Abstract
The voltage-dependent anion channel (VDAC), also known as mitochondrial porin, is the most abundant protein in the mitochondrial outer membrane (MOM). VDAC is the channel known to guide the metabolic flux across the MOM and plays a key role in mitochondrially induced apoptosis. Here, we present the 3D structure of human VDAC1, which was solved conjointly by NMR spectroscopy and x-ray crystallography. Human VDAC1 (hVDAC1) adopts a β-barrel architecture composed of 19 β-strands with an α-helix located horizontally midway within the pore. Bioinformatic analysis indicates that this channel architecture is common to all VDAC proteins and is adopted by the general import pore TOM40 of mammals, which is also located in the MOM.
Keywords: membrane protein, NMR, porin, structure, x-ray
The outer membrane of mitochondria (MOM) contains three integral membrane protein families, two of which form channels as part of larger protein complexes (for review, see ref. 1). These two MOM complexes, the general import pore TOM and the SAM insertase, allow for the entire translocation and insertion of nearly all newly synthesized proteins destined to the mitochondrial organelle (2, 3). The third protein family of typically high abundance (≈10,000 copies per mitochondrion) is termed voltage-dependent anion channels (VDACs), because of the voltage sensitivity of its open probability (4, 5). Together, this small number of protein families is sufficient for full communication between mitochondria with their cellular environment (1).
The VDAC channel was initially described as being reminiscent of bacterial porins and primarily responsible for the exchange of chemical energy equivalents between the cytosol and the mitochondrion (4, 6). Indeed, a variety of structural features (like barrel geometry and dimension) known from the bacterial precursors are maintained (7, 8). By contrast, a variety of functions have been ascribed to the VDAC isoforms among which the direct coupling of the mitochondrial matrix to the energy maintenance of the cytosol seems to be the most general function (9). The structure of VDAC is of interest because of a substantial body of evidence connecting VDAC to apoptosis. It is suggested that VDAC is a critical player in the release of apoptogenic factors from mitochondria of mammalian cells, and consequently several hypotheses describing the mechanism of mitochondria-mediated apoptosis involving VDAC have been proposed (for review, see ref. 10).
Results and Discussion
Structure Determination of hVDAC1: Combining NMR Spectroscopy and X-Ray Crystallography.
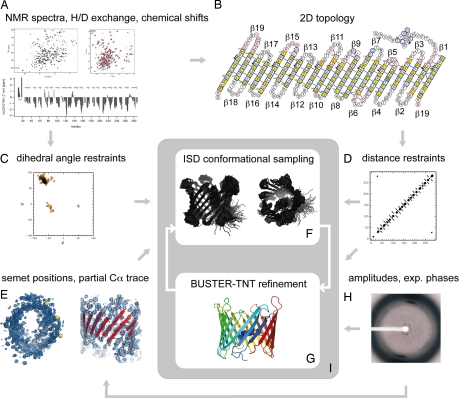
In a parallel structural biology approach, we set out to characterize the structure of hVDAC1, the major isoform of this channel in mammalian tissues, by a combination of NMR spectroscopy and x-ray crystallography. The idea behind this project was to gain complementary structural information to have a solid basis for future studies, e.g., analysis of protein heterocomplex formation by NMR and crystal structures as a basis for drug target design. Only information derived from both methods and the application of an iterative structure calculation procedure allowed the structure determination of the protein (see Fig. 1).
Fig. 1.
Flow chart of the conjoint NMR and x-ray structure determination. The 3D structure of VDAC was determined by integrating NMR and x-ray data in an iterative structure calculation protocol: (A) The NMR data (H/D exchange, chemical shifts) served as a basis to determine the secondary structure of hVDAC1. In addition, 95 NOEs could be assigned, 65 of which are interstrand NOEs that framed the register of neighboring β-strands. (B) A 2D topology model was derived from the secondary structure, the interstrand NOEs and paramagnetic relaxation enhancements (PRE) of 18 spin-labeled single-cysteine mutants (β19 is shown twice to indicate that the barrel is closed). (C) From the chemical shifts, main-chain dihedral angle restraints for 184 φ/ψ-pairs were predicted with the program TALOS (14). The SHIFTOR server predicted 170 additional side-chain χ-angle restraints (15). (D) The NOEs, the inter strand hydrogen bonding pattern and the strand assignments were converted into distance restraints. The NOEs provided 95 distance restraints between amide protons. These were supplemented with 150 hydrogen bonds inferred from the topology model. In addition, we imposed 370 distance restraints that restrain the intrastrand Cα-distances to multiples of 3.4 Å. (E) Positional restraints were provided by four seleno-methionines (M10 (L in WT), M129, M155, M184 (V in WT), and an eight-stranded partial Cα-trace that was built manually into the well defined regions of the electron density. The Se-Met positions were used to dock the model into the density map and to structurally align the model and the Cα-trace. (F) Using the distance, dihedral angle and positional restraints as input an initial structure ensemble was generated with the ISD software (16). (G) The highest-probability conformation served as a starting structure in a subsequent reciprocal space refinement with BUSTER-TNT (17). In addition to the structure factor amplitudes and experimental phases from SHARP (18) (H), secondary structure distance restraints (D) were imposed in all BUSTER-TNT refinements. (I) The structure calculation proceeded by cycling between ensemble generation and crystallographic refinement. In the conformational sampling, the positional restraints (E) were used only in the initial iteration. In subsequent calculations, the last structure obtained by crystallographic refinement served as a template (positional restraints to all Cα-coordinates). The distance and dihedral angle restraints were imposed in all ISD structure calculations.
