Abstract
Resonance Raman spectra are reported for substrate-free and camphor-bound cytochrome P450cam and its isotopically labeled analogues that have been reconstituted with protoheme derivatives that bear -CD3 groups at the 1,3,5 and 8-positions (d12-protoheme) or deuterated methine carbons (d4-protoheme). In agreement with previous studies of this and similar enzymes, substrate binding induces changes in the high frequency and low frequency spectral regions, with the most dramatic effect in the low frequency region being activation of a new mode near 367 cm−1. This substrate-activated mode had been previously assigned as a second “propionate bending” mode (Chen, Z.; Ost, T. W. B.; Schelvis, J. P. M. Biochemistry 2004, 43, 1798−1808), arising in addition to the single propionate bending mode observed for the substrate-free form at 380 cm−1. In the present work, this newly activated mode is observed to shift by 8 cm−1 to lower frequency in the d12-protoheme reconstituted enzyme (i.e., the same shift as that observed for the higher frequency “propionate bending” mode) and is therefore consistent with the suggested assignment. However, the newly acquired data for the d4-protoheme substituted analogue also support an earlier alternate suggestion (Deng, T. J.; Proniewicz, L. M.; Kincaid, J. R.; Yeom, H.; Macdonald, I. D. G.; Sligar, S. G. Biochemistry 1999 38, 13699−13706) that substrate binding activates several heme out-of-plane modes, one of which ( γ6) is accidentally degenerate with the 367 cm−1 propionate bending mode. Finally, the study of the enzyme reconstituted with the protoheme-d4, which shifts the macrocycle ν10 mode, has now allowed a definitive identification of the vinyl C=C stretching modes.
Introduction
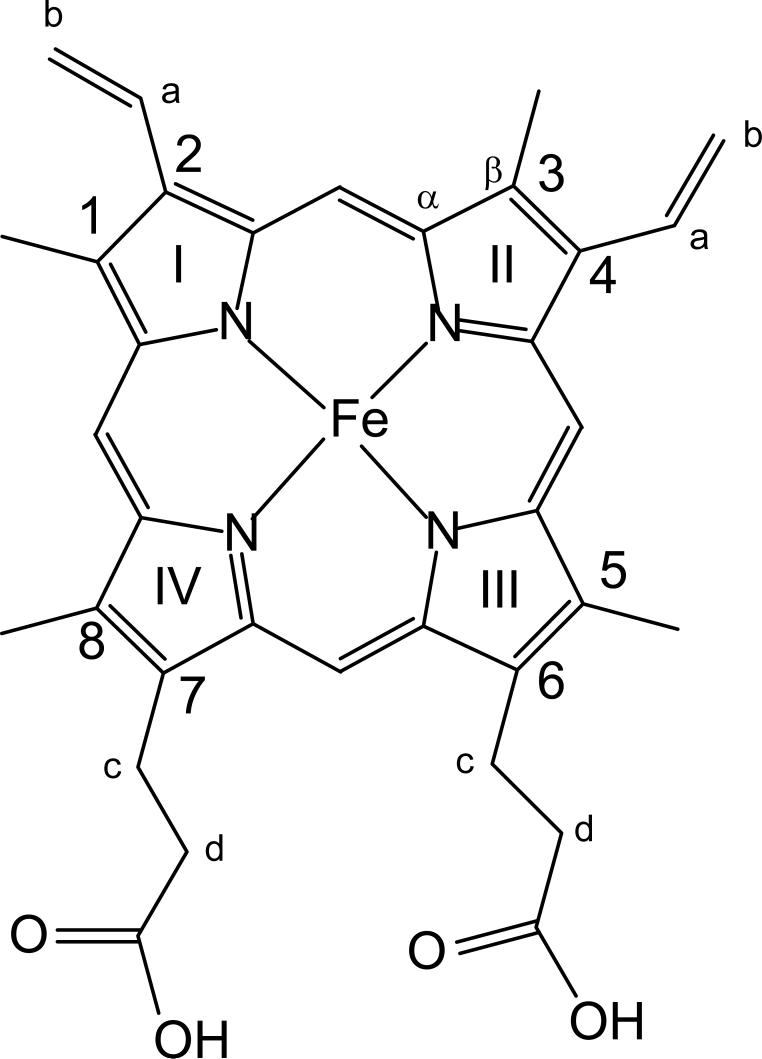
Resonance Raman spectroscopy has long been established as an exquisite probe of active site structure for a wide range of heme proteins and enzymes.1-3 Most applications have focused on the so-called marker bands in the high frequency region, which effectively document oxidation and spin-state of the central iron, in addition to providing a reliable indicator of heme core size.1,2 In recent years it has become increasingly apparent that low frequency modes in the RR spectra of heme proteins are quite sensitive to active site structure changes that can induce out-of-plane distortions of the macrocycle or alter the orientation of the heme peripheral substituents4-11 (see Figure 1 for protoheme labeling scheme).
Figure 1.
