Abstract
Nucleoside diphoshate kinases (NDKs), an evolutionarily conserved family of proteins, synthesize nucleoside triphosphates from nucleoside diphosphates and ATP. Here, we have characterized the kinase activity and DNA processing functions of eight human proteins that contain at least one domain homologous to E. coli NDK. Not all human proteins with NDK-like domains exhibited NDK activity when expressed as recombinant proteins in E. coli. Human NDK1 (NM23-H1) has been reported to have 3' to 5' exonuclease activity. In addition to human NDK1, we also find that human NDK5, NDK7 and NDK8 contain 3' to 5' exonuclease activity. Site-directed mutagenesis, competition assays between wild type and mutant NDK proteins, and NMR studies confirmed that the DNA binding and 3' to 5' exonuclease activity of human NDK1 is an intrinsic activity of the protein. Using double-stranded DNA substrates containing modified bases, human NDK1 efficiently excised nucleotides from the single strand break produced by APE1 or Nth1. When human cells were treated with various DNA damaging agents, human NDK1 translocated from the cytoplasm to the nucleus. These results suggest that, in addition to maintenance of nucleotide pool balance, the human NDK-like proteins may have previously unrecognized roles in DNA nucleolytic processing.
Nucleoside diphosphate kinases (NDKs) have been characterized as a large family of conserved proteins that synthesize nucleoside triphosphates from nucleoside diphosphates and ATP (1). One of the main functions of the NDKs is the maintenance of the intracellular nucleoside triphosphate pools. However, it has been shown that the disruption of the single NDK genes in E. coli, Saccharomyces cerevisiae, and Schizosaccharomyces pombe has no effect on cell growth or morphology (2-4). Some microorganisms such as mycoplasma species and Thermotoga maritima lack an NDK gene altogether (1). E. coli strains with a disrupted NDK gene show a mutator phenotype which is thought to be due to imbalances in nucleotide pools (4, 5).
It has become recognized that some NDK proteins have additional or different roles, in particular in mammals. These genes and their corresponding proteins are referred to here as NDKs, but the mammalian homologues have also been named NM23-H or NME genes based on the first discovery of a mouse gene that showed strongly reduced expression in metastatic mouse melanoma cell lines (NM23 stands for “nonmetastatic clone 23”) (6). To date, eight human NDK homologues have been identified (1). NDK1 (NM23-H1) seems to be involved in suppression of tumor metastasis since low expression of NDK1 has been linked to increased metastatic potential in many cell lines and tumors (6-9). The NDK proteins are usually expressed ubiquitously but NDK5, NDK7, and NDK8 are mainly found in testis (1, 10, 11). NDK4 and NDK6 are localized predominantly in mitochondria (12, 13).
There is increasing evidence that some NDK proteins have DNA processing functions. In particular, NDK2 has been shown to be involved in regulation of transcription, by binding to and activating a nuclease-hypersensitive element in the c-myc promoter (14). Certain NDK proteins, such as NDK1, NDK2, and E. coli NDK can cleave DNA sequences with unusual structures in vitro (15-17). A recent study has shown that NDK1 is involved as the DNA cleavage component in a complex that promotes cytotoxic T-lymhocyte-mediated apoptosis (18) and this protein has been characterized biochemically as a 3' to 5' exonuclease (19).
Recently, it has been reported that E. coli NDK possesses multifunctional base excision repair activities, namely uracil-DNA glycosylase, AP-lyase and 3'-phosphodiesterase activity, in vitro (20). However, subsequently, two different groups have reported that E. coli NDK does not possess uracil-DNA glycosylase (21, 22) and AP-lyase activity (21).
Here, we have characterized the NDP kinase activity and DNA processing functions of eight human proteins that contain at least one domain homologous to E. coli NDK. We report that only human NDK1, NDK2 and NDK4 contain kinase activity and that human NDK1, NDK5, NDK7 and NDK8 retain 3' to 5' exonuclease activity.
MATERIALS AND METHODS
Reagents and oligonucleotides
Oligonucleotides for PCR primers and substrates containing uracil (U) were purchased from IDT (Coralville, IA). Oligonucleotides containing thymine glycol (Tg) were kindly provided by Dr. Shigenori Iwai (Osaka University) and were synthesized as previously described (23). E. coli uracil DNA glycosylase (UDG), and restriction enzymes were purchased from New England Biolabs (Beverly, MA). All chemicals and reagents were from Sigma (St. Louis, MO). E. coli Fpg and Nth proteins, and human APE1 were purified as previously described (24, 25).
Construction of plasmids containing E.coli NDK, human NDKs and human NDK1 mutants
A PCR fragment of the E. coli NDK gene was prepared with E. coli genomic DNA, Pfu turbo DNA polymerase (Stratagene; La Jolla, CA) and two primers, forward primer 5'GGTCGGGATCCGATGGCTATTGAACGT (BamHI site underlined) and reverse primer 5'GTGCTCGAGTTAACGGGTGCGCGG (XhoI site underlined). To construct plasmids containing human NDKs, human NDK cDNA clones or EST clones were obtained from the ATCC (Manassas, VA). Human NDK1 and NDK2 cDNAs were directly subcloned into plasmid pET-28a(+) (Novagen; Madison, WI). For all other human NDKs, PCR products containing appropriate restriction sites were prepared from cDNA clones or EST clones using Pfu turbo DNA polymerase and specific primers for in frame insertion. The PCR products were cloned into plasmid pET-28b(+). The ligated plasmids were transformed into bacterial competent cells BL21 (DE3) (Stratagene; La Jolla, CA). To construct the pET-28a(+) vectors containing point mutations of human NDK1, the Quikchange™ site-directed mutagenesis kit (Stratagene; La Jolla, CA) was used. The plasmids were isolated and sequenced to confirm the cDNA sequence. These expression vectors generate an N-terminal 6x-histidine-tagged NDK open reading frame.
Purification of recombinant human NDK proteins
Recombinant 6x-His-tagged NDK proteins were purified from E. coli under native conditions using the QIAexpressionist kit (Qiagen; Valencia, CA). The bound proteins were released from the Ni-NTA-agarose column with elution buffer (50 mM sodium phosphate, pH 8.0, 0.1% Triton X-100, 500 mM NaCl and each of 100 mM, 150 mM and 250 mM imidazole, respectively). Purified NDKs were dialyzed against storage buffer (20 mM HEPES, pH 7.9, 20 mM KCl, 5 mM beta-mercaptoethanol and 40% glycerol).
DEAE sepharose (Amersham Pharmacia Biotech) chromatography was used for additional purification of the NDK proteins. The 250 mM imidazole fractions of human NDKs were dialyzed overnight at 4°C against loading buffer (20 mM Tris-HCl, pH 8.2, 0.5 mM EDTA, 5 mM beta-mercaptoethanol and 5% glycerol). Bound proteins were eluted with a linear gradient of 0-500 mM NaCl in loading buffer. The eluted human NDK proteins were dialyzed against storage buffer. For human NDK1, additional gel filtration chromatography was carried out with ACA54 columns (Amersham Pharmacia Biotech).
Phosphoenzyme and enzyme coupled kinase assay
Purified E. coli NDK, human NDKs and human NDK1 mutants were tested for kinase activity. For phosphoenzyme formation, 500 ng of each NDK was used in an autophosphorylation reaction with 1 μM gamma-32P-ATP for 10 min at 37°C in 10 μl reaction buffer (20 mM HEPES, pH 7.9, 20 mM KCl, 2 mM MgCl2 and 20% glycerol). The reaction was stopped by adding SDS gel loading buffer and the samples were loaded onto a 15% SDS-polyacrylamide gel. After electrophoresis, the wet gel was autoradiographed and then stained with Coomassie blue.
The kinase activity of NDKs and human NDK1 mutants was determined by an enzyme coupled assay as previously described (16, 17, 20, 26). Briefly, one milliliter reactions in buffer containing 50 mM Tris-HCI, pH 7.4, 50 mM KCI, 6 mM MgCl2, 0.1 mM phosphoenolpyruvate, 0.1 mg/ml NADH, 2 units of pyruvate kinase, 2.5 units of lactate dehydrogenase, and 1 mg/ml BSA (bovine serum albumin) in plastic disposable cuvettes were incubated at room temperature in the presence of ATP as the donor of phosphate and dTDP as the phosphate acceptor nucleotides. The reaction was initiated by the addition of 200 ng of purified NDKs. NADH oxidation, which produced a decrease in absorbance at 340 nm was measured by UV spectrophotometry. Nucleoside diphosphate kinase activity of the NDKs was determined by calculation of the decreasing value of absorbance per minute at the initial rates where a typical reaction rate is produced.
