Abstract
Salicylic acid (SA) is a primary factor responsible for exerting diverse immune responses in plants and is synthesized in response to attack by a wide range of pathogens. The Arabidopsis (Arabidopsis thaliana) sid2 mutant is defective in a SA biosynthetic pathway involving ISOCHORISMATE SYNTHASE1 (ICS1) and consequently contains reduced levels of SA. However, the sid2 mutant as well as ICS-suppressed tobacco (Nicotiana benthamiana) still accumulate a small but significant level of SA. These observations along with previous studies suggest that SA might also be synthesized by another pathway involving benzoic acid (BA). Here we isolated a benzoic acid hypersensitive1-Dominant (bah1-D) mutant that excessively accumulated SA after application of BA from activation-tagged lines. This mutant also accumulated higher levels of SA after inoculation with Pseudomonas syringae pv tomato DC3000. Analysis of the bah1-D sid2 double mutant suggested that the bah1-D mutation caused both ICS1-dependent and -independent accumulation. In addition, the bah1-D mutant showed SA-dependent localized cell death in response to P. syringae pv tomato DC3000. The T-DNA insertional mutation that caused the bah1-D phenotypes resulted in the suppression of expression of the NLA gene, which encodes a RING-type ubiquitin E3 ligase. These results suggest that BAH1/NLA plays crucial roles in the ubiquitination-mediated regulation of immune responses, including BA- and pathogen-induced SA accumulation, and control of cell death.
Plants sense contacts with microbial pathogens and exert diverse immune responses, including defense responses triggered by the recognition of pathogen-associated molecular patterns (PAMPs) or microbial-associated molecular patterns, hypersensitive responses (HRs) mediated by resistance (R) proteins, and subsequent systemic acquired resistance. Salicylic acid (SA) plays crucial roles as a signaling molecule in acquiring resistance. The accumulation of SA is induced after infection with both virulent and avirulent pathogens (Spoel et al., 2003). A recent report showed that a PAMP derived from a bacterial pathogen, Pseudomonas syringae pv tomato (Pst) DC3000, induces SA accumulation (Tsuda et al., 2008). Pathogens eluding the PAMP-triggered immune responses dispatch effector proteins to the insides of plant cells through a type III secretion system to acquire virulence effects. Some effectors, called avirulence proteins, are recognized by R proteins and lead to the HR. SA is involved in the regulation of HR including the oxidative burst and programmed cell death (Draper, 1997; Mur et al., 1997; Shirasu et al., 1997). Endogenous and exogenous SA can activate signal transduction pathways to induce systemic acquired resistance and the expression of defense-related genes, for example, PR1 (Durrant and Dong, 2004). SA-deficient plants are compromised in all of these immune responses and accordingly become hypersusceptible to both avirulent and virulent pathogens (Delaney et al., 1994; Mur et al., 1997; Nawrath and Métraux, 1999; Dewdney et al., 2000; Wildermuth et al., 2001; Tsuda et al., 2008). Moreover, emerging evidence suggests that some effectors target SA-mediated signaling and SA biosynthesis to disturb host immune responses (DebRoy et al., 2004; Jelenska et al., 2007). Given these findings, SA is a fundamental factor in plant-pathogen interactions and provides a backbone to immune responses.
SA is assumed to be synthesized through the Phe ammonia-lyase (PAL) pathway in which Phe is converted to benzoic acid (BA) by several enzymes including PAL and subsequently BA is converted to SA by a putative enzyme BA 2-hydroxylase (BA2H; Leon et al., 1993; Mauch-Mani and Slusarenko, 1996; Shirasu et al., 1997; Coquoz et al., 1998; Ferrari et al., 2003; Ogawa et al., 2005; Yaeno et al., 2006). However, at present there is no clear evidence that the PAL pathway is responsible for the regulation of SA biosynthesis because neither the gene encoding BA2H nor relevant mutants have been found in any plant species. An alternative pathway involving ISOCHORISMATE SYNTHASE1 (ICS1) was found in Arabidopsis (Arabidopsis thaliana; Wildermuth et al., 2001). A mutation in the ICS1 gene (the sid2 mutation) causes a substantial reduction of SA accumulation (Nawrath and Métraux, 1999). Although the ICS1 pathway is now thought to be the predominant pathway responsible for SA synthesis in immune responses, its regulatory mechanisms are poorly understood. A reasonable hypothesis is the so-called amplification loop model, in which SA induces the expression of the PAD4 gene and then the PAD4 protein activates SA synthesis after pathogen infection (Jirage et al., 1999). An issue is how the primary SA in the loop is synthesized. Furthermore, the sid2 mutant and ICS-suppressed tobacco (Nicotiana benthamiana) plants still accumulate small but significant levels of SA (Wildermuth et al., 2001; Catinot et al., 2008). Therefore, it is possible that the PAL pathway might be involved in the basal synthesis of SA for the amplification loop.
In this study, we isolated and characterized the benzoic acid hypersensitive1-Dominant (bah1-D) mutant, an activation-tagged line carrying a mutation in a RING-type ubiquitin E3 ligase, which accumulated excess amounts of SA. The bah1-D mutant was allelic to the nla mutant, which showed early senescence under low nitrogen condition (Peng et al., 2007). Our results suggest that the wild-type version of this gene may play a role as a negative regulator during both ICS1-independent and ICS1-mediated SA production in immune responses.