Structure and labeling scheme for protohemin IX
One of the most interesting and widely studied classes of heme proteins are the cytochromes P450, which bind and cleave molecular oxygen to generate a potent intermediate capable of hydroxylating even relatively inert hydrocarbon substrates.12 Within the catalytic cycle there are several important functional consequences of structural changes induced by binding of substrates or redox partners of these cytochrome P450 enzymes.2,3,12 Thus, considering cytochrome P450cam from Pseudomonas putida, for example, binding of camphor or other substrate analogue induces a change in spin state from a relatively planar 6-coordinate, low spin (6CLS) aquo complex to a 5-coordinate, high spin (5CHS) complex that is then susceptible to reduction by an appropriate electron donor, such as the natural putidaredoxin redox partner.12
In an earlier RR study of another bacterial P450, the so-called P450 BM3, distinctive changes were observed in the low frequency RR spectrum upon binding of natural substrate fatty acids; specifically, a new mode appeared near 365 cm−1.13 Based on the fact that the known conversion from LS to HS upon substrate binding is expected to cause out-of-plane distortions of the heme macrocycle, this newly activated mode was tentatively assigned to γ6, an out-of-plane heme macrocycle mode known to be activated upon non-planar distortions of the heme macrocycle.5,7,11,14 In two later studies, similar behavior was also demonstrated for cytochrome P450cam upon binding of camphor.15, 16 While the report by Reipa and coworkers16 concurred with the initial assignment13 of the new 367 cm−1 mode to γ6, in the work by Zhang, and in another independent work,17 the possibility was considered that this substrate-sensitive low frequency mode might be attributable to changes in orientation of the peripheral substituents (e.g., the 6,7-propionate groups) and preliminary efforts in our laboratory,15 employing specifically labeled cytochrome P450cam, were made to further elucidate the origin of this substrate-activated mode. In following up on these initial studies we here report RR spectral data for isotopically labeled derivatives of cytochrome P450cam, which contain either of two deuterated protohemes; d12-protoheme, which bears four -CD3 groups on the heme periphery, a substitution that induces shifts of propionate bending modes, and d4-protoheme, which contains deuterium atoms at the four methine positions inducing substantial shifts in low-frequency out-of-plane heme modes. Interestingly, the newly acquired data for these two labeled analogues provide evidence in support of both of the previously suggested assignments; i.e., binding of substrate definitely gives rise to a new propionate bending mode near 367 cm−1, as suggested by Schelvis and coworkers,17 but also activates the accidently degenerate γ6 out-of-plane mode, along with several other out-of plane modes.13,16
Experimental
Adequate supplies of the enzyme were obtained by expression and purification of cytochrome P450cam (C334A) from bacteria according to established procedures.18 A measured purity index, RZ value,19 of 1.6 was indicative of high purity enzyme. The d12-protoheme was prepared by base catalyzed exchange of the 1,3,5,8-methyl groups (Figure 1) of natural protoheme in perdeuterated dimethyl sulfoxide by methods described previously9,20 and d4-protoheme by using procedures published previously.8,21
The labeled cytochrome P450cam (d4- or d12- P450cam) was obtained from reconstitution of the apoprotein using methods similar to those originally reported by Wagner and Gunsalus.22, 23 Specifically, the apoprotein was generated by butanone extraction of an acidic solution of the enzyme. The separated aqueous protein layer was dialyzed in 0.01 % (w/v) solution of bicarbonate and the precipitated apoprotein then dissolved in a histidine buffer, pH 8.0, containing 20% glycerol and 0.5 mM camphor. This solution was treated with 1.2 equivalents of labeled protoheme dissolved in a minimal amount (∼1 ml) of 0.10 M KOH. The mixture was allowed to equilibrate under an inert atmosphere, while periodically checking the preparation for the presence of the reconstituted enzyme; this was accomplished by taking a small aliquot of the mixture (approximately 10 μL), adding it to a solution containing CO-saturated buffer and treating it with excess dithionite so as to generate the CO adduct of cytochrome P450cam. When the characteristic absorbance at 446 nm had maximized (typically about 50 hours) the reconstitution was considered to be complete. The excess heme was removed by passage over a Sephdex G25 column, equilibrated with 50 mM phosphate buffer pH 7.0 containing 1 mM camphor. The eluted protein was then purified on a DEAE-52 column equilibrated with 20 mM phosphate buffer pH 7.0 by eluting with a gradient of 50−200 mM KCl; a total volume of 100 mL of buffer was used at 4 mL/hr flow rate. Fractions with Rz > 1.4 were combined and concentrated. The solution of purified protein was stored in liquid nitrogen. To prepare the substrate-free forms of the native P450cam, a solution of the camphor-bound form was passed through a Sephadex G25 column equilibrated with 50 mM MOPS (morpholino-propane sulfonic acid), pH 7.4.23 The eluent was checked by UV-Vis spectrophotometry; for substrate free P450cam, a symmetrical peak at 416 nm is expected. The process was repeated if there were still traces of the 390 nm absorption peak, as determined by UV-Vis spectrophotometry. The MOPS was removed by passing the protein solution over a separate Sephadex G25 column equilibrated with 20 mM phosphate buffer (pH 7.4), containing 100 mM KCl (without camphor). The substrate-free sample was then concentrated to 0.2 mM by ultrafiltration using Centricon PM30 devices. The samples of substrate free P450's were used for Raman measurement within approximately 1 hr after camphor removal.
Resonance Raman spectra were obtained using a Spex 1269 spectrometer equipped with Andor Newton EMCCD detector (Model DU971, Andor Technologies). The excitation lines employed were the 406.7 nm line from a Kr+ laser (Coherent Innova 100). Fenchone was used to calibrate all spectra, which were processed with Grams 32/AI (Galactic Industries, Salem, NH). Rayleigh scattering was removed by use of an appropriate Notch filter from Kaiser Optical. The power at the sample was approximately 15 mW. The NMR tube containing the sample was spun and the RR spectra were collected at room temperature using 180° geometry in combination with a cylindrical lens which focuses the laser beam on the sample as a line image24,25 to avoid local heating. The spectra were accumulated for 30 min for low frequency region and 10 min for the high frequency region, the spectral resolution was about 5 cm−1. The UV-Vis spectra of the reduced, CO-forms, were taken before and after measurements to confirm the integrity of the sample. UV-Vis spectra were obtained using a Hewlett-Packard Model 8452 Diode Array Spectrophotometer.
The low frequency spectra, somewhat congested with a relatively large number of overlapping bands, were analyzed with Grams 32/AI software in an attempt to extract reliable vibrational parameters. This was done by imposing certain restrictions, as described here. First, in fitting the three traces (natural abundance and two deuterated samples), the same number of bands were included in each trace; i.e., it was not necessary to postulate the appearance or disappearance of any band upon deuteration. The bandwidths (assuming a 50/50 % Lorentzian/Gaussian function)26 employed here were held to within 10−13 cm−1 for the native and were only allowed to increase slightly for the deuterated analogues; i.e., for bands that experience moderate (5−15 cm−1) shifts the bandwidth was allowed to increase by 1 cm−1, while those with relatively large isotope shifts (> 15 cm−1) were allowed to slightly expand by 2 cm−1, modest increases that effectively account for incomplete deuteration (e.g., 90−95%). In general, efforts were made to prevent large changes in intensities in proceeding from one isotopomer to another; e.g., the intensities of the three vinyl modes between 400 and 430 cm−1 remain approximately the same, as do the relative intensities of the ν8 and ν50 modes and the propionate bending modes seen near 365−380 cm−1.