Exonuclease assays
The exonuclease activities of purified recombinant E.coli and human NDKs were tested with a standard 30-mer single-stranded oligonucleotide, 5'-CTCGTCAGCATCATGATCATACAGTCAGTG-3'. To verify the substrate specificity, various oligonucleotide substrates were tested (27). The oligonucleotide sequences were 5'TTGAGGCAGAGTCC (O-1), 5'GGACTCTGCCTCAA (O-2), 5'GGACTCT-GCCTCAAG (O-3), 5'GGACTCTGCCTCAAGACG (O-4), 5'CACGTTGACTA-CCGTC (O-5), 5'GGACTCTGCCTCAAGACGGTAGTCAA-CGTG (O-6), and 5'GATGTCAAG-CAGTCCTAAGTTTGAGGCAGAGTCC (O-7). The 5' end-labeling of each top strand was carried out with T4 polynucleotide kinase and 32P-ATP. The end-labeled oligonucleotides were annealed to their complementary bottom strands in annealing buffer (10 mM Tris-HCl, pH 8.0, 1 mM EDTA, 30 mM NaCl and 40 mM KCl). The standard assay was performed in a 30 μl volume with reaction buffer (20 mM HEPES, pH 7.9, 20 mM KCl and 2 mM MgCl2), 5 pmoles DNA, and E. coli or human NDKs (100 ng protein) at 37°C for 1 hour. The exonuclease activity of human NDK1 mutants was tested with the 5' end-labeled single-stranded 30-mer in standard assay as described above. For competition assays between wild type and the catalytically inactive mutants, 0.5-5 μg of human NDK1 mutants E5A, E5Q, E129A, and E129Q, were preincubated with the 5'end-labeled single-stranded 30mer in reaction buffer (20 mM HEPES, pH 7.9, 20 mM KCl and 2 mM MgCl2) at room temperature for 20 min. After adding 0.5 μg of wild type protein, an additional reaction was carried out at 37°C for 30 min. The reaction was terminated by phenol/chloroform extraction and ethanol precipitation. The DNA pellet was dissolved in formamide loading dye. The cleavage of the oligomers was analyzed by 16% denaturing polyacrylamide gel electrophoresis.
To test the processing of substrates containing modified bases, double stranded oligonucleotides containing U/A or Tg/A pairs were used. The 30-mer top strand oligonucleotides were 5'-CTCGTCAGCATCA-(U-or-Tg)GATCATACAGTCAGTG-3', where “U or Tg” stands for uracil and thymine glycol (Tg), respectively. The 5'-labeled top strands were annealed to the bottom strands and the double stranded oligonucleotides containing uracil were first incubated with E. coli UDG (10 units) or human UDG (50 ng) for 20 min at 37°C. After directly adding human APE1 (50 ng), the mixtures were incubated for an additional 20 min at 37°C. The reaction mixtures were split into two further reactions with hNDK1, direct incubation with hNDK1 or incubation with hNDK1 after removing UDG and hAPE1 by phenol/chloroform extraction and ethanol precipitation. The samples were incubated for an additional 30 min at 37°C. The cleaved DNA was analyzed as described above.
Cell lines and antibodies
HeLa cervical carcinoma and ARPE-19 retinal epithelial cell lines were purchased from ATCC and grown in DMEM with 10% fetal bovine serum, 2 mM glutamine, 100 units/ml penicillin, and 100 mg/ml streptomycin. Rabbit antisera against human NDK1, HDAC2, beta-tubulin, H-CAM, normal rabbit IgG and HRP-conjugated goat anti-rabbit IgG, mouse mAb against hNDK1, horseradish peroxidase (HRP) conjugated donkey anti-mouse IgG, and normal mouse IgG, and protein A and G plus-agarose were purchased from Santa Cruz Biotechnology (Santa Cruz, CA). Rabbit anti-human UDG (Imgenex; San Diego, CA), mouse anti-human APE1 (Trevigen; Gaithersburg, MD), rabbit anti-human NTH1 (Novus Biologicals; Littleton, CO) were used for Western blot analysis.
Immunohistochemistry and Western blot analysis
HeLa cells were grown on glass coverslips (40-50% confluence) and treated with various DNA damaging agents such as H2O2 (1 or 5 mM, 20 min at 37°C), cisplatin (5-20 μM, 20 min at 37°C), solar simulated UV radiation for 5 or 10 min emitted from a 1000 W solar UV simulator (Oriel Inc.), UVB light emitted from sunlamps (556 J/m2 per minute), and UVA light emitted from two 360 nm black lights (Sylvania 15W F15T8; 1.13 kJ/m2 per minute). The UV dose emitted from the UV lamps was measured with a UVX radiometer (Ultraviolet Products; Upland, CA). For DNA damaging agent treatment, the medium was removed and cells were treated in PBS. After treatment, the DMEM culture medium was replaced and cells were incubated for an additional 20 min or 24 hours. Cells were fixed in 3.7% (v/v) paraformaldehyde in PBS for 10 min at room temperature and were then washed with PBS. Cells were permeabilized with 0.2% Triton X-100 in PBS for 15 min at room temperature and incubated in blocking buffer (PBS containing 1% bovine serum albumin and 0.2% Tween 20) for 1 hour at room temperature. Rabbit anti-human NDK1 (0.2 μg/ml) was added in blocking buffer and incubated for 1 hour at room temperature. Cells were washed 3 times with PBS containing 0.2% Tween 20 for 5 min each. Alexa Fluor 488-conjugated anti-rabbit secondary antibody (1 μg/ml; Molecular Probes; Eugene, OR) was added to the cells and incubated in blocking buffer for 1 hour at room temperature followed by 3 washes as described above. Nuclear counterstaining was performed with 0.2 μg/ml 4′, 6′-diamidino-2-phenylindole (DAPI) in PBS for 20 min. The cells were washed five times with PBS, mounted with Fluoromount G (Southern Biotechnology Associates; Birmingham, AL), and visualized with a fluorescence microscope (Olympus IX81).
For Western blot analysis, HeLa and ARPE-19 cells (1-2 × 106 cells) were treated with various DNA damaging agents such as solar-simulated UV irradiation, H2O2, cisplatin, UVB irradiation and UVA irradiation. Using a nuclear extract preparation kit (Chemicon; Temecula, CA), cellular and nuclear extracts were prepared from DNA damaging agent treated cells. Equivalent amounts (approximately 10 μg) of prepared cellular and nuclear extracts were mixed with SDS loading buffer and heated at 95°C for 5 min. The proteins were separated on a 15% SDS-polyacrylamide gel and transferred to a PVDF membrane. The blots were probed with polyclonal antibodies against human NDK1, HDAC2, and β-tubulin followed by appropriate secondary antibodies conjugated with horseradish peroxidase. The signals were detected using ECL-Plus (Amersham).
NMR
The 15N-labled NDK1 protein sample was dissolved in 90% H2O/10% D2O containing 10 mM phosphate buffer at pH 7.0 (final concentration of the monomer protein was approximately 0.5 mM). Then a sample of 20 mM unlabeled DNA was titrated to the protein to a molar ratio of 2:1. The DNA molecule was a single stranded 30-oligomer, which was the same as the one used as standard substrate in the exonuclease activity assays. The NMR spectra were acquired at 25°C on a Brucker Avance 600 spectrometer equipped with a cryoprobe. The NMR data were processed with NMRPIPE (28) and analyzed with NMRVIEW (29).
RESULTS
Purified human NDK1, NDK5, NDK7 and NDK8 contain 3' to 5' exonuclease activity
Genome database searches and review of the literature (1) indicated that the human genome contains at least eight homologues of E. coli NDK. The eight human genes were obtained from cDNA or EST clones and were sequenced. Figure 1A indicates the degree of homology between the NDK domains. Human NDKs 1-4 are most closely related to the E. coli protein. NDK7 has a duplicated NDK domain (Fig. 1B). NDK8 is a fusion protein composed of a thioredoxin domain and two and a half tandemly repeated NDK domains and has also been described as SPTRX2 (11).
Figure 1. Human proteins with domains homologous to E. coli NDK.
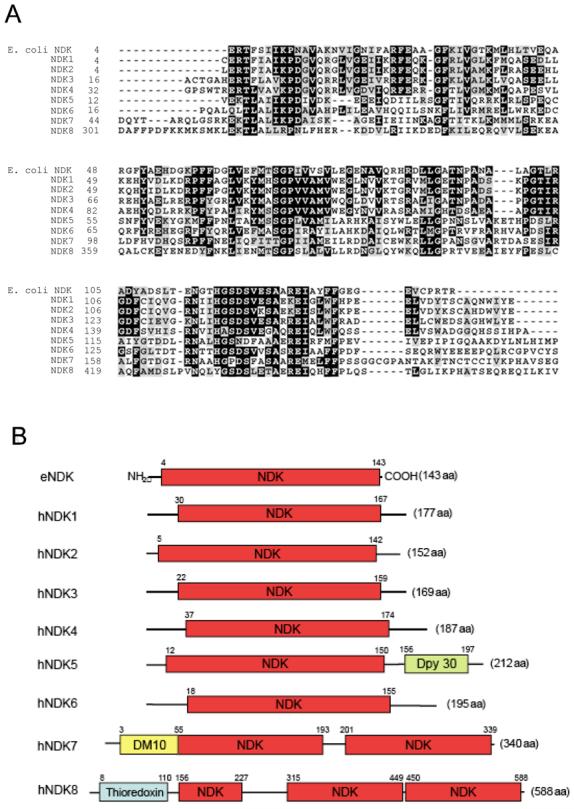
A. The sequence alignment of E. coli NDK protein and the NDK domains found in eight human homologues is shown. Alignment was performed with the Clustal W program. For NDK7, the N-terminal NDK domain was used for alignment and for NDK8, the central domain was used. B. Domain structures of E. coli NDK and the human NDK domain proteins. Additional domains indicated are a Dpy30 motif (probably formed of two alpha-helices) in NDK5, a DM10 motif (unknown function) in NDK7, and a thioredoxin domain in NDK8.
