Abstract
Chromatin modification is important for virtually all aspects of DNA metabolism but little is known about the consequences of such modification in trypanosomatids, early branching protozoa of significant medical and veterinary importance. MYST-family histone acetyltransferases in other species function in transcription regulation, DNA replication, recombination and repair. Trypanosoma brucei HAT3 was recently shown to acetylate histone H4K4 and we now report characterization of all three T. brucei MYST acetyltransferases (HAT1–3). First, GFP-tagged HAT1–3 all localize to the trypanosome nucleus. While HAT3 is dispensable, both HAT1 and HAT2 are essential for growth. Strains with HAT1 knock-down display mitosis without nuclear DNA replication and also specific de-repression of a telomeric reporter gene, a rare example of transcription control in an organism with widespread and constitutive polycistronic transcription. Finally, we show that HAT2 is responsible for H4K10 acetylation. By analogy to the situation in Saccharomyces cerevisiae, we discuss low-level redundancy of acetyltransferase function in T. brucei and suggest that two MYST-family acetyltransferases are essential due to the absence of a Gcn5 homologue. The results are also consistent with the idea that HAT1 contributes to establishing boundaries between transcriptionally active and repressed telomeric domains in T. brucei.
Introduction
Histone N-terminal tails extend from the core of the nucleosome and can be post-translationally modified with a range of chemical groups. These combined modifications may constitute a code (Jenuwein and Allis, 2001) that regulates access to DNA, possibly during all chromatin-templated processes (Millar and Grunstein, 2006). For example, the transfer of an acetyl group from Ac-CoA to specific Lys ε-amino groups on histone H4 can modulate chromosome structure and interaction (Shogren-Knaak et al., 2006) or provide binding platforms for regulatory factors (de la Cruz et al., 2005). For identical DNA sequences to exhibit distinct properties within chromatin, cells are equipped with the enzymes that specifically and reversibly modify the histones in this way. Acetylation of histones H3 and H4 has been characterized in some detail in Saccharomyces cerevisiae and in human cells (Rice and Allis, 2001; Lachner and Jenuwein, 2002) but comparison among organisms ‘argue against there being a universal histone code and underscore the need to avoid general conclusions obtained from one organism’ (Garcia et al., 2007).
The MYST family is a group of related histone acetyltransferases that appears to have members in all eukaryotes (Yang, 2004). The acronym MYST is from the founding members, human MOZ (monocytic leukaemia zinc finger protein) (Borrow et al., 1996), yeast Ybf2 (renamed Sas3, for Something about silencing 3) and Sas2 (Reifsnyder et al., 1996) and mammalian TIP60 (HIV Tat-interacting protein 60 kDa) (Kamine et al., 1996). A third MYST protein in S. cerevisiae is Esa1 (Essential sas2-related acetyltransferase 1) (Smith et al., 1998; Clarke et al., 1999). Besides MOZ and TIP60 in humans are hMOF (orthologue of Drosophila Mof, males-absent-on-the-first) (Taipale et al., 2005), HBO1 (HAT bound to ORC1, replication origin recognition complex 1) (Iizuka and Stillman, 1999) and MORF (MOZ-related factor) (Champagne et al., 1999).
MYST acetyltransferases have been shown to impact upon transcription, DNA replication, recombination and repair (Carrozza et al., 2003). The enzymes function in multi-protein complexes in vivo (Lee and Workman, 2007). For example, S. cerevisiae Esa1, Sas2 and Sas3 are the catalytic components of the NuA4 (Nucleosomal H2A/H4) (Eisen et al., 2001; Doyon and Cote, 2004), SAS (Sutton et al., 2003) and NuA3 (Nucleosomal Acetyltransferase of histone H3) (John et al., 2000) complexes respectively. Other components of the complexes may be required for activity (Sutton et al., 2003) or substrate recruitment (Lee and Workman, 2007) while there is also an interplay between histone acetylation and methylation. Indeed, MYST acetyltransferase complexes may require histone methylation for substrate recruitment (Martin et al., 2006) and MYST acetyltransferases can function in the same complex as a methyltransferase (Dou et al., 2005).
Trypanosomatids, including Trypanosoma brucei, T. cruzi and Leishmania spp., branched early in the eukaryotic lineage and trypanosomes are established, ‘differently evolved’ model organisms. Although trypanosome histone tails are divergent, acetylation has been identified on histone H4K2 (2% of sites modified), K4 (73%), K5 (7%), K10 (7%) and K14 (1%) (da Cunha et al., 2006; Janzen et al., 2006a; Mandava et al., 2007). These mono-flagellated protozoa have a devastating impact on the world's poor, causing African trypanosomiasis, Chagas disease and leishmaniasis (http://www.who.int/tdr/diseases/). The consequences of this range of human and animal diseases are hundreds of thousands of deaths each year, ∼1.5 million cases a year of the disfiguring lesions associated with cutaneous leishmaniasis and severely curtailed agricultural development throughout sub-Saharan Africa. Human African trypanosomaiasis is fatal if untreated and represents the leading cause of mortality in some areas. T. brucei also causes Nagana in cattle, rendering 10 million square kilometres of land unsuitable for livestock.
In trypanosomatids, almost all genes are transcribed as long polycistrons (Clayton, 2002). No RNA polymerase II (pol II) promoters have been identified for protein-coding genes but certain protein-coding genes are transcribed by RNA polymerase I (pol I) (Palenchar and Bellofatto, 2006). Although gene expression in trypanosomatids is predominantly regulated post-transcriptionally (Clayton, 2002; Horn, 2008), several lines of evidence point to important roles for chromatin structure and modification in gene expression, cell cycle control and differentiation (Ersfeld et al., 1999; Belli, 2000; Horn, 2001). More recently, acetylated and methylated histones were shown to be enriched at regions where the polycistronic transcription units diverge in T. cruzi (Respuela et al., 2008). One of the four putative class I/II histone deacetylases characterized in T. brucei is required for normal cell cycle progression (Ingram and Horn, 2002) while the only nuclear class III, sirtuin-type deacetylase (Kowieski et al., 2008) is involved in DNA repair (Garcia-Salcedo et al., 2003) and telomeric gene silencing (Alsford et al., 2007). Trypanosomatid genome sequencing revealed six putative histone acetyltransferases in T. brucei: three related to the MYST family, two of the elongator type (see table S4 in Ivens et al., 2005) and a PHD-finger protein (D. Horn, unpublished). HAT3, one of the MYST-family proteins, was recently shown to be the histone H4K4 acetyltransferase (Siegel et al., 2008). We now report characterization of all three MYST-family proteins (HAT1–3). While HAT3, the H4K4 acetyltransferase, is dispensable for growth, HAT1 is required for telomeric silencing and growth, and possibly, for DNA replication and HAT2 is required for H4K10 acetylation and growth.
Results
Three MYST-family acetyltransferases expressed in T. brucei
Four proteins were identified encoded by T. cruzi and Leishmania major and three by T. brucei related to the MYST-family acetyltransferases (Ivens et al., 2005). For comparison, the human genome encodes five MYST-family proteins: S. cerevisiae, three and Schizosaccharomyces pombe, two (Yang, 2004). The trypanosomatid proteins were designated HAT1–4 for putative Histone AcetylTransferase and it is HAT4 that is not present in the corresponding syntenic region or elsewhere in the T. brucei genome. Phylogenetic analysis indicated that the trypanosomatid proteins are remarkably divergent relative to other MYST-family members (Fig. 1). Trypanosomatid MYST proteins cluster into distinct groups but can all be traced to the root of the tree through a branch not shared with any other species. This suggests that the MYST proteins may have diversified in a common trypanosomatid ancestor. Thus, shared phylogeny does not allow us to identify putative homologues for the trypanosomatid proteins in model organisms.
Fig. 1.

Phylogenetic analysis. The trypanosomatid HATs were compared with other MYST-family proteins. The unrooted neighbour-joining tree was generated using clustal 1.8X and TreeView. Where excellent (≥ 99.9%), branching confidence is indicated (open circles). Tb, T. brucei; Tc, T. cruzi; Lm, L. major; Hs, Homo sapiens; Sc, S. cerevisiae; Gl, Giardia lamblia; Eh, Entamoeba histolytica; Sp, Schizosaccharomyces pombe; Pf, Plasmodium falciparum; Tg, Toxoplasma gondii. All accession numbers are indicated. The GeneDB IDs for the T. brucei proteins are: HAT1, Tb927.7.4560; HAT2, Tb11.01.3380; and HAT3, Tb10.6k15.2190.