Results and Discussion
Sample Integrity
In the present work P450cam was reconstituted with d12-protoheme and d4-protoheme. The d12-protoheme was prepared according to the method originally reported by Goff and coworkers,20 while d4-protoheme was obtained using well know procedures employing CH3OD as the deuterium source21 and as used previously in our laboratory.7,8,27,28 The effective deuteration at all four methyl groups in d12- and all four methine protons in d4-protohemin was checked by comparison of the 1H NMR spectrum of the bis-cyano derivative of the deuterated analogue and the natural abundance starting material, as reported earlier.9,29 There is no significant exchange at the 6,7-d-CH2 groups in d12-protoheme; thus, these peaks are used as a reference in calculating the extent of deuteration of the methyl groups. For d12-protoheme the deuteration of 1,3 methyl group was complete (> 98 %) and the exchange of the 5- and 8-CH3 groups was calculated to be about 90 %, while the extent of methine deuteration in d4-protoheme was about 95 %. The chemical integrity of the deuterated protohemin samples was further confirmed by TLC30,31 and comparison of the pyridine hemochromogen spectra of natural hemin with those of the d12- and d4-protohemin samples;30,32 these are virtually identical, with the Soret, β and α bands being observed at 418 nm, 524 nm and 556 nm.30,32 It is also expected that the ratio of αmaximum/ βmaximum is equal to 1.97 (the value obtained in this work was 1.98).30,32 Both the native and reconstituted protein samples used in this work were checked for the presence of P420 form by measuring the spectra of the CO-saturated reduced forms and in all cases were found to contain less than 10% contribution from the P420 form.
For the Resonance Raman studies conducted here, substrate-free samples were required. The removal of substrate was monitored by the disappearance of the 646 nm band, as well as the shifting of the Soret band by 24 nm, from 392 nm for the camphor-bound form to 416 nm for the substrate-free form, reflecting the spin state changes from 5CHS to 6CLS after removal of substrate.
Resonance Raman Spectra
High Frequency Region
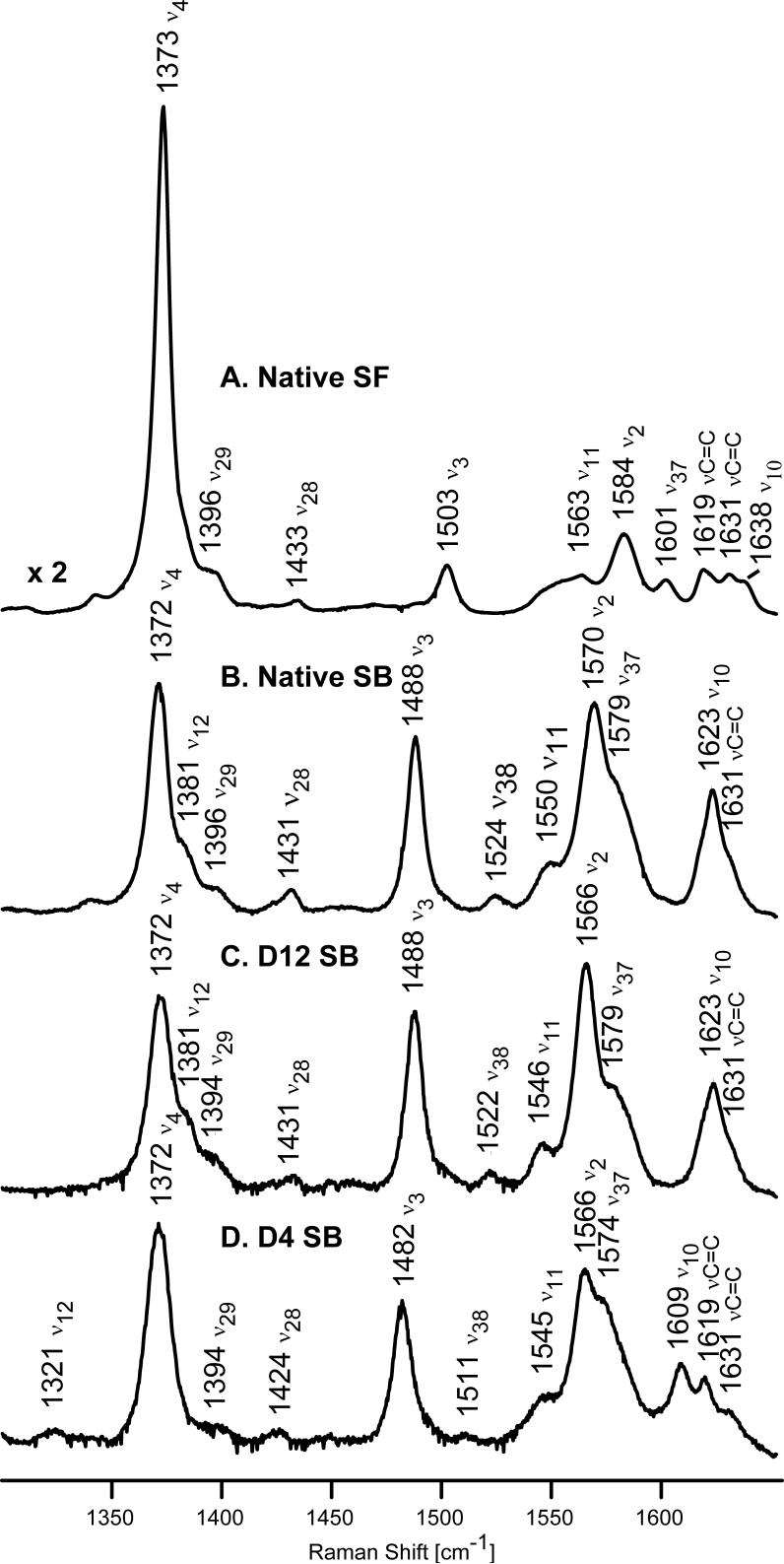
The present work represents the first reported resonance Raman study of deuterated analogues of a cytochrome P450 and provides a new opportunity to clarify RR spectral response to substrate binding, not only in the low frequency region (vide infra), but also in the high frequency region, where rather substantial isotopic shifts effectively eliminate annoying overlaps in the spectrally crowded region between 1600−1640 cm−1. Thus, the high frequency spectra of the substrate-free and substrate-bound native forms of P450cam (traces A and B) are shown in Figure 2 along with those of the substrate-bound forms of d12-P450cam and d4-P450cam (traces C and D, respectively). The modes in this region are sensitive to oxidation and spin state changes or alterations to heme core size. For the substrate-free and camphor-bound forms, the oxidation state marker, ν4, appears at 1372−1373 cm−1 for both the native and reconstituted proteins, as expected for a ferric oxidation