Abstract
The keratin intermediate filament network is abundant in epithelial cells, but its function in the establishment and maintenance of cell polarity is unclear. Here, we show that Albatross complexes with Par3 to regulate formation of the apical junctional complex (AJC) and maintain lateral membrane identity. In nonpolarized epithelial cells, Albatross localizes with keratin filaments, whereas in polarized epithelial cells, Albatross is primarily localized in the vicinity of the AJC. Knockdown of Albatross in polarized cells causes a disappearance of key components of the AJC at cell–cell borders and keratin filament reorganization. Lateral proteins E-cadherin and desmoglein 2 were mislocalized even on the apical side. Although Albatross promotes localization of Par3 to the AJC, Par3 and ezrin are still retained at the apical surface in Albatross knockdown cells, which retain intact microvilli. Analysis of keratin-deficient epithelial cells revealed that keratins are required to stabilize the Albatross protein, thus promoting the formation of AJC. We propose that keratins and the keratin-binding protein Albatross are important for epithelial cell polarization.
Introduction
Keratin filaments are epithelial-specific intermediate filaments (IFs) whose major function is apparently to provide mechanical strength (Fuchs and Cleveland, 1998; Coulombe and Wong, 2004). However, the expression of components of IFs is strictly regulated in cell-, tissue-, and differentiation-specific fashions, though other major cytoskeletal proteins, actin and tubulin, are highly conserved through different cell types. The keratin subfamily has >20 members. In simple epithelia, keratin 8 and 18 (K8/18) are the major subtypes and form hetero-oligomeric filaments. The apparent redundancy of keratins has for a long time obscured the analysis of keratin-specific functions, but recently, several novel keratin functions have been described. For example, they may modulate apoptosis (Caulin et al., 2000; Gilbert et al., 2001; Inada et al., 2001) and control cell growth through interaction with 14-3-3 proteins (Kim et al., 2006). It is likely that keratins have additional physiological roles.
To address this issue, we have performed searches for keratin-binding proteins. Here, we report findings for a novel protein involved in regulation of the apical junctional complex (AJC) and lateral domains in epithelial cells. In the past, we described the chaperoning protein of Mrj (Izawa et al., 2000), the TNF receptor 1–associated protein of TRADD (Inada et al., 2001) and a novel protein of trichoplein (Nishizawa et al., 2005), to be keratin-binding proteins. Among these proteins, trichoplein has a unique domain, trichohyalin and plectin homology domain (TPHD), which has similarities with trichohyalin and plectin of classical keratin-binding proteins (Izawa and Inagaki, 2006).
The functions of the AJC are believed to: (1) act as a mechanical link between adjacent cells via adherens junctions (AJ) and desmosomes (DS); (2) provide a fence between apical and lateral plasma membrane domains to establish cell polarity by tight junctions (TJ), though TJ are actually not necessary for fence functions (Umeda et al., 2006); and (3) selectively block paracellular pathways via TJ (Tsukita et al., 2001). It is a key character of polarized epithelial cells (Rodriguez-Boulan and Nelson, 1989). In this paper, we document that Albatross (also known as Fas-binding factor-1; Schmidt et al., 2000), a keratin-binding protein containing TPHD, regulates the AJC and lateral domains in epithelial cells. In addition, keratins stabilize Albatross and play a role in its AJC localization.
Results and discussion
Albatross was initially noted as one TPHD-containing protein in a basic local alignment search tool (BLAST) search. To characterize this protein, we generated anti-Albatross antibodies that detected a 130-kD form upon immunoblotting (Fig. 1 A). This putative molecular size was calculated from cloned human Albatross (Fig. S1, A to C, available at http://www.jcb.org/cgi/content/full/jcb.200803133/DC1). Using the antibody, we found Albatross to be expressed in various epithelial cells (Fig. 1 B). In immunoprecipitation assays with an anti-Albatross antibody, K8/18 coimmunoprecipitated (Fig. 1 C). In a yeast hybrid system, Albatross was further found to bind to these keratins, and in a cosedimentation assay, they cosedimented with Albatross, in contrast to desmin and vimentin (Fig. S1, D and E). Albatross has the unique TPHD domain (Fig. S1, A and B) and directly binds keratin 18 via this domain in a yeast two-hybrid system (Fig. S1 D). In addition, immunofluorescence revealed Albatross to be discontinuously associated with the K8/18 filament network (Fig. 1 D). The results thus strongly indicate that Albatross is a keratin-binding protein.