To have large quantities of the channel available in manipulable form, we overexpressed the protein in Escherichia coli and refolded the channel to produce labeled protein for NMR and x-ray studies (11). For NMR studies, 15N-labeled hVDAC1 was refolded in detergents (LDAO or Cymal-5) and qualitatively analyzed by 15N-1H TROSY experiments. hVDAC1 refolded in LDAO showed ≈250 well dispersed cross-peaks [supporting information (SI) Fig. S1A]. Using a combination of TROSY-type triple-resonance experiments, seven different amino acid specific 15N and 15N/13C-labeled samples, several 15N-resolved NOESY spectra and 20 different point mutations combined with paramagnetic spin labeling (SI Text, Fig. S2), the backbone resonances of 192 of the 282 residues of hVDAC1 (excluding the N-terminal methionine) were assigned. Cα secondary chemical shifts and slow hydrogen-deuterium exchange of amide protons revealed the topology of 19 β-strands with 65 interstrand NOEs unambiguously determining the register of all 19 β-strands (Fig. 1B and Fig. S3). At the N terminus, a combination of NMR parameters identified an α-helix that binds to the barrel wall (Fig. 1 A and B and Table S1).
To obtain the crystal structure by x-ray diffraction, hVDAC1 was subjected to an extensive detergent screening. Native crystals were obtained with the hanging drop method by mixing 1.5-μl protein-Cymal-5 detergent solution containing 5–15 mg/ml protein with 0.75 μl of reservoir solution (30% PEG 400; 0.1 M Na Hepes, pH 7.5; 0.2 M magnesium chloride). The trigonal crystals (space group P321) diffracted up to 4 Å (12). Multiwavelength anomalous diffraction phasing with data from Se-Met crystals was used to approach the phase problem. Together with phases from a Pt-derivative and after careful density modification, an initial low-resolution electron density map was obtained that clearly showed the protein envelope and the overall barrel dimensions. This initial density was further improved through enforcing of a barrel-like solvent envelope leading to an electron density that allowed the tracing of eight β-strands and the N-terminal helix (see Fig. 1E). Both normal and B-factor sharpened (13) density maps were used for model building. In general, the B-factor sharpened density maps (after density modification) gave a clearer indication of the position and direction for the β-strands. Although the electron density of the strands in the N- and C-terminal parts was readily interpretable, the quality of the intermediate part remained problematic. This could be due to various reasons: (i) the lack of very good low-resolution reflections because of overloads or beamstop shadow; (ii) the general high nonisomorphism between different datasets from different crystals, making it difficult to combine phase information from different datasets in SHARP (19); (iii) the relatively high anisotropy of the reflection data (see footnote, Table S2); and (iv) the low resolution of the x-ray data (Fig. 1E).
The quality of the electron density was not sufficient to allow the determination of the hVDAC1 structure by standard crystallographic methods alone. Therefore, we pursued a hybrid approach in which we integrated complementary information from NMR spectroscopy and x-ray crystallography. These calculations were done iteratively by cycling between real space ensemble generation using the Inferential Structure Determination (ISD) program (16) and reciprocal space refinement using the BUSTER-TNT (17) program (compare with Fig. 1).
In the ISD real space calculations, the crystallographic data are introduced as positional restraints. To generate the initial structure ensemble, positional restraints were derived from the four SeMet atoms that could be assigned unambiguously and from eight β-strands that were built manually into the electron density (see Fig. 1F). In later iterations, the positional restraints were derived from the structure obtained by reciprocal space refinement. The four SeMet sites in the mutant L10M, V184M are at positions 10, 129, 155, and 184.
The positional restraints were combined with standard NMR restraints: 95 distance restraints were obtained from the NOEs, 368 backbone and 170 side chain dihedral angle restraints were predicted from the chemical shifts. Moreover, H/D exchange rates in combination with the topology model were used to derive 150 hydrogen bonds that were also introduced as distance restraints into the ensemble calculation. Finally, we applied additional Cα-Cα distance restraints within the β-strand regions. These restraints improved the convergence of the calculation but were not essential for the overall structure. Test calculations showed that virtually the same ensemble is obtained without imposing the Cα intrastrand restraints.