state. As had been documented earlier by us and others,3,13,16,33 the core size marker bands, ν3, ν2 and ν10, which are sensitive to spin state and coordination number, exhibit anticipated changes upon binding of substrate. Thus, for the substrate-free form of the native protein (Figure 2, trace A), these bands appear at 1503, 1584 and 1638 cm−1. The two vinyl ν(C=C) stretching modes appearing at 1619 cm−1 and 1631 cm−1 confirm the presence of at least two distinct vinyl conformers in the substrate-free form. Specifically, based on the elegant analysis by Smulevich and coworkers34 and later extended by others,35 which defines the relationship between ν(C=C) vinyl stretching modes and the orientation of vinyl groups with respect to the pyrrole ring, the 1619 cm−1 mode implies a torsional angle of between |τ| = 100−180° or |τ| = 0−10° degrees, while that at 1631 cm−1 corresponds to values between |τ| = 20−90°.
Figure 2.
High frequency resonance Raman spectra of native P450cam substrate-free (A) and substrate-bound (B); and substrate-bound deuterated analogues of P450cam, d12-P450cam (C) and d4-P450cam (D). Excitation line 406.7 nm, enzyme concentration 50 μM.
Consistent with previous reports,3,13,16,33 upon binding of camphor, the ν3 and ν2 marker modes are observed at 1488 and 1570 cm−1, this behavior being indicative of a five coordinate, high spin complex (5CHS) arising in response to a water molecule being displaced by binding of substrate. The unresolved envelope of three overlapping bands appearing in the region of 1620−1635 cm−1 is attributable to the ν10 spin state marker and the two vinyl stretches mentioned above. The maximum of this envelope at 1623 cm−1 is reasonably assigned to the ν10 mode, based on the 15 cm−1 downshift from its position in the spectrum of substrate-free P450cam. A similar downshift of this ν10 mode was previously observed for the low- to high-spin transition in P450cam3,33 consistent with present results.
The deuteration of all four methyl groups (Figure 2, trace C) causes shifts of the ν2 marker mode to lower frequency by 4 cm−1, a shift that is not unexpected inasmuch as reliable normal mode calculations36-38 indicate this mode contains major contributions of Cβ-Cβ stretch, and the labeled (CD3) groups are attached to the Cβ carbons. The only other high frequency mode that shows sensitivity to the methyl group deuteration is the ν11 mode observed at 1550 cm−1 in the spectrum of native substrate-bound (Figure 2, trace B), which shifts to 1546 cm−1 in the spectrum of d12-P450cam; it is noted that this mode also contains substantial contributions from the Cβ-Cβ stretch.36-38 On the other hand, deuteration of methine protons causes more significant changes in the spectra (Figure 2, trace D). Now all the spin state markers are shifted to lower frequency, results consistent with the RR study of model compounds and their deuterated analogues2,36-37 as well as deuterated myoglobins;5 this shift of the ν10 mode from 1623 cm−1 (trace B) to 1609 cm−1 (trace D) reveals the precise frequency of the lower frequency ν(C=C) vinyl stretch to be 1619 cm−1, unshifted from it's position in the substrate-free form (trace A).