A schematic representation of the T. brucei MYST proteins is shown in Fig. 2A. Alignment and sequence comparison revealed a number of features (see Fig. 2B). Motif A (Q/Rx2GxG/A) is involved in binding Coenzyme A (Roth et al., 2001) and this motif is present in HAT1 (Qx2GxG in all three trypanosomatids) and HAT3 (Rx2GxG in all three trypanosomatids) and can also be found in HAT4 in T. cruzi and Leishmania (not shown). Key residues found to be critical for catalytic activity in Esa1 are Glu338 and Cys304 (Yan et al., 2000; 2002) and these strictly conserved residues are also found in HAT1–3 in all three trypanosomatids. In Esa1, these residues participate in a ping-pong catalytic mechanism involving a Cys304 self-acetylation intermediate (Yan et al., 2002). Like several MYST-type acetyltransferases, HAT1 and HAT3 in all three trypanosomatids (see T. brucei proteins in Fig. 2) and HAT4 in T. cruzi and Leishmania have a C2HC (Cx2Cx12Hx3−5C) zinc-binding motif thought to be involved in catalytic activity and/or substrate recognition (Akhtar and Becker, 2001). Motif A and the C2HC motif are absent from all three trypanosomatid HAT2s. Relative to Esa1, HAT1 and HAT2 have insertions within the MYST-homology domain (one in HAT1 and two in HAT2, see Fig. 2) and similarly located, but variable-size, insertions are also found in these proteins in T. cruzi and Leishmania. Several MYST-family proteins contain an N-terminal chromo (chromosome organization modifier) domain thought to deliver these regulators to their sites of action on chromatin (Jones et al., 2000) by mediating binding to methyl-lysine (Nielsen et al., 2002). The chromodomain of HP1 (heterochromatin protein 1) recognizes histone H3 methylated on Lys9 for example (Bannister et al., 2001) but these domains may also interact with RNA (Akhtar et al., 2000). Sequence alignments suggest the presence of chromodomains towards the N-terminus of both HAT1 and HAT2. The evidence is weaker for HAT2, but HAT1 alignment with Esa1 (Fig. 2A, boxed) indicates the presence of a MOF-like chromo-barrel domain (Nielsen et al., 2005) suggesting a link between acetylation and methylation in trypanosomatids. Taken together, our analysis indicates that each of the HATs has similar features regardless of the trypanosomatid under consideration. We have focused on the three T. brucei HATs (1–3) found on chromosomes 7, 11 and 10, encoding proteins with predicted molecular mass of approximately 53.5, 67.7 and 32.4 kDa respectively.
Fig. 2.
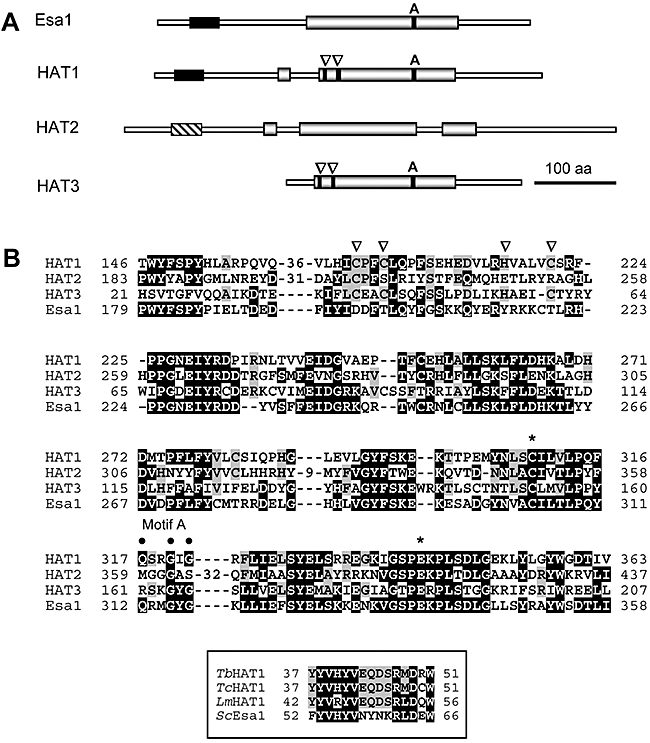
MYST-family acetyltransferases in T. brucei.
A. Schematic representation of the predicted T. brucei MYST-family proteins compared with S. cerevisiae Esa1. The core acetyltransferase domains (grey boxes) and N-terminal chromodomains (black boxes and cross-hatched box; see the text) are indicated. Arrowheads indicate a C2HC zinc-finger motif. ‘A’ indicates motif A.
B. Sequence alignment. The core acetyltransferase domains from the T. brucei HATs were aligned with the equivalent region from Esa1 using clustalw followed by manual adjustment. Residues that are shared between Esa1 and any of the T. brucei proteins are white on a black background. Other residues shared among the T. brucei proteins are on a grey background. Motif A and the zinc-finger motif are indicated (see A). Asterisks indicate a glutamate residue [E] and a cysteine residue [C] required for Esa1 catalytic activity. Insertions within the HAT1 and HAT2 MYST-homology domains have been removed to optimize the alignment. The box shows the trypanosomatid chromodomain from HAT1 aligned with the corresponding domain from S. cerevisiae Esa1.
We analysed expression and mapped RNA processing signals using reverse transcription polymerase chain reaction (RT-PCR). This technique exploits the fact that all mature T. brucei mRNAs possess an identical 5′-spliced leader (SL) sequence added at a specific AG dinucleotide, splice acceptor. DNA sequencing identified acceptor sites 84, 64 and 5 nt upstream of the start codons for HAT1–3 respectively. Using Northern blotting, we detected cognate mRNA transcripts for HAT1–3 of approximately 2.1 kb, 2.1 kb and 1.3 kb respectively. All three genes are expressed in bloodstream and insect-stage cells, and mRNA abundance revealed no evidence for differential expression in these major life cycle stages (data not shown).
All three acetyltransferases localize to the T. brucei nucleus
Eukaryotic MYST acetyltransferases typically localize to the nucleus. Nuclear localization signals are not well defined in trypanosomatids and we obtained no clear prediction for subcellular localization from bioinformatic analysis so we determined subcellular localization in bloodstream-form strains engineered for Tet-inducible expression (Alsford et al., 2005) of GFP-tagged versions of each gene. GFPHAT1–3 peptides are predicted to be 80.6, 94.7 and 59.5 kDa, respectively, and proteins of the predicted size were conditionally expressed in each strain as demonstrated by Western blotting (Fig. 3). Microscopic analysis of these strains did not reveal any significant GFP signal in un-induced cultures (data not shown). In contrast, fluorescence or immunofluorescence analysis of each induced GFPHAT strain indicated specific accumulation in the nucleus (Fig. 3). These results were confirmed with strains expressing epitope (myc)-tagged versions of each acetyltransferase (data not shown).
Fig. 3.
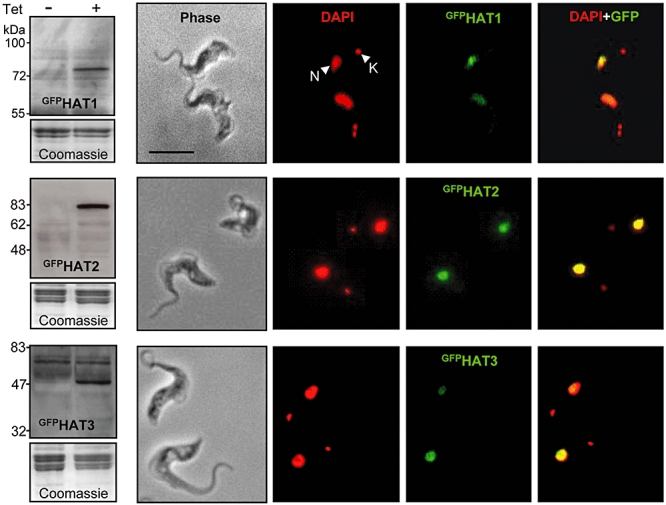
MYST-family proteins localize to the T. brucei nucleus. Strains were cultured without Tet (−) or with Tet (1 μg ml−1) for 24 h (+) to induce GFPHAT expression. Fusion proteins were detected by Western blotting using α-GFP (left). Coomassie-stained gels are shown as loading controls. Representative examples of immunofluorescence (HAT1 and HAT3) or direct fluorescence (HAT2) images of induced cultures are shown. HAT localization is indicated in green. DAPI, the DNA counterstain (4′,6-diamino-2-phenylindole), is false-coloured red and reveals the nucleus (N) and the smaller mitochondrial, kinetoplast DNA (K). Merged images indicate nuclear colocalization in yellow. Scale bar: 10 μm.
HAT3 is dispensable while HAT1 and HAT2 are essential for growth
To examine MYST-family acetyltransferase function in bloodstream-form T. brucei, we initially attempted gene disruption or knockout (Fig. 4A). The HAT genes are ‘single copy’ and T. brucei is diploid, so we assembled a pair of constructs with different selectable markers for each gene. All six constructs targeted the correct loci as determined using PCR assays but we were unable to disrupt both alleles of either HAT1 or HAT2 (data not shown). In contrast, HAT3 null strains were obtained and confirmed by Southern analysis (Fig. 4C). The results indicate that HAT3 is dispensable and suggest that HAT1 and HAT2 are essential for growth. Although it was proposed that more abundant histone acetylation in lower eukaryotes reflects a higher proportion of transcriptionally competent chromatin, HAT3 knockout suggests that 73% H4K4 acetylation (Mandava et al., 2007; Siegel et al., 2008) has little impact on transcription.
Fig. 4.
