the 5′ terminus of the GFP gene in frame, under the control of the cauliflower mosaic virus 35S promoter. Cells in the epidermal layers of onion (Allium cepa) bulbs were transformed by particle bombardment as described previously (Han et al., 2005), with gold particles (Bio-Rad) coated with pGFP-OsRAA1 or pGFP-221 as control plasmid DNA. Twenty-four hours after bombardment, epidermal peels were visualized by fluorescence microscopy (Zeiss).
BiFC
The interaction of OsRAA1 and OsRPT4 identified by BiFC was analyzed as described (Walter et al., 2004). The ORF excluding the termination code of OsRAA1 was amplified with the forward primer 5′-GGACTAGTATGTCAGGGGTTTGGGTG-3′ (SpeI site underlined) and the reverse primer 5′-CCATCGATGGCGTCGACGACGC-3′ (ClaI site underlined). The constructed vector was termed pUCSPYNE-OsRAA1. Correspondingly, OsRPT4 was generated by PCR with the primers 5′-CGGGATCCATGGCCGACGGG-3′ (BamHI site underlined) and 5′-GGGGTACCCTCTTTGCCGAAGTC-3′ (KpnI site underlined), subsequently constructed in pUCSPYCE, termed pUCSPYCE-OsRPT4. pUCSPYNE-OsRAA1 and pUCSPYCE-OsRPT4 were mixed at 1:1 with gold particles (Bio-Rad). Cells in the epidermal layers of Chinese cabbage (Brassica campestris pekinensis) were transformed by particle bombardment as described previously (Varagona et al., 1992), with pUCSPYNE-OsRAA1 and pUCSPYCE, pUCSPYNE and pUCSPYCE-OsRPT4, and pUCSPYNE and pUCSPYCE as controls. YFP fluorescence in Chinese cabbage epidermal cells was visualized with a Zeiss fluorescence microscope at 24 h after bombardment.
Immunofluorescence of Rice Root Tips
The seeds of wild-type and OsRAA1-overexpressed transgenic rice plants were generated and grown for 3 to 4 d at 28°C. In each treatment, 60 root tips of 1 to 2 mm were fixed in 4% paraformaldehyde in PEM buffer (50 mm PIPES, 5 mm EGTA, 0.1 mm EDTA, and 5 mm MgSO4, pH 6.9) for 1 h, rinsed three times with PEM buffer for 10 min each, followed by 1% cellulase and 0.25% pectolyase digestion at room temperature for 50 min. After being washed with PEM and twice with 1× PBS, cells were fixed on coverslips with 1% polylysine and squashed with the use of a pencil eraser between two coverslips, then rested at −20°C for 5 min and at room temperature for 2 min. The coverslips were blocked by 1% BSA for 20 min, then incubated with 1% Triton X-100 for 30 min and rinsed three times with 1× PBS for 10 min each. The primary antibody anti-β-tubulin and the secondary antibody anti-mouse IgG fluorescein isothiocyanate conjugate were applied at room temperature for 2 h, then cells were washed three times with 1× PBS. Cells were labeled with the DNA dye 4,6-diamidino-2-phenylindole (DAPI) at 0.25 μg mL−1 in 1× PBS for 5 min. The coverslips were then sealed with nail polish. Cells were viewed with a Zeiss microscope.
Colocalization of OsRAA1 and Tubulin in the BY-2 Cell Line
BY-2 cells were maintained in Murashige and Skoog (MS) medium containing 0.5 mg L−1 2,4-dichlorophenoxyacetic acid and 30 g L−1 Suc, pH 5.8. Cells were incubated at 28°C in a shaker set at 130 rpm and subcultured weekly. The construct p35S∷GFP or p35S∷OsRAA1-GFP within a binary vector (pBI121) was introduced into Agrobacterium tumefaciens (strain C58) by electroporation and used to transform BY-2 cells. Transfected BY-2 cells were transferred onto MS medium containing kanamycin (100 μg mL−1) and cefotaxim (200 μg mL−1) for 3 to 4 weeks until transformed colonies were formed. Resistant cell colonies were identified by GFP fluorescence with a fluorescence microscope, and more than five positive cell lines were further transferred into MS liquid medium containing the antibiotics mentioned to initiate suspension culture and used for subsequent analysis.