Low Frequency Region
The low frequency region is particularly useful in providing information about the interaction between heme vinyl and propionate groups with the protein environment.4-11,13,16,17 In other heme proteins, such as myoglobin,9 hemoglobin39,40 and its isolated subunits,27 the modes traditionally associated with the propionate and vinyl groups exhibited shifts ranging from 9 to 13 cm−1 upon reconstitution of these proteins with d12-protoheme. As mentioned above, a new mode appears near 367 cm−1 upon substrate binding to cytochrome P450 BM313,17 and cytochrome P450cam.15,16 This mode was originally attributed to an out of plane γ6 mode,13 with the initial assignment being supported by Riepa and coworkers in their carefully conducted studies of P450cam,16 but questioned by Zhang15 and also Schelvis and coworkers,17 who suggested that this mode is more reasonably associated with substrate-induced activation of a “second” propionate mode. To investigate this issue further, the present work employs both d12-protohemeand d4-protoheme-reconstituted cytochrome P450cam in order to extend the earlier preliminary work completed in this laboratory by Zhang.15
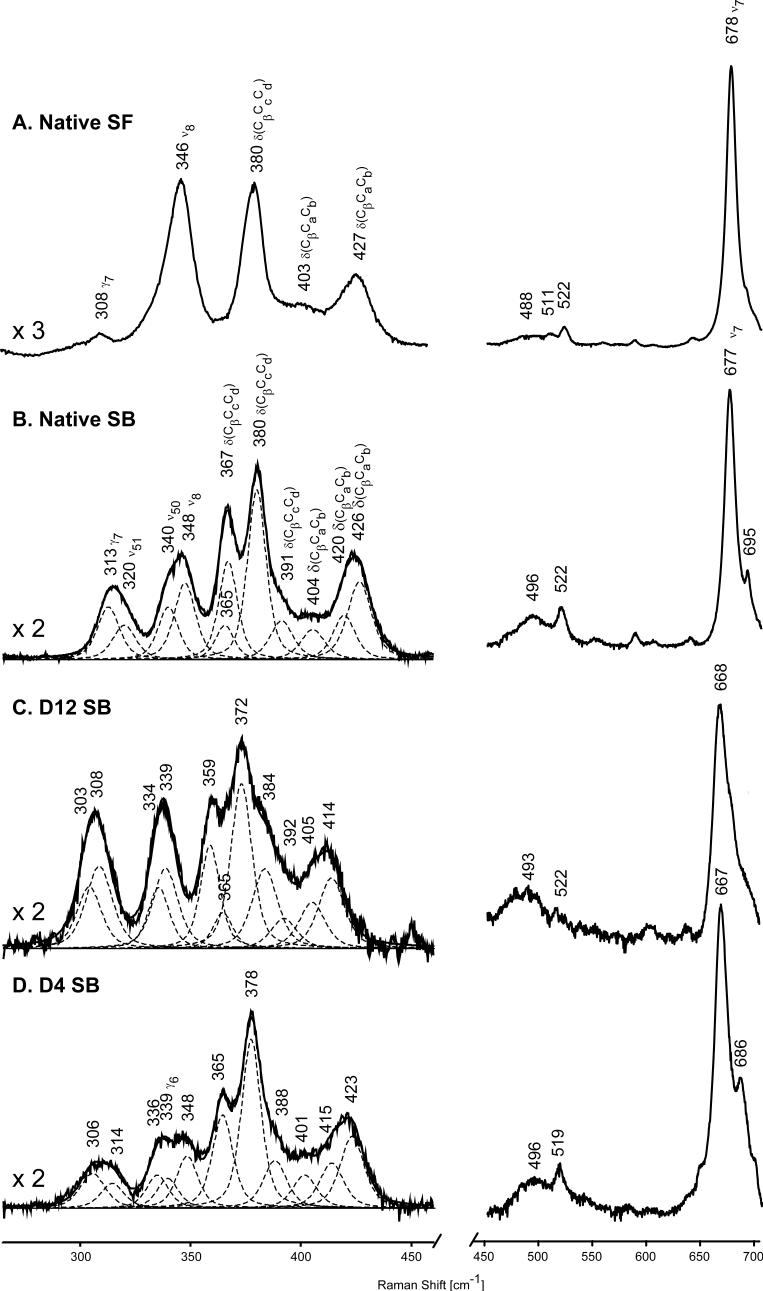
The low frequency spectra of the relevant cytochrome P450cam samples are shown in Figure 3, noting that for the substrate-bound samples of interest here, both the raw spectra and the bands used in simulating the spectral traces are given, with the procedure and restrictions used to deconvolute the traces being summarized in the Experimental section. It is noted that, in agreement with the discussion below, three component bands were included in this region to account for slight asymmetry in the band near 425 cm−1 and to restrict bandwidths to < 13 cm−1.The spectral parameters extracted from this procedure are presented in Table I. Also, it is noted here that the alternative deconvolution scheme presented in Supporting Information reproduces the raw spectra, but violates some of these restrictions.
Figure 3.
Low frequency resonance Raman spectra of native P450cam substrate-free (A) and substrate-bound (B); and substrate-bound deuterated analogues of P450cam, d12-P450cam (C) and d4-P450cam (D). Excitation line 406.7 nm, enzyme concentration 50 μM.
Table I.
The frequencies, bandwidths and isotope shifts of extracted resonance Raman bands of substrate-bound P450cam and its isotopicaly substituted analogues.
| positions of RR bands [band width] for SB P450cam in cm−1 |
isotopic shifts for d12 /d4 analogues in cm−1 | |||
|---|---|---|---|---|
| native | d12 | d4 | ||
| γ7 |
313 [12] |
308 [13] |
306 [13] |
5 / 7 |
| ν51 |
320 [12] |
303 [14] |
314 [13] |
17 / 6 |
| ν50 |
340 [11] |
334 [12] |
336 [11] |
6 / 4 |
| ν8 |
348 [11] |
339[12] |
348 [11] |
9 / 0 |
| γ6 |
365 [10] |
365 [10] |
339 [12] |
0 / 26 |
| δ(CβCcCd) |
367 [10] |
359 [11] |
365 [10] |
8 / 2 |
| δ(CβCcCd) |
380 [10] |
372 [11] |
378 [10] |
8 / 2 |
| δ(CβCcCd)a |
391 [12] |
384 [13] |
388 [12] |
7 / 3 |
| δ(CβCaCb) |
404 [13] |
392 [14] |
401 [13] |
12 / 3 |
| δ(CβCaCb) |
420 [13] |
405 [14] |
415 [13] |
15 / 5 |
| δ(CβCaCb) | 426 [13] | 414 [14] | 423 [13] | 12 / 3 |
Normal mode calculations for NiOEP, including all seven atoms for each ethyl group (C2h or S4 molecular symmetry),14,37 indicate the possibility for several modes between 350 and 450 cm−1, designated as δ(CβC1C2) or γ2 (A1u), whose d4 and d16 shifts are reasonably consistent with those observed here.