RESULTS
Isolation and Characterization of the bah1-D Mutant
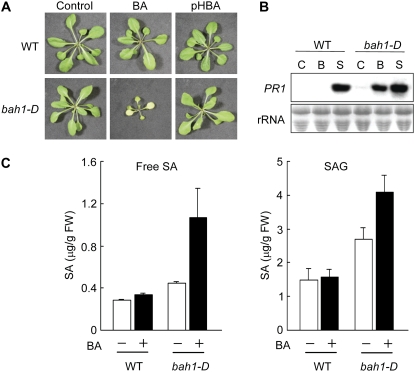
Several SA-accumulating mutants show the dwarf phenotype, and the reduction of SA in these plants can restore the wild-type phenotype (Bowling et al., 1997; Shah et al., 1999; Mauch et al., 2001), indicating that SA has an inhibitory effect on plant growth. In fact, the growth of wild-type Arabidopsis plants was inhibited when grown on Murashige and Skoog medium containing SA. On the other hand, BA, a putative precursor of SA, did not inhibit growth (Fig. 1A). We took advantage of this difference to screen for a mutant related to SA biosynthesis in which BA was involved. The screening strategy was based on the following assumptions. First, if the screening is performed using activation tagging, a dominant mutant, in which the biosynthesis of SA is activated, would be efficiently selected. Second, if the biosynthesis is activated, the mutant would overproduce SA when grown on a medium containing abundant BA. Therefore, we screened activation-tagged lines grown on a medium containing 50 μm BA for those showing growth inhibition. After screening approximately 20,000 seedlings, 121 such lines were selected and transplanted to normal medium to rescue them. In the subsequent screening of the next generation, 23 lines that grow normally on a medium without BA were selected and the most BA-sensitive mutant, bah1-D was identified (Fig. 1A). The growth of the bah1-D mutant was inhibited in the presence of BA, but it grew normally on the control medium. Although p-hydroxybenzoic acid, an isomer of SA, is similar in chemical formula to BA, the growth of the bah1-D mutant was not inhibited by it, suggesting that the growth inhibition effect is specific for BA.
Figure 1.
Sensitivity of the bah1-D mutant to BA. A, Growth of bah1-D and wild-type (WT) plants on normal medium (Control), or in the presence of 50 μm BA or p-hydroxybenzoic acid (pHBA). B, RNA gel-blot analysis of PR1 expression in bah1-D and wild-type plants grown on normal medium (C) or medium containing 50 μm BA (B) or SA (S). rRNA staining was used as a loading control. C, Endogenous levels of SAG in bah1-D and wild-type plants treated with 1 mm BA. SAG, SA glucoside.
To analyze expression of the SA-inducible PR1 gene, leaves were harvested from wild-type and bah1-D plants grown on a media containing 50 μm BA. As expected, PR1 transcripts accumulated in both the wild-type and the bah1-D plants grown on the SA-containing medium. On the other hand, PR1 transcripts accumulated in the bah1-D plants but not the wild-type plants grown on the BA-containing medium (Fig. 1B). These results suggested that the accumulation of SA may be activated in bah1-D plants grown in the presence of BA.
Free SA and conjugated SA levels were examined in leaves of 2-week-old seedlings that had been treated by spraying with 1 mm BA. The bah1-D plants showed slightly higher levels of free SA than wild-type plants without BA treatment (Fig. 1C). After BA treatment, the levels of free SA increased significantly in the bah1-D plants but only very slightly in the wild-type plants (Fig. 1C). This suggested that exogenously applied BA may infiltrate into the leaves of the bah1-D plants and then be converted to SA, or may stimulate the biosynthesis of SA. Both the basal level and the level after BA treatment of conjugated SA were significantly higher in the mutant than the wild-type plants (Fig. 1C). Possibly, basal SA biosynthesis might be enhanced in the bah1-D plants, resulting in the accumulation of conjugated SA, which is less toxic than free SA. This is supported by the observations that a small amount of PR1 transcript was detected in bah1-D plants grown on normal Murashige and Skoog medium, and a cDNA microarray analysis showed that PR1 expression was slightly up-regulated in bah1-D plants grown on normal medium (Fig. 1B; Supplemental Table S1).
The bah1-D Mutant Accumulates Higher Levels of SA in Response to Inoculation with Pst DC3000
The enhanced accumulation of basal SA in the bah1-D plants raises the possibility that basal disease resistance may be activated in the mutant. To study this possibility, the bah1-D plants were inoculated with Pst DC3000 strains and bacterial growth 3 d after inoculation was measured (Fig. 2A). Bacterial growth in the bah1-D plants inoculated with Pst DC3000 (avrRpm1) was almost the same as that in the wild-type plants, indicating that the bah1-D plants are as resistant to Pst DC3000 (avrRpm1) as the wild-type plants. On the other hand, bacterial growth in the bah1-D plants inoculated with Pst DC3000 was significantly reduced compared with that in the wild-type plants. This enhanced resistance to Pst DC3000 may be attributable to high basal accumulation of SA and/or to Pst DC3000-inducible accumulation of SA in the bah1-D plants. Therefore, we examined the levels of SA in the bah1-D plants after inoculation with Pst DC3000. As previously reported, the levels of both free and conjugated SA increased in the wild-type plants 3 d after inoculation with Pst DC3000 (Spoel et al., 2003). The levels of both forms of SA in the bah1-D plants were much higher than those in the wild-type plants, indicating that the bah1-D mutant was responsive to infection with Pst DC3000 (Fig. 2B). These results suggest that the enhanced resistance in the mutant is also conferred by an additional increase in SA levels after infection. The increase in free SA levels may result from a release from the conjugates. Alternatively, the free SA accumulated in the bah1-D plants may be derived from de novo synthesis because the conjugated SA also accumulated substantially in the bah1-D plants inoculated with Pst DC3000. Incidentally, the bah1-D plants accumulated higher levels of SA than the wild-type plants after inoculation with Pst DC3000 (avrRpm1), suggesting that the increased responsiveness to Pst was independent of R-mediated responses (Supplemental Fig. S1).
Figure 2.
Responses of the bah1-D mutant to Pst DC3000. A, Growth of Pst DC3000 strains 0 or 3 d after inoculation (106 cfu/mL) in bah1-D and wild-type plants. B, Endogenous levels of SAG in leaves of bah1-D and wild-type plants inoculated with Pst DC3000. Leaves were harvested 3 d after syringe inoculation with Pst DC3000 (106 cfu/mL) or syringe injection with 10 mm MgCl2 (Mock). dpi, Days postinoculation.