Figure 1.
Albatross is a keratin-binding protein. (A) Immunoblotting with affinity-purified rabbit anti-Albatross antibodies. Specific recognition of a polypeptide with a relative molecular mass of ∼130 kD, the putative size of Albatross, in HeLa and Caco-2 epithelial cells (middle). Preincubation of the antibody with the antigen selectively inhibited the immunoreactivity (right). CBB, a loading control (left). (B) Lysates prepared from the indicated cells were subjected to immunoblot analysis for Albatross. (C) Anti-Albatross antibody immunoprecipitants were subsequently immunoblotted with the indicated antibodies. Left and right lanes show immunoprecipitates with control rabbit IgG and anti-Albatross antibodies, respectively. With the indicated epithelial cells, Albatross, keratin 8, and keratin 18 immunoprecipitated as a complex. (D) Double staining of keratin 18 and Albatross. Albatross is discontinuously associated with the K8/18 filament network in the indicated epithelial cells. Mag., hyper-magnification of regions indicated by arrowheads. (E) Immunoblotting for keratin 8 and 18, and the indicated junctional and cell-polarity proteins in control and Albatross knockdown A549 (A1050 and A1160) cells. Albatross knockdown cells were produced successfully in which the expression level of keratin 8 and 18, the indicated AJC-specific transmembrane and undercoat proteins, lateral proteins, and apical proteins were not reduced. α-tubulin was included as a loading control. (F) Double staining for Albatross and keratin 18 in control and Albatross knockdown A549 (Albatross KD) cells. In control cells, Albatross is localized at the apical side of cell–cell border, and keratin filaments form apical bundles, whereas in Albatross KD cells, keratin filaments were apparently dissociated from cell–cell borders (arrowheads). Top and bottom columns illustrate projections of x-y planes and z sections, respectively. Bars, 10 μm.
When epithelial tissues were double-stained for Albatross and keratin 8/18, Albatross was localized not only on keratin filaments but also in the vicinity of the AJC, especially in polarized epithelial cells (Fig. S2, available at http://www.jcb.org/cgi/content/full/jcb.200803133/DC1). A549 lung adenocarcinoma cells form distinct surface domains with well-developed AJCs (Fig. 1 F). Using this cell line, we generated Albatross knockdown A549 cells (Fig. 1 E). Immunostaining revealed that in the apical region of control cells, keratin filaments were assembled into thick bundles along the cell–cell borders. However, in Albatross knockdown A549 cells, the attachment of keratin filaments to intercellular junctions was apparently weakened (Fig. 1 F). As it is has been found that keratin is anchored on the AJC via DS in polarized epithelial cells (Jones and Green, 1991; Bornslaeger et al., 1996; Perez-Moreno et al., 2003), we assumed that this phenotype was caused either by affecting the anchoring of keratin filaments on DS or by affecting the AJC itself.