Coimmunolocalization of OsRAA1 and tubulin in transgenic BY-2 cell lines was as described (Li et al., 2006). Transgenic BY-2 cells were double labeled. The primary antibodies mouse anti-β-tubulin monoclonal antibody (Sigma) and rabbit anti-GFP polyclonal antibody were incubated together at 4°C overnight followed by three 10-min washes with PBS. The secondary antibodies TRITC-conjugated goat anti-mouse IgG (Jackson) shown in red and Alexa Fluor 488-conjugated goat anti-rabbit IgG (Molecular Probes) shown in green were added and incubated at 37°C for 2 h. Fluorescence images were collected with a Zeiss LSM 510 META confocal laser-scanning microscope with a 40× oil-immersion objective. Images were processed for publication by the use of Photoshop 7.0 (Adobe Systems).
Determination of the Half-Life of OsRAA1
Rice seedlings at 3 d old were treated with a solution of 100 μm CHX in the presence or absence of 100 μm MG132. Proteins were then extracted from the seedlings at 0, 5, 10, 15, 20, and 23 h followed by western-blot analysis. Probes were antibodies to OsRAA1, and the antibodies also cross-reacted with a smaller nonspecific band, which served as a loading control. Western blots were quantified by densitometry using the BioCapt program. OsRAA1 band intensity was normalized to the loading control band and then normalized to the time zero controls. Each decrease of 1 unit of log2 (band intensity) is equivalent to one half-life (Stommel and Wahl, 2004).
Protein Degradation Assay in Vitro
The procedure was as described (Yamamoto et al., 2004). The protein mixture of buds (150 μg) was extracted in TEMG buffer (50 mm Tris buffer [pH 7.5], 1 mm EDTA, 5 mm mercaptoethanol, 15% glycerol, and 400 mm NaCl) and incubated with 10 μg of ubiquitin (Sigma) and 1 mm ATP in the presence or absence of 100 μm MG132 or MG115 (Sigma) at 25°C for 8 h. The mixture was separated by 16.5% T, 3% C gel and subjected to western-blot analysis with the antiserum of OsRAA1 (1:200). For quantification of proteins, band intensity was detected by densitometry using the BioCapt program. The loading sample was normalized, and the relative level of OsRAA1 was the intensity ratio of the western band to the specific loading band.
Morphological Examination of Transformed S. pombe Cells
The ORF of OsRAA1 was constructed in a vector of pESPM with an NMT1 promoter, which was inducible with thiamine removal. The transformants were selected on plates containing minimal medium with thiamine at 30°C. Cells from the selected clones were grown to midexponential phase in minimal medium containing thiamine at 30°C, washed three times with the minimal medium without thiamine, and then incubated at 30°C for 22 h. Then, cells were examined with a light microscope. The transformant cells of pESPM plasmid were used as a control. Cells were fixed with methanol or formaldehyde and stained with DAPI dissolved in 1× PBS (1 μg mL−1) to visualize the DNA, then examined with a Zeiss microscope.
Protein Extraction from S. pombe and in Vitro Degradation Assay
Protein was extracted by the use of the Yeast Buster reagent (Novagen). The yeast cells were harvested from 100 mL of liquid culture (optical density at 600 nm up to 0.8) by centrifugation at 3,000g for 10 min at 4°C. Yeast Buster reagent and THP solution were added to the cell pellet and incubated on a shaking platform at 45°C for 40 min. After centrifugation at 16,000g for 20 min at 4°C, the supernatant was obtained as a protein mixture. The degradation assay of OsRAA1 in yeast followed that of rice as described above.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession number AY659938.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Alignment of RPT4 homologs.
Supplemental Figure S2. Expression pattern analysis of OsRPT4 by RT-PCR.
Supplemental Figure S3. RT-PCR analysis of the OsRAA1 transgenic yeast lines.
Supplementary Material
Acknowledgments
We thank Dr. Stephen Albert Johnston (Center for Biomedical Inventions, University of Texas-Southwestern Medical Center) for the generous gifts of antibodies against RPT4 and Dr. Jorg Kudla (Institut fur Botanik und Botanischer Garten, Universitat Munster) for the kind gifts of plasmids for BiFC assay.