After each ensemble generation, a joint NMR/x-ray refinement was carried out by using BUSTER-TNT. The highest-probability model of the ensemble was docked into the electron-density envelope generated by the x-ray crystallographic phases and refined against the spatial constraints given by the diffraction data (Fig. 1G). Distance restraints based on the NMR secondary structure assignment were used for all β-sheet residues and for the N-terminal helix (21). Cyclic rebuilding and addition of further restraints into the model calculation improved the initial phases to yield an electron density map that allowed tracing of the entire model against the 4-Å resolution data (Fig. 1I, crystallographic data: Tables S2 and S3). The improvement of the electron density from the initial experimental map after refinement with BUSTER-TNT is shown in Fig. S4.
3D Structure of hVDAC1.
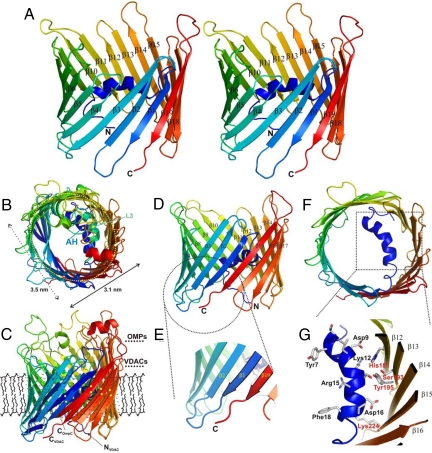
The channel pore of slightly concave shape (Fig. 2A) has dimensions of ≈3.5 × 3.1 nm in the horizontal and ≈4 nm in the vertical directions (Fig. 2B). Similar dimensions for VDAC proteins in the native state have also been obtained by high resolution AFM investigations (3.8 × 2.7-nm diameter) (22) and electron microscopy studies (diameter of ≈3 nm) (7), both of which derived from Saccharomyces cerevisiae in natural membrane composition. hVDAC1 reconstituted in artificial membranes showed identical dimensions (3.7-nm diameter × 4.3-nm height) (23). The inner diameter of the pore is ≈1.5 × 1 nm and therefore leaves space for diffusion of small metabolites (Fig. 2 B and F).
Fig. 2.
3D structure of hVDAC1. (A) Stereo representation of the hVDAC1 structure in a ribbon presentation color coded from the N to the C terminus (N and C). Some β-strands are marked with their respective numbers. All images were prepared using PYMOL (20). (B, C) Structure superposition of OmpC from E. coli (PDB ID code 2J1N) onto the refined hVDAC1 (115 Cα-atoms were superposed with an rmsd of 2.5 Å) viewed from top (B) and side (C), respectively. The view shown in B indicates a similar cork architecture of two different structural elements (AH, N-terminal helix of hVDAC; L3, constricting loop of porin). The horizontal dimensions of hVDAC1 are given (3.5 × 3.1 nm). Residues in the C-terminal half (β10–β19) of the two barrel proteins (in C marked with NVDAC, CVDAC, and COmpC) match well (matching elements carry yellow points for better visibility), whereas the N-terminal region differs in the relative inclination. The approximate membrane dimension surrounding the protein in the MOM is indicated by a “theoretical” lipid layer. (D) Side view of hVDAC1 in a ribbon presentation color coded from the N to the C terminus (N and C). The 19 β-strands are marked (β1–β19), whereas the N-terminal helix is folded into the pore interior. (E) Close-up of the C terminus (C) with the two terminal β-strands (β1 and β19) forming the parallel arrangement. (F) Top view onto the hVDAC1 barrel with the α-helix enclosed by and attached to the barrel walling.(G) Closeup of the N-terminally located α-helical element. Residues contacting the barrel wall or pointing toward the interior of the pore are marked (in black: Tyr7-Phe18) as well as residues on the barrel wall (in red). The secondary structure of the barrel is marked β12–β16.
The final protein structure contained all residues with the N and C termini of the protein oriented to the same side of the barrel (Fig. 2 A, C, and D). In contrast to bacterial outer membrane proteins, which archetypically show structures with an even number of strands and a distribution of long loops pointing to the extracellular and short turns to the periplasmic side (24), hVDAC1 shows an uneven number of 19 strands and 18 loop-like connections with a less distinct distribution of shortened loop structures to one side of the membrane (see Fig. 2C). Adjacent to the loops, which are dynamic on the pico- to nanosecond time scale according to NMR spectroscopic relaxation measurements (Fig. S1C), the formation of two incomplete aromatic girdles separated by a distance of only ≈1.5 nm on an axis parallel to the membrane normal is visible (data not shown). The unequal number of strands in the barrel requires one parallel interaction of two adjacent and slightly twisted terminal β-strands ending on the same side (β1 and β19) of the membrane (Fig. 2 D and E). The average inclination of the β-strands relative to the barrel axis is 37°, varies between 27 and 46°, and forms a barrel of almost circular shape (see Fig. 2F), similar, e.g., to the monomeric porin OmpG (25, 26).