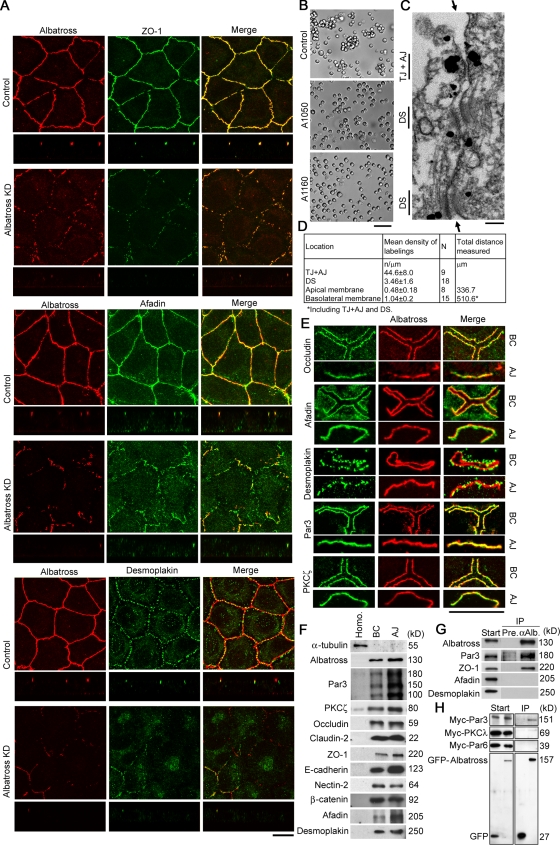
To distinguish between these possibilities, immunostaining was performed (Fig. 2 A and Fig. S3 A, available at http://www.jcb.org/cgi/content/full/jcb.200803133/DC1). In Albatross knockdown cells, AJC components no longer accumulated on the AJC, except in regions where residual Albatross was present: TJ, ZO-1; AJ, afadin; DS, desmoplakin (Fig. 2 A). Their transmembrane proteins were similarly affected (Fig. S3 A), which strongly suggested impaired cell–cell adhesion. We therefore executed an aggregation assay (Fig. 2 B; Takeichi, 1977). In Albatross knockdown A549 cells, the formation of cell aggregates was indeed reduced, and single cells were increased. Thus, the cell–cell adhesive properties were weakened. Furthermore, immunoelectron microscopy revealed that Albatross was predominantly localized in the vicinity of AJC (Fig. 2, C and D; and Fig. S3 B). From these data, we concluded that Albatross is crucial for AJC formation with respect to both components and physiological functions in A549 cells.
Figure 2.
Impaired AJC formation in Albatross knockdown cells. (A) Double staining for Albatross (red) and the undercoat proteins (green) for each AJC component: TJ, ZO-1; AJ, afadin; DS, desmoplakin. Top and bottom columns show projections of x-y planes and z sections, respectively. Albatross knockdown A549 (Albatross KD) cells lack accumulation of these proteins at the cell–cell borders except in regions where residual Albatross is present. (B) Cell–cell adhesive properties evaluated by a cell aggregation assay. In the differential interference contrast images, control cells show cell aggregation. With Albatross knockdown A549 (A1050 and A1160) cells, the aggregated cell population is reduced and free cells are increased. The percentages of single cells in total cells (mean ± SD) are: control, 36.1 ± 3.9; A1050, 52.4 ± 2.8; A1160 cells, 59.4 ± 10.2. n = 4 and P < 0.01. (C) Immunoelectron microscopy of A549 cells with anti-Albatross antibodies. Note that the cytoplasm in the vicinity of AJCs is labeled. TJ, AJ, and DS are indicated. Arrows indicate cell–cell contacts. (D) Quantitative data from C. (E) BC fraction and AJ fraction were immunostained for Albatross with the indicated AJC proteins, PKCζ or Par3. Note that Albatross is well colocalized with them. (F) Immunoblotting of fractions derived from mouse liver: homogenates (left), BC (middle), and AJ (right). Not only Albatross but also Par3 is enriched in line with the concentrations of the indicated AJC components. (G) Immunoprecipitation of A549 cells with anti-Albatross antibodies. Start and IP indicate starting lysates and immunoprecipitates with preimmune (Pre.) and anti-Albatross (αAlb.) antibodies, respectively. Note the Par3 precipitation with Albatross. Among AJC components, ZO-1 also coprecipitated. (H) Immunoprecipitation analysis with tagged Albatross and Par3. Start and IP indicate starting lysates and immunoprecipitates with anti-GFP antibodies, respectively. Left lanes show results for negative controls expressing GFP alone. Par3 was the most precipitated with GFP-Albatross among coexpressed myc-Par3, -Par6, and -PKCλ. Bars: (A) 10 μm; (B) 100 μm; (C) 0.1 μm; (E, BC) 13 μm; (E, AJ) 10 μm.