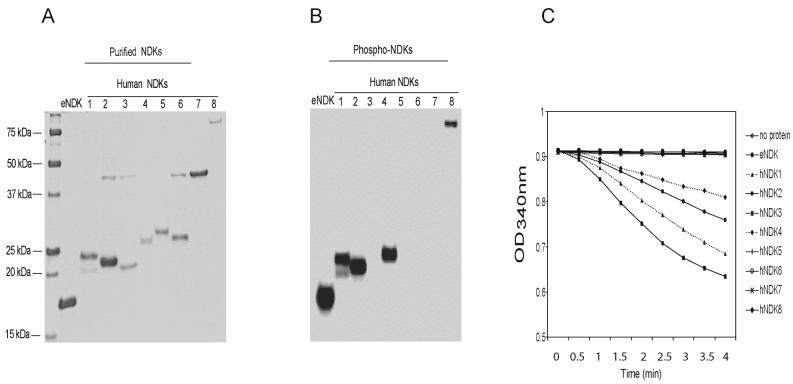
E. coli NDK and the eight human proteins were expressed and purified as recombinant histidine-tagged proteins from E. coli (Fig. 2A). We tested the recombinant NDK proteins for potential nucleoside diphosphate kinase activity, both by gel-based detection of a phosphohistidine intermediate (Fig. 2B) and by enzymatic assays (Fig. 2C). Only E. coli NDK, and human NDK1, NDK2, and NDK4 displayed measurable nucleoside diphosphate kinase activities. NDK8 showed autophosphorylation but was inactive in the standard dNDP substrate assay. The lack of NDP kinase activity in the other recombinant NDKs indicates that these proteins intrinsically lack such activity or improper folding or lack of posttranslational modifications precluded its detection.
Figure 2. Purified recombinant human NDK proteins and nucleoside diphosphate kinase assays.
A. The recombinant proteins, obtained after Ni-NTA-agarose and DEAE sepharose chromatography, were resolved on an SDS-polyacrylamide gel and stained with Coomassie Blue. Preparations of NDK2, NDK3, and NDK6 contained a contaminant of 45 kDa, which could not be separated from the NDK proteins. B. Phosphoenzyme essay. Catalytically active NDKs contain a histidine phosphotransferase activity. This could be demonstrated for E. coli NDK, and human NDK1, NDK2, NDK4, and NDK8. C. Enzymatic assay for nucleoside diphosphate kinase activity. A decrease in absorbance at 340 nm is indicative of the presence of enzymatic activity. This could be demonstrated for E. coli NDK, and human NDK1, NDK2, and NDK4.
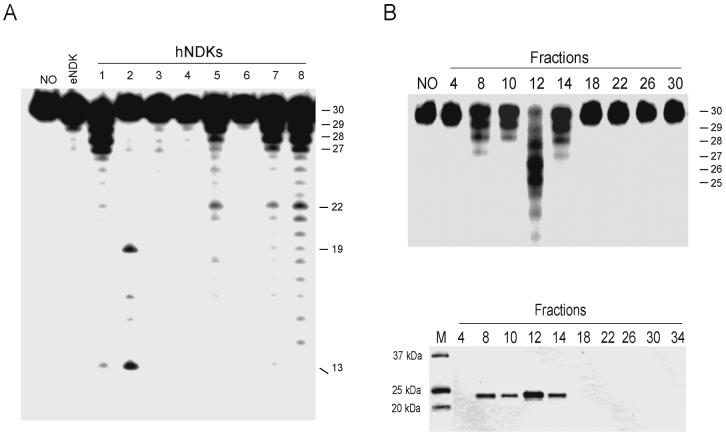
To search for exonuclease activity in the recombinant proteins, we used a 30-mer single stranded oligonucleotide substrate that was labeled at its 5' end. Interestingly, only human NDK1, NDK5, NDK7, and NDK8 showed pronounced 3' to 5' exonuclease activity (Fig. 3A). The DNA laddering observed is suggestive of removal of one nucleotide at a time by these enzymes. Human NDK2 showed evidence for internal cleavage of the oligonucleotide. However, E. coli NDK and other human NDKs exhibited little or no exonuclease activity (Fig. 3A). We found that human NDK1 required 1 to 2 mM Mg2+ for optimal reaction. Exonucleolytic cleavage increased as a function of time and NDK1 concentration and a relatively high amount of hNDK1 (50 ng or more per reaction with 5 picomoles of DNA) was required to observe exonucleolytic cleavage of single stranded DNA substrates (data not shown; see also below, Fig. 7C).
Figure 3. Exonuclease activity of purified human NDK proteins and co-elution of human NDK 1 with exonuclease activity.
A. The single-stranded 30-mer oligonucleotide (5 pmoles) was labeled at the 5' end. Purified recombinant E. coli NDK and human NDKs (100 ng each) were tested for exonuclease activity. No, no enzyme. B. Ni-NTA column-purified human NDK1 was eluted from a DEAE sepharose column and the fractions were assayed for exonuclease activity (top panel) and protein content by SDS-PAGE (bottom panel).
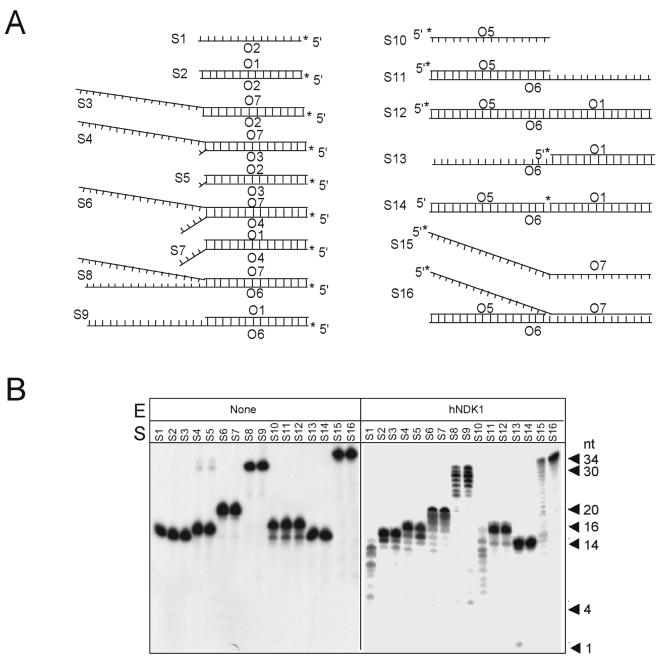
Figure 7. Exonuclease activity of human NDK 1 protein with various substrates.
Oligonucleotide substrates with single stranded, double stranded, or partially single stranded configurations were incubated with no protein (control) or with human NDK1 (200 ng). The cleavage products were separated on polyacrylamide gels. A. Composition of the substrates. The asterisks indicate the positions of the label. B. Cleavage assays. Exonuclease activity of human NDK1 was most pronounced with single-stranded substrates or with substrates containing single-stranded 3' overhangs. Similar data were obtained with human NDK5 and NDK7 (data not shown).
Since the proteins were purified as recombinants from E. coli, we were concerned that the enzyme preparations were potentially contaminated with E. coli proteins having exonuclease activity. To exclude this possibility, we purified human NDK1, NDK5, and NDK7 through subsequent steps consisting of Ni-NTA-agarose, DEAE sepharose, and gel filtration columns. In all cases, the NDK proteins, detected on SDS gels, exactly co-purified with the 3' to 5' exonuclease activities (Fig. 3B for human NDK1; human NDK5 and NDK7 data not shown).