The bah1-D sid2 Double Mutant Still Accumulates SA in Response to BA and Pst DC3000
The accumulation of total SA was examined in bah1-D sid2 double-mutant plants. SA levels in bah1-D sid2 plants were much lower than those in the bah1-D plants after treatment with BA (Fig. 3A). Therefore, the high levels of SA in bah1-D plants may be synthesized by ICS1. However, detectable levels of total SA did accumulate in both sid2 and bah1-D sid2 plants, and the levels in the bah1-D sid2 plants were significantly higher than those in the sid2 plants (Fig. 3B). These results suggest that the majority of SA accumulation in the bah1-D plants is synthesized by the ICS1 pathway, but a residual amount of SA in both sid2 and bah1-D sid2 plants may be derived from an ICS1-independent pathway. Likewise, SA levels increased in both sid2 and bah1-D sid2 plants after inoculation with Pst DC3000 (Fig. 3C). This indicates that the sid2 mutant is still able to accumulate SA in response to Pst DC3000. SA levels in the bah1-D sid2 plants were higher than those in the sid2 plants after both mock inoculation and inoculation with Pst DC3000, suggesting that the bah1-D mutation causes basal and induced accumulation of SA independently of ICS1. While bah1-D plants accumulated approximately 3.6-fold higher levels of SA than the wild-type plants, bah1-D sid2 plants accumulated only approximately 1.5-fold higher levels of SA than sid2 plants (Figs. 2B and 3C). Therefore, the bah1-D mutation amplifies the accumulation of SA drastically if ICS1 exists. Taken together, BAH1 may be involved in inhibitory mechanisms underlying the ICS1-independent basal accumulation of SA and the ICS1-mediated amplification of SA.
Figure 3.
SA levels and the cell death phenotypes of bah1-D sid2 double mutants. A, Total SA levels in wild-type (WT), bah1-D, sid2, and bah1-D sid2 leaves treated with 1 mm BA. B, A more precise analysis of total SA levels in sid2 and bah1-D sid2 leaves treated with 1 mm BA. The asterisk indicates a significant difference between sid2 and bah1-D sid2 mutants by the Student's t test (*P < 0.03). C, Total SA levels in sid2 and bah1-D sid2 plants inoculated with Pst DC3000. Leaves were harvested 3 d after syringe inoculation with Pst DC3000 (106 cfu/mL) or mock inoculation (10 mm MgCl2). Asterisks indicate values significantly different (**, P < 0.02; ***, P < 0.01) from those of mock inoculation. D, Localized cell death occurred at the inoculation site (arrow) in bah1-D leaves. Photographs were taken 3 d after inoculation with Pst DC3000 (106 cfu/mL). E, Age-related cell death occurred in older leaves (shown on the right in each photograph of the top section) of bah1-D but not wild-type plants after bolting. The leaves on the left in each of the top photographs are comparatively young leaves without cell death. The leaves from the right in top photographs were stained with trypan blue (bottom section). Insets show stained cells at 200× magnification. F, Age-related cell death occurred in bah1-D and bah1-D sid2 plants, but not in wild-type and sid2 plants.
The bah1-D Mutation Causes SA-Dependent Pathogen-Induced Localized Cell Death and SA-Independent Age-Related Cell Death
Notably, localized cell death was observed in the bah1-D plants 2 d after inoculation with Pst DC3000 and resulted in a restriction of the spread of disease symptoms (Fig. 3D). On the other hand, characteristic chlorotic disease symptoms developed in the wild-type and the sid2 plants. The localized cell death was similar to, but occurred more slowly than, the R-gene-mediated HR cell death that occurred in response to Pst DC3000 (avrRpm1) inoculation (data not shown). In fact, the bah1-D mutant also accumulated higher levels of SA and was resistant to Pst DC3000 as if R-gene-mediated signals induced the resistance (Fig. 2). The localized cell death was abolished and disease symptoms developed in bah1-D sid2 plants, indicating that cell death required the ICS1-dependent accumulation of SA (Fig. 3D).
The bah1-D plants also exhibited spontaneous cell death in leaves of bolted plants, but not in leaves of young plants before bolting (Fig. 3E). Once the senescence-like response started, it did not stop until the whole leaf was withered. Clusters of dead cells were visualized by trypan blue staining of bah1-D leaves (Fig. 3E). The age-related cell death was not abolished in the bah1-D sid2 plants (Fig. 3F). SA accumulated in the dying leaves of bah1-D plants without BA treatment and in BA-treated leaves of bah1-D plants that were not undergoing cell death (Fig. 1C; data not shown). Therefore, BA-related biosynthesis may be activated in the bah1-D plants irrespective of cell death. These results suggest that BAH1 regulates pathogen-induced localized cell death and age-related cell death in SA-dependent and SA-independent manners, respectively.
The BAH1 Gene Encodes a Ubiquitin E3 Ligase with RING and SPX Domains
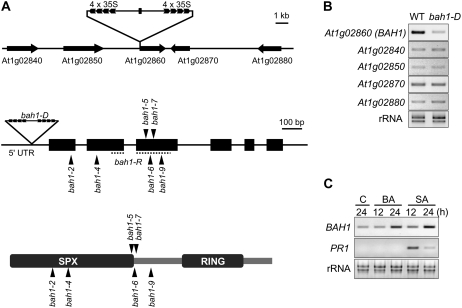
The sequences flanking the T-DNA insertion site were identified by plasmid rescue (Fig. 4A). The insertion consisted of at least two T-DNA fragments connected inversely, with 4× 35S elements at each end, and was located in the 5′-untranslated region of the At1g02860 gene. Unexpectedly, reverse transcription (RT)-PCR analysis revealed that the expression of this gene was suppressed in the bah1-D mutant (Fig. 4B). No other neighboring genes were affected by this insertion. These results suggested that the bah1-D mutation may be caused by suppression of the At1g02860 gene. To confirm this, we generated RNAi plants (named bah1-R) in which the At1g02860 gene was suppressed (Fig. 5). The bah1-R plants showed similar phenotypes to those of the bah1-D mutant, including growth inhibition in the presence of BA, accumulation of SA after treatment with BA, and age-related cell death. As a further confirmation, TILLING analysis was performed to screen for more bah1 alleles carrying point mutations. Mutant alleles with substituted amino acids in BAH1 were obtained and these plants showed bah1-like phenotypes (Fig. 4A; Supplemental Table S2; data not shown). The expression of the BAH1 gene was induced 24 h after application of BA and SA (Fig. 4C). SA accumulation coincided with suppressed expression of the BAH1 gene in the bah1-D mutant, suggesting that the BAH1 gene may play a role in SA-mediated negative feedback regulation of SA accumulation. BAH1 is predicted to contain a SPX (SYG1/Pho81/XPR1) domain and a RING-finger domain (Fig. 4A). The function of the SPX domain in plants is not yet known. The RING domain is a feature of the RING-type ubiquitin E3 ligases, but E3 ligase activity has not yet been confirmed in the BAH1 protein (Stone et al., 2005). Age-related cell death was also observed in bah1-4 plants, indicating that a mutation in the SPX domain can cause a malfunction of the BAH1 protein (Fig. 4A; data not shown). Recently, the nla mutant that is also defective in this gene was isolated (Peng et al., 2007). The nla mutation is caused by a deletion of the RING domain that is responsible for binding to the ubiquitin E2 conjugase UBC8. This suggests that the BAH1/NLA protein may function as an E3 ligase.