Then, to examine the colocalization of Albatross with other components in AJC, we prepared a “junctional complex–enriched” fraction, as described previously (Tsukita and Tsukita, 1989). Immunostaining of the bile canaliculi (BC) fraction and the AJ fraction revealed that Albatross was colocalized with Par3 and other AJC proteins like occludin, afadin, and desmoplakin (Fig. 2 E). On immunoblotting of BC and AJ, not only Albatross but also Par3 was enriched, in line with the concentrations of other AJC components (Fig. 2 F). In immunoprecipitates of A549 cells with an anti-Albatross antibody, Par3 and ZO-1 were found to be present (Fig. 2 G). The association with Par3 was further confirmed by immunoprecipitation with tagged proteins (Figs. 2 H and S3 C). These immunoprecipitation results indicate that Albatross also binds with ZO-1, which suggests that Albatross may have other binding partners in addition to Par3.
In Albatross knockdown cells, Par3 at AJC was certainly abolished, but Par3 at the apical domains remained (Fig. 3 A). It is well known that Par3 regulates apical cell polarity (Nelson, 2003; Hirose et al., 2006). In our cells, ezrin was also found to be still localized on apical domains. Microvilli were also intact, presumably because of the retention of these apical proteins (Fig. 3, B and C). In Albatross knockdown A549 cells, the lateral domain components of E-cadherin and desmoglein 2 no longer accumulated on the AJC and at lateral domains, and were mislocalized even at the apical membranes (Fig. 3, D and E). The cortical actin bundles along the cell–cell borders were not organized (Fig. 3 F). These data suggest that Albatross with Par3 regulates the integrity of the AJC and maintenance of lateral membrane identity but does not affect Par3 at the apical surface.
Figure 3.
Albatross knockdown affects the localization of Par3 at AJCs and lateral proteins without affecting apical components. (A, B, and D–F) Double staining for Albatross and the indicated proteins. The top two rows indicate projections of x-y planes, and the bottom rows are for merged images of z sections. (A) Compared with control cells, Par3 is reduced in AJCs of Albatross knockdown A549 (Albatross KD) cells, though Par3 in the apical domains remains. (B) Ezrin is still localized in the apical domains of Albatross KD cells. (C) Scanning electron micrograph showing that microvilli on the apical domains are not impaired in Albatross KD (A1050 and A1160) cells. (D) In control cells, E-cadherin is localized on lateral domains and in AJCs. In Albatross KD cells, it is no longer accumulated on AJCs and lateral domains, and is mislocalized even at apical membranes. (E) Desmoglein 2 demonstrates similar alteration. (F) In control cells, cortical actin bundles are organized along the cell–cell border. In Albatross KD cells, they are not organized. Bars: (A, B, and D–F) 10 μm; (C) 1 μm.
To confirm the phenotype described above, we introduced GFP-mouse Albatross into cloned Albatross knockdown A549 cells and developed a stable transformant (Fig. 4 A). In these cells, the localization of Albatross at the cell–cell border was promoted as compared with GFP-introduced cells. Other AJC components and Par3 were also localized at the AJC.
Figure 4.
Albatross–Par3 complex functions. (A) Rescue experiments. Double staining for Albatross and GFP, AJC components, or Par3 was performed with Albatross knockdown A549 cells in which GFP-Albatross or GFP (negative control) were introduced. Note that the GFP-Albatross–introduced cells show staining of GFP at cell–cell borders and recovery of the localization of Albatross, AJC components, and Par3. (B) Immunoblotting for the indicated junctional and cell-polarity proteins in control and Par3 knockdown A549 (A1555 and A1556) cells. Par3 knockdown cells have normal expression levels of the indicated AJC-specific proteins, lateral proteins, and apical proteins. GAPDH is included as a loading control. (C–E) Double staining for Par3 and the indicated proteins in Par3 knockdown A549 (Par3 KD) cells. The top two rows show projections of x-y planes and the bottom rows show merged images of z sections. (C) Compared with control cells, Albatross is reduced in AJCs of Par3 KD cells. (D) As in Albatross knockdown cells, desmoglein 2 no longer accumulated on AJCs and lateral domains, and was mislocalized even on the apical membranes in Par3 KD cells. (E) Par3 KD cells show impairment of apical localization of ezrin, in contrast to Albatross knockdown cells. Bars, 10 μm.