VDACs within a certain species, e.g., mammals or fungi, share a high sequence identity (typically >80%) (data not shown), suggesting that all VDAC proteins share the same fold. In addition to the VDAC isoforms, the translocase of the outer mitochondrial membrane (human TOM40) (2) shows a striking homology to hVDAC1 in terms of both sequence and predicted secondary structure (data not shown), suggesting that hTOM40 also folds into a 19-stranded β-barrel.
N-Terminal Helix Is Located Inside the Pore.
The N-terminal helix comprising residues Tyr-7 to Val-17 is folded horizontally inside the barrel wall approximately at the midpoint of the hydrophobic portion of the membrane (Fig. 2 F and G). Although this arrangement has no “precursors” in bacterial porins, the position and the influence of the helix onto the overall barrel architecture is reminiscent of the long loop L3 present in bacterial porins (24), which folds in a similar position halfway perpendicular to the membrane normal into the barrel lumen (Fig. 2B). Whereas the preceding N-terminal residues (Val-3–Tyr-7) are rather loosely attached to the barrel wall and point toward the pore outlet, the N terminus with an overall charge of zero is amphiphatic and more strongly connected to the barrel by a short stretch of residues running almost in parallel to the angle of the β-sheets. Positively charged residues face the barrel interior, whereas negatively charged aspartates interact with the barrel wall. The counter residues on the barrel wall (His-181, Ser 193, Tyr-195, Lys-224) are predominantly hydrophilic and are conserved among most VDAC proteins (data not shown).
Dimerization of VDAC via a Small Hydrophobic Interface.
Previous studies have indicated that VDAC from several species can exist in different oligomerization states from monomers to dimers, trimers, tetramers, hexamers, and higher oligomers (27–29). A transient dimerization is in agreement with chemical cross-linking experiments where dimers and higher-order oligomers were found in diluted solutions of hVDAC1. NMR relaxation measurements of VDAC in LDAO, which correspond to a much lower local concentration than that of VDAC in the mitochondrial membrane indicate a global correlation time of the VDAC-micelle assembly of 60 ns, corresponding to a monomer/dimer equilibrium (see SI Text).
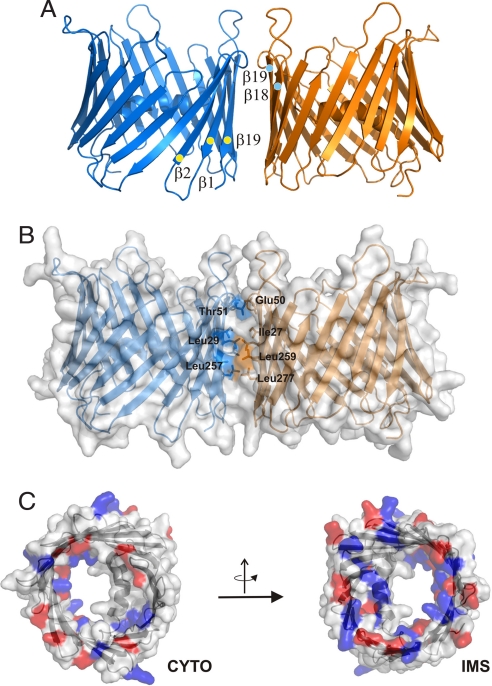
Using crystallographic symmetry operators, a dimer was constructed that coincides with electron micrographs and implies the formation of a parallel dimer (Fig. 3A) (23). In analogy to OMPs, this interface is formed by the four β-strands (β18, β19, β1, β2) with the lowest inclination values. The interface provided by residues in β-strands β1 (Ile-27, Leu-29), β2 (Glu-50, Thr-51), β18 (Leu-257, Leu-259), and β19 (Leu-277) covers 550 Å2, suggesting that it might be much more dynamic than in bacterial porins (Fig. 3B). It should be noted that residues showing both enhanced (β1, β2) and slow exchange rates (β18, β19) with water (see below) are involved in the dimer interface excluding transient dimerization as the reason of broadening of NH resonances of strands β1 and β2.
Fig. 3.
Distribution of charged residues and dimer model of hVDAC1. (A) Ribbon representation of the protein dimer and strands involved in dimerization are marked. (B) Surface representation of the dimer with residues at the dimer interface color coded accordingly and marked. (C) Surface representations of hVDAC1 visualized from both sides of the membrane with the charged residues colored in red (Glu, Asp) and blue (Lys, Arg). CYTO, cytoplasmic side; IMS, intermembrane space side.