This work was supported by the Major State Basic Research Program of the People's Republic of China (grant no. 2005CB120806) and the National Science Foundation of China for Distinguished Young Scholars (grant no. 30525026), as well as by innovation grants from the Chinese Academy of Sciences.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Kang Chong (chongk@ibcas.ac.cn).
The online version of this article contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Ambrose JC, Li W, Marcus A, Ma H, Cyr R (2005) A minus-end-directed kinesin with plus-end tracking protein activity is involved in spindle morphogenesis. Mol Biol Cell 16 1584–1592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blilou I, Frugier F, Folmer S, Serralbo O, Willemsen V, Wolkenfelt H, Eloy NB, Ferreira PC, Weisbeek P, Scheres B (2002) The Arabidopsis HOBBIT gene encodes a CDC27 homolog that links the plant cell cycle to progression of cell differentiation. Genes Dev 16 2566–2575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandeis M, Hunt T (1996) The proteolysis of mitotic cyclins in mammalian cells persists from the end of mitosis until the onset of S phase. EMBO J 15 5280–5289 [PMC free article] [PubMed] [Google Scholar]
- Cohen-Fix O, Peters JM, Kirschner MW, Koshland D (1996) Anaphase initiation in Saccharomyces cerevisiae is controlled by the APC-dependent degradation of the anaphase inhibitor Pds1p. Genes Dev 10 3081–3093 [DOI] [PubMed] [Google Scholar]
- Dawson SP, Arnold JE, Mayer NJ, Reynolds SE, Billett MA, Gordon C, Colleaux L, Kloetzel PM, Tanaka K, Mayer RJ (1995) Developmental changes of the 26S proteasome in abdominal intersegmental muscles of Manduca sexta during programmed cell death. J Biol Chem 270 1850–1858 [DOI] [PubMed] [Google Scholar]
- Di Laurenzio L, Wysocka-Diller J, Malamy JE, Pysh L, Helariutta Y, Freshour G, Hahn MG, Feldmann KA, Benfey PN (1996) The SCARECROW gene regulates an asymmetric cell division that is essential for generating the radial organization of the Arabidopsis root. Cell 86 423–433 [DOI] [PubMed] [Google Scholar]
- Evans T, Rosenthal ET, Youngblom J, Distel D, Hunt T (1983) Cyclin: a protein specified by maternal mRNA in sea urchin eggs that is destroyed at each cleavage division. Cell 33 389–396 [DOI] [PubMed] [Google Scholar]
- Fang G, Yu H, Kirschner MW (1998) Direct binding of CDC20 protein family members activates the anaphase-promoting complex in mitosis and G1. Mol Cell 2 163–171 [DOI] [PubMed] [Google Scholar]
- Ferrell K, Wilkinson CR, Dubiel W, Gordon C (2000) Regulatory subunit interactions of the 26S proteasome, a complex problem. Trends Biochem Sci 25 83–88 [DOI] [PubMed] [Google Scholar]
- Fleury D, Himanen K, Cnops G, Nelissen H, Boccardi TM, Maere S, Beemster GT, Neyt P, Anami S, Robles P, et al (2007) The Arabidopsis thaliana homolog of yeast BRE1 has a function in cell cycle regulation during early leaf and root growth. Plant Cell 19 417–432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu H, Doelling JH, Rubin DM, Vierstra RD (1999) Structural and functional analysis of the six regulatory particle triple-A ATPase subunits from the Arabidopsis 26S proteasome. Plant J 18 529–539 [DOI] [PubMed] [Google Scholar]
- Fu H, Reis N, Lee Y, Glickman MH, Vierstra RD (2001) Subunit interaction maps for the regulatory particle of the 26S proteasome and the COP9 signalosome. EMBO J 20 7096–7107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funabiki H, Yamano H, Kumada K, Nagao K, Hunt T, Yanagida M (1996) Cut2 proteolysis required for sister-chromatid separation in fission yeast. Nature 381 438–441 [DOI] [PubMed] [Google Scholar]