It has been found that Par3 controls the assembly of TJ and AJ in AJCs (Mertens et al., 2005; Chen and Macara, 2005; Mertens et al., 2006; Ooshio et al., 2007). In Par3 knockdown A549 cells (Fig. 4 B), Albatross was not concentrated at cell–cell borders (Fig. 4 C). ZO-1, afadin, and desmoplakin at AJCs were also abolished, and the lateral protein of desmoglein 2 was mislocalized even on the apical side, in accordance with the phenotype of Albatross knockdown cells (Figs. 4 D and S3 D). As described previously (Hirose et al., 2006), Par3 knockdown distinctively caused reduction of ezrin at the apical surface (Fig. 4 E). Collectively with the results in Fig. 3, these findings indicate that there are two types of Par complex, based on whether they associate with or without Albatross.
To investigate potential common functions of keratins and Albatross in cell polarity, we introduced K8/18 into keratin-deficient, SW13 epithelial cells (Fig. 5, A–D). The amount of Albatross was elevated according to the increase of keratins (Fig. 5, A and B). Furthermore, stable introduction of K8/18 in SW13 cells caused elevation of cytosolic Albatross and promoted its localization to cell–cell borders, as assessed by immunostaining (Fig. 5 D). However, the level of Albatross mRNA was not changed (Fig. 5 C). These results indicate that keratins stabilize Albatross without altering the expression level, which results in the prompt localization of AJC components to cell–cell borders (Fig. 5 D). This was confirmed by Albatross knockdown in K8/18-introduced SW13 cells, which exhibit disturbed AJC formation (Fig. 5 E), indicating that K8/18-induced junction formation is dependent on Albatross. In other words, keratins function as dynamic and multipurpose scaffolds. As an explanation, disruption of the keratin cytoskeleton may be the mechanism of AJC disruption through Albatross function.
Figure 5.
Functions of keratins and Albatross–Par3 complexes. (A–C) The amounts of Albatross protein and mRNA were analyzed in both keratin 8 and keratin 18 (K8/18)-introduced SW13 cells. As a control, an empty vector was transfected. As loading controls, α-tubulin and GAPDH were used. Two independent experiments were performed. (A) Immunoblotting. In transiently K8/18-introduced SW13 cells, the amount of Albatross protein is elevated, along with the amount of keratin 18. (B) With stable lines, the same results were obtained. (C) RT-PCR. In K8/18-introduced SW13 cells, the mRNA level of K18 is elevated, but not that of Albatross. β-actin is included as an internal control. (D) Double staining for K8/18 and the indicated proteins: Albatross, AJC components of ZO-1 and afadin, and Par3. (top) In control cells, K8/18 is absent and only limited amounts of Albatross are apparent at cell–cell junctions. In stably K8/18-introduced SW13 cells, Albatross is well localized in cell–cell junctions compared with control cells. (middle and bottom) ZO-1, afadin, and Par3 similarly accumulated at the cell–cell borders in stably K8/18-introduced SW13 cells. (E) Immunostaining of stably K8/18-introduced SW13 cells transfected with control or Albatross siRNA. Note that ZO-1, afadin, and Par3 are reduced at cell–cell borders with knockdown of Albatross. (F) A model for the regulation of AJC and lateral domains with the Albatross–Par3 complex and keratins. Albatross–Par3 complexes regulate the formation of AJC and maintain lateral membrane identity. However, Par3 without Albatross regulates apical structures. Keratins stabilize Albatross, promoting the formation of AJC. Knockdown effects are also indicated. Bars, 10 μm.
Here, we propose a model for the regulation of AJC and lateral domains with an Albatross–Par3 complex and keratins (Fig. 5 F). The Albatross–Par3 complex regulates AJC formation and maintains lateral membrane identity. However, Par3 without Albatross regulates apical membrane formation. Keratins stabilize Albatross, which promotes formation of the AJC. Thus, keratins and the keratin-binding protein Albatross may play critical roles in the polarization of epithelial cells. These findings add a new dimension to the biological significance of these proteins in polarized epithelial cells within tissues.