Figure 4.
BAH1 encodes a RING-type ubiquitin E3 ligase with an SPX domain. A, Physical map of the BAH1 locus, showing the T-DNA insertion site (top). Structure of the BAH1 gene showing the T-DNA insertion in the 5′ untranslated region (middle). The dashed line indicates the region used for RNAi. Arrowheads indicate bah1 alleles isolated by the TILLING method. Exons are shown as black rectangles. Scheme of the BAH1 protein, showing the putative SPX and RING domains, and sites of the bah1 mutations (bottom). SPX, SYG1/Pho81/XPR1. B, RT-PCR analysis of expression of the At1g02860 (BAH1), At1g02840, At1g02850, At1g02870, and At1g02880 genes in wild-type (WT) and bah1-D plants grown on normal Murashige and Skoog medium. rRNA staining was used as a loading control. C, Induction of BAH1 and PR1 expression in wild-type plants that were untreated (Control, C) or treated with 1 mm BA or SA. rRNA staining was used as a loading control. Samples were taken for RT-PCR analysis at 12 and 24 h after treatment.
Figure 5.
The knockdown of BAH1 by RNAi causes bah1-D-like phenotypes. A, Growth of bah1-D and BAH1-suppressed plants (bah1-R) was inhibited in the presence of 50 μm BA. B, Total SA levels in wild-type, bah1-D, and bah1-R plants treated with 1 mm BA. The asterisk indicates that the SA level in bah1-R plants was not significantly different from that in bah1-D plants by the Student's t test (*, P = 0.19). C, Age-related cell death occurred in bah1-D and bah1-R plants. D, RT-PCR analysis of BAH1 expression in wild-type, bah1-D, and bah1-R plants. rRNA staining was used as a loading control.
Nitrogen Limitation Induces the Accumulation of SA in Both Wild Type and bah1-D Mutant
The nla mutant shows age-related cell death under conditions of nitrogen limitation (Peng et al., 2007). As with the nla mutant, age-related cell death in the bah1-D and bah1-D sid2 plants was suppressed by supplying sufficient nitrate (Supplemental Fig. S2A). Cell death-associated SA accumulation was also suppressed in bah1-D plants supplied with nitrate (Fig. 6A). To examine the dose-dependent effects of nitrate on cell death and SA accumulation, wild-type and bah1-D plants were grown in vermiculite supplied with 1, 3, or 10 mm nitrate. Cell death was observed in bah1-D plants supplied with 1 mm nitrate at 28 d after germination but this was alleviated by 3 or 10 mm nitrate (Supplemental Fig. S2B). The accumulation of SA in bah1-D plants decreased with the increase in the supply of nitrate (Fig. 6B). The wild-type plants also accumulated SA in a nitrate-dependent manner even though there was no cell death (Fig. 6B; Supplemental Fig. S2B). These findings suggest that the regulation of SA accumulation may be closely related to mechanisms controlling the adaptation to nitrate concentrations in soil. Nitrogen limitation induces the accumulation of phenylpropanoids including BA (Fritz et al., 2006), and this was confirmed for both in the wild-type and bah1-D plants (Fig. 6). Unlike SA, the bah1-D plants accumulated less BA than the wild-type plants under lower nitrate conditions (Fig. 6B). However, the decrease in BA levels was not equivalent to the increase in SA levels in the bah1-D plants supplied with 1 mm nitrate. BA is possibly synthesized from a derivative of chorismate (Wildermuth, 2006; Strawn et al., 2007), and if so, the synthesis of BA may decrease due to activation of ICS1. Alternatively, trans-cinnamic acid may be used for the synthesis of other phenylpropanoids. In fact, genes involved in phenylpropanoid biosynthesis were up-regulated in the bah1-D mutant (Supplemental Table S1). BAH1/NLA may be involved in the overall regulation of SA, BA, and phenylpropanoid biosynthesis.
Figure 6.
SA and BA accumulation induced by nitrogen limitation. A, Total SA and BA levels in bah1-D and bah1-D sid2 plants grown on peat pellets supplied with or without 10 mm nitrate. B, Total SA and BA levels in wild-type (WT) and bah1-D plants grown on vermiculite irrigated with nutrient solution containing 1, 3, or 10 mm nitrate.
DISCUSSION
The levels of SA were substantially reduced by the sid2 mutation in Arabidopsis. However, detectable levels remained, and increased in the bah1-D sid2 double mutant after application of BA and after inoculation with Pst DC3000 (Fig. 3, B and C). These results demonstrate that the bah1-D mutation can cause the accumulation of SA independently of ICS1. Because the sid2 mutant has another functional ICS gene, ICS2, there is a possibility that ICS2 gene contributed to the accumulation of SA in the bah1-D sid2 mutant. According to the microarray analysis, ICS2 gene expression levels were almost the same in the bah1-D mutant and wild-type plants (Supplemental Table S1). Unlike ICS1, the expression of ICS2 gene is not induced after infection with the biotrophic fungus Erysiphe orontii (Wildermuth et al., 2001). Taken together, ICS2 induction may not be responsible for the accumulation of SA in defense responses or in the bah1-D mutant, but the ICS2 protein may be posttranscriptionally modulated to make a slight contribution to the accumulation of SA. More recently, Garcion et al. (2008) demonstrated the existence of an ICS-independent pathway using the mutant that lacks both the ICS1 and ICS2 genes.