The 3' to 5' exonuclease activity of human NDK1 is confirmed by site-directed mutagenesis, competition, and NMR studies
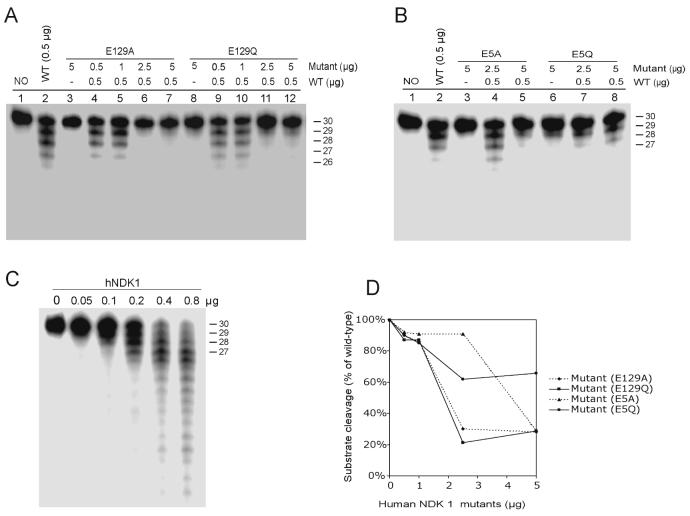
To confirm the exonuclease activity of human NDKs, we created and purified several human NDK1 mutants and tested their exonuclease activity. Since acidic amino acids, in particular Asp (D) and Glu (E), having carboxylic acid residues commonly associated with divalent cation binding, are often essential for nuclease activity (27, 30), we chose the four acidic amino acid residues, E5, D54, D121, and E129, which are the most highly conserved in the NDK domain (Fig. 1). We also converted three additional amino acids, F33, K12, and H118. F33 is also a highly conserved amino acid in the NDK domain. In previous studies, K12 was critical for both the kinase activity and DNA cleaving activity of human NDK2 (17) and the H118F mutation prevented the formation of a phosphorylated intermediate required for kinase activity (26). A total of nine human NDK1 mutants along with the wild-type protein were employed to test the exonuclease activity and kinase activity. The purified mutants are shown in Figure 4A. Several of these mutants almost completely lost the 3' to 5' exonuclease activity (Fig. 4B). The E5Q and H118F mutants showed approximately 20% of wild type enzyme activity but other mutants contained less than 10% of wild type activity. However, the NDK activity among these mutants was differently affected by the mutations compared to the exonuclease activity (Fig. 4C). Whereas some mutants such as K12Q, H118F, E129A and E129Q completely lost kinase activity, D54A, E5A and E5Q still retained significant NDP kinase activity. These data confirm that the 3' to 5' exonuclease activity of human NDK1 is not due to contamination by bacterial nucleases but is an intrinsic activity of this protein. Moreover, the active site and critical amino acids for the kinase and exonuclease activities appear to be different (Table 1). For example, D54A showed kinase activity close to that of the wild type protein but lost almost completely the exonuclease activity.
Figure 4. Exonuclease and NDP kinase activity of purified human NDK1 mutants.
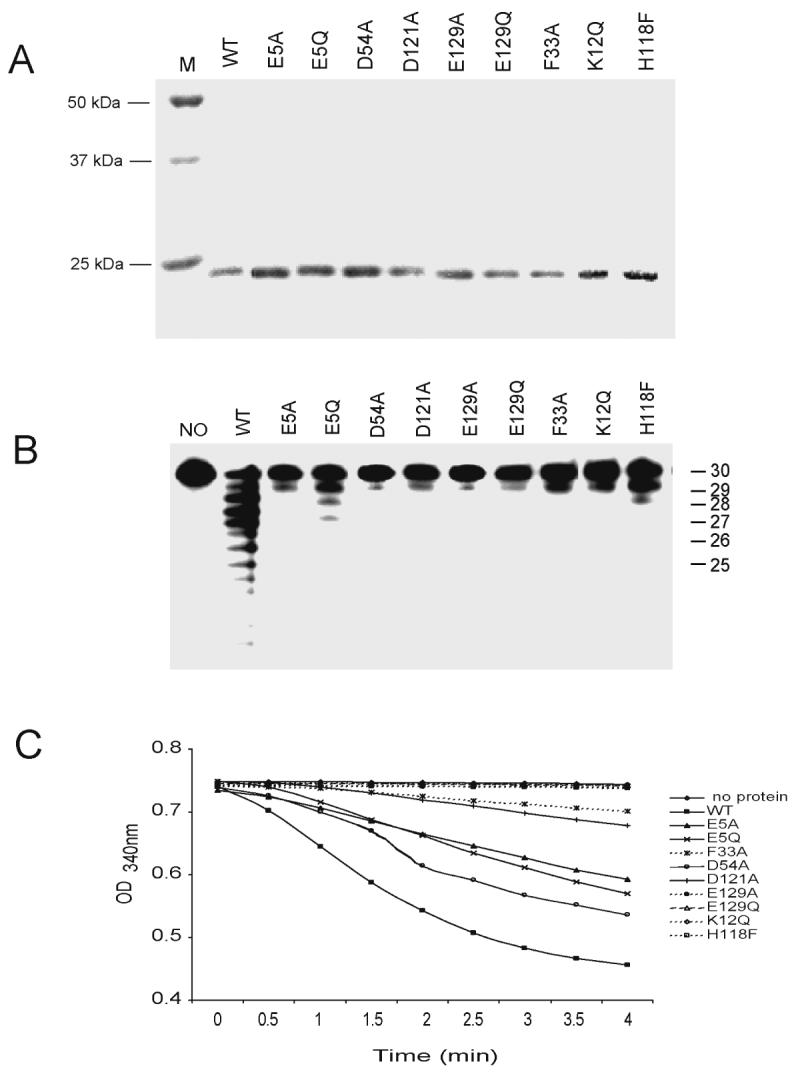
A. Ni-NTA column-purified human wild type and mutant NDK1 proteins on an SDS-PAGE gel. B. Exonuclease activity of purified human wild type and mutant NDK1 proteins was tested with a single-stranded 30-mer oligonucleotide and equal amounts (100 ng) of purified proteins. C. Enzymatic assay for nucleoside diphosphate kinase activity. D54A, E5Q and E5A mutants retained relatively high kinase activity.
Table 1.
Activities of human NDK1 mutants
Since inactive mutants are suspected to lose their activity in either of two ways, a defect in substrate binding, loss of cleavage activity or both (30), we performed a competition assay between four mutants (E5A, E5Q, E129A and E129Q) and wild type human NDK1 with single stranded oligonucleotide substrates. If the mutation only affects catalysis but not substrate binding, then it may inhibit wild-type enzyme activity at relatively high concentrations of the mutant. Various amounts (0.5-5 μg) of human NDK1 mutants were preincubated with DNA substrates to allow them to bind to the substrates and then wild type protein was added to the mixture. As shown in Figure 5A and 5B, at a lower concentration of mutants (0.5-1 μg), there is little inhibitory effect on the wild type reaction so that DNA cleavage was similar to the level of the wild type alone reaction. Although there is different inhibitory capacity among the mutants (Fig. 5C), at high concentrations (10-fold excess), all the mutants severely inhibited the wild type activity. These results also provide good evidence to support the intrinsic exonuclease activity of human NDK1.
Figure 5. Competition assay between human NDK1 wild type and mutants with a single stranded oligonulceotide substrate.
A. The competition effect of E129A and E129Q mutants on wild type NDK1 exonuclease activity is shown. Various amounts of E129A and E129Q proteins were preincubated with the single-stranded oligonucleotide substrate followed by incubation with 0.5 μg of wild type human NDK1. B. E5A and E5Q mutants were used for the competition assay. C. Increasing amounts of wild type NDK1 were used with the same substrate. D. Quantified data of the competition effects by human NDK1 mutants. The cleaved substrates were quantified by phophoimager analysis and converted to percentage values relative to wild type NDK1.
To investigate if hNDK1 specifically binds to a DNA substrate, an NMR study was performed. The interaction between human NDK1 and a single-stranded oligonucleotide substrate was characterized by NMR chemical shift perturbation (Fig. 6). Uniformly 15N-labeled human NDK1 and unlabeled DNA were used in this study. When a protein forms specific interactions with another molecule, such as DNA, the amino acid residues at the binding interface will inevitably experience changes of their environments. Such changes of environment would result in specific changes of chemical shifts of the nuclei of the amino acid residues at the binding interface. The amino acid residues that are not located at the binding interface would not experience such changes of environment, and thus would not exhibit any changes of chemical shifts. 1H-15N heteronuclear single-quantum coherence (HSQC) experiments were used to investigate whether human NDK1 binds specifically to the DNA substrates. Before the titration, both the protein and DNA were in an identical buffer at identical pH, in order to ensure that the pH of the solution remains unchanged during the titration. The resonances in the 1H-15N HSQC spectra of human NDK1 in the presence or absence of DNA were very broad and not well resolved in the central region, which is consistent with previous findings that human NDK1 forms an oligomer in solution (31). However, regions of the spectra contain well-resolved resonances. Figure 6 shows such a region of the superimposed 1H-15N HSQC spectra of human NDK1, free and when bound to DNA. Most of the resonances of human NDK1 did not show any changes of chemical shifts, as shown by the good overlay of most cross peaks. This suggests that binding the DNA does not cause a global conformational change in the protein. However, specific chemical shift changes occurred upon the titration with DNA, and the changes are linearly related to the amount of DNA titrated to the protein sample. Thus, these chemical shift changes, as shown in Figure 6, indicate that human NDK1 binds specifically to the DNA substrate. The NMR data lend support to the notion that DNA binding of human NDK1 is an intrinsic activity of the protein.
Figure 6. NMR study with purified human NDK1 and a single stranded oligonucleotide substrate.