Materials and methods
Cloning of full-length Albatross
Full-length mouse Albatross cDNA was correctly generated from integrated molecular analysis of genomes and their expression (IMAGE) cDNA clones (available from GenBank/EMBL/DDBJ under accession nos. BC043126 and CB519635) according to the cDNA sequence deposited in GenBank (accession no. AF241249) and the genomic sequence of mouse Albatross in the University of California, Santa Cruz, Genome Browser (http://www.genome.ucsc.edu/index.html). Human Albatross cDNA was cloned from A549 cells by RT-PCR with a set of primers designed according to the predicted cDNA sequence deposited in GenBank (accession no. NM_001080542). Then, the sequence was determined (GenBank/EMBL/DDBJ accession no. AB354594).
RT-PCR
Semiquantitative RT-PCR was performed using a poly-dT primer and SuperScript III transcriptase (Invitrogen), followed by PCR with human Albatross-specific primers (5′-CCGGAATTCGCACCAAAAACCAAGAAAGGA-3′ and 5′-GCTCTAGATCAGGCTGAATGAGATGTC-3′), human keratin 18–specific primers (5′-ATGAGCTTCACCACTCGCTC-3′ and 5′-TTAATGCCTCAGAACTTTGGTGT-3′), or β-actin–specific primers (5′-TGACCCAGATCATGTTTGAGA-3′ and 5′-ACTCCATGCCCAGGAAGGA-3′).
Yeast hybrid assay
Yeast hybrid assays were performed by the regular method (James et al., 1996) or with a three-hybrid approach (Van Criekinge et al., 1998). In the latter case, two plasmids containing the GAL4 DNA-binding domain and nutritionally selective genes of TRP1 or URA3 (pGBD or pGBDU, respectively) fused with keratin 18 or keratin 8, or without them, were transformed into Saccharomyces cerevisiae PJ69-4A and selected. Subsequently, plasmids containing the LEU2 and GAL4 DNA-activation domains (pGAD) fused with or without Albatross were independently introduced into the yeast and assayed under appropriate selective conditions.
Generation of anti-Albatross antibodies
The GST-fused mouse Albatross fragment (amino acid residues 774–954) expressed in Escherichia coli was purified with glutathione Sepharose 4B beads (GE Healthcare) and used as the antigen in rabbits. Then, the generated anti-Albatross antibodies were preabsorbed with GST and affinity-purified.
Immunoblotting
For immunoblotting, proteins were separated by SDS-PAGE and electrophoretically transferred onto polyvinylidene fluoride sheets (Immobilon-P; Millipore), which were then incubated with primary antibodies. Binding was detected with HRP-conjugated secondary antibodies and Western blotting detection reagents (Western lightning chemiluminescence reagent plus; PerkinElmer; or SuperSignal West Femto Maximum Sensitivity Substrate; Thermo Fisher Scientific).
Immunoprecipitation
For Fig. 1C, cells were lysed on ice for 20 min in lysis buffer consisting of 1% Triton X-100, 20 mM Tris/HCl, pH 7.5, 150 mM NaCl, 1 mM EDTA, 10 μM PMSF, and 10 μg/ml leupeptin. Lysates were clarified by centrifugation at 15 krpm for 30 min. Endogenous Albatross was immunoprecipitated from cell lysates with Albatross antibody or control rabbit IgG, and protein G–agarose beads. Then, they were washed four times with lysis buffer. Immunoprecipitates were analyzed by immunoblotting. For Fig. 2 G, the following modifications were made. Lysis buffer containing 0.1% NP-40, 25 mM Tris/Cl, pH 8.0, 150 mM NaCl, 1 mM EDTA, 1 mM EGTA, 1 mM DTT, 1 mM PMSF, and 10 μg/ml leupeptin was used, and lysates were sonicated. They were clarified by centrifugation at 15 krpm for 10 min.
Immunoprecipitations with tagged proteins
Tagged proteins were ectopically expressed in COS7 cells and immunoprecipitated with anti-GFP antibodies as described previously (Nishimura et al., 2005).