An ICS-independent pathway is possibly activated by bah1-D mutation and may involve the BA2H. Actually, BA levels in the bah1-D mutant were less than those in the wild type (Fig. 6B), suggesting that some BA may be used for the synthesis of SA in the bah1-D mutant. Although the gene encoding BA2H has not yet been identified in any plant species, the BA2H protein has been studied in tobacco (Nicotiana tabacum). The tobacco BA2H is thought to be a 160-kD cytochrome P450 protein (Leon et al., 1995), whose activity is increased in tobacco leaves by the application of BA (Leon et al., 1993). Arabidopsis may also have a BA2H enzyme, however, the molecular masses of almost all the annotated P450 proteins in Arabidopsis are not more than 70 kD. Thus, a protein with BA2H activity in Arabidopsis may be considerably different in size from that in tobacco, or it may not be a P450 protein. Conceivably, the putative BA2H protein and some regulatory factors may be controlled by BAH1/NLA.
In the bah1-D sid2 double mutant, the bah1-D mutation raised both the basal and Pst DC3000-inducible levels of SA, despite the lack of ICS1 (Fig. 3). In the bah1-D mutant with a functional ICS1, SA accumulated at an accelerated pace (Figs. 2B and 3A). These results suggest that BAH1/NLA is involved in the negative regulation of both the basal and the Pst DC3000-inducible accumulation of SA. The bah1-D sid2 mutation may be equivalent to a runaway car with a gear-change failure. The bah1-D mutation may cause the continuation of signals promoting SA production, despite the lack of ICS1 in the bah1-D sid2 mutant, as if an accelerator is kept pressed down in a car that is stuck in first gear. SA can overaccumulate in the bah1-D mutant, due to the presence of a functional ICS1, like a car with the accelerator pressed down in top gear. This raises the possibility that the basal accumulation may control the ICS1-dependent pathogen-inducible accumulation. Similarly, a low level of SA activates PAD4 expression and PAD4 stimulates SA accumulation, which further induces PAD4 expression, resulting in an amplifying loop (Jirage et al., 1999). PAD4 positively regulates SA accumulation in response to Pst DC3000, but not to Pst DC3000 (avrRpm1) (Feys et al., 2001). On the other hand, BAH1/NLA is involved in the negative regulation of SA accumulation in response to both Pst DC3000 and Pst DC3000 (avrRpm1) (Fig. 2B; Supplemental Fig. S1). It might be necessary to study whether PAD4 is involved in SA accumulation in the bah1-D mutant, although the microarray data indicated that the expression of PAD4 was not affected in the aseptically grown bah1-D mutant (Supplemental Table S1).
It was reported recently that ICS1-dependent SA accumulation is induced by a PAMP from Pst DC3000 hrcC, which lacks the type III secretion system (Tsuda et al., 2008). SA accumulates excessively in the bah1-D mutant presumably in response to PAMPs from Pst DC3000. In addition, HR-like localized cell death may also occur in response to PAMPs in the bah1-D mutant inoculated with Pst DC3000. This cell death is suppressed by the sid2 mutation, suggesting that the PAMP-induced accumulation of SA is required for the induction of this HR-like cell death in the bah1-D mutant. Because SA per se cannot induce cell death, both the PAMP-induced SA accumulation and the bah1-D mutation might be required for the induction of this HR-like cell death. BAH1/NLA may play a role in the negative regulation of these PAMP-induced responses. To verify whether the bah1-D mutant is responsive to PAMPs, it may be necessary to study SA accumulation and cell death in the mutant plants inoculated with Pst DC3000 hrcC, or treated with a PAMP such as flg22.
Unlike the HR-like localized cell death, age-related cell death was not suppressed in the bah1-D sid2 mutant (Fig. 3F), suggesting that the induction of age-related cell death does not require SA. Even though SA levels in the bah1-D sid2 mutant were slightly higher than those in the sid2 mutant (Fig. 3B), it is unlikely that age-related cell death was affected by this difference, because it was not suppressed in the double mutant even though its SA levels were much lower than those in the bah1-D mutant (Fig. 3A). Therefore, the mechanisms leading to age-related cell death may be considerably different from those leading to HR-like cell death. When age-related cell death occurs in leaves, the regions in which cell death has yet to occur turn yellow simultaneously. Isochorismate is the major precursor for the biosynthesis of both SA and phylloquinone (Gross et al., 2006). The absence of phylloquinone, which can occur due to defects in ICS proteins, causes a reduction in PSI activity and a reduction in chlorophyll accumulation (Gross et al., 2006). The bah1-D mutant may run out of isochorismate to synthesize phylloquinone, resulting in a reduction in chlorophyll accumulation with increasing age. It is reasonable to assume that the age-related cell death was not be suppressed in the bah1-D sid2 mutant, because residual isochorismate may be depleted by the bah1-D mutation, while the synthesis of isochorismate is suppressed by the sid2 mutation. ICS2 contributes slightly to the supply of phylloquinone (Gross et al., 2006; Garcion et al., 2008). The slightly increased SA levels in the bah1-D sid2 mutant after application of BA or inoculation with Pst DC3000 suggest the possibility that the bah1-D mutation also activates ICS2 dependent, or possibly independently of both ICS proteins, SA accumulation in the double mutant (Fig. 3).
Leaf senescence is a crucial strategy for recycling the degradation products of chlorophyll as nitrogen nutrients under conditions of nitrogen starvation (Smart, 1994; Diaz et al., 2006). The depletion of isochorismate also results in the degradation of chlorophyll (Gross et al., 2006). The bah1-D mutation causes the accumulation of excessive levels of SA, resulting in the depletion of isochorismate, and cell death occurs as a consequence of senescence induced by nitrogen starvation (Fig. 6; Supplemental Fig. S2). In addition, SA regulates the expression of genes during senescence (Morris et al., 2000). Interestingly, SAG101, a protein encoded by a senescence-associated gene, acts together with EDS1, which is a partner of PAD4 in defense responses against pathogens (Feys et al., 2005). Thus, several lines of evidence suggest that SA is closely related to senescence, which controls the nitrogen nutrition status, and BAH1/NLA may play a role in the regulation of the ratio between the levels of isochorismate and SA under conditions of nitrogen starvation.