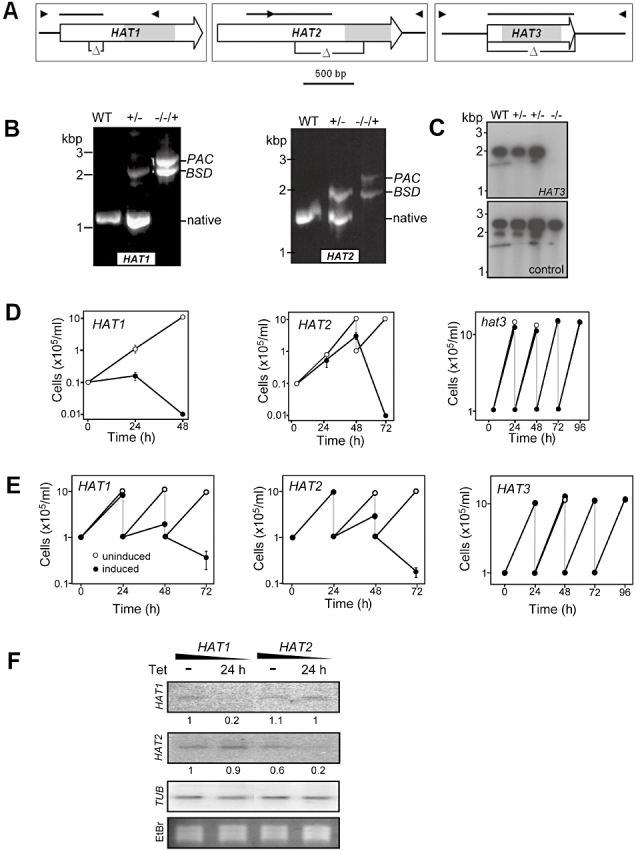
HAT3 is dispensable while HAT1 and HAT2 are essential for growth.
A. Schematics illustrating the gene targeting strategies. The symbols ‘Δ’ indicate regions targeted for deletion. Arrowheads represent the positions of the primers used for PCR assays; 29M45/3, 18M145/3 and H31/4 for HAT1–3 respectively; bars represent probes used for Southern and Northern blot analysis and the grey boxes indicate the regions targeted for RNAi by dsRNA.
B. Genomic DNA from clones with both native alleles disrupted in the presence of conditionally expressed GFPHAT1 and GFPHAT2 were analysed by PCR and the products were visualized in agarose gels. The BSD and PAC cassettes are 1 and 1.3 kb respectively. The recombinant GFPHAT genes are not detected using this assay (see primer locations in A).
C. Southern blot analysis of hat3 null T. brucei. Genomic DNA was digested with AccI and the blot was sequentially hybridized with the probes indicated (the control probe is from HAT1, see bars in A).
D. Growth analysis of strains expressing conditional copies of GFPHAT1 or GFPHAT2 and a hat3 null strain. For GFPHAT1 and GFPHAT2: GFPHAT expressed (+Tet), open circles; GFPHAT expression inactivated (−Tet), closed circles. For HAT3: wild type, open circles; hat3 null, closed circles. Cells were split back to 1 × 105 ml−1 every 24 h (grey lines). Error bars, ± one standard deviation.
E. Growth analysis of three independent RNAi strains during knock-down of each HAT (target regions indicated in A). Un-induced, open circles; RNAi induced (+Tet), closed circles. Cells were split back to 1 × 105 ml−1 every 24 h (grey lines). Error bars, ± one standard deviation.
F. Northern analysis during RNAi knock-down. Membranes were probed with HAT1 and HAT2 gene fragments (see bars in A). Tubulin (TUB) was used as a loading control. Relative HAT1 or HAT2 mRNA signals, as determined by phosphorimager analysis and corrected for loading, are indicated. EtBr, ethidium bromide.
We next generated conditional deficient strains for HAT1 and HAT2. We were able to disrupt both native alleles in T. brucei conditionally expressing GFPHAT1 or GFPHAT2 (Fig. 4B and see Fig. 3) indicating that both GFPHAT1 and GFPHAT2 complement the HAT defects. Downregulation of GFPHAT1 and GFPHAT2 expression produced a severe growth defect in both cases (Fig. 4D). In contrast, hat3 null mutants grew at the same rate as wild type (Fig. 4D). This verified our assignment of HAT1 and HAT2 as essential for growth but the conditional growth defects were not stable. Phenotype instability was found to be due to the outgrowth of cells in which intact native HAT alleles were regenerated through recombination between the ectopic gene and portions of the coding region that remained intact in these strains (see Fig. 4A, data not shown). We therefore turned to RNA interference (RNAi) to generate stable knock-down strains. We used an RNAi system for conditional expression of long, intramolecular, ‘stem-loop’ (Durand-Dubief et al., 2003) double-stranded RNA (dsRNA) (517, 459 and 568 bp for HAT1–3 respectively) checked for specificity (Redmond et al., 2003). Expression of dsRNA specific for either HAT1 or HAT2 induced growth defects similar to those observed following downregulation of GFPHAT1 and GFPHAT2 (compare Fig. 4D and E). Northern analysis indicated specific knock-down of HAT1 and HAT2 mRNA respectively (Fig. 4F) and the growth defects following RNAi induction were highly reproducible in multiple independent strains (Fig. 4E) and at multiple times after prolonged culture (data not shown). HAT3 RNAi strains provided controls for Tet induction in cells harbouring the RNAi vector system and, as expected, showed no growth defect (Fig. 4E).
Cell cycle analysis following HAT1 or HAT2 knock-down
The histone H4 acetyltransferase, Esa1, is required for G2/M progression and is the only essential MYST-type HAT in S. cerevisiae (Clarke et al., 1999). In addition, the T. brucei deacetylase, DAC4, is required for normal G2/M progression (Ingram and Horn, 2002). No method is available to synchronize bloodstream-form T. brucei, but nuclear and mitochondrial (kinetoplast) DNA, stained with DAPI, provides excellent cytological markers that define position in the cell cycle (Woodward and Gull, 1990). During normal growth, approximately 80% of cells display a single nucleus and a single kinetoplast (1N1K) corresponding to earlier phases of the cell cycle (G1/S). A single nucleus and two kinetoplasts (1N2K) correspond to nuclear G2 and two nuclei and two kinetoplasts (2N2K) indicate completion of mitosis.
We used the HAT RNAi strains to explore the role of MYST-type HATs in cell cycle progression in T. brucei. After 24 h of RNAi induction, we saw very little or no growth defect compared with un-induced strains. Between 24 and 48 h of induction, the HAT1 and HAT2 knock-down strains showed approximately five- and twofold reduction in cell density respectively (Fig. 4E). To examine primary rather than secondary defects, we analysed cell cycle position at 24 and 48 h, but focused on trends established by 24 h, prior to detectable growth defect. Within the G2/M population, in both HAT1 and HAT2 knock-down strains, we saw a reduction in the number of pre-mitotic (1N) cells and an increase in the number of post-mitotic (2N) cells (Fig. 5A) indicating accumulation pre-cytokinesis. A HAT3 RNAi strain provided a control as neither this (Fig. 5A) nor a hat3 null strain (data not shown) displayed evidence of cell cycle aberration.
Fig. 5.
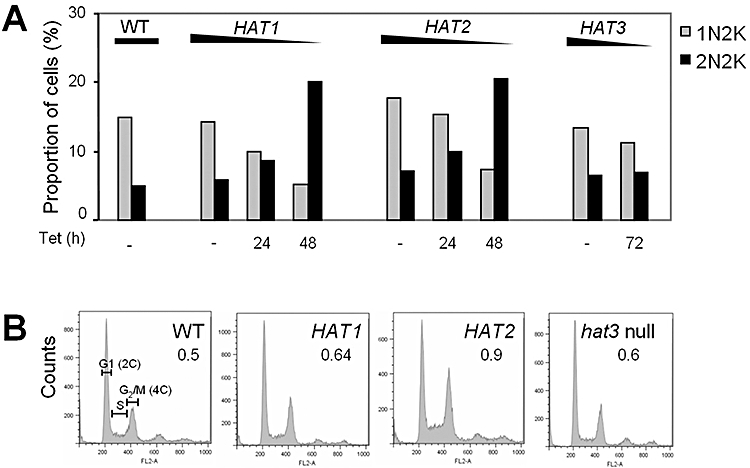
Cell cycle analysis following HAT knock-down.
A. Analysis of DAPI-stained T. brucei. 1N2K indicates cells with a single nucleus and two kinetoplasts and corresponds to nuclear G2. 2N2K indicates cells with two nuclei and two kinetoplasts and corresponds to completion of mitosis. n > 300 cells for each sample per time point.
B. DNA content analysis using flow cytometry. HAT1- and HAT2-depleted cells were analysed following 48 h +Tet. A hat3 null strain was analysed in parallel. 2C, cells with a diploid nuclear content; 4C, cells that have passed through S-phase and replicated their DNA. The proportion of 4C relative to 2C is indicated. n = 20 000 cells.
We next analysed cellular DNA content by flow cytometry (Fig. 5B). A hat3 null strain was not significantly different from wild type and, consistent with accumulation pre-cytokinesis, we saw a substantial increase in 4C (4× haploid nuclear DNA content) cells following HAT2 knock-down. Interestingly, a similar increase in 4C cells was not seen following HAT1 knock-down. DNA synthesis is normally complete before mitosis but the increased number of 2N2K cells in the absence of a corresponding increase in 4C cells indicates progression through mitosis without nuclear DNA replication.