Expression and purification of recombinant proteins
Recombinant Albatross was expressed in E. coli as maltose-binding protein (MBP)-tagged protein a using pMAL vector (QIAGEN). MBP-tagged Albatross was expressed and purified according to the manufacturer's instructions. Recombinant human keratin 8, keratin 18, desmin, and mouse vimentin expressed in E. coli were prepared as described previously (Izawa et al., 2000).
IF assembly and cosedimentation assay
In vitro assembly and cosedimentation assays were performed as described previously (Izawa et al., 2000). Polymerized IF proteins, heterotypic complexes of K8/18, vimentin, and desmin, were incubated with or without MBP-Albatross. The assembled complexes were subjected to centrifugation at 15,000 rpm for 30 min followed by SDS-PAGE.
Cells
HeLa cells and T24 human bladder cell carcinoma cells were grown in DME supplemented with 10% FBS. Human mammary epithelial cells (HMEC) were immortalized by retroviral gene transfer of a cDNA encoding the catalytic component of human telomerase (pLXSN-fhTERT), provided by D.A. Galloway (Fres Hutchinson Cancer Research Center, Seattle, WA), and grown as described previously (Kiyono et al., 1998). DJM-1 human trichilemmal cyst carcinoma cells were grown in Eagle's minimum essential medium supplemented with 10% FBS, 20 ng/ml EGF, 0.4 μg/ml hydrocortisone, and 84 ng/ml cholera toxin. Caco-2 colorectal adenocarcinoma cells were grown in DME with 10% FBS and 10 μg/ml human transfelin. The SW13 human adenocarcinoma is devoid of both keratins and vimentin. Epstein-Barr virus nuclear antigen–expressing SW13 cells (Inada et al., 2001) were grown in DME with 10% FBS. A549 lung adenocarcinoma cells were grown in RPMI medium supplemented with 1% FBS. For polarization, A549 cells were cultured after confluence for several days. In each experiment, growth conditions were equalized for control and treated cells. All cells were cultured in an air–5% CO2 atmosphere with constant humidity.
Immunofluorescence microscopy
Cells were grown on glass coverslips or as cell culture inserts with 0.4-μm pores (BD Biosciences). They were fixed with 100% methanol at −20°C for 10 min followed by 100% acetone at room temperature for 1 min, then washed with PBS. For F-actin, they were fixed with 1% formaldehyde in PBS at room temperature for 15 min and similarly washed. After soaking in PBS containing 1% BSA, the samples were incubated with primary antibodies for 1 h. Then they were washed with PBS, followed by incubation for 30 min with the appropriate secondary antibodies. For double-staining with two different rabbit or mouse antibodies, Zenon rabbit or mouse IgG labeling kits (Invitrogen) were used. Mouse tissues were excised and then frozen in liquid nitrogen. Sections were cut at ∼6 μm on a cryostat, mounted on coverslips, air dried, and fixed in 95% ethanol at 4°C for 30 min followed by 100% acetone at room temperature for 1 min. After soaking in PBS containing 1% BSA, primary and second antibodies were applied as described above. Fluorescence images of alveoli, liver, kidney, and brain were recorded with a fluorescence microscope (BX60; Olympus) with a Plan Apo 60×/1.40 NA oil immersion lens and a charge-coupled device camera (DP50; Olympus) controlled by Viewfinder Lite software (Pixera Corporation). Other images were recorded with a confocal microscope system (Radiance; Bio-Rad Laboratories) equipped on a fluorescence microscope (BX50; Olympus) with a Plan Apo 60×/1.40 NA oil immersion lens and a UPlan Apo 100×/1.35 NA oil immersion lens (Olympus) controlled by LaserSharp2000 software (Bio-Rad), or a confocal laser-scanning microscope (LSM510 META; Carl Zeiss, Inc.) equipped with a Plan-Apochromat 100×/1.4 NA oil immersion lens and LSM Image Browser software (Carl Zeiss, Inc). Images were taken at room temperature and further processed using Photoshop Elements 5.0 (Adobe) according to Journal of Cell Biology guidelines.
Northern blot analysis
Detection of mRNA for Albatross was performed with a multiple-choice northern RNA blot (OriGene Technologies, Inc.) containing 2 μg of Poly A+ RNA per lane from mouse tissue. Hybridization and washing were performed according to