Rice (Oryza sativa) and Medicago truncatula have orthologs of BAH1/NLA (Supplemental Fig. S3). A reduction in nitrogen fertilizer application has been established as a traditional way to control disease in these agricultural plants. In line with this, we found that nitrogen limitation could induce the accumulation of SA even in wild-type Arabidopsis plants (Fig. 6B). Therefore, it will be important to elucidate the roles of BAH1/NLA orthologs in SA accumulation and disease resistance in various agricultural plants, and to understand the role that nitrogen plays in the action of these genes.
Similar to the bah1-D mutant, the npr1 mutant also accumulates higher levels of SA after inoculation with a pathogen (Delaney et al., 1995; Shah et al., 1997). NPR1 negatively regulates the expression of the ICS1 gene (Wildermuth et al., 2001). Since SA accumulation was largely dependent on the ICS1 protein, and expression of both ICS genes was not altered in the bah1-D mutant, BAH1/NLA might be involved in mechanisms regulating activation of the ICS enzymes rather than transcription of the ICS genes. Therefore, BAH1/NLA might not collaborate with NPR1 in transcriptional regulation even though both are localized in the nucleus (Kinkema et al., 2000; Peng et al., 2007). It is possible that expression of the BAH1/NLA gene is regulated via NPR1-mediated SA signaling.
If BAH1/NLA acts as a RING-type E3 ligase, the SPX domain might be a binding site for substrate proteins. In fact, the bah1-D phenotypes were also observed in the bah1-4 allele that has an amino acid change in the SPX domain, demonstrating the importance of this domain in the BAH1/NLA protein. Because the function of the SPX domain remains unclear, it will be crucial to identify proteins that bind to it to better understand the function of this unknown E3 ligase in plant immune responses.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
Arabidopsis (Arabidopsis thaliana) plants were grown on Jiffy-7 peat pellets (Jiffy Products International AS) under continuous light in a growth chamber at 23°C. Seeds of the Weigel activation-tagged collection were obtained from the Arabidopsis Biological Resource Center (Weigel et al., 2000). Col-7 accession was used as the wild type. For screening of BA sensitivity, plants were grown on Murashige and Skoog medium containing 1% Suc, 0.5% 2-morpholinoethanesulfonic acid, 0.4% gellan gum (Wako), and 50 μm BA. Additional bah1 alleles were obtained through the TILLING project (http://tilling.fhcrc.org/). For the experiments on nitrogen limitation, plants were grown on vermiculite irrigated with nutrient solution (1, 3, or 10 mm KNO3−, 10 mm KH2PO4, 2 mm MgSO4, 1 mm CaCl2, 0.1 mm Fe-EDTA, 50 μm H3BO4, 12 μm MnSO4, 1 μm ZnCl2, 1 μm CuSO4, 0.2 μm Na2MoO4; Peng et al., 2007).
Pathological Analyses
Leaves of 17-d-old plants were syringe inoculated with Pst DC3000 (empty vector or avrRpm1) at a titer of 106 colony forming units (cfu) per mL. Bacterial growth assays and trypan blue staining were carried out as described previously (Yaeno et al., 2004).
Measurement of SA and BA
Leaves of 1.5- to 2-week-old plants were harvested 1 week after treatment with 1 mm BA solution containing 0.01% Tween 20. Leaves of 17-d-old plants were harvested 3 d after syringe inoculation with Pst (106 cfu/mL). Approximately 0.2 g of tissue was used for analyses of the wild-type and bah1-D plants, and 1 g of tissue was used for analyses of the sid2 and bah1-D sid2 plants. Extraction and measurement of SA was carried out as described previously (Yaeno et al., 2006). For BA, methanol extracts were dried and then suspended in 5% trichloroacetic acid, then saponified with NaOH for 30 min at room temperature. After neutralization with HCl, BA was extracted with ethylacetate:cyclohexane (1:1, v/v). BA was detected by UV spectrophotometry at 290 nm using a photodiode array detector (SPD-M10A, Shimadzu).
Gene Expression Analyses
RNA was extracted from 2-week-old plants grown on Murashige and Skoog medium containing 50 μm BA or SA. RNA gel-blot analysis of the PR1 expression was performed as described previously (Yaeno et al., 2006). To detect the BAH1 transcripts in the wild-type and bah1-D plants, RT-PCR was performed. RNA was extracted from 17-d-old soil-grown plants treated with 1 mm BA or SA. Expression of the BAH1, At1g02840, At1g02850, At1g02870, At1g02880, and PR1 genes were monitored using the specific primer sets, 5′-AATGCCCAGTTTGTGATGGGAC-3′/5′-AAACACCGTGTCCAGGCATATTGAAC-3′, 5′-GCAAGATCTCAAGGATCACATGCG-3′/5′-CTTGAACGCGATCTTCTTGGAGAC-3′, 5′-GGCTTTTACCAAGTGGAAGAGGAC-3′/5′-TCTAGCTGTTGCCTTGTACTGTTG-3′, 5′-ATGGCGAGGTCGAGAAGAAAGTAC-3′/5′-TCGGTACATGCCCTCAATGTCATC-3′, 5′-TAAACTTGGGAACTAAGGTTATAGATG-3′/5′-CTAAGGTGTATGGTCTTGTATGG-3′, and 5′-GCTCTTGTAGGTGCTCTTGT-3′/5′-TTACACCTCACTTTGGCACA-3′, respectively.
Genetic and DNA Analysis of the bah1-D Mutant
The bah1-D mutant was backcrossed three times with the wild-type Col-7. The T-DNA contains the Basta resistance gene as a selectable marker. A homozygous mutant line was established by analyzing the segregation of Basta resistance and BA sensitivity. DNA gel-blot analysis along with the segregation analyses revealed that there was one T-DNA insertion in the bah1-D mutant. The restriction enzymes EcoRI and XhoI were used for plasmid rescue. Three independent plasmids were obtained and sequenced by primer walking.