HAT1 modulates telomeric silencing
Many MYST-family HATs regulate transcription. In T. brucei, pol I transcription is repressed immediately adjacent to telomeres (Glover and Horn, 2006) in a telomere- (Glover et al., 2007) and sirtuin deacetylase- (Alsford et al., 2007) dependent manner. In S. cerevisiae, telomeric silencing is similarly dependent on a sirtuin and also the MYST-family HATs, Sas2 (Reifsnyder et al., 1996) and Esa1 (Clarke et al., 2006). The S. pombe MYST-family HAT mst2+ appears to perform a similar function (Gomez et al., 2005). To explore the role of MYST-HATs in telomeric silencing in T. brucei, we placed a reporter cassette driven by a ribosomal RNA promoter within 2 kb of a de novo telomere in the HAT RNAi strains (Fig. 6A); the reporter is repressed by telomere position effect in these strains. We first analysed NPT reporter expression before and after HAT knock-down by Western blotting with α-NPT. The analysis indicated increased telomeric reporter expression following HAT1 knock-down with no change in expression following HAT2 or HAT3 knock-down (data not shown). To confirm these results, we analysed NPT reporter mRNA by Northern blotting before and after HAT1 and HAT2 knock-down (Fig. 6A). The analysis confirmed increased reporter expression following HAT1 knock-down (twofold compared with un-induced control and fourfold compared with HAT2 control) with no change in reporter expression following HAT2 knock-down. Increased reporter expression was not simply due to loss of viability, because extracts prepared from the HAT2 RNAi strain (Fig. 6A) and a histone variant RNAi strain (data not shown), which also suffered substantial loss of viability, had unchanged levels of NPT expression. Thus, telomeric silencing is specifically compromised following T. brucei HAT1 knock-down.
Fig. 6.
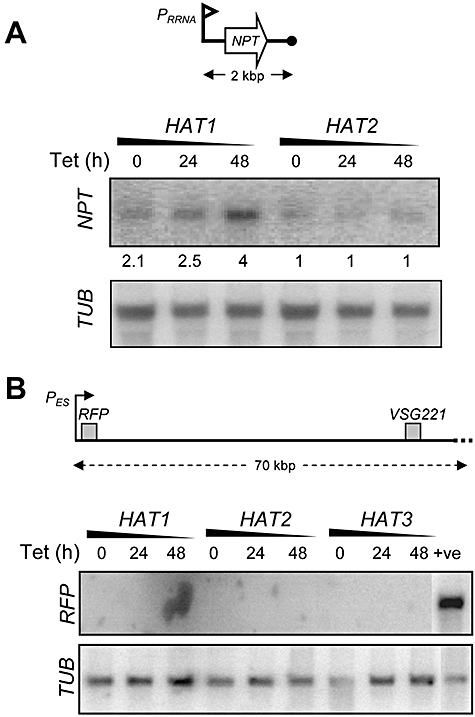
HAT1 modulates telomeric silencing but not VSG expression site repression.
A. A ribosomal RNA promoter (PRRNA) driving the expression of an NPT gene was placed 2 kb from a telomere in HAT RNAi strains. NPT expression was assessed before and after knock-down by Northern blot. TUB was used as a loading control. Relative NPT expression, determined by phosphorimager analysis and corrected for loading, is indicated.
B. HAT1–3 knock-down was induced in strains with an RFP reporter immediately downstream of a repressed VSG221 expression site promoter (PES). RFP expression was assessed before and after knock-down using Northern blotting. The positive control is a similar strain but expressing VSG221. Tubulin (TUB) was used as a loading control. All of the knock-down samples expressed RFP at < 1% relative to the control as determined by phosphorimager analysis.
Most genes appear to be constitutively transcribed by RNA pol II in T. brucei, but antigenic variation, unusually, relies on regulated pol I transcription. Although multiple variant surface glycoprotein expression site promoters are found close to telomeres, all are repressed but one in the bloodstream form. These promoters are usually approximately 50 kb from telomeres and recruit RNA pol I despite being distinct in sequence compared with the ribosomal RNA promoter. We therefore examined the expression of a reporter placed immediately downstream of a repressed expression site promoter (Fig. 6B) following HAT knock-down. None of the HATs appear to play a primary role in expression site promoter repression as determined by Northern blot analysis of reporter expression (Fig. 6B). The data are consistent with previous reports of mechanistically distinct VSG expression site promoter silencing and telomeric silencing (Alsford et al., 2007; Hughes et al., 2007).
HAT2 acetylates histone H4K10
The key residues involved in MYST factor catalysis are Glu338 and Cys304 (Yan et al., 2002) but motif A and the C2HC motif are also conserved and involved in Ac-CoA binding (Sternglanz and Schindelin, 1999) and HAT activity in some MYST-family proteins respectively (Takechi and Nakayama, 1999; Akhtar and Becker, 2001). Both motif A and the C2HC motif are absent from all three trypanosomatid HAT2s but neither motif is absolutely required for activity as demonstrated by Esa1 which complements esa1 cell cycle defects when motif A is mutated (Smith et al., 1998) and displays a TFIIIA-type zinc finger fold in place of the C2HC motif (Yan et al., 2000). To determine whether HAT2 displays acetyltransferase activity, we first asked whether the protein could transfer acetyl groups to a T. brucei histone H4 peptide substrate in vitro. As several recombinant MYST-family acetyltransferases lack activity in the absence of cofactors (Iizuka and Stillman, 1999; Sutton et al., 2003), we used GFPHAT2 purified from T. brucei for this analysis (see Fig. 3). Importantly, we previously demonstrated that GFPHAT2 complements the hat2 defect (Fig. 4E) indicating association with any additional factors required for activity. Despite the absence of a canonical motif A or C2HC motif within HAT2, affinity-purified GFPHAT2 (complex) displays acetyltransferase activity in vitro (Fig. 7A). We also assembled a mutant GFPHAT2(C351A) (see Fig. 2) ORF but the protein was expressed at a level insufficient for an in vitro assay, possibly due to selection against dominant-negative replacement of native HAT2 complexes (data not shown). This, and the finding that ESA1(E338Q) was dominant negative (Yan et al., 2000), is consistent with the hypothesis that HAT activity is essential for MYST function.
Fig. 7.
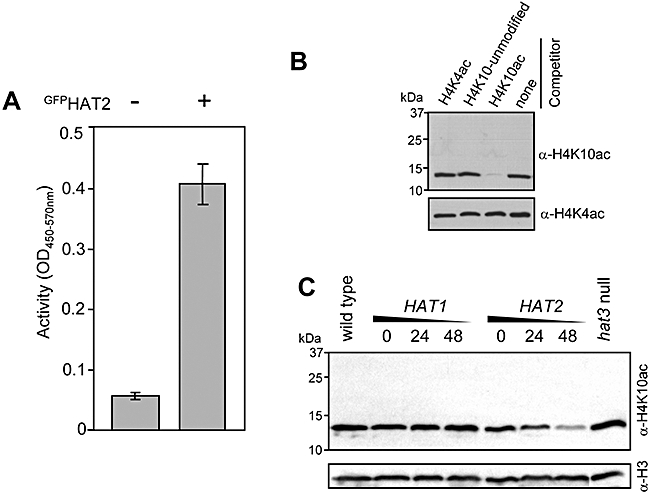
HAT2 acetylates histone H4K10.
A. In vitro acetyltransferase activity assay using eluates from T. brucei lacking (−) or expressing (+) GFPHAT2. The substrate was a T. brucei histone H4 tail peptide (H4A1-L20). n = 3. Error bars, ± one standard deviation.
B. Characterization of α-histone H4K10ac. A Western blot of whole trypanosome extracts (2 × 106 cells lane−1) was probed using α-H4K10ac. Peptide competitors used are shown above. To confirm equal loading, the blot was stripped and reprobed with α-H4K4ac (Siegel et al., 2008).
C. Specific depletion of the H4K10ac signal following HAT2 knock-down by RNAi. Western blotting was carried out as for (B) but without peptide competitor. Note that Siegel et al. (2008) demonstrated that total H4 levels are not depleted during HAT2 knock-down. To confirm equal loading, the blot was stripped and reprobed with α-histone H3.
Importantly, acetylation of a T. brucei histone H4 tail peptide does not necessarily reflect the nature of the substrate in vivo as substrate specificity can be lost or compromised in vitro. As substrate specificity underpins the epigenetic code, we sought to link acetyl modifications to specific HATs in vivo. The highly divergent sequence of the trypanosome histone H4 N-terminal tail prohibits the use of commercially available antibodies to specific modifications but T. brucei histone-specific antibodies were recently used to demonstrate that HAT3, but not HAT1 or HAT2, acetylates H4K4 (Siegel et al., 2008). A similar approach was used to generate antibodies to acetylated H4K10 and specificity was tested by pre-incubation with peptide competitors before Western blotting. This demonstrated α-H4K10ac affinity only for the corresponding peptide and no cross-reactivity to other modified or unmodified sites (Fig. 7B). We then used α-H4K10 to screen a series of T. brucei protein extracts harvested from wild-type, HAT1 and HAT2 RNAi strains induced for different periods and a hat3 null strain. This demonstrated a reduction in signal specific to the HAT2 knock-down strain (Fig. 7C). We conclude that trypanosomes use distinct MYST-family acetyltransferases to acetylate K4 and K10 on the N-terminal tail of histone H4; H4K4 (73% of sites modified, see above) is acetylated by the dispensable HAT3 while H4K10 (7%) is acetylated by the essential HAT2. The data also show that loss of H4K4 acetylation has no discernible effect on H4K10 acetylation (Fig. 7C) and vice versa (Siegel et al., 2008).