Conformational Instability of the N-Terminal Part of hVDAC1.
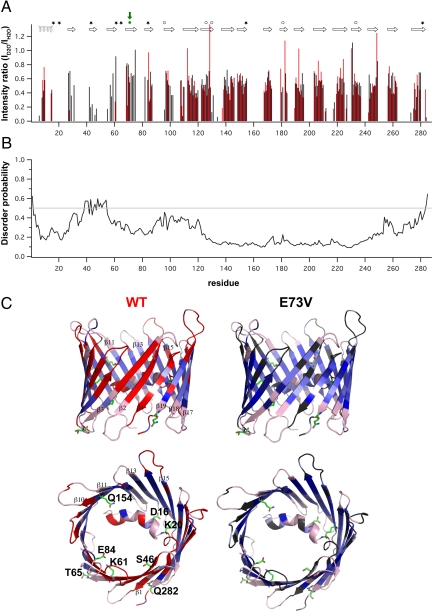
To probe the conformational stability and the influence of particular residues, we performed hydrogen/deuterium exchange coupled to NMR spectroscopy. Surprisingly, residues in β1- β4 of WT hVDAC1 solubilized in LDAO or in Cymal-5 showed rapid amide proton exchange (Fig. 4 A and C and Fig. S5), whereas this was not observed for the other β-strands. This indicates that the N-terminal four β-strands are less stable than other regions of the VDAC barrel and switch between different conformations. As part of this conformational exchange, hydrogen bonds within the four N-terminal strands of the barrel are transiently broken. This is consistent with computational analyses that predict an increased flexibility for the N-terminal region (Fig. 4B). Importantly, mutants that affected the voltage sensitivity of Saccharomyces cerevisiae VDAC (scVDAC) are mainly located in the N-terminal domain: the α-helix (conserved Asp-16), the linker (conserved Lys-20), and β-strands β1- β5 (Lys-46, Lys 61, Lys 65, and Lys 84 in scVDAC) (Fig. 4C) (30). This suggests that the voltage gating of VDAC requires conformational flexibility.
Fig. 4.
Conformational instability of the N-terminal region of the hVDAC1 barrel. (A) Comparison of amide proton exchange in wt (red) and E73V (black) hVDAC1 at 37°C, pH 6.8. Shown are signal intensity ratios of hVDAC1 in D2O and H2O. The NMR measurements were completed 2 hours after transfer of the proteins into 100% D2O. The location of the 19 β-strands and the helix are shown on top. Mutants that affect the voltage gating of scVDAC (Asp-16, Lys-20, Lys-46, Lys-61, Lys-65, Lys-84, Q154, Q282) are indicated by stars (30). Residues, which on the basis of channel conductance measurements were proposed to remain in the wall of the closed-channel pore of VDAC (31), are indicated by circles. The position of E73V is highlighted by a green square and arrow. (B) Probability for disorder in hVDAC1 as predicted by the DisProt web server (32). Based on the primary sequence of hVDAC1 a higher disorder propensity is predicted for the N-terminal part of the β-barrel including strands β1-β4 that NMR shows to be conformationally unstable. (C) Ribbon representation of WT hVDAC1 (Left) and E73V hVDAC1 (Right) in a side view and rotated by 90° resulting in a top view. Absolute signal intensities in the H/D exchange TROSY were visualized by using a continuous blue scale (light blue, <105; dark blue, >4×105). Residues without detectable signal in the exchange TROSY are shown in red for wt hVDAC1 and gray for E73V hVDAC1. Unassigned residues are colored in light pink. Residues that affected the voltage gating of scVDAC (30) are shown with side chains. They are labeled by the one letter amino acid code in the top view of hVDAC1.
The structural instability of the N-terminal part is particularly influenced by Glu-73. This residue is conserved in different isoforms of human VDAC and in VDAC from mammals and fungi (data not shown). In the 3D structure, E73 points to the membrane and interrupts the amphipathic pattern in strand β4 (Fig. 1B and Fig. S1). We replaced Glu-73 by valine, a substitution that resulted in slower amide proton exchange in β-strands β1-β4 (Fig. 4 A and C and Fig. S5), indicating that the conformation of the N-terminal part of the barrel is stabilized. Importantly, substitution of Glu-73 by glutamine has important consequences for the function of VDAC. This mutation abolished ruthenium red and hexokinase I-mediated inhibition of VDAC channel activity and ruthenium red and hexokinase I-mediated protection against apoptosis (33). In both E73V and E73Q, the charged residue that breaks the alternating pattern of hydrophobic and hydrophilic residues in strand β4 is removed. Thus, the β-barrel might also become more stable by the E73Q mutation. This is consistent with the finding that ruthenium red is no longer able to close the pore of E73Q VDAC and the channel gating occurs at higher voltages for E73Q VDAC compared with WT murine VDAC (34). We conclude that the conformational flexibility of β-strands β1–β4 is, in addition to the N-terminal helix, important for the voltage gating and for protein–protein interactions of VDAC in vitro and in vivo.