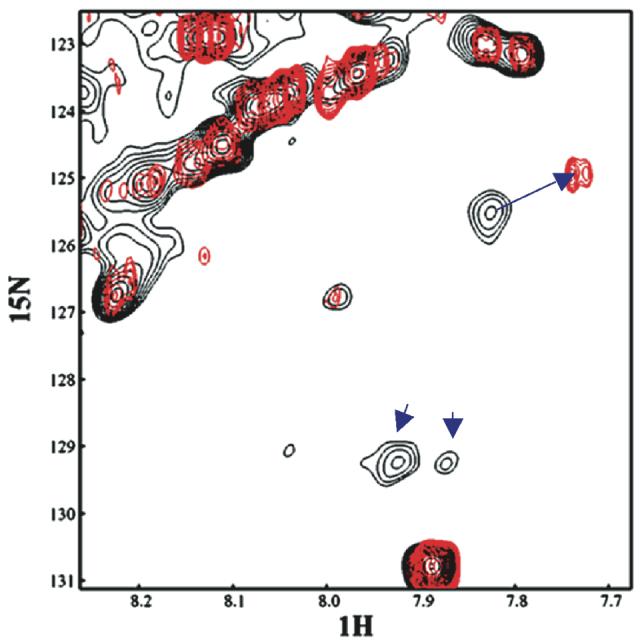
Overlay of a well-resolved region of the 1H-15N HSQC spectra of NDK1, free and bound to DNA. Resonances in black correspond to the spectrum of free NDK1 while those in red correspond to the DNA-bound protein when the protein to DNA ratio is 1:2. The cross peak that shows a specific chemical shift change in this region is indicated by a blue arrow. Two other cross peaks in this region disappeared upon binding to DNA, suggesting that chemical perturbation also occurs in these resonances.
In order to further substantiate the exonuclease activity present in human NDK proteins, we used a series of oligonucleotide substrates containing double-stranded, single-stranded or partially single-stranded DNA. Human NDK1, NDK5, and NDK7 were tested in these assays and they all exhibited significant exonuclease activity (see Fig. 7 for data with no protein control and NDK1; NDK5 and NDK7 produced very similar cleavage patterns, data not shown). The 3' to 5' exonuclease activity was most pronounced with single-stranded DNA substrates or substrates containing 3' single strand overhangs. We noticed that a substrate with a 3'-terminal mismatch (S4) was cleaved only slightly better than a substrate without mismatch (S3) (Fig. 7B) suggesting that this exonuclease probably does not have a major role in supporting proofreading activity.
Human NDK1, NDK5 and NDK7 show highly specific exonuclease activity in concert with UDG, human APE1 and NTH proteins
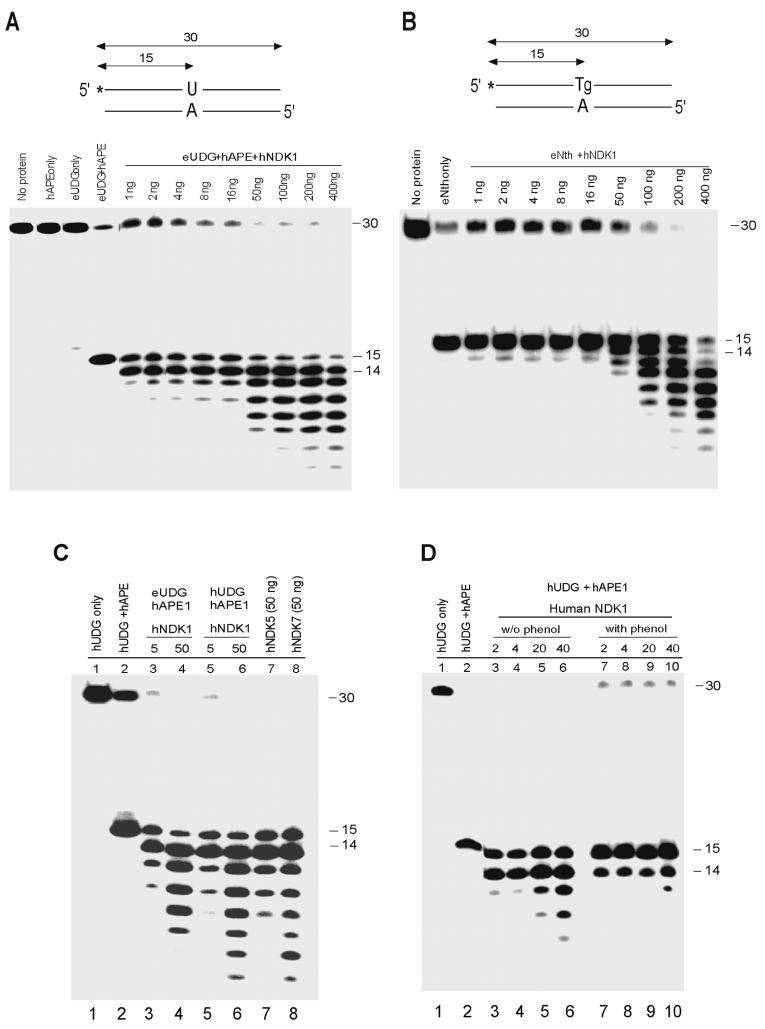
Previous studies indicated that the purified E. coli NDK protein may possess AP site cleavage activity although this activity was not fully characterized and is controversial (20, 21). A recent study has also reported that human NDK1 is involved as the DNA cleavage component in a protein complex (SET complex) that promotes cytotoxic T-lymphocyte-mediated apoptosis (18). Interestingly, human APE1 has also been found as one of the components of the SET complex (18). On the basis of these findings, we hypothesized that the 3' to 5' exonuclease activity of human NDK proteins might be involved in DNA processing events in concert with other repair proteins, particularly with human APE1 functioning in base excision repair. To test this hypothesis, we investigated human NDK1 activity in the presence of DNA glycosylase or AP lyase enzymes and hAPE1. First, we found that human NDK1 by itself has very weak or no activity towards cleaving abasic sites (data not shown). We used a double-stranded oligonucleotide containing U/A or thymine glycol (Tg)/A base pairs as base excision substrates (Fig. 8). Human or E. coli UDG was incubated with oligonucleotide substrates containing U/A to produce an abasic site (Fig. 8A). Human APE1 was then added to the mixture to cleave the abasic site followed by the addition of human NDKs. In the other reaction (Fig. 8B), E. coli Nth protein was incubated with an oligomer substrate containing Tg/A followed by the addition of human NDKs. Human NDK1 efficiently cleaved the double-stranded oligonucleotide substrate to produce a 3' to 5' exonuclease cleavage pattern initiated from the AP cleavage site produced by hAPE1 or Nth (Fig. 8A,B). The substrate incised in the presence of APE1 was more efficiently processed than the Nth cleavage product. Even at very low concentrations (2-4 ng of protein per 30 μl reaction), human NDK1 appeared to be sufficient to initiate cleavage of this substrate. The use of E. coli or human UDG made no difference (Fig. 8C). Human NDK5 and NDK7 proteins also showed efficient exonuclease activity in this concerted reaction (Fig. 8C). The activity in the concerted reaction was much more efficient than when human NDK1 was used alone in reactions with single-stranded or double-stranded oligonucleotide substrates (Fig. 3A and 7). With NDKs alone, 50 to 100 ng of purified NDK proteins were required to detect a significant cleavage with both single- and double-stranded substrates. A substrate with a simple nick (substrate S12 in Figure 7) was cleaved far less efficiently than the base excision repair intermediates, which have a single strand break bracketed by 3'-hydroxyl and 5'-deoxyribose-5-phosphate (5'-dRp) termini (Fig. 8). In addition, when UDG and hAPE1 proteins were removed by phenol/chloroform extraction, the exonuclease activity of human NDK1 was significantly lower than that in the presence of UDG and hAPE1 (Fig. 8D). These findings suggest that for optimal exonuclease activity, human NDKs might require the coordination with other proteins such as a DNA glycosylase and human APE1.
Figure 8. Enzymatic activity of human NDK1 protein in presence of base excision repair proteins.
A. Cleavage of an abasic site-containing oligonucleotide by combined reaction with UDG, human APE1 and human NDK1 proteins. The abasic site was created by eUDG that excised uracil from a double stranded oligonucleotide containing a U/A mispair. The abasic site was then cleaved by human APE1 followed by reaction with human NDK1 protein. Human NDK1 showed specific exonuclease activity emanating from the cleaved abasic site. B. Cleavage of an abasic site-containing oligonucleotide by combined reaction with eNth and human NDK1 proteins. The double stranded oligonucleotide containing a Tg/A mispair was preincubated with eNth followed by the addition of human NDK1 protein. C. Cleavage of an abasic site-containing oligonucleotide by combined reaction with E. coli UDG or human UDG, human APE1 and human NDK1, NDK5 or NDK7 proteins. eUDG (lane 3-4) or hUDG (lane 1-2 and 5-8) were preincubated with double stranded oligonucleotides containing U/A followed by adding hAPE1. Human NDKs were then added to the reaction mixture. D. The exonuclease activity of human NDK1 in the presence or absence of UDG and hAPE1. The reaction conditions were as described in panel A. Preincubated hUDG and hAPE1 were removed by phenol/chloroform extraction and then human NDK1 protein was added (lanes 7-10). The exonuclease activity of human NDK1 was much higher in the presence of UDG and hAPE1 (lanes 3-6).