- Ge L, Chen H, Jiang JF, Zhao Y, Xu ML, Xu YY, Tan KH, Xu ZH, Chong K (2004) Overexpression of OsRAA1 causes pleiotropic phenotypes in transgenic rice plants, including altered leaf, flower, and root development and root response to gravity. Plant Physiol 135 1502–1513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genschik P, Criqui MC, Parmentier Y, Derevier A, Fleck J (1998) Cell cycle-dependent proteolysis in plants: identification of the destruction box pathway and metaphase arrest produced by the proteasome inhibitor mg132. Plant Cell 10 2063–2076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez F, Delahodde A, Kodadek T, Johnston SA (2002) Recruitment of a 19S proteasome subcomplex to an activated promoter. Science 296 548–550 [DOI] [PubMed] [Google Scholar]
- Hames RS, Wattam SL, Yamano H, Bacchieri R, Fry AM (2001) APC/C-mediated destruction of the centrosomal kinase Nek2A occurs in early mitosis and depends upon a cyclin A-type D-box. EMBO J 20 7117–7127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Y, Jiang JF, Liu HL, Ma QB, Xu WZ, Xu YY, Xu ZH, Chong K (2005) Overexpression of OsSIN, encoding a novel small protein, causes short internodes in Oryza sativa. Plant Sci 169 487–495 [Google Scholar]
- Han Y, Wang X, Jiang J, Xu Y, Xu Z, Chong K (2005) Biochemical character of the purified OsRAA1, a novel rice protein with GTP-binding activity, and its expression pattern in Oryza sativa. J Plant Physiol 162 1057–1063 [DOI] [PubMed] [Google Scholar]
- Hochstrasser M (1995) Ubiquitin, proteasomes, and the regulation of intracellular protein degradation. Curr Opin Cell Biol 7 215–223 [DOI] [PubMed] [Google Scholar]
- Hoyt MA (1997) Eliminating all obstacles: regulated proteolysis in the eukaryotic cell cycle. Cell 91 149–151 [DOI] [PubMed] [Google Scholar]
- Ichii M, Ishikawa M (1997) Genetic analysis of newly induced short-root mutants in rice (Oryza sativa L.). Breed Sci 47 121–125 [Google Scholar]
- Inukai Y, Miwa M, Nagato Y, Kitano H, Yamauchi A (2001) RRL1, RRL2 and CRL2 loci regulating root elongation in rice. Breed Sci 51 123–129 [Google Scholar]
- Inze D, De Veylder L (2006) Cell cycle regulation in plant development. Annu Rev Genet 40 77–105 [DOI] [PubMed] [Google Scholar]
- Jiang H, Wang S, Dang L, Wang S, Chen H, Wu Y, Jiang X, Wu P (2005) A novel short-root gene encodes a glucosamine-6-phosphate acetyltransferase required for maintaining normal root cell shape in rice. Plant Physiol 138 232–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juang YL, Huang J, Peters JM, McLaughlin ME, Tai CY, Pellman D (1997) APC-mediated proteolysis of Ase1 and the morphogenesis of the mitotic spindle. Science 275 1311–1314 [DOI] [PubMed] [Google Scholar]
- King RW, Deshaies RJ, Peters JM, Kirschner MW (1996) How proteolysis drives the cell cycle. Science 274 1652–1659 [DOI] [PubMed] [Google Scholar]
- Koepp DM, Harper JW, Elledge SJ (1999) How the cyclin became a cyclin: regulated proteolysis in the cell cycle. Cell 97 431–434 [DOI] [PubMed] [Google Scholar]
- Kominami K, DeMartino GN, Moomaw CR, Slaughter CA, Shimbara N, Fujimuro M, Yokosawa H, Hisamatsu H, Tanahashi N, Shimizu Y, et al (1995) Nin1p, a regulatory subunit of the 26S proteasome, is necessary for activation of Cdc28p kinase of Saccharomyces cerevisiae. EMBO J 14 3105–3115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YR, Liu B (2004) Cytoskeletal motors in Arabidopsis: sixty-one kinesins and seventeen myosins. Plant Physiol 136 3877–3883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li CL, Chen ZL, Yuan M (2006) Actomyosin is involved in the organization of the microtubule preprophase band in Arabidopsis suspension cultured cells. J Integr Plant Biol 48 53–61 [Google Scholar]