While the bands appearing between 400 and 440 cm−1 have been (and will probably continue to be) designated as the “vinyl bending” modes, because of previously observed shifts upon full or partial deuteration of the vinyl groups,4,5,41,42 they are perhaps more accurately described as deformations of pyrrole rings I or II that contain significant vinyl bending contributions, as more fully discussed in recent work using several methyl deuterated protohemes; i.e., it is noted there that these so-called vinyl bending modes shift significantly upon deuteration of the adjacent methyl groups.8,9,15 Given the apparently complex nature of these modes, it is then not surprising that more than two “vinyl bending modes” can be observed in this region; the degree to which an actual vinyl deformation motion contributes to any given mode in this region can not be readily assessed without acquiring data for protohemes bearing (2H- or 13C-) labeled vinyl groups. However, only a small number of reports on the RR spectra of such vinyl-labeled heme proteins have appeared and only for myoglobin4,5,41 or cytochrome c peroxidase42; although the spectra acquired in those studies were of generally lower resolution, there was no clear evidence obtained for the presence of three modes in this region. In the present case, and especially in the earlier study of cytochrome P450 BM3 by Schelvis and coworkers,17 the presence of three modes in this region is more evident. While further studies, employing vinyl-labeled protohemes, will be needed to clarify the precise nature of the three “vinyl bending” modes seen for P450s in this region, this issue is not of great importance for the present work; i.e., in agreement with the behavior of the vinyl stretching modes discussed above, substrate binding has little or no effect on vinyl group dispositions, as judged by the lack of significant changes for the vinyl mode envelopes observed near 403 and 427 cm−1 in traces A and B. All three components seen here in the spectrum of native substrate-bound P450cam experience 12−15 cm−1 shifts for d12-P450cam, similar to the shifts observed for d12-oxyMb,9 but only 3−5 cm−1 downshifts upon d4 substitution, also consistent with results from the deuterated Mb and Hb studies.5,9,27,39,40
In considering the utility of future work with labeled vinyl groups, it is pointed out that early work with heme model compounds by Bocian and coworkers,43 as well as the more recent studies by Schelvis and coworkers on cytochrome P450 BM3,17 suggest that the lower frequency bending modes and the lower frequency ν(C=C) stretching modes correspond to a nearly in-plane vinyl conformation, while higher frequency values are associated with more out-of-plane orientations, a suggestion that is consistent with the extensive correlations of ν(C=C) stretching modes with documented vinyl group orientations, as initially reported by Smulevich and coworkers34 and later by others.35 Although, it seems reasonable to assign the lower frequency bending and stretching modes to an in- plane vinyl conformer and the higher frequency ones to out-of-plane conformers, in line with previous studies, it remains true that without labeling specific vinyl groups, it is not possible to definitively associate a given feature with a particular vinyl group; i.e., as first pointed out by Bocian and coworkers,43 a given vinyl group actually may contribute to both features, depending upon whether or not it can assume different conformations.
The most striking change that occurs in the low frequency region upon substrate binding is the appearance of the new band at 367 cm−1. Inasmuch as substrate binding also led to a shift and reasonably significant increase in the intensity of the out-of-plane γ7 mode appearing at 308 and 313 cm−1 in the substrate-free and substrate-bound forms (traces A and B in Figure 3), the 367 cm−1 feature was originally attributed in the earlier works13,16 to the out-of-plane γ6 mode, presumably activated by substrate binding in analogy with the γ7 counterpart. However, the expectation5,7-9 that γ6 normally occurs at frequencies near 340 cm−1 led Zhang15 and Schelvis and coworkers17 to argue for assignment of the substrate-activated mode to a new propionate associated vibration, probably the propionate at the 7-position, this suggestion also being supported by crystallographic data44 showing that binding of substrate causes breaking of the hydrogen bond between 7-propionate and an amino acid Lysine, (Lys69) for P450 BM3. Obviously, further work on isotopically labeled derivatives was needed to clarify this issue. Now, the new RR spectral results for P450cam containing d12-protoheme and d4-protoheme, reported here for the first time, do in fact support this assignment of the 367 cm−1 feature to a mode associated with propionate motion; however, as is explained below, corresponding experiments with d4-P450cam samples also provide evidence for activation of the out-of-plane γ6 mode occurring at almost the same frequency.
The 367 cm−1mode observed here for the native substrate-bound form shifts down by 8 cm−1 for the d12-labeled P450cam sample, exhibiting a shift identical to that obtained for the higher frequency propionate mode seen at 380 cm−1. Furthermore, as can be seen in trace D, a feature is observed at 365 cm−1 for the d4-labeled protein, demonstrating that a strong band persists at this frequency upon methine deuteration. The persistence of this feature rules out the possibility that the 367 cm−1 band is associated solely with a γ6 assignment, because it is well known that γ6 exhibits large (∼25−30 cm−1) downshifts for methine deuterated hemes.5,9,37 Thus, the present results provide definitive evidence for the assignment of this new 367 cm−1 feature to a second propionate bending mode, as was indeed proposed by Schelvis and coworkers.17 On the other hand, as explained below, the spectra observed for the d4-protoheme substituted sample are not entirely explainable without also invoking the presence of an activated γ6 mode in the substrate-bound form. Thus, in the spectrum of substrate-bound d4-P450cam, shown in trace D of Figure 3, two modes are observed at 365 cm−1 and 378 cm−1, both being 2 cm−1 lower than those observed for native substrate-bound P450cam (Figure 3, trace B). This slight sensitivity of propionate modes to methine deuteration is in excellent agreement with similar studies on other heme proteins.5,9,27,39,40 However, it is also notable that the relative intensity ratio of those two modes is significantly different than that observed in the spectrum of native protein (Figure 3, trace B), with the 365 cm−1 component being significantly less intense. As explained in the Experimental section, the relative component intensities of multi-component envelopes, including the three vinyl bends, two propionate bending modes and the ν8/ν50 modes, were held constant in the absence of any valid arguments for variations. Thus, this loss of intensity of the 365 cm−1 component, coupled with the apparent observation of a new band appearing within the 335−350 cm−1 envelope of bands in trace D, is most reasonably attributable to the activation of the γ6 mode at a frequency near 365 cm−1; i.e., given the 25−30 cm−1 shift expected for γ6.5,9,37 While it is noted that unjustifiable relaxation of certain restrictions within the deconvolution procedure does allow one to generate fits that do not demand the presence of the γ6 mode (Supporting Information), the most satisfying fit is obtained by including it.
The appearance of a second propionate mode appearing at 367 cm−1 upon binding of substrate is most reasonably interpreted to arise from rupture of the propionate's H-bonding interactions with associated peptide residues,6,7,17,45; i.e., upon substrate binding one of the propionate residues retains these interactions and associated ∼380 cm−1 bending mode that is still observable in the substrate-bound form. Inasmuch as the analysis of crystallographic structure of camphor-bound P450cam (2CPP, Protein Data Bank) 46 reveals that camphor binds nearer to the 7-propionate group, the new 367 cm−1 band is tentatively associated with that substituent.