Nuclear translocation of human NDK1
If human NDK1 has exonuclease activities related to apoptosis and/or DNA repair, this protein should be localized in the nucleus. What is puzzling in this regard is that a predominantly cytoplasmic localization has been reported for several of the mammalian NDKs (1). However, it is possible that the NDKs are transported to the nucleus upon a stimulus and carry out repair and/or other DNA processing reactions only under specific circumstances. Arabidopsis thaliana NDK1a is present in the nucleus and in the cytosol and is induced by UVB irradiation (32). Human NDK1/NM23-H1 is found in an ER-associated complex with hAPE1 and other proteins and is translocated to the nucleus upon a proapoptotic stimulus triggered by cytotoxic T lymphocytes (18) where it carries out a DNA cleavage reaction.
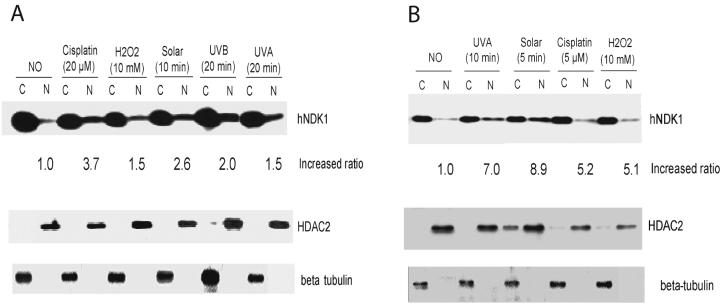
On the basis of these findings and the data described above, we hypothesized that when cells are treated with DNA damaging agents, human NDKs may be translocated from the cytosol to the nucleus. To investigate NDK1 nuclear localization, we carried out Western blot and immunohistochemistry studies for human NDK1. HeLa cervical carcinoma and ARPE-19 retinal epithelial cells were treated with various DNA damaging agents including cisplatin, H2O2, and different sources of UV radiation. Cytoplasmic and nuclear extracts were prepared from DNA damaging agent treated cells. Nuclear translocation of NDK1 was analyzed by immunoblot of cytosolic and nuclear fractions with anti-hNDK1 antibody and control antibodies (HDAC2 for the nuclear fraction and beta-tubulin for the cytoplasmic fraction). Figure 9 shows that most of the NDK1 protein was predominantly, but not exclusively, in the cytoplasm before treatment in both cell types. Interestingly, nuclear localization of NDK1 was significantly increased when cells were treated with DNA damaging agents. Depending on cell type and DNA damaging agent used, the nuclear translocation levels were different. The nuclear localization was more significantly increased in ARPE-19 cells than in HeLa cells (up to 8.9-fold relative to untreated cells with solar simulated UV). While simulated solar UV and UVA irradiation were relatively more effective in ARPE-19 cells (Fig. 9B), cisplatin and solar UV light were most effective in HeLa cells (Fig. 9A).
Figure 9. Western blot analysis of human NDK1 in HeLa and ARPE-19 cells.
A. Cytoplasmic and nuclear extracts from DNA damaging agent-treated HeLa cells were separated on SDS-PAGE. Endogenous human NDK1 in both cytoplasmic (C) and nuclear extracts (N) was detected with anti-human NDK1 antibody. HDAC2 was used as a control for the nuclear protein fraction and as a loading standard. β-tubulin was used as a cytoplasmic specific and loading control. Cells were lysed 20 min after DNA damage induction. The “increased ratio” refers to the relative increase in the percentage of nuclear localized protein in damaging agent-treated cells relative to untreated cells, corrected for the values of the loading controls. B. Localization analysis of human NDK1 in untreated and DNA damaging agents-treated ARPE-19 cells.
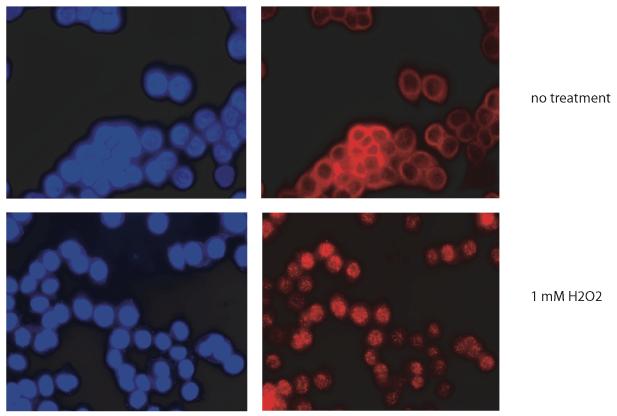
The nuclear translocation of NDK1 was also detected by immunohistochemistry with anti-hNDK1 antibody and fluorescence microscopy. Consistent with the Western blot analysis, in untreated cells, most human NDK1 is localized in the cytoplasm (Fig. 10, no treatment). However, when cells were treated with DNA damaging agents, the anti-hNDK1 antibody stained both cytoplasmic and nuclear regions (Fig. 10). In some cells treated with damaging agents, a focal or dot signal in the nucleus was detected. Time course experiments indicated that NDK1 was translocated to the nucleus within 20 min after DNA damage treatment and persisted there for at least up to 24 hours (Figs. 10 and 11). These results indicate that in response to DNA damage, human NDK1 might participate in DNA processing reactions in the nucleus. However, the treatment conditions used did not produce any evidence for apoptotic DNA cleavage, either in the form of nuclear fragmentation detected by DAPI staining (Figs. 10 and 11) or by DNA laddering detected by agarose gel analysis (not shown).
Figure 10. Immunohistochemistry of human NDK1 in HeLa cells.
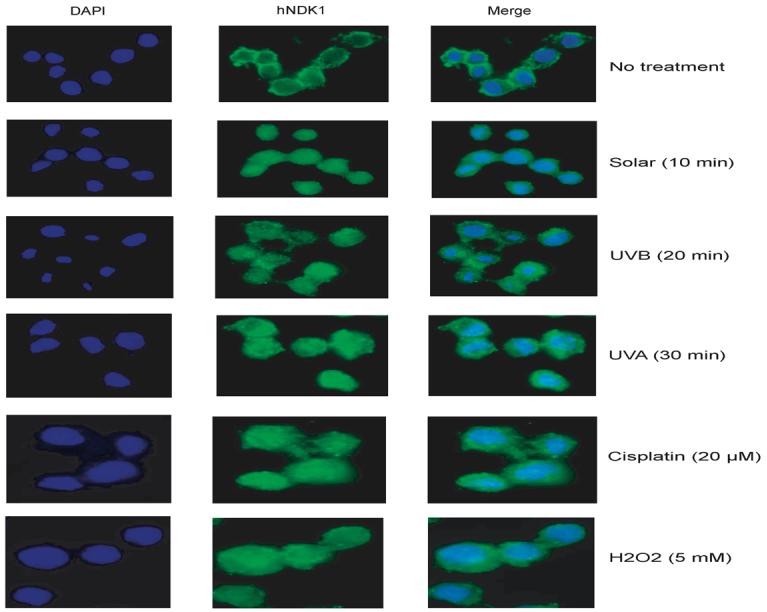
Untreated and DNA damaging agents-treated HeLa cells were fixed and stained with anti-human NDK1 antibody 20 min after DNA damage induction. The stained signals were visualized by Alexa Fluor 488-conjugated secondary antibody (middle). Control nuclear staining was performed with DAPI (left). Merged images are also shown (right).
Figure 11. Persistence of human NDK1 in the nuclei of HeLa cells after DNA damage.
Untreated and H2O2-treated HeLa cells were fixed and stained with anti-human NDK1 antibody 24 hours after DNA damage induction. The NDK1 signals were visualized in the right panels. Control nuclear staining was performed with DAPI (left panels).
DISCUSSION
We show here that several human proteins that contain NDK domains are in fact devoid of nucleoside diphosphate kinase activity, at least when tested as recombinant proteins in vitro. In mammalian cells, NDK1 (NM23-H1) and NDK2 (NM23-H2) have been reported to possess nuclease activities (14-17). Moreover, human NDK1 has been implicated in cytotoxic T lymphocyte-initiated apoptotic DNA fragmentation (18). However, the detailed reaction mechanism of the nuclease activity of NDK proteins is still unclear.
In this report we have demonstrated that the 3' to 5' exonuclease activity of human NDK1 is intrinsic and is unlikely a contamination by bacterial nucleases. This conclusion is based on the following findings: (1) Human NDK1 tightly co-purifies with the 3' to 5' exonuclease activity through several column chromatographic procedures. (2) A series of NDK1 catalysis-defective mutants show no exonuclease activity (Table 1). Competition assays with wild type and mutant NDK1 proteins indicate that mutant proteins can compete with the wild type. (4) NMR studies indicate specific binding of NDK1 protein to DNA. (5) The exonuclease activity of NDK1 is particularly pronounced when the reaction is part of a base excision repair step involving a DNA glycosylase and APE1.
The specific activity of NDK1 is much lower than that of DNA polymerase associated exonuclease activities. It is consistent, however, with that of other autonomous 3′-5′ exonucleases, which require higher enzyme concentrations for activity. For example, WRN exhibits significant DNA cleavage activity only when analyzed at concentrations in the nanomolar range (33, 34). One possible explanation is that these exonucleases normally function in concert with other proteins that may modulate their activity. In case of NDK1, we have shown that the presence of base excision repair proteins enhances the exonuclease activity of NDK1 substantially (Fig. 8).