RNAi Construction
For the RNAi construction, antisense and sense fragments were prepared using the primer sets, 5′-TCTAGACCAGATTCCTTCTTAGATTC-3′/5′-GGATCCTGCTCAGAAGCTTCTTGAGC-3′ and 5′-GGCGCGCCTGCTCAGAAGCTTCTTGAGC-3′/5′-GAGCTCCCAGATTCCTTCTTAGATTC-3′. These fragments were cleaved at prepared restriction sites and inserted along with a linker fragment and the 35S promoter into the vector pBIH101 as described previously (Yaeno et al., 2006; Yara et al., 2007).
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers NM_100167, NM_129450, NM_001057323, NM_001067141, and ABE85047.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Endogenous levels of free and conjugated SA (SAG) in leaves of bah1-D plants inoculated with Pst DC3000 (avrRpm1).
Supplemental Figure S2. Nitrogen supplementation suppresses age-related cell death.
Supplemental Figure S3. Alignment of the predicted amino acid sequences of BAH1/NLA orthologs from Arabidopsis, rice, and Medicago.
Supplemental Table S1. Expression of defense-related genes in the bah1-D mutant.
Supplemental Table S2. bah1 alleles.
Supplementary Material
Acknowledgments
We are grateful to Dr. Jeff Dangl and Dr. Ken Shirasu for useful advice and discussions on the manuscript.
This work was supported by Core Research for Evolutional Science and Technology of the Japan Science and Technology Agency and by the Japan Society of the Promotion of Science (grant no. 17370019).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Koh Iba (koibascb@mbox.nc.kyushu-u.ac.jp).
The online version of this article contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Bowling SA, Clarke JD, Liu Y, Klessig DF, Dong X (1997) The cpr5 mutant of Arabidopsis expresses both NPR1-dependent and NPR1-independent resistance. Plant Cell 9 1573–1584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catinot J, Buchala A, Abou-Mansour E, Métraux JP (2008) Salicylic acid production in response to biotic and abiotic stress depends on isochorismate in Nicotiana benthamiana. FEBS Lett 582 473–478 [DOI] [PubMed] [Google Scholar]
- Coquoz JL, Buchala A, Métraux JP (1998) The biosynthesis of salicylic acid in potato plants. Plant Physiol 117 1095–1101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DebRoy S, Thilmony R, Kwack YB, Nomura K, He SY (2004) A family of conserved bacterial effectors inhibits salicylic acid-mediated basal immunity and promotes disease necrosis in plants. Proc Natl Acad Sci USA 101 9927–9932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaney TP, Friedrich L, Ryals JA (1995) Arabidopsis signal transduction mutant defective in chemically and biologically induced disease resistance. Proc Natl Acad Sci USA 92 6602–6606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaney TP, Uknes S, Vernooij B, Friedrich L, Weymann K, Negrotto D, Gaffney T, Gut-Rella M, Kessmann H, Ward E, et al (1994) A central role of salicylic acid in plant disease resistance. Science 266 1247–1250 [DOI] [PubMed] [Google Scholar]
- Dewdney J, Reuber TL, Wildermuth MC, Devoto A, Cui J, Stutius LM, Drummond EP, Ausubel FM (2000) Three unique mutants of Arabidopsis identify eds loci required for limiting growth of a biotrophic fungal pathogen. Plant J 24 205–218 [DOI] [PubMed] [Google Scholar]
- Diaz C, Saliba-Colombani V, Loudet O, Belluomo P, Moreau L, Daniel-Vedele F, Morot-Gaudry JF, Masclaux-Daubresse C (2006) Leaf yellowing and anthocyanin accumulation are two genetically independent strategies in response to nitrogen limitation in Arabidopsis thaliana. Plant Cell Physiol 47 74–83 [DOI] [PubMed] [Google Scholar]
- Draper J (1997) Salicylate, superoxide synthesis and cell suicide in plant defence. Trends Plant Sci 2 162–165 [Google Scholar]
- Durrant WE, Dong X (2004) Systemic acquired resistance. Annu Rev Phytopathol 42 185–209 [DOI] [PubMed] [Google Scholar]
- Ferrari S, Plotnikova JM, De Lorenzo G, Ausubel FM (2003) Arabidopsis local resistance to Botrytis cinerea involves salicylic acid and camalexin and requires EDS4 and PAD2, but not SID2, EDS5 or PAD4. Plant J 35 193–205 [DOI] [PubMed] [Google Scholar]
- Feys BJ, Moisan LJ, Newman MA, Parker JE (2001) Direct interaction between the Arabidopsis disease resistance signaling proteins, EDS1 and PAD4. EMBO J 20 5400–5411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feys BJ, Wiermer M, Bhat RA, Moisan LJ, Medina-Escobar N, Neu C, Cabral A, Parker JE (2005) Arabidopsis SENESCENCE-ASSOCIATED GENE101 stabilizes and signals within an ENHANCED DISEASE SUSCEPTIBILITY1 complex in plant innate immunity. Plant Cell 17 2601–2613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritz C, Palacios-Rojas N, Feil R, Stitt M (2006) Regulation of secondary metabolism by the carbon-nitrogen status in tobacco: nitrate inhibits large sectors of phenylpropanoid metabolism. Plant J 46 533–548 [DOI] [PubMed] [Google Scholar]
- Garcion C, Lohmann A, Lamodière E, Catinot J, Buchala A, Doermann P, Métraux JP (2008) Characterization and biological function of the ISOCHORISMATE SYNTHASE2 gene of Arabidopsis. Plant Physiol 147 1279–1287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross J, Cho WK, Lezhneva L, Falk J, Krupinska K, Shinozaki K, Seki M, Herrmann RG, Meurer J (2006) A plant locus essential for phylloquinone (vitamin K1) biosynthesis originated from a fusion of four eubacterial genes. J Biol Chem 281 17189–17196 [DOI] [PubMed] [Google Scholar]