Finally, in commenting on the appearance of the γ6 mode, as was pointed out in one of the earlier works dealing with this issue,13 the enhancement of this mode in the spectra of the substrate-bound form of P450cam is accompanied by an increase in the intensity of a second A2u mode in this region, namely the γ7 mode seen at 313 cm−1. Generally, the enhancement of these and other out-of-plane modes are expected to attend out-of-plane distortions of the heme macrocycle, but the systematics of such enhancements are somewhat complex. This issue has been clearly outlined in the discussion of comprehensive and elegant studies of horsereadish peroxidase by Schweitzer-Stenner.47 Applying the so-called Normal Coordinate Structural Decomposition method developed by Shelnutt and coworkers,48,49 it is shown that, generally, actual protein-induced out-of-plane distortions can be decomposed into combinations of distortions of various possible symmetries; i.e., A2u(“doming”), B1u (“ruffling”), B2u (“saddling”) and Eg (“waving”). As explained by Schweitzer-Stenner,47 in the absence of complications, it is anticipated that the out-of-plane distortions of a given symmetry type will lead to enhancement of out-of-plane vibrational modes of the same symmetry class; i.e. a purely A2u distortion should enhance only the A2u out-of-plane modes. Inasmuch as the γ6 and γ7 , both of A2u symmetry, are most strongly enhanced upon substrate binding, these acquired RR spectra suggest a more domed heme macrocycle, although it is noted that a weak broadened band is seen near 480−540 cm−1 in the substrate bound form, near the region where the γ12 (B1u, ruffled) and γ21, γ22 (Eg, waving) modes are expected to occur.
Conclusion
In summary, the RR data acquired here for deuterated analogues of cytochrome P450cam provide an important clarification of the response of the active site to substrate binding. Thus, the present results, employing two different isotopically labeled cytochrome P450cam samples, resolve an apparent controversy13,15-17 in the interpretation of the changes observed in the RR spectra upon substrate binding. While the new results obtained here for both deuterated samples definitely confirm the assignment of the newly observed 367 cm−1 feature to a second conformation of the propionate fragments, as originally suggested by Schelvis and coworkers,17 the data obtained here for the d4-protoheme labeled enzyme are most reasonably interpreted to also support the previous suggestion13,16 that the binding of substrate activates several out-of-plane heme modes, including γ7 and γ6, whose frequency is close to that of the newly activated propionate mode at 367 cm−1; the enhancement of these A2u out-of-plane modes is most consistent with a substrate-induced doming distortion of the heme macrocycle.
Supplementary Material
Acknowledgments
This work was supported by a grant from the National Institutes of Health (DK35153 to J.R.K.) and by some additional funding provided by the Pfletschinger Habermann Fund of Marquette University. The authors thank Dr. Daniel Sem and Dr. Chris McCollough of Marquette University for advice and assistance with the expression of cytochrome P450cam.
References
- 1.Li X-Y, Spiro TG. In: Biological Applications of Raman Spectroscopy. Spiro TG, editor. Vol. 3. Wiley; New York: 1988. pp. 1–37. [Google Scholar]
- 2.Kincaid JR. In: The Porphyrin Handbook. Kadish KM, Smith KM, Guilard R, editors. Vol. 7. Academic Press; New York: 1999. pp. 225–291. [Google Scholar]
- 3.Champion PM. In: Biological Applications of Raman Spectroscopy. Spiro TG, editor. Vol. 3. Wiley; New York: 1988. pp. 249–292. [Google Scholar]
- 4.Choi S, Spiro TG, Langry KC, Smith KM. J Am Chem Soc. 1982;104:4337–4344. [Google Scholar]
- 5.Hu S, Smith KM, Spiro TG. J Am Chem Soc. 1996;118:12638–12646. [Google Scholar]
- 6.Cerda-Colon JF, Silfa E, Lopez-Garriga J. J Am Chem Soc. 1998;120:9312–9317. [Google Scholar]
- 7.Peterson ES, Friedman JM, Chien EYT, Sligar SG. Biochemistry. 1988;37:12301–12319. doi: 10.1021/bi980752u. [DOI] [PubMed] [Google Scholar]
- 8.Podstawka E, Rajani C, Kincaid JR, Proniewicz LM. Biopolymers (Biospectroscopy) 2000;57:201–207. doi: 10.1002/1097-0282(2000)57:4<201::AID-BIP1>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- 9.Mak PJ, Podstawka E, Kincaid JR, Proniewicz LM. Biopolymers. 2004;75:217–228. doi: 10.1002/bip.20115. [DOI] [PubMed] [Google Scholar]
- 10.Nagai M, Aki M, Li R, Jin Y, Sakai H, Nagatomo S, Kitagawa T. Biochemistry. 2000;39:13093–13105. doi: 10.1021/bi001029i. [DOI] [PubMed] [Google Scholar]
- 11.Zbylut SD, Kincaid JR. J Am Chem Soc. 2002;124:6751–6758. doi: 10.1021/ja012578u. [DOI] [PubMed] [Google Scholar]