VDAC Retains the Sidedness of Bacterial Porins.
In mitochondria, hVDAC1 faces the cytoplasm and the IMS by hydrophilic residue exposure (Fig. 3C). In the structure of hVDAC1, an asymmetry in the length of the loop regions is visible (Fig. 3), in agreement with AFM measurements that showed a rough and a flat side of VDAC (27). In addition, tyrosine residues are found almost exclusively at the water–membrane interface at the C-terminal end of every second strand (Fig. 1B) (35). Moreover, antibodies raised against the N terminus of scVDAC were able to bind to these membranes only when mitochondrial envelopes were disrupted (36). The combined data suggest that the C terminus of VDAC extends into the IMS, resembling the orientation found in all bacterial OMPs (24).
Summary and Perspectives.
Our combined approach using data of both NMR spectroscopy and x-ray crystallography enabled the determination of the 3D structure of the first MOM protein, hVDAC1. The finding of a 19-stranded β-barrel is in strong contrast to evolutionary theories predicting bacterial β-barrels and related proteins to be formed by an ancient β-hairpin motif (37). The new channel architecture is likely to be a consequence of the differences in membrane environment, sorting signal and partner proteins experienced by integral membrane proteins of the outer membrane of bacteria and those of the MOM and most likely adopted by other MOM proteins. On the basis of the NMR resonance assignment and the 3D structure of the hVDAC1 channel, a variety of experiments and structural studies of other VDAC isoforms will be possible. The functional difference of the E73 mutant compared with WT could be rationalized based on the structure described in this work.
Materials and Methods
Expression, refolding, and purification of hVDAC1 were done as described. NMR spectra were recorded from a perdeuterated 15N and 15N/13C labeled samples containing 0.6 mM HVDAC, 25 mM BisTris, ≈250 mM Lauryldimethylamine-oxide (LDAO), and 10% D2O, pH 6.8. Assignment of backbone resonance of hVDAC1 was achieved through six independent types of NMR data: Cα chemical-shift connectivity, intrastrand and sequential HN-HN NOEs, amino acid-specific labeling, 20 point mutations, paramagnetic broadening induced by spin labels, and hydrogen/deuterium exchange experiments. Native crystals were obtained with the hanging-drop method by mixing 1.5 μl of protein-Cymal-5 detergent solution containing 5–15 mg/ml protein with 0.75 μl of reservoir solution (30% PEG 400; 0.1 M Na Hepes, pH 7.5; 0.2 M magnesium chloride). The trigonal crystals (space group P321) diffracted up to 4 Å. Structure calculations were done iteratively by cycling between real space ensemble generation using the ISD program and reciprocal space refinement using the BUSTER-TNT program. Detailed information on methodological aspects is provided as SI Text.
Supplementary Material
Acknowledgments.
We thank Vinesh Vijayan for help with NMR measurements in the initial stages of the project and Jegannath Korukottu for useful discussions. This work was supported by the Max Planck Society, the Fonds der Chemischen Industrie, the German science foundation Deutsche Forschungsgemeinschaft (DFG) (ZE 522/2-3 scholarship, to T.M.), and through a DFG Heisenberg scholarship (ZW 71/2-1 and 3-1, to M.Z.). The help of Dr. Johannes Söding (bioinformatics) and Christel Weyrauch (protein purification, crystallization) in the beginning of this project is appreciated. The help of people from beamlines PXII at the Swiss Light Source and beamlines ID29/ID23 at the European Synchrotron Radiation Facility is highly appreciated.
Footnotes
The authors declare no conflict of interest.
Data deposition: The coordinates and structure factors have been deposited in the Protein Data Bank (PDB ID code 2jk4).
This article contains supporting information online at www.pnas.org/cgi/content/full/0808115105/DCSupplemental.