Discussion
Histone modification is under study in a range of parasitic protozoa (Ramakrishnan et al., 2004; Smith et al., 2005; Merrick and Duraisingh, 2006; Sullivan et al., 2006). Histone acetylation and methylation have been described in T. brucei (Janzen et al., 2006a) and T. cruzi (da Cunha et al., 2006) and it is thought that trypanosomatids employ a relatively simple histone code to regulate access to DNA (Horn, 2007). T. brucei expresses three distinct MYST-family members, all of which have homologues in T. cruzi and Leishmania and we describe non-redundant roles for each of these histone acetyltransferases in bloodstream-form T. brucei. HAT1 modulates telomeric silencing and is required for growth, and possibly, for DNA replication; HAT2 is required for H4K10 acetylation and growth; and, HAT3 is required for H4K4 acetylation and is dispensable for growth. The orthologues in other trypanosomatids likely have similar roles. The non-redundant functions for T. brucei HAT1–3 appear to reflect unique substrates for each acetyltransferase and further support the idea of a simplified, non-redundant histone code in these divergent parasites.
Both HAT1 and HAT2 are essential for growth in bloodstream-form T. brucei, as demonstrated using both conditional expression in a null background and knock-down using RNAi. To our knowledge, this is the first example of a unicellular eukaryote with two essential MYST-family acetyltransferases. For comparison, S. cerevisiae (Howe et al., 2001) and S. pombe (Gomez et al., 2005) each express a single essential MYST acetyltransferase. Esa1 is essential in S. cerevisiae (Smith et al., 1998; Clarke et al., 1999) while the other, non-essential, MYST-family members, Sas2 and Sas3, also function in transcription regulation and cell cycle control (Reifsnyder et al., 1996). Interestingly, S. cerevisiae Sas3 displays redundancy with Gcn5, a non-MYST acetyltransferase. In the absence of Gcn5, Sas3 is also essential (Howe et al., 2001). This is thought to be because Esa1 favours histone H4 as substrate while Sas3 and Gcn5 both favour histone H3. Thus, two MYST-family acetyltransferases may be essential in T. brucei due to the absence of a Gcn5 homologue (see table S4 in Ivens et al., 2005). Further work will be required to determine whether the MYST-family HATs acetylate histone H3 lysine residues or other additional sites in T. brucei.
In S. cerevisiae, histone acetylation is essential for cell cycle progression (Clarke et al., 1999), and cells lacking either histone H3 or H4 tails are unable to progress through G2/M (Ling et al., 1996). Some conventional cell cycle checkpoints may be absent from T. brucei (Hammarton, 2007) but the T. brucei deacetylase, DAC4, is required for normal G2/M cell cycle progression (Ingram and Horn, 2002). Cells accumulate pre-cytokinesis following HAT1 or HAT2 knock-down and the absence of a parallel increase in tetraploid cells may reflect a DNA replication defect upon HAT1 knock-down. Indeed, yeast Sas2 is involved in chromatin assembly associated with DNA replication (Meijsing and Ehrenhofer-Murray, 2001; Osada et al., 2001). A similar defect was reported in cells defective in H3K79 methylation but in that case the cells progressed through cytokinesis to produce cells with a haploid DNA content (Janzen et al., 2006b).
Histone acetylation is predominantly linked to increased transcription as opposed to repression and can increase transcription by all three major eukaryotic RNA polymerases (Ura et al., 1997; Hirschler-Laszkiewicz et al., 2001). Acetylation may influence transcription by altering promoter accessibility or by facilitating elongation. The MYST complexes tend to exert their effects over domains rather than by targeting promoters. Most genes are constitutively transcribed by RNA pol II in T. brucei and conventional pol II promoters have not been identified for protein-coding genes (Palenchar and Bellofatto, 2006). Unusually, in T. brucei, the variant surface glycoprotein genes are transcribed by RNA pol I and regulation here appears to be at the level of transcription elongation (Vanhamme et al., 2000) while transcription mediated by the canonical ribosomal RNA pol I promoter is repressed immediately adjacent to telomeres in a sirtuin (SIR2rp1) deacetylase-dependent manner (Alsford et al., 2007). We demonstrate that HAT1 modulates telomeric silencing in T. brucei but neither SIR2rp1 (Alsford et al., 2007) nor HAT1–3 (this study) appear to have a role in variant surface glycoprotein expression site repression. The role in telomeric silencing is reminiscent of S. cerevisiae Sas2 (Reifsnyder et al., 1996) and Esa1 (Clarke et al., 2006) and S. pombe mst2+ function (Gomez et al., 2005). Sas2 is a histone H4K16-specific acetyltransferase (Meijsing and Ehrenhofer-Murray, 2001; Sutton et al., 2003; Shia et al., 2005) that opposes the action of the Sir2 histone deacetylase to establish boundaries between telomeric heterochromatin and euchromatin (Kimura et al., 2002; Suka et al., 2002). Deletion of Sas2 allows Sir2-dependent heterochromatin to extend further into subtelomeric DNA (Kristjuhan et al., 2003) and this redistribution of a limiting pool of Sir2 dilutes the effect immediately adjacent to the telomere. Our results therefore are consistent with a conserved role in telomeric silencing from yeast to trypanosomatids. Other MYST-family acetyltransferases have been shown to be required for repression but these effects are also now thought to be indirect. For example, Sas3 was originally identified as a regulator of gene silencing (Reifsnyder et al., 1996), but is now thought to stimulate transcription and replication (John et al., 2000). We cannot rule out a direct role for acetylation in telomeric silencing (Braunstein et al., 1996) but, like Sas2/Esa1 and Sir2 in S. cerevisiae, we propose that HAT1 and the sole nuclear sirtuin deacetylase, SIR2rp1, establish boundaries between transcriptionally active and repressed telomeric domains in T. brucei.
Although histone N-terminal sequences are highly divergent in trypanosomes, a number of specific lysine residues are acetylated within the N-terminal tail of T. brucei histone H4 (Mandava et al., 2007). Antisera against human and yeast histone modifications do not recognize specific modification on T. brucei histones so specific antibodies against trypanosome modifications are required to facilitate both identification of the relevant modifiers and further studies on HAT function. Specific antibodies have been used to show that HAT3 acetylates H4K4 (Siegel et al., 2008) and we now show that HAT2 acetylates H4K10. In humans (Sobel et al., 1995), flies (Sobel et al., 1994) and Tetrahymena (Chicoine et al., 1986), histone H4K5 and H4K12 are diacetylated by a cytosolic type-B HAT (Parthun et al., 1996). The role of these modifications is poorly understood but they may function in histone deposition (Sobel et al., 1995). Trypanosome H4K4, acetylated at > 70% of sites, does not appear to be analogous to H4K5 in other eukaryotes (Siegel et al., 2008) raising the possibility that trypanosome H4K5 and K10, each acetylated at about 7% of sites (Mandava et al., 2007), could be analogous to H4K5 and H4K12. Whatever the precise function of H4K4ac and H4K10ac, the presence of a pair of residues within the first 10 N-terminal residues of histone H4 independently targeted by specific MYST-family proteins is unprecedented and has implications for the evolution of the epigenetic code and for the interpretation of the code in different organisms.
Trypanosomatid genomes encode relatively few acetyltransferases, methyltransferases and cognate binding modules (see table S4 in Ivens et al., 2005), and the histones also display fewer modifications (da Cunha et al., 2006; Janzen et al., 2006a; Mandava et al., 2007), compared with other eukaryotes amenable to experimental analysis. Low-level redundancy should facilitate further functional dissection of the processes enacted by the T. brucei acetyltransferases.
Experimental procedures
T. brucei growth and manipulation
All strains were derived from Lister 427 bloodstream-form MITat1.2 (clone 221a). T. brucei were grown in HMI-11 and transformed with linear DNA constructs as previously described (Alsford et al., 2005). For expression of tagged proteins, and for RNAi, recombinant vectors were integrated at a marked RRNA locus in 2T1 T. brucei that express the tetracycline repressor (TetR) (Alsford et al., 2005). Drugs were added ∼6 h post transfection. Recombinant protein expression and RNAi were induced by growing T. brucei in 1 μg ml−1 tetracycline (Sigma) for 24 h unless stated otherwise. For growth analysis, T. brucei were seeded at a density of 1 × 105 ml−1 and split back to 1 × 105 ml−1 every 24 h where necessary. Counts were carried out using a haemocytometer.
Plasmid constructs
DNA fragments were amplified by PCR from genomic DNA using Phusion high-fidelity DNA polymerase (New England Biolabs) and T. brucei Lister 427 genomic DNA in conjunction with specific primer pairs (relevant restriction enzyme sites are indicated in italics and stop codons in lower-case below). For N-terminal eGFP-tagging constructs, pGFPHAT1–3, we used the Tet-responsive expression vector pRPaGFP (Alsford et al., 2005) and primers were as follows: for HAT1, HAT15N (GCTCTAGAGTAATGTTTGAGGTGCGGC) and HAT13N (GAAGATCTttaTACTTTCTCAGCTCTACG); for HAT2, HAT25N (GCTCTAGAGCGTCGTTAGCAGCAAAAAA) and HAT23N (CGGGATCCtcaGCTCTGGTGCTGATTG); and for HAT3, H35N (GCTCTAGAAAACGACAGAGGTCGGGA) and H33N (CGGGATCCttaTGATATCGGCACCCATAG).