- Liu W, Xu ZH, Luo D, Xue HW (2003) Roles of OsCKI1, a rice casein kinase I, in root development and plant hormone sensitivity. Plant J 36 189–202 [DOI] [PubMed] [Google Scholar]
- McDonald HB, Byers B (1997) A proteasome cap subunit required for spindle pole body duplication in yeast. J Cell Biol 137 539–553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald HB, Helfant AH, Mahony EM, Khosla SK, Goetsch L (2002) Mutational analysis reveals a role for the C terminus of the proteasome subunit Rpt4p in spindle pole body duplication in Saccharomyces cerevisiae. Genetics 162 705–720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaelis C, Ciosk R, Nasmyth K (1997) Cohesins: chromosomal proteins that prevent premature separation of sister chromatids. Cell 91 35–45 [DOI] [PubMed] [Google Scholar]
- Moreno-Romero J, Carme Espunya M, Platara M, Arino J, Carmen Martinez M (2008) A role for protein kinase CK2 in plant development: evidence obtained using a dominant-negative mutant. Plant J 55 118–130 [DOI] [PubMed] [Google Scholar]
- Murray A (1995) Cyclin ubiquitination: the destructive end of mitosis. Cell 81 149–152 [DOI] [PubMed] [Google Scholar]
- Nagata T, Nemoto Y, Hasezawa S (1992) Tobacco BY-2 cell line as the ‘HeLa’ cell in the cell biology of higher plants. Int Rev Cytol 132 1–30 [Google Scholar]
- Paquette AJ, Benfey PN (2005) Maturation of the ground tissue of the root is regulated by gibberellin and SCARECROW and requires SHORT-ROOT. Plant Physiol 138 636–640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Perez JM, Serralbo O, Vanstraelen M, Gonzalez C, Criqui MC, Genschik P, Kondorosi E, Scheres B (2008) Specialization of CDC27 function in the Arabidopsis thaliana anaphase-promoting complex (APC/C). Plant J 53 78–89 [DOI] [PubMed] [Google Scholar]
- Peter M, Herskowitz I (1994) Joining the complex: cyclin-dependent kinase inhibitory proteins and the cell cycle. Cell 79 181–184 [DOI] [PubMed] [Google Scholar]
- Peters JM, Franke WW, Kleinschmidt JA (1994) Distinct 19 S and 20 S subcomplexes of the 26 S proteasome and their distribution in the nucleus and the cytoplasm. J Biol Chem 269 7709–7718 [PubMed] [Google Scholar]
- Plesse B, Fleck J, Genschik P (1998) The ubiquitin dependent proteolytic pathway and cell cycle control. In D Francis, D Oudits, D Inzé, eds, Plant Cell Division. Portland, Colchester, UK, pp 145–163
- Rechsteiner M, Hoffman L, Dubiel W (1993) The multicatalytic and 26 S proteases. J Biol Chem 268 6065–6068 [PubMed] [Google Scholar]
- Ren H, Santner A, del Pozo JC, Murray JA, Estelle M (2008) Degradation of the cyclin-dependent kinase inhibitor KRP1 is regulated by two different ubiquitin E3 ligases. Plant J 53 705–716 [DOI] [PubMed] [Google Scholar]
- Renaudin JP, Doonan JH, Freeman D, Hashimoto J, Hirt H, Inze D, Jacobs T, Kouchi H, Rouze P, Sauter M, et al (1996) Plant cyclins: a unified nomenclature for plant A-, B- and D-type cyclins based on sequence organization. Plant Mol Biol 32 1003–1018 [DOI] [PubMed] [Google Scholar]
- Renaudin JP, Savouré A, Philippe H, Van Montagu M, Inzé D, Rouzé P (1998) Characterization and classification of plant cyclin sequences related to A- and B-type cyclins. In D Francis, D Oudits, D Inzé, eds, Plant Cell Division. Portland, Colchester, UK, pp 97–98
- Russell SJ, Gonzalez F, Joshua-Tor L, Johnston SA (2001) Selective chemical inactivation of AAA proteins reveals distinct functions of proteasomal ATPases. Chem Biol 8 941–950 [DOI] [PubMed] [Google Scholar]
- Russell SJ, Reed SH, Huang W, Friedberg EC, Johnston SA (1999) The 19S regulatory complex of the proteasome functions independently of proteolysis in nucleotide excision repair. Mol Cell 3 687–695 [DOI] [PubMed] [Google Scholar]