- Jelenska J, Yao N, Vinatzer BA, Wright CM, Brodsky JL, Greenberg JT (2007) A J domain virulence effector of Pseudomonas syringae remodels host chloroplasts and suppresses defenses. Curr Biol 17 499–508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jirage D, Tootle TL, Reuber TL, Frost LN, Feys BJ, Parker JE, Ausubel FM, Glazebrook J (1999) Arabidopsis thaliana PAD4 encodes a lipase-like gene that is important for salicylic acid signaling. Proc Natl Acad Sci USA 96 13583–13588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinkema M, Fan W, Dong X (2000) Nuclear localization of NPR1 is required for activation of PR gene expression. Plant Cell 12 2339–2350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leon J, Shulaev V, Yalpani N, Lawton MA, Raskin I (1995) Benzoic acid 2-hydroxylase, a soluble oxygenase from tobacco, catalyzes salicylic acid biosynthesis. Proc Natl Acad Sci USA 92 10413–10417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leon J, Yalpani N, Raskin I, Lawton MA (1993) Induction of benzoic acid 2-hydroxylase in virus-inoculated tobacco. Plant Physiol 103 323–328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mauch F, Mauch-Mani B, Gaille C, Kull B, Haas D, Reimmann C (2001) Manipulation of salicylate content in Arabidopsis thaliana by the expression of an engineered bacterial salicylate synthase. Plant J 25 67–77 [DOI] [PubMed] [Google Scholar]
- Mauch-Mani B, Slusarenko AJ (1996) Production of salicylic acid precursors is a major function of phenylalanine ammonia-lyase in the resistance of Arabidopsis to Peronospora parasitica. Plant Cell 8 203–212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris K, Mackerness SA, Page T, John CF, Murphy AM, Carr JP, Buchanan-Wollaston V (2000) Salicylic acid has a role in regulating gene expression during leaf senescence. Plant J 23 677–685 [DOI] [PubMed] [Google Scholar]
- Mur LA, Bi YM, Darby RM, Firek S, Draper J (1997) Compromising early salicylic acid accumulation delays the hypersensitive response and increases viral dispersal during lesion establishment in TMV-infected tobacco. Plant J 12 1113–1126 [DOI] [PubMed] [Google Scholar]
- Nawrath C, Métraux JP (1999) Salicylic acid induction-deficient mutants of Arabidopsis express PR-2 and PR-5 and accumulate high levels of camalexin after pathogen inoculation. Plant Cell 11 1393–1404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa D, Nakajima N, Sano T, Tamaoki M, Aono M, Kubo A, Kanna M, Ioki M, Kamada H, Saji H (2005) Salicylic acid accumulation under O3 exposure is regulated by ethylene in tobacco plants. Plant Cell Physiol 46 1062–1072 [DOI] [PubMed] [Google Scholar]
- Peng M, Hannam C, Gu H, Bi YM, Rothstein SJ (2007) A mutation in NLA, which encodes a RING-type ubiquitin ligase, disrupts the adaptability of Arabidopsis to nitrogen limitation. Plant J 50 320–337 [DOI] [PubMed] [Google Scholar]
- Shah J, Kachroo P, Klessig DF (1999) The Arabidopsis ssi1 mutation restores pathogenesis-related gene expression in npr1 plants and renders defensin gene expression salicylic acid dependent. Plant Cell 11 191–206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah J, Tsui F, Klessig DF (1997) Characterization of a salicylic acid-insensitive mutant (sai1) of Arabidopsis thaliana, identified in a selective screen utilizing the SA-inducible expression of the tms2 gene. Mol Plant Microbe Interact 10 69–78 [DOI] [PubMed] [Google Scholar]
- Shirasu K, Nakajima H, Rajasekhar VK, Dixon RA, Lamb C (1997) Salicylic acid potentiates an agonist-dependent gain control that amplifies pathogen signals in the activation of defense mechanisms. Plant Cell 9 261–270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smart CM (1994) Gene expression during leaf senescence. New Phytol 126 419–448 [DOI] [PubMed] [Google Scholar]
- Spoel SH, Koornneef A, Claessens SM, Korzelius JP, Van Pelt JA, Mueller MJ, Buchala AJ, Métraux JP, Brown R, Kazan K, et al (2003) NPR1 modulates cross-talk between salicylate- and jasmonate-dependent defense pathways through a novel function in the cytosol. Plant Cell 15 760–770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone SL, Hauksdottir H, Troy A, Herschleb J, Kraft E, Callis J (2005) Functional analysis of the RING-type ubiquitin ligase family of Arabidopsis. Plant Physiol 137 13–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strawn MA, Marr SK, Inoue K, Inada N, Zubieta C, Wildermuth MC (2007) Arabidopsis isochorismate synthase functional in pathogen-induced salicylate biosynthesis exhibits properties consistent with a role in diverse stress responses. J Biol Chem 282 5919–5933 [DOI] [PubMed] [Google Scholar]
- Tsuda K, Sato M, Glazebrook J, Cohen JD, Katagiri F (2008) Interplay between MAMP-triggered and SA-mediated defense responses. Plant J 53 763–775 [DOI] [PubMed] [Google Scholar]
- Weigel D, Ahn JH, Blazquez MA, Borevitz JO, Christensen SK, Fankhauser C, Ferrandiz C, Kardailsky I, Malancharuvil EJ, Neff MM, et al (2000) Activation tagging in Arabidopsis. Plant Physiol 122 1003–1013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wildermuth MC (2006) Variations on a theme: synthesis and modification of plant benzoic acids. Curr Opin Plant Biol 9 288–296 [DOI] [PubMed] [Google Scholar]
- Wildermuth MC, Dewdney J, Wu G, Ausubel FM (2001) Isochorismate synthase is required to synthesize salicylic acid for plant defence. Nature 414 562–565 [DOI] [PubMed] [Google Scholar]
- Yaeno T, Matsuda O, Iba K (2004) Role of chloroplast trienoic fatty acids in plant disease defense responses. Plant J 40 931–941 [DOI] [PubMed] [Google Scholar]
- Yaeno T, Saito B, Katsuki T, Iba K (2006) Ozone-induced expression of the Arabidopsis FAD7 gene requires salicylic acid, but not NPR1 and SID2. Plant Cell Physiol 47 355–362 [DOI] [PubMed] [Google Scholar]
- Yara A, Yaeno T, Hasegawa M, Seto H, Montillet JL, Kusumi K, Seo S, Iba K (2007) Disease resistance against Magnaporthe grisea is enhanced in transgenic rice with suppression of ω-3 fatty acid desaturases. Plant Cell Physiol 48 1263–1274 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.