References
- 1.Neupert W, Herrmann JM. Translocation of proteins into mitochondria. Annu Rev Biochem. 2007;76:723–749. doi: 10.1146/annurev.biochem.76.052705.163409. [DOI] [PubMed] [Google Scholar]
- 2.Hill K, et al. Tom40 forms the hydrophilic channel of the mitochondrial import pore for preproteins [see comment] Nature. 1998;395:516–521. doi: 10.1038/26780. [DOI] [PubMed] [Google Scholar]
- 3.Wiedemann N, et al. Machinery for protein sorting and assembly in the mitochondrial outer membrane. Nature. 2003;424:565–571. doi: 10.1038/nature01753. [DOI] [PubMed] [Google Scholar]
- 4.Schein SJ, Colombini M, Finkelstein A. Reconstitution in planar lipid bilayers of a voltage-dependent anion-selective channel obtained from paramecium mitochondria. J Membr Biol. 1976;30:99–120. doi: 10.1007/BF01869662. [DOI] [PubMed] [Google Scholar]
- 5.Blachly-Dyson E, Forte M. VDAC channels. IUBMB Life. 2001;52:113–118. doi: 10.1080/15216540152845902. [DOI] [PubMed] [Google Scholar]
- 6.Benz R. Permeation of hydrophilic solutes through mitochondrial outer membranes: Review on mitochondrial porins. Biochim Biophys Acta. 1994;1197:167–196. doi: 10.1016/0304-4157(94)90004-3. [DOI] [PubMed] [Google Scholar]
- 7.Guo XW, Mannella CA. Conformational change in the mitochondrial channel, VDAC, detected by electron cryo-microscopy. Biophys J. 1993;64:545–549. doi: 10.1016/S0006-3495(93)81399-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shao L, Kinnally KW, Mannella CA. Circular dichroism studies of the mitochondrial channel, VDAC, from Neurospora crassa. Biophys J. 1996;71:778–786. doi: 10.1016/S0006-3495(96)79277-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Grimm S, Brdiczka D. The permeability transition pore in cell death. Apoptosis. 2007;12:841–855. doi: 10.1007/s10495-007-0747-3. [DOI] [PubMed] [Google Scholar]
- 10.Shoshan-Barmatz V, Israelson A, Brdiczka D, Sheu SS. The voltage-dependent anion channel (VDAC): Function in intracellular signalling, cell life and cell death. Curr Pharmacol Des. 2006;12:2249–2270. doi: 10.2174/138161206777585111. [DOI] [PubMed] [Google Scholar]
- 11.Engelhardt H, et al. High-level expression, refolding and probing the natural fold of the human voltage-dependent anion channel isoforms I and II. J Membr Biol. 2007;216:93–105. doi: 10.1007/s00232-007-9038-8. [DOI] [PubMed] [Google Scholar]
- 12.Meins T, Vonrhein C, Zeth K. Crystallization and preliminary X-ray crystallographic studies of human voltage-dependent anion channel isoform I (HVDAC1) Acta Crystallogr F. 2008;64:651–655. doi: 10.1107/S174430910801676X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.DeLaBarre B, Brunger AT. Considerations for the refinement of low-resolution crystal structures. Acta Crystallogr D. 2006;62:923–932. doi: 10.1107/S0907444906012650. [DOI] [PubMed] [Google Scholar]
- 14.Cornilescu G, Delaglio F, Bax A. Protein backbone angle restraints from searching a database for chemical shift and sequence homology. J Biomol NMR. 1999;13:289–302. doi: 10.1023/a:1008392405740. [DOI] [PubMed] [Google Scholar]
- 15.Neal S, Berjanskii M, Zhang H, Wishart DS. Accurate prediction of protein torsion angles using chemical shifts and sequence homology. Magn Reson Chem. 2006;44 Spec No:S158–167. doi: 10.1002/mrc.1832. [DOI] [PubMed] [Google Scholar]
- 16.Rieping W, Habeck M, Nilges M. Inferential structure determination. Science. 2005;309:303–306. doi: 10.1126/science.1110428. [DOI] [PubMed] [Google Scholar]
- 17.Roversi P, Blanc E, Vonrhein C, Evans G, Bricogne G. Modelling prior distributions of atoms for macromolecular refinement and completion. Acta Crystallogr D. 2000;56:1316–1323. doi: 10.1107/s0907444900008490. [DOI] [PubMed] [Google Scholar]
- 18.Vonrhein C, Blanc E, Roversi P, Bricogne G. Automated structure solution with autoSHARP. Methods Mol Biol. 2007;364:215–230. doi: 10.1385/1-59745-266-1:215. [DOI] [PubMed] [Google Scholar]
- 19.Bricogne G, Vonrhein C, Flensburg C, Schiltz M, Paciorek W. Generation, representation and flow of phase information in structure determination: Recent developments in and around SHARP 2.0. Acta Crystallogr D. 2003;59:2023–2030. doi: 10.1107/s0907444903017694. [DOI] [PubMed] [Google Scholar]