- Russell SJ, Sathyanarayana UG, Johnston SA (1996) Isolation and characterization of SUG2: a novel ATPase family component of the yeast 26 S proteasome. J Biol Chem 271 32810–32817 [DOI] [PubMed] [Google Scholar]
- Schagger H, von Jagow G (1987) Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal Biochem 166 368–379 [DOI] [PubMed] [Google Scholar]
- Scheres B, Laurenzio L, Willemsen V, Hanser MT, Janmaat K, Weisbeek P, Benfey PN (1995) Mutations affecting the radical organization of the Arabidopsis root display specific defects throughout the radical axis. Development 121 53–62 [Google Scholar]
- Shibahara T, Kawasaki H, Hirano H (2002) Identification of the 19S regulatory particle subunits from the rice 26S proteasome. Eur J Biochem 269 1474–1483 [DOI] [PubMed] [Google Scholar]
- Smalle J, Kurepa J, Yang P, Babiychuk E, Kushnir S, Durski A, Vierstra RD (2002) Cytokinin growth responses in Arabidopsis involve the 26S proteasome subunit RPN12. Plant Cell 14 17–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smalle J, Kurepa J, Yang P, Emborg TJ, Babiychuk E, Kushnir S, Vierstra RD (2003) The pleiotropic role of the 26S proteasome subunit RPN10 in Arabidopsis growth and development supports a substrate-specific function in abscisic acid signaling. Plant Cell 15 965–980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stommel JM, Wahl GM (2004) Accelerated MDM2 auto-degradation induced by DNA-damage kinases is required for p53 activation. EMBO J 23 1547–1556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Townsley FM, Ruderman JV (1998) Proteolytic ratchets that control progression through mitosis. Trends Cell Biol 8 238–244 [DOI] [PubMed] [Google Scholar]
- Tyers M, Jorgensen P (2000) Proteolysis and the cell cycle: with this RING I do thee destroy. Curr Opin Genet Dev 10 54–64 [DOI] [PubMed] [Google Scholar]
- Ueda M, Matsui K, Ishiguro S, Sano R, Wada T, Paponov I, Palme K, Okada K (2004) The HALTED ROOT gene encoding the 26S proteasome subunit RPT2a is essential for the maintenance of Arabidopsis meristems. Development 131 2101–2111 [DOI] [PubMed] [Google Scholar]
- Varagona MJ, Schmidt RJ, Raikhel NV (1992) Nuclear localization signal(s) required for nuclear targeting of the maize regulatory protein Opaque-2. Plant Cell 4 1213–1227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter M, Chaban C, Schutze K, Batistic O, Weckermann K, Nake C, Blazevic D, Grefen C, Schumacher K, Oecking C, et al (2004) Visualization of protein interactions in living plant cells using bimolecular fluorescence complementation. Plant J 40 428–438 [DOI] [PubMed] [Google Scholar]
- Wang W, Chevray PM, Nathans D (1996) Mammalian Sug1 and c-Fos in the nuclear 26S proteasome. Proc Natl Acad Sci USA 93 8236–8240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Q, Guo HS, Dallman G, Fang S, Weissman AM, Chua NH (2002) SINAT5 promotes ubiquitin-related degradation of NAC1 to attenuate auxin signals. Nature 419 167–170 [DOI] [PubMed] [Google Scholar]
- Xu ML, Jiang JF, Ge L, Xu YY, Chen H, Zhao Y, Bi YR, Wen JQ, Chong K (2005) FPF1 transgene leads to altered flowering time and root development in rice. Plant Cell Rep 24 79–85 [DOI] [PubMed] [Google Scholar]
- Yamamoto T, Kimura S, Mori Y, Oka M, Ishibashi T, Yanagawa Y, Nara T, Nakagawa H, Hashimoto J, Sakaguchi K (2004) Degradation of proliferating cell nuclear antigen by 26S proteasome in rice (Oryza sativa L.). Planta 218 640–646 [DOI] [PubMed] [Google Scholar]
- Zur A, Brandeis M (2002) Timing of APC/C substrate degradation is determined by fzy/fzr specificity of destruction boxes. EMBO J 21 4500–4510 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.