For HAT1 and HAT2 gene disruption constructs, we amplified and cloned DNA fragments using the following primer pairs: for HAT1, 29M45 (TCCCCGCGGTCCTGCGACGTTAGGACTTA) and 29M43 (GGGGTACCGGTGTAGTCTTCTCTTTG) and for HAT2, 18M145 (TCCCCGCGGTACTATGCGCCTTATGGTA) and 18M143 (GGGGTACCGAATAGAAGAGTGAGGTAG). A 130 bp BamHI–Bsp120I fragment from HAT1 and a 679 bp EcoRV–SmaI fragment from HAT2 were replaced by blasticidin S deaminase (BSD) or puromycin N-acetyltransferase (PAC) selectable marker gene cassettes. For HAT3 knockout we amplified upstream and downstream targets using the H31 (CCACTAGTACCCCAGTAGA), H32 (ATCCTGCAGTCCACCTATACGGAAGGGA) and H33 (GGGGTACCAGTGTTTTTCACCTGTTT), H34 (CTGGTACCTCAGAAACAGG) primer pairs respectively. The targets were assembled such that they flanked a PAC or neomycin phospotransferase (NPT) selectable marker.
We used a stem-loop vector (pRPaiSL) to generate intramolecular dsRNA for RNAi. The RNAi trigger fragments were amplified using the following primers: for HAT1, H1R5SL (GATCGGGCCCGGTACCCCCTTTTCGGAACATGAAGA) and H1R3SL (GATCTCTAGAGGATCCAGTGTGACGCAGTGACTT); for HAT2, H2R5SL (GATCGGGCCCGGTACCACTCACAGACTTGGGAGCA) and H2R3SL (GATCTCTAGAGGATCCTCATGTCATGTGCCCACTTT); and for HAT3, H3R5SL (GATCGGGCCCGGTACCTGACCTCATTAAGCACGCTG) and H3R3SL (GATCTCTAGAGGATCCAAGGGCAAGGGCAGTATCTT).
DNA and RNA analysis
PCR, RT-PCR, Southern and Northern analysis were all carried out according to standard protocols. Signals on Northern blots were quantified using a Phosphorimager (Amersham).
Protein analysis
SDS-polyacrylamide gel electrophoresis and Western blotting were performed according to standard protocols (Ausubel et al., 1998). Western blots were developed using rabbit α-GFP RF795PB (kind gift from M. Rout) or rabbit α-GFP 11E5 (Molecular Probes) according to the manufacturer's instructions and tagged proteins were detected using enhanced chemiluminescence (Amersham). Western blot analysis using α-H4K10ac and α-H3 (Abcam, 1791) were carried out as described (Siegel et al., 2008). Indirect immunofluorescence microscopy was carried out as described (Alsford et al., 2007).
Cell cycle analysis and flow cytometry
Nuclear and kinetoplast DNA, counterstained with DAPI, were analysed as described (Ingram and Horn, 2002). For flow cytometry, 1 × 106 cells were fixed in 70% methanol, 30% PBS and incubated at 4°C overnight. Cells were washed in PBS then re-suspended in PBS containing 10 μg ml−1 propidium iodide and 10 μg ml−1 RNase A and incubated at 37°C for 45 min. A Becton Dickinson FACScalibur was used to analyse samples with cell quest software and detector FL2-A with an Amp gain value of 1.75. Data analysis was carried out using FlowJo (Treestar).
Histone acetyltransferase assay
Cells (1.5 × 108) expressing GFPHAT2 or the wild-type strain were lysed in RIPA buffer (Upstate) containing protease inhibitor cocktail (Sigma) at 4°C. Tagged protein (complex) was enriched using Catch and Release columns (Upstate) according to the manufacturers' instructions. Briefly, 9 μl of rabbit α-GFP serum (M. Rout, Rockefeller University) and 430 μl of lysate were applied to each column and incubated for 6 h at 4°C with continuous rotation followed by washing and elution with 70 μl of 1× non-denaturing buffer. An indirect ELISA (Upstate) was used to measure HAT activity according to the manufacturers' instructions. Briefly, streptavidin-coated plates were loaded with 1 μg ml−1T. brucei histone H4 peptide (H4A1-L20), biotin conjugate. Eluate (20 μl) was added followed by incubation at 30°C for 30 min followed by incubation with rabbit α-Ac-Lys and goat α-rabbit HRP conjugate both at 0.4 ng ml−1. Signals were quantified on a plate reader at 450 and 570 nm.
Acknowledgments
This work was supported by a Research Career Development Fellowship (to D.H., 052323) and a Project Grant (069909) from The Wellcome Trust, by the National Institutes of Health (AI 21729), a Medical Research Council studentship to T.K. and fellowships and travel grants to T.N.S. from Boehringer Ingelheim Fonds and the David Rockefeller Graduate Program. We thank John Kelly and Martin Taylor (LSHTM) for critical reading of the manuscript; Najib El-Sayed (TIGR, USA), Sara Melville and Vanessa Leech (University of Cambridge, UK) for sheared DNA clones (41a7 and 5a08); Farhana Chishti for assistance with sequencing and Mike Rout (Rockefeller University, USA) for α-GFP.
References
- Akhtar A, Becker PB. The histone H4 acetyltransferase MOF uses a C2HC zinc finger for substrate recognition. EMBO Rep. 2001;2:113–118. doi: 10.1093/embo-reports/kve022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akhtar A, Zink D, Becker PB. Chromodomains are protein–RNA interaction modules. Nature. 2000;407:405–409. doi: 10.1038/35030169. [DOI] [PubMed] [Google Scholar]
- Alsford S, Kawahara T, Glover L, Horn D. Tagging a T. brucei RRNA locus improves stable transfection efficiency and circumvents inducible expression position effects. Mol Biochem Parasitol. 2005;144:142–148. doi: 10.1016/j.molbiopara.2005.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alsford S, Kawahara T, Isamah C, Horn D. A sirtuin in the African trypanosome is involved in both DNA repair and telomeric gene silencing but is not required for antigenic variation. Mol Microbiol. 2007;63:724–736. doi: 10.1111/j.1365-2958.2006.05553.x. [DOI] [PubMed] [Google Scholar]
- Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K. Current Protocols in Molecular Biology. New York, NY: John Wiley and Sons; 1998. [Google Scholar]
- Bannister AJ, Zegerman P, Partridge JF, Miska EA, Thomas JO, Allshire RC, Kouzarides T. Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature. 2001;410:120–124. doi: 10.1038/35065138. [DOI] [PubMed] [Google Scholar]
- Belli SI. Chromatin remodelling during the life cycle of trypanosmatids. Int J Parasitol. 2000;30:679–687. doi: 10.1016/s0020-7519(00)00052-7. [DOI] [PubMed] [Google Scholar]
- Borrow J, Stanton VP, Andresen JM, Becher R, Behm FG, Chaganti RS, et al. The translocation t(8;16)(p11;p13) of acute myeloid leukaemia fuses a putative acetyltransferase to the CREB-binding protein. Nat Genet. 1996;14:33–41. doi: 10.1038/ng0996-33. [DOI] [PubMed] [Google Scholar]
- Braunstein M, Sobel RE, Allis CD, Turner BM, Broach JR. Efficient transcriptional silencing in Saccharomyces cerevisiae requires a heterochromatin histone acetylation pattern. Mol Cell Biol. 1996;16:4349–4356. doi: 10.1128/mcb.16.8.4349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrozza MJ, Utley RT, Workman JL, Cote J. The diverse functions of histone acetyltransferase complexes. Trends Genet. 2003;19:321–329. doi: 10.1016/S0168-9525(03)00115-X. [DOI] [PubMed] [Google Scholar]
- Champagne N, Bertos NR, Pelletier N, Wang AH, Vezmar M, Yang Y, et al. Identification of a human histone acetyltransferase related to monocytic leukemia zinc finger protein. J Biol Chem. 1999;274:28528–28536. doi: 10.1074/jbc.274.40.28528. [DOI] [PubMed] [Google Scholar]
- Chicoine LG, Schulman IG, Richman R, Cook RG, Allis CD. Nonrandom utilization of acetylation sites in histones isolated from Tetrahymena. Evidence for functionally distinct H4 acetylation sites. J Biol Chem. 1986;261:1071–1076. [PubMed] [Google Scholar]
- Clarke AS, Lowell JE, Jacobson SJ, Pillus L. Esa1p is an essential histone acetyltransferase required for cell cycle progression. Mol Cell Biol. 1999;19:2515–2526. doi: 10.1128/mcb.19.4.2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke AS, Samal E, Pillus L. Distinct roles for the essential MYST family HAT Esa1p in transcriptional silencing. Mol Biol Cell. 2006;17:1744–1757. doi: 10.1091/mbc.E05-07-0613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clayton C. Life without transcriptional control? From fly to man and back again. EMBO J. 2002;21:1881–1888. doi: 10.1093/emboj/21.8.1881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Cruz X, Lois S, Sanchez-Molina S, Martinez-Balbas MA. Do protein motifs read the histone code? Bioessays. 2005;27:164–175. doi: 10.1002/bies.20176. [DOI] [PubMed] [Google Scholar]
- da Cunha JP, Nakayasu ES, de Almeida IC, Schenkman S. Post-translational modifications of Trypanosoma cruzi histone H4. Mol Biochem Parasitol. 2006;150:268–277. doi: 10.1016/j.molbiopara.2006.08.012. [DOI] [PubMed] [Google Scholar]
- Dou Y, Milne TA, Tackett AJ, Smith ER, Fukuda A, Wysocka J, et al. Physical association and coordinate function of the H3, K4 methyltransferase MLL1 and the H4, K16 acetyltransferase MOF. Cell. 2005;121:873–885. doi: 10.1016/j.cell.2005.04.031. [DOI] [PubMed] [Google Scholar]
- Doyon Y, Cote J. The highly conserved and multifunctional NuA4 HAT complex. Curr Opin Genet Dev. 2004;14:147–154. doi: 10.1016/j.gde.2004.02.009. [DOI] [PubMed] [Google Scholar]
- Durand-Dubief M, Kohl L, Bastin P. Efficiency and specificity of RNA interference generated by intra- and intermolecular double stranded RNA in Trypanosoma brucei. Mol Biochem Parasitol. 2003;129:11–21. doi: 10.1016/s0166-6851(03)00071-9. [DOI] [PubMed] [Google Scholar]
- Eisen A, Utley RT, Nourani A, Allard S, Schmidt P, Lane WS, et al. The yeast NuA4 and Drosophila MSL complexes contain homologous subunits important for transcription regulation. J Biol Chem. 2001;276:3484–3491. doi: 10.1074/jbc.M008159200. [DOI] [PubMed] [Google Scholar]
- Ersfeld K, Melville SE, Gull K. Nuclear and genome organization of Trypanosoma brucei. Parasitol Today. 1999;15:58–63. doi: 10.1016/s0169-4758(98)01378-7. [DOI] [PubMed] [Google Scholar]
- Garcia BA, Hake SB, Diaz RL, Kauer M, Morris SA, Recht J, et al. Organismal differences in post-translational modifications in histones H3 and H4. J Biol Chem. 2007;282:7641–7655. doi: 10.1074/jbc.M607900200. [DOI] [PubMed] [Google Scholar]
- Garcia-Salcedo JA, Gijon P, Nolan DP, Tebabi P, Pays E. A chromosomal SIR2 homologue with both histone NAD-dependent ADP-ribosyltransferase and deacetylase activities is involved in DNA repair in Trypanosoma brucei. EMBO J. 2003;22:5851–5862. doi: 10.1093/emboj/cdg553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glover L, Horn D. Repression of polymerase I-mediated gene expression at Trypanosoma brucei telomeres. EMBO Rep. 2006;7:93–99. doi: 10.1038/sj.embor.7400575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glover L, Alsford S, Beattie C, Horn D. Deletion of a trypanosome telomere leads to loss of silencing and progressive loss of terminal DNA in the absence of cell cycle arrest. Nucleic Acids Res. 2007;35:872–880. doi: 10.1093/nar/gkl1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez EB, Espinosa JM, Forsburg SL. Schizosaccharomyces pombe mst2+ encodes a MYST family histone acetyltransferase that negatively regulates telomere silencing. Mol Cell Biol. 2005;25:8887–8903. doi: 10.1128/MCB.25.20.8887-8903.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammarton TC. Cell cycle regulation in Trypanosoma brucei. Mol Biochem Parasitol. 2007;153:1–8. doi: 10.1016/j.molbiopara.2007.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirschler-Laszkiewicz I, Cavanaugh A, Hu Q, Catania J, Avantaggiati ML, Rothblum LI. The role of acetylation in rDNA transcription. Nucleic Acids Res. 2001;29:4114–4124. doi: 10.1093/nar/29.20.4114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horn D. Nuclear gene transcription and chromatin in Trypanosoma brucei. Int J Parasitol. 2001;31:1157–1165. doi: 10.1016/s0020-7519(01)00264-8. [DOI] [PubMed] [Google Scholar]
- Horn D. Introducing histone modification in trypanosomes. Trends Parasitol. 2007;23:239–242. doi: 10.1016/j.pt.2007.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horn D. Codon usage suggests that translational selection has a major impact on protein expression in trypanosomatids. BMC Genomics. 2008;9:2. doi: 10.1186/1471-2164-9-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howe L, Auston D, Grant P, John S, Cook RG, Workman JL, Pillus L. Histone H3 specific acetyltransferases are essential for cell cycle progression. Genes Dev. 2001;15:3144–3154. doi: 10.1101/gad.931401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes K, Wand M, Foulston L, Young R, Harley K, Terry S, et al. A novel ISWI is involved in VSG expression site downregulation in African trypanosomes. EMBO J. 2007;26:2400–2410. doi: 10.1038/sj.emboj.7601678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iizuka M, Stillman B. Histone acetyltransferase HBO1 interacts with the ORC1 subunit of the human initiator protein. J Biol Chem. 1999;274:23027–23034. doi: 10.1074/jbc.274.33.23027. [DOI] [PubMed] [Google Scholar]
- Ingram AK, Horn D. Histone deacetylases in T. brucei: two are essential and another is required for normal cell-cycle progression. Mol Microbiol. 2002;45:89–97. doi: 10.1046/j.1365-2958.2002.03018.x. [DOI] [PubMed] [Google Scholar]
- Ivens AC, Peacock CS, Worthey EA, Murphy L, Aggarwal G, Berriman M, et al. The genome of the kinetoplastid parasite, Leishmania major. Science. 2005;309:436–442. doi: 10.1126/science.1112680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janzen CJ, Fernandez JP, Deng H, Diaz R, Hake SB, Cross GA. Unusual histone modifications in Trypanosoma brucei. FEBS Lett. 2006a;580:2306–2310. doi: 10.1016/j.febslet.2006.03.044. [DOI] [PubMed] [Google Scholar]
- Janzen CJ, Hake SB, Lowell JE, Cross GA. Selective di- or trimethylation of histone H3 lysine 76 by two DOT1 homologs is important for cell cycle regulation in Trypanosoma brucei. Mol Cell. 2006b;23:497–507. doi: 10.1016/j.molcel.2006.06.027. [DOI] [PubMed] [Google Scholar]
- Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- John S, Howe L, Tafrov ST, Grant PA, Sternglanz R, Workman JL. The something about silencing protein, Sas3, is the catalytic subunit of NuA3, a yTAF(II)30-containing HAT complex that interacts with the Spt16 subunit of the yeast CP (Cdc68/Pob3)–FACT complex. Genes Dev. 2000;14:1196–1208. [PMC free article] [PubMed] [Google Scholar]
- Jones DO, Cowell IG, Singh PB. Mammalian chromodomain proteins: their role in genome organisation and expression. Bioessays. 2000;22:124–137. doi: 10.1002/(SICI)1521-1878(200002)22:2<124::AID-BIES4>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- Kamine J, Elangovan B, Subramanian T, Coleman D, Chinnadurai G. Identification of a cellular protein that specifically interacts with the essential cysteine region of the HIV-1 Tat transactivator. Virology. 1996;216:357–366. doi: 10.1006/viro.1996.0071. [DOI] [PubMed] [Google Scholar]
- Kimura A, Umehara T, Horikoshi M. Chromosomal gradient of histone acetylation established by Sas2p and Sir2p functions as a shield against gene silencing. Nat Genet. 2002;32:370–377. doi: 10.1038/ng993. [DOI] [PubMed] [Google Scholar]
- Kowieski TM, Lee S, Denu JM. Acetylation-dependent ADP-ribosylation by Trypanosoma brucei Sir2. J Biol Chem. 2008;283:5317–5326. doi: 10.1074/jbc.M707613200. [DOI] [PubMed] [Google Scholar]
- Kristjuhan A, Wittschieben BO, Walker J, Roberts D, Cairns BR, Svejstrup JQ. Spreading of Sir3 protein in cells with severe histone H3 hypoacetylation. Proc Natl Acad Sci USA. 2003;100:7551–7556. doi: 10.1073/pnas.1332299100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lachner M, Jenuwein T. The many faces of histone lysine methylation. Curr Opin Cell Biol. 2002;14:286–298. doi: 10.1016/s0955-0674(02)00335-6. [DOI] [PubMed] [Google Scholar]
- Lee KK, Workman JL. Histone acetyltransferase complexes: one size doesn't fit all. Nat Rev Mol Cell Biol. 2007;8:284–295. doi: 10.1038/nrm2145. [DOI] [PubMed] [Google Scholar]
- Ling X, Harkness TA, Schultz MC, Fisher-Adams G, Grunstein M. Yeast histone H3 and H4 amino termini are important for nucleosome assembly in vivo and in vitro: redundant and position-independent functions in assembly but not in gene regulation. Genes Dev. 1996;10:686–699. doi: 10.1101/gad.10.6.686. [DOI] [PubMed] [Google Scholar]
- Mandava V, Fernandez JP, Deng H, Janzen CJ, Hake SB, Cross GA. Histone modifications in Trypanosoma brucei. Mol Biochem Parasitol. 2007;156:41–50. doi: 10.1016/j.molbiopara.2007.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin DG, Grimes DE, Baetz K, Howe L. Methylation of histone H3 mediates the association of the NuA3 histone acetyltransferase with chromatin. Mol Cell Biol. 2006;26:3018–3028. doi: 10.1128/MCB.26.8.3018-3028.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meijsing SH, Ehrenhofer-Murray AE. The silencing complex SAS-I links histone acetylation to the assembly of repressed chromatin by CAF-I and Asf1 in Saccharomyces cerevisiae. Genes Dev. 2001;15:3169–3182. doi: 10.1101/gad.929001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merrick CJ, Duraisingh MT. Heterochromatin-mediated control of virulence gene expression. Mol Microbiol. 2006;62:612–620. doi: 10.1111/j.1365-2958.2006.05397.x. [DOI] [PubMed] [Google Scholar]
- Millar CB, Grunstein M. Genome-wide patterns of histone modifications in yeast. Nat Rev Mol Cell Biol. 2006;7:657–666. doi: 10.1038/nrm1986. [DOI] [PubMed] [Google Scholar]
- Nielsen PR, Nietlispach D, Mott HR, Callaghan J, Bannister A, Kouzarides T, et al. Structure of the HP1 chromodomain bound to histone H3 methylated at lysine 9. Nature. 2002;416:103–107. doi: 10.1038/nature722. [DOI] [PubMed] [Google Scholar]
- Nielsen PR, Nietlispach D, Buscaino A, Warner RJ, Akhtar A, Murzin AG, et al. Structure of the chromo barrel domain from the MOF acetyltransferase. J Biol Chem. 2005;280:32326–32331. doi: 10.1074/jbc.M501347200. [DOI] [PubMed] [Google Scholar]
- Osada S, Sutton A, Muster N, Brown CE, Yates JR, Sternglanz R, Workman JL. The yeast SAS (something about silencing) protein complex contains a MYST-type putative acetyltransferase and functions with chromatin assembly factor ASF1. Genes Dev. 2001;15:3155–3168. doi: 10.1101/gad.907201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palenchar JB, Bellofatto V. Gene transcription in trypanosomes. Mol Biochem Parasitol. 2006;146:135–141. doi: 10.1016/j.molbiopara.2005.12.008. [DOI] [PubMed] [Google Scholar]
- Parthun MR, Widom J, Gottschling DE. The major cytoplasmic histone acetyltransferase in yeast: links to chromatin replication and histone metabolism. Cell. 1996;87:85–94. doi: 10.1016/s0092-8674(00)81325-2. [DOI] [PubMed] [Google Scholar]
- Ramakrishnan G, Gilchrist CA, Musa H, Torok MS, Grant PA, Mann BJ, Petri WA., Jr Histone acetyltransferases and deacetylase in Entamoeba histolytica. Mol Biochem Parasitol. 2004;138:205–216. doi: 10.1016/j.molbiopara.2004.09.002. [DOI] [PubMed] [Google Scholar]
- Redmond S, Vadivelu J, Field MC. RNAit: an automated web-based tool for the selection of RNAi targets in Trypanosoma brucei. Mol Biochem Parasitol. 2003;128:115–118. doi: 10.1016/s0166-6851(03)00045-8. [DOI] [PubMed] [Google Scholar]
- Reifsnyder C, Lowell J, Clarke A, Pillus L. Yeast SAS silencing genes and human genes associated with AML and HIV-1 Tat interactions are homologous with acetyltransferases. Nat Genet. 1996;14:42–49. doi: 10.1038/ng0996-42. [DOI] [PubMed] [Google Scholar]
- Respuela P, Ferella M, Rada-Iglesias A, Åslund L. Histone acetylation and methylation at sites initiating divergent polycistronic transcription in Trypanosoma cruzi. J Biol Chem. 2008;283:15884–15892. doi: 10.1074/jbc.M802081200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice JC, Allis CD. Histone methylation versus histone acetylation: new insights into epigenetic regulation. Curr Opin Cell Biol. 2001;13:263–273. doi: 10.1016/s0955-0674(00)00208-8. [DOI] [PubMed] [Google Scholar]
- Roth SY, Denu JM, Allis CD. Histone acetyltransferases. Ann Rev Biochem. 2001;70:81–120. doi: 10.1146/annurev.biochem.70.1.81. [DOI] [PubMed] [Google Scholar]
- Shia WJ, Osada S, Florens L, Swanson SK, Washburn MP, Workman JL. Characterization of the yeast trimeric-SAS acetyltransferase complex. J Biol Chem. 2005;280:11987–11994. doi: 10.1074/jbc.M500276200. [DOI] [PubMed] [Google Scholar]
- Shogren-Knaak M, Ishii H, Sun JM, Pazin MJ, Davie JR, Peterson CL. Histone H4-K16 acetylation controls chromatin structure and protein interactions. Science. 2006;311:844–847. doi: 10.1126/science.1124000. [DOI] [PubMed] [Google Scholar]
- Siegel TN, Kawahara T, Degrasse JA, Janzen CJ, Horn D, Cross GA. Acetylation of histone H4K4 is cell cycle regulated and mediated by HAT3 in Trypanosoma brucei. Mol Microbiol. 2008;67:762–771. doi: 10.1111/j.1365-2958.2007.06079.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith ER, Eisen A, Gu W, Sattah M, Pannuti A, Zhou J, et al. ESA1 is a histone acetyltransferase that is essential for growth in yeast. Proc Natl Acad Sci USA. 1998;95:3561–3565. doi: 10.1073/pnas.95.7.3561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith AT, Tucker-Samaras SD, Fairlamb AH, Sullivan WJ., Jr MYST family histone acetyltransferases in the protozoan parasite Toxoplasma gondii. Eukaryot Cell. 2005;4:2057–2065. doi: 10.1128/EC.4.12.2057-2065.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobel RE, Cook RG, Allis CD. Non-random acetylation of histone H4 by a cytoplasmic histone acetyltransferase as determined by novel methodology. J Biol Chem. 1994;269:18576–18582. [PubMed] [Google Scholar]
- Sobel RE, Cook RG, Perry CA, Annunziato AT, Allis CD. Conservation of deposition-related acetylation sites in newly synthesized histones H3 and H4. Proc Natl Acad Sci USA. 1995;92:1237–1241. doi: 10.1073/pnas.92.4.1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sternglanz R, Schindelin H. Structure and mechanism of action of the histone acetyltransferase Gcn5 and similarity to other N-acetyltransferases. Proc Natl Acad Sci USA. 1999;96:8807–8808. doi: 10.1073/pnas.96.16.8807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suka N, Luo K, Grunstein M. Sir2p and Sas2p opposingly regulate acetylation of yeast histone H4 lysine16 and spreading of heterochromatin. Nat Genet. 2002;32:378–383. doi: 10.1038/ng1017. [DOI] [PubMed] [Google Scholar]
- Sullivan WJ, Naguleswaran A, Angel SO. Histones and histone modifications in protozoan parasites. Cell Microbiol. 2006;8:1850–1861. doi: 10.1111/j.1462-5822.2006.00818.x. [DOI] [PubMed] [Google Scholar]
- Sutton A, Shia WJ, Band D, Kaufman PD, Osada S, Workman JL, Sternglanz R. Sas4 and Sas5 are required for the histone acetyltransferase activity of Sas2 in the SAS complex. J Biol Chem. 2003;278:16887–16892. doi: 10.1074/jbc.M210709200. [DOI] [PubMed] [Google Scholar]
- Taipale M, Rea S, Richter K, Vilar A, Lichter P, Imhof A, Akhtar A. hMOF histone acetyltransferase is required for histone H4 lysine 16 acetylation in mammalian cells. Mol Cell Biol. 2005;25:6798–6810. doi: 10.1128/MCB.25.15.6798-6810.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takechi S, Nakayama T. Sas3 is a histone acetyltransferase and requires a zinc finger motif. Biochem Biophys Res Comm. 1999;266:405–410. doi: 10.1006/bbrc.1999.1836. [DOI] [PubMed] [Google Scholar]
- Ura K, Kurumizaka H, Dimitrov S, Almouzni G, Wolffe AP. Histone acetylation: influence on transcription, nucleosome mobility and positioning, and linker histone-dependent transcriptional repression. EMBO J. 1997;16:2096–2107. doi: 10.1093/emboj/16.8.2096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanhamme L, Poelvoorde P, Pays A, Tebabi P, Van Xong H, Pays E. Differential RNA elongation controls the variant surface glycoprotein gene expression sites of Trypanosoma brucei. Mol Microbiol. 2000;36:328–340. doi: 10.1046/j.1365-2958.2000.01844.x. [DOI] [PubMed] [Google Scholar]
- Woodward R, Gull K. Timing of nuclear and kinetoplast DNA replication and early morphological events in the cell cycle of Trypanosoma brucei. J Cell Sci. 1990;95:49–57. doi: 10.1242/jcs.95.1.49. [DOI] [PubMed] [Google Scholar]
- Yan Y, Barlev NA, Haley RH, Berger SL, Marmorstein R. Crystal structure of yeast Esa1 suggests a unified mechanism for catalysis and substrate binding by histone acetyltransferases. Mol Cell. 2000;6:1195–1205. doi: 10.1016/s1097-2765(00)00116-7. [DOI] [PubMed] [Google Scholar]
- Yan Y, Harper S, Speicher DW, Marmorstein R. The catalytic mechanism of the ESA1 histone acetyltransferase involves a self-acetylated intermediate. Nat Struct Biol. 2002;9:862–869. doi: 10.1038/nsb849. [DOI] [PubMed] [Google Scholar]
- Yang XJ. The diverse superfamily of lysine acetyltransferases and their roles in leukemia and other diseases. Nucleic Acids Res. 2004;32:959–976. doi: 10.1093/nar/gkh252. [DOI] [PMC free article] [PubMed] [Google Scholar]