- 20.DeLano WL. PyMOL Reference Manual. San Carlos, CA: DeLano Scientific; 2003. [Google Scholar]
- 21.Fabiola GF, Krishnaswamy S, Nagarajan V, Pattabhi V. C-H. O hydrogen bonds in beta-sheets. Acta Crystallogr D. 1997;53:316–320. doi: 10.1107/S0907444997000383. [DOI] [PubMed] [Google Scholar]
- 22.Goncalves RP, Buzhynskyy N, Prima V, Sturgis JN, Scheuring S. Supramolecular assembly of VDAC in native mitochondrial outer membranes. J Mol Biol. 2007;369:413–418. doi: 10.1016/j.jmb.2007.03.063. [DOI] [PubMed] [Google Scholar]
- 23.Dolder M, et al. Crystallization of the human, mitochondrial voltage-dependent anion-selective channel in the presence of phospholipids. J Struct Biol. 1999;127:64–71. doi: 10.1006/jsbi.1999.4141. [DOI] [PubMed] [Google Scholar]
- 24.Schulz GE. The structure of bacterial outer membrane proteins. Biochim Biophys Acta. 2002;1565:308–317. doi: 10.1016/s0005-2736(02)00577-1. [DOI] [PubMed] [Google Scholar]
- 25.Yildiz O, Vinothkumar KR, Goswami P, Kuhlbrandt W. Structure of the monomeric outer-membrane porin OmpG in the open and closed conformation. EMBO J. 2006;25:3702–3713. doi: 10.1038/sj.emboj.7601237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Subbarao GV, van den Berg B. Crystal structure of the monomeric porin OmpG. J Mol Biol. 2006;360:750–759. doi: 10.1016/j.jmb.2006.05.045. [DOI] [PubMed] [Google Scholar]
- 27.Hoogenboom BW, Suda K, Engel A, Fotiadis D. The supramolecular assemblies of voltage-dependent anion channels in the native membrane. J Mol Biol. 2007;370:246–255. doi: 10.1016/j.jmb.2007.04.073. [DOI] [PubMed] [Google Scholar]
- 28.Zalk R, Israelson A, Garty ES, Azoulay-Zohar H, Shoshan-Barmatz V. Oligomeric states of the voltage-dependent anion channel and cytochrome c release from mitochondria. Biochem J. 2005;386:73–83. doi: 10.1042/BJ20041356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Malia TJ, Wagner G. NMR structural investigation of the mitochondrial outer membrane protein VDAC and its interaction with antiapoptotic Bcl-xL. Biochemistry. 2007;46:514–525. doi: 10.1021/bi061577h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Thomas L, Blachly-Dyson E, Colombini M, Forte M. Mapping of residues forming the voltage sensor of the voltage-dependent anion-selective channel. Proc Natl Acad Sci USA. 1993;90:5446–5449. doi: 10.1073/pnas.90.12.5446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Peng S, Blachly-Dyson E, Forte M, Colombini M. Large scale rearrangement of protein domains is associated with voltage gating of the VDAC channel. Biophys J. 1992;62:123–131. doi: 10.1016/S0006-3495(92)81799-X. discussion 131–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Peng K, et al. Optimizing long intrinsic disorder predictors with protein evolutionary information. J Bioinform Comput Biol. 2005;3:35–60. doi: 10.1142/s0219720005000886. [DOI] [PubMed] [Google Scholar]
- 33.Abu-Hamad S, Zaid H, Israelson A, Nahon E, Shoshan-Barmatz V. Hexokinase-I protection against apoptotic cell death is mediated via interaction with the voltage-dependent anion channel-1: Mapping the site of binding. J Biol Chem. 2008;283:13482–13490. doi: 10.1074/jbc.M708216200. [DOI] [PubMed] [Google Scholar]
- 34.Zaid H, Abu-Hamad S, Israelson A, Nathan I, Shoshan-Barmatz V. The voltage-dependent anion channel-1 modulates apoptotic cell death. Cell Death Differ. 2005;12:751–760. doi: 10.1038/sj.cdd.4401599. [DOI] [PubMed] [Google Scholar]
- 35.Jackups R, Jr, Liang J. Interstrand pairing patterns in beta-barrel membrane proteins: The positive-outside rule, aromatic rescue, and strand registration prediction. J Mol Biol. 2005;354:979–993. doi: 10.1016/j.jmb.2005.09.094. [DOI] [PubMed] [Google Scholar]
- 36.Guo XW, et al. Molecular design of the voltage-dependent, anion-selective channel in the mitochondrial outer membrane. J Struct Biol. 1995;114:41–59. doi: 10.1006/jsbi.1995.1004. [DOI] [PubMed] [Google Scholar]
- 37.Arnold T, Poynor M, Nussberger S, Lupas AN, Linke D. Gene duplication of the eight-stranded beta-barrel OmpX produces a functional pore: A scenario for the evolution of transmembrane beta-barrels. J Mol Biol. 2007;366:1174–1184. doi: 10.1016/j.jmb.2006.12.029. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.