The data obtained so far suggest that NDK1 (and possibly other NDK proteins) may be involved in DNA repair processes. The in vitro data point to a role of NDK1 in base excision repair, although an involvement of NDK1 in base excision repair in vivo remains to be proven, a task that may be difficult due to the expected redundancy of NDK exonucleases. A prevalent 3' to 5' exonucleolytic excision step in base excision repair has not been described. However, this may be due to limitations in the sensitivity of the assays employed and because of the predominant use of cell extracts rather than in vivo assays. Such excision extending 5' to the incised damage site could provide a facilitated entry point for DNA polymerases sealing the gap. There are other 3' to 5' exonucleases that may participate in base excision repair processes such as TREX (35) and WRN (36). APE1 has a 3' to 5' exonuclease activity itself (37-41) and can potentially degrade DNA substrates that mimic BER intermediates. However, the exonuclease activity of APE1 is inhibited by 5′-dRP at nicks and gaps and is enhanced only after the 5′-dRP is removed by Pol beta (38, 39).
The strongest activity of NDK1 for substrate cleavage was observed with single-stranded substrates or substrates having 3' single-stranded overhangs. This raises the possibility that NDK proteins may potentially function in double-strand break repair processes where they may be involved in trimming 3' single-stranded overhangs before ligation can occur. This possibility requires further investigation.
Although we have not detected apoptotic cells under the conditions we used (where NDK1 is translocated to the nucleus; Figs. 10 and 11), we cannot currently exclude the possibility that NDK1, under certain circumstances, may function as an apoptotic nuclease in response to DNA damage. Human NDK1 has been implicated in cytotoxic T lymphocyte-initiated apoptotic DNA fragmentation (18). Cytotoxic T lymphocytes release the serine protease granzyme A which triggers a caspase-independent apoptotic pathway in which DNA is damaged by single strand nicks, not oligosomal fragmentation (42, 43). Granzyme A targets a 270-420 kDa ER-associated complex (SET complex) which contains APE1 and NDK1/NM23-H1 (18). In this process, APE1 is cleaved proteolytically after Lys31 (44). In addition it has been shown that both APE1 (45) and NDK1 (this study) move to the nucleus upon DNA damage. Others have reported that an N-terminally truncated form of APE1 (AN34) arises in apoptotic cells and has endonuclease as well as 3' to 5' exonuclease activity (46). Thus, it can be speculated that APE1 and NDK1, acting together, can provide a functional module in DNA damage-induced DNA fragmentation. In this case, the DNA lesions themselves would provide the signal for apoptotic processing via recruitment of a base excision repair and nuclease complex including NDK1.
Additional studies are warranted to further characterize the role of mammalian NDK proteins in DNA repair and in DNA damage-induced apoptosis pathways and to determine whether they function in one or both pathways perhaps dependent on the level of DNA damage.
TOC Figure.

ACKNOWLEDGEMENTS
We thank Steven Bates for assistance with cell culture.
This work was supported by grants from the National Cancer Institute (CA84469) and National Institute of Environmental Health Sciences (ES06070) to G.P.P and by a grant from the National Cancer Institute (CA85344) to B.S.
Abbreviations
- NDK
nucleoside diphosphate kinase
- APE1
apurinic/apyrimidinic endonuclease 1
- UDG
uracil DNA glycosylase
- Tg
thymine glycol
- 5′-dRp
5′-deoxyribose-5-phosphate
REFERENCES
- 1.Lacombe ML, Milon L, Munier A, Mehus JG, Lambeth DO. The human Nm23/nucleoside diphosphate kinases. J. Bioenerg. Biomembr. 2000;32:247–258. doi: 10.1023/a:1005584929050. [DOI] [PubMed] [Google Scholar]
- 2.Fukuchi T, Nikawa J, Kimura N, Watanabe K. Isolation, overexpression and disruption of a Saccharomyces cerevisiae YNK gene encoding nucleoside diphosphate kinase. Gene. 1993;129:141–146. doi: 10.1016/0378-1119(93)90710-k. [DOI] [PubMed] [Google Scholar]
- 3.Izumiya H, Yamamoto M. Cloning and functional analysis of the ndk1 gene encoding nucleoside-diphosphate kinase in Schizosaccharomyces pombe. J. Biol. Chem. 1995;270:27859–27864. doi: 10.1074/jbc.270.46.27859. [DOI] [PubMed] [Google Scholar]
- 4.Lu Q, Zhang X, Almaula N, Mathews CK, Inouye M. The gene for nucleoside diphosphate kinase functions as a mutator gene in Escherichia coli. J. Mol. Biol. 1995;254:337–341. doi: 10.1006/jmbi.1995.0620. [DOI] [PubMed] [Google Scholar]
- 5.Miller JH, Funchain P, Clendenin W, Huang T, Nguyen A, Wolff E, Yeung A, Chiang JH, Garibyan L, Slupska MM, Yang H. Escherichia coli strains (ndk) lacking nucleoside diphosphate kinase are powerful mutators for base substitutions and frameshifts in mismatch-repair-deficient strains. Genetics. 2002;162:5–13. doi: 10.1093/genetics/162.1.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Steeg PS, Bevilacqua G, Kopper L, Thorgeirsson UP, Talmadge JE, Liotta LA, Sobel ME. Evidence for a novel gene associated with low tumor metastatic potential. J. Natl. Cancer Inst. 1988;80:200–204. doi: 10.1093/jnci/80.3.200. [DOI] [PubMed] [Google Scholar]
- 7.Chang CL, Zhu XX, Thoraval DH, Ungar D, Rawwas J, Hora N, Strahler JR, Hanash SM, Radany E. Nm23-H1 mutation in neuroblastoma. Nature. 1994;370:335–336. doi: 10.1038/370335a0. [DOI] [PubMed] [Google Scholar]
- 8.Leone A, Flatow U, King CR, Sandeen MA, Margulies IM, Liotta LA, Steeg PS. Reduced tumor incidence, metastatic potential, and cytokine responsiveness of nm23-transfected melanoma cells. Cell. 1991;65:25–35. doi: 10.1016/0092-8674(91)90404-m. [DOI] [PubMed] [Google Scholar]
- 9.MacDonald NJ, Freije JM, Stracke ML, Manrow RE, Steeg PS. Site-directed mutagenesis of nm23-H1. Mutation of proline 96 or serine 120 abrogates its motility inhibitory activity upon transfection into human breast carcinoma cells. J. Biol. Chem. 1996;271:25107–25116. doi: 10.1074/jbc.271.41.25107. [DOI] [PubMed] [Google Scholar]
- 10.Munier A, Feral C, Milon L, Pinon VP, Gyapay G, Capeau J, Guellaen G, Lacombe ML. A new human nm23 homologue (nm23-H5) specifically expressed in testis germinal cells. FEBS Lett. 1998;434:289–294. doi: 10.1016/s0014-5793(98)00996-x. [DOI] [PubMed] [Google Scholar]
- 11.Sadek CM, Damdimopoulos AE, Pelto-Huikko M, Gustafsson JA, Spyrou G, Miranda-Vizuete A. Sptrx-2, a fusion protein composed of one thioredoxin and three tandemly repeated NDP-kinase domains is expressed in human testis germ cells. Genes Cells. 2001;6:1077–1090. doi: 10.1046/j.1365-2443.2001.00484.x. [DOI] [PubMed] [Google Scholar]
- 12.Mehus JG, Deloukas P, Lambeth DO. NME6: a new member of the nm23/nucleoside diphosphate kinase gene family located on human chromosome 3p21.3. Hum. Genet. 1999;104:454–459. doi: 10.1007/s004390050987. [DOI] [PubMed] [Google Scholar]
- 13.Milon L, Meyer P, Chiadmi M, Munier A, Johansson M, Karlsson A, Lascu I, Capeau J, Janin J, Lacombe ML. The human nm23-H4 gene product is a mitochondrial nucleoside diphosphate kinase. J. Biol. Chem. 2000;275:14264–14272. doi: 10.1074/jbc.275.19.14264. [DOI] [PubMed] [Google Scholar]
- 14.Postel EH, Berberich SJ, Flint SJ, Ferrone CA. Human c-myc transcription factor PuF identified as nm23-H2 nucleoside diphosphate kinase, a candidate suppressor of tumor metastasis. Science. 1993;261:478–480. doi: 10.1126/science.8392752. [DOI] [PubMed] [Google Scholar]
- 15.Levit MN, Abramczyk BM, Stock JB, Postel EH. Interactions between Escherichia coli nucleoside-diphosphate kinase and DNA. J. Biol. Chem. 2002;277:5163–5167. doi: 10.1074/jbc.M111170200. [DOI] [PubMed] [Google Scholar]