- 12.Ortiz de Montellano PR. Cytochrome P450: Structure, Mechanism and Biochemistry. Kluwer Academic/ Plenum Publishers; New York: 2005. [Google Scholar]
- 13.Deng TJ, Proniewicz LM, Kincaid JR, Yeom H, Macdonald IDG, Sligar SG. Biochemistry. 1999;38:13699–13706. doi: 10.1021/bi991287j. [DOI] [PubMed] [Google Scholar]
- 14.Li XY, Czernuszewicz RS, Kincaid JR, Spiro TG. J Am Chem Soc. 1989;111:7012–7023. [Google Scholar]
- 15.Zhang HMS. Thesis, Marquette University. 1999. [Google Scholar]
- 16.Niaura G, Reipa V, Mayhew MP, Holden M, Vilker VL. Arch Biochem Biophys. 2003;409:102–112. doi: 10.1016/s0003-9861(02)00581-7. [DOI] [PubMed] [Google Scholar]
- 17.Chen Z, Ost TWB, Schelvis JPM. Biochemistry. 2004;43:1798–1808. doi: 10.1021/bi034920g. [DOI] [PubMed] [Google Scholar]
- 18.Sibbesen O, Voss JJ, Ortiz de Montellano PR. J Biol Chem. 1996;271:22462–22469. doi: 10.1074/jbc.271.37.22462. [DOI] [PubMed] [Google Scholar]
- 19.O'Keeffe DH, Ebel RE, Peterson JA. Methods in Enzymology. 1978;52:151–156. doi: 10.1016/s0076-6879(78)52017-x. [DOI] [PubMed] [Google Scholar]
- 20.Godziela GM, Kraner SK, Goff HM. Inorg Chem. 1986;25:4286–4288. [Google Scholar]
- 21.Kenner GW, Smith KM, Sutton MJ. Tetrahedron Lett. 1973;16:1303–1306. [Google Scholar]
- 22.Wagner GC, Gunsalus IC, Wang M-YR, Hoffman BM. J Biol Chem. 1981;256:6266–6273. [PubMed] [Google Scholar]
- 23.Wagner GC, Perez M, Toscano WA, Jr., Gunsalus IC. J Biol Chem. 1981;256:6262–6265. [PubMed] [Google Scholar]
- 24.Shriver DF, Dunn JBR. Appl Spectrosc. 1974;28:319–323. [Google Scholar]
- 25.Wu Q, Balakrishnan G, Pevsner A, Spiro TG. J Phys Chem A. 2003;107:8047–8051. [Google Scholar]
- 26.Rodgers KR, Su C, Subramaniam S, Spiro TG. J Am Chem Soc. 1992;114:3697–3709. [Google Scholar]
- 27.Podstawka E, Mak PJ, Kincaid JR, Proniewicz LM. Biopolymers. 2006;83:455–466. doi: 10.1002/bip.20573. [DOI] [PubMed] [Google Scholar]
- 28.Rwere F, Mak PJ, Kincaid JR. Biopolymers. 2008;89:179–186. doi: 10.1002/bip.20887. [DOI] [PubMed] [Google Scholar]
- 29.Barbush M, Dixon DW. Biochem Biophys Res Comm. 1985;129:70–75. doi: 10.1016/0006-291x(85)91404-4. [DOI] [PubMed] [Google Scholar]
- Fuhrhop J-H, Smith KM. Laboratory methods. In: Smith KM, editor. Porphyrin and Metalloporphyrins. Elsevier/North Holland; Amsterdam: 1975. pp. 757–869. [Google Scholar]
- 31.DiNello RK, Dolphin DH. Anal Biochem. 1975;64:444–449. doi: 10.1016/0003-2697(75)90452-2. [DOI] [PubMed] [Google Scholar]
- 32.Buchler JW. The Porphyrins. In: Dolphin D, editor. Structure and Synthesis. Part A. I. Academic Press; New York: 1978. pp. 389–483. [Google Scholar]
- 33.Unno M, Christian JF, Benson DE, Gerber NC, Sligar SG, Champion PM. J Am Chem Soc. 1997;119:6614–6620. [Google Scholar]
- 34.Marzocchi MP, Smulevich G. J Raman Spectrosc. 2003;34:725–736. [Google Scholar]
- 35.Hudecek J, Hodek P, Anzenbacherova E, Anzenbacher P. Biochim Biophys Acta. 2007;1770:413–419. doi: 10.1016/j.bbagen.2006.10.008. [DOI] [PubMed] [Google Scholar]
- 36.Abe M, Kitagawa T, Kyogoku Y. J Chem Phys. 1978;69:4526–4534. [Google Scholar]
- 37.Li XY, Czernuszewicz RS, Kincaid JR, Stein PB, Spiro TG. J Phys Chem. 1990;94:47–61. [Google Scholar]
- 38.Stoll LK, Zgierski MZ, Kozlowski PM. J Phys Chem A. 2003;107:4165–4171. [Google Scholar]
- 39.Podstawka E, Kincaid JR, Proniewicz LM. J Mol Struct. 2001;596:157–162. [Google Scholar]
- 40.Rajani C, Kincaid JR. J Am Chem Soc. 1998;120:7278–7285. [Google Scholar]
- 41.Uchida K, Susai Y, Hirotani E, Kimura T, Yoneya T, Takeuchi H, Harada I. J Biochem. 1988;103:979–985. doi: 10.1093/oxfordjournals.jbchem.a122397. [DOI] [PubMed] [Google Scholar]
- 42.Smulevich G, Hu S, Rodgers KR, Goodin DB, Smith KM, Spiro TG. Biospectroscopy. 1996;2:365–376. [Google Scholar]
- 43.Kalsbeck WA, Ghosh A, Pandey RK, Smith KM, Bocian DF. J Am Chem Soc. 1995;117:10959–10968. [Google Scholar]
- 44.Li H, Poulos TL. Nat Struct Biol. 1997;4:140–146. doi: 10.1038/nsb0297-140. [DOI] [PubMed] [Google Scholar]
- 45.Chen Z, Wang L-H, Schelvis JPM. Biochemistry. 2003;42:2542–2551. doi: 10.1021/bi027206s. [DOI] [PubMed] [Google Scholar]
- 46.Poulos TL, Finzel BC, Howard AJ. J Mol Biol. 1987;195:687–700. doi: 10.1016/0022-2836(87)90190-2. [DOI] [PubMed] [Google Scholar]
- 47.Huang Q, Schweitzer-Stenner R. J Ram Spect. 2005;36:363–375. [Google Scholar]
- 48.Jentzen W, Ma J-G, Shelnutt JA. Biophys J. 1998;74:7653. doi: 10.1016/S0006-3495(98)74000-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shelnutt JA, Song X-Z, Ma J-G, Jia S-L, Jentzen W, Medforth CJ. Chem Soc Rev. 1998;27:31–42. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.