- 16.Ma D, Xing Z, Liu B, Pedigo NG, Zimmer SG, Bai Z, Postel EH, Kaetzel DM. NM23-H1 and NM23-H2 repress transcriptional activities of nuclease-hypersensitive elements in the platelet-derived growth factor-A promoter. J. Biol. Chem. 2002;277:1560–1567. doi: 10.1074/jbc.M108359200. [DOI] [PubMed] [Google Scholar]
- 17.Postel EH, Abramczyk BM, Levit MN, Kyin S. Catalysis of DNA cleavage and nucleoside triphosphate synthesis by NM23-H2/NDP kinase share an active site that implies a DNA repair function. Proc. Natl. Acad. Sci. U.S.A. 2000;97:14194–14199. doi: 10.1073/pnas.97.26.14194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fan Z, Beresford PJ, Oh DY, Zhang D, Lieberman J. Tumor suppressor NM23-H1 is a granzyme A-activated DNase during CTL-mediated apoptosis, and the nucleosome assembly protein SET is its inhibitor. Cell. 2003;112:659–672. doi: 10.1016/s0092-8674(03)00150-8. [DOI] [PubMed] [Google Scholar]
- 19.Ma D, McCorkle JR, Kaetzel DM. The metastasis suppressor NM23-H1 possesses 3′-5′ exonuclease activity. J. Biol. Chem. 2004;279:18073–18084. doi: 10.1074/jbc.M400185200. [DOI] [PubMed] [Google Scholar]
- 20.Postel EH, Abramczyk BM. Escherichia coli nucleoside diphosphate kinase is a uracil-processing DNA repair nuclease. Proc. Natl. Acad. Sci. U.S.A. 2003;100:13247–13252. doi: 10.1073/pnas.2333230100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kumar P, Krishna K, Srinivasan R, Ajitkumar P, Varshney U. Mycobacterium tuberculosis and Escherichia coli nucleoside diphosphate kinases lack multifunctional activities to process uracil containing DNA. DNA Repair (Amst) 2004;3:1483–1492. doi: 10.1016/j.dnarep.2004.06.007. [DOI] [PubMed] [Google Scholar]
- 22.Bennett SE, Chen CY, Mosbaugh DW. Escherichia coli nucleoside diphosphate kinase does not act as a uracil-processing DNA repair nuclease. Proc. Natl. Acad. Sci. USA. 2004;101:6391–6396. doi: 10.1073/pnas.0401031101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yoon JH, Iwai S, O'Connor TR, Pfeifer GP. Human thymine DNA glycosylase (TDG) and methyl-CpG-binding protein 4 (MBD4) excise thymine glycol (Tg) from a Tg:G mispair. Nucleic Acids Res. 2003;31:5399–5404. doi: 10.1093/nar/gkg730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Graves RJ, Felzenszwalb I, Laval J, O'Connor TR. Excision of 5′-terminal deoxyribose phosphate from damaged DNA is catalyzed by the Fpg protein of Escherichia coli. J. Biol. Chem. 1992;267:14429–14435. [PubMed] [Google Scholar]
- 25.O'Connor T. The use of DNA glycosylases to detect DNA damage. In: Pfeifer GP, editor. Technologies for Detection of DNA damage and mutations. Plenum Press; New York: 1996. pp. 155–167. [Google Scholar]
- 26.Postel EH, Ferrone CA. Nucleoside diphosphate kinase enzyme activity of NM23-H2/PuF is not required for its DNA binding and in vitro transcriptional functions. J. Biol. Chem. 1994;269:8627–8630. [PubMed] [Google Scholar]
- 27.Qiu J, Yoon JH, Shen B. Search for apoptotic nucleases in yeast: Role of Tat-D nuclease in apoptotic DNA degradation. J. Biol. Chem. 2005;280:15370–15379. doi: 10.1074/jbc.M413547200. [DOI] [PubMed] [Google Scholar]
- 28.Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J Biomol. NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 29.Johnson BA. Using NMRView to visualize and analyze the NMR spectra of macromolecules. Methods Mol. Biol. 2004;278:313–352. doi: 10.1385/1-59259-809-9:313. [DOI] [PubMed] [Google Scholar]
- 30.Shen B, Nolan JP, Sklar LA, Park MS. Essential amino acids for substrate binding and catalysis of human flap endonuclease 1. J. Biol. Chem. 1996;271:9173–9176. doi: 10.1074/jbc.271.16.9173. [DOI] [PubMed] [Google Scholar]
- 31.Gilles AM, Presecan E, Vonica A, Lascu I. Nucleoside diphosphate kinase from human erythrocytes. Structural characterization of the two polypeptide chains responsible for heterogeneity of the hexameric enzyme. J. Biol. Chem. 1991;266:8784–8789. [PubMed] [Google Scholar]
- 32.Zimmermann S, Baumann A, Jaekel K, Marbach I, Engelberg D, Frohnmeyer H. UV-responsive genes of arabidopsis revealed by similarity to the Gcn4-mediated UV response in yeast. J. Biol. Chem. 1999;274:17017–17024. doi: 10.1074/jbc.274.24.17017. [DOI] [PubMed] [Google Scholar]
- 33.Huang S, Li B, Gray MD, Oshima J, Mian IS, Campisi J. The premature ageing syndrome protein, WRN, is a 3′-->5′ exonuclease. Nat. Genet. 1998;20:114–116. doi: 10.1038/2410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Brosh RM, Jr., Karmakar P, Sommers JA, Yang Q, Wang XW, Spillare EA, Harris CC, Bohr VA. p53 Modulates the exonuclease activity of Werner syndrome protein. J. Biol. Chem. 2001;276:35093–35102. doi: 10.1074/jbc.M103332200. [DOI] [PubMed] [Google Scholar]
- 35.Mazur DJ, Perrino FW. Excision of 3′ termini by the Trex1 and TREX2 3′-->5′ exonucleases. Characterization of the recombinant proteins. J. Biol. Chem. 2001;276:17022–17029. doi: 10.1074/jbc.M100623200. [DOI] [PubMed] [Google Scholar]
- 36.Ahn B, Harrigan JA, Indig FE, Wilson DM, 3rd, Bohr VA. Regulation of WRN helicase activity in human base excision repair. J. Biol. Chem. 2004;279:53465–53474. doi: 10.1074/jbc.M409624200. [DOI] [PubMed] [Google Scholar]
- 37.Cistulli C, Lavrik OI, Prasad R, Hou E, Wilson SH. AP endonuclease and poly(ADP-ribose) polymerase-1 interact with the same base excision repair intermediate. DNA Repair (Amst) 2004;3:581–591. doi: 10.1016/j.dnarep.2003.09.012. [DOI] [PubMed] [Google Scholar]
- 38.Wilson DM., 3rd Properties of and substrate determinants for the exonuclease activity of human apurinic endonuclease Ape1. J. Mol. Biol. 2003;330:1027–1037. doi: 10.1016/s0022-2836(03)00712-5. [DOI] [PubMed] [Google Scholar]
- 39.Wong D, DeMott MS, Demple B. Modulation of the 3′-->5′-exonuclease activity of human apurinic endonuclease (Ape1) by its 5′-incised Abasic DNA product. J. Biol. Chem. 2003;278:36242–36249. doi: 10.1074/jbc.M306065200. [DOI] [PubMed] [Google Scholar]
- 40.Chou KM, Cheng YC. An exonucleolytic activity of human apurinic/apyrimidinic endonuclease on 3′ mispaired DNA. Nature. 2002;415:655–659. doi: 10.1038/415655a. [DOI] [PubMed] [Google Scholar]
- 41.Ishchenko AA, Yang X, Ramotar D, Saparbaev M. The 3′->5′ exonuclease of Apn1 provides an alternative pathway to repair 7,8-dihydro-8-oxodeoxyguanosine in Saccharomyces cerevisiae. Mol. Cell. Biol. 2005;25:6380–6390. doi: 10.1128/MCB.25.15.6380-6390.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Beresford PJ, Kam CM, Powers JC, Lieberman J. Recombinant human granzyme A binds to two putative HLA-associated proteins and cleaves one of them. Proc. Natl. Acad. Sci. USA. 1997;94:9285–9290. doi: 10.1073/pnas.94.17.9285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shresta S, Graubert TA, Thomas DA, Raptis SZ, Ley TJ. Granzyme A initiates an alternative pathway for granule-mediated apoptosis. Immunity. 1999;10:595–605. doi: 10.1016/s1074-7613(00)80059-x. [DOI] [PubMed] [Google Scholar]
- 44.Fan Z, Beresford PJ, Zhang D, Xu Z, Novina CD, Yoshida A, Pommier Y, Lieberman J. Cleaving the oxidative repair protein Ape1 enhances cell death mediated by granzyme A. Nat. Immunol. 2003;4:145–153. doi: 10.1038/ni885. [DOI] [PubMed] [Google Scholar]
- 45.Ramana CV, Boldogh I, Izumi T, Mitra S. Activation of apurinic/apyrimidinic endonuclease in human cells by reactive oxygen species and its correlation with their adaptive response to genotoxicity of free radicals. Proc. Natl. Acad. Sci. USA. 1998;95:5061–5066. doi: 10.1073/pnas.95.9.5061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yoshida A, Urasaki Y, Waltham M, Bergman AC, Pourquier P, Rothwell DG, Inuzuka M, Weinstein JN, Ueda T, Appella E, Hickson ID, Pommier Y. Human apurinic/apyrimidinic endonuclease (Ape1) and its N-terminal truncated form (AN34) are involved in DNA fragmentation during apoptosis. J. Biol. Chem. 2003;278:37768–37776. doi: 10.1074/jbc.M304914200. [DOI] [PubMed] [Google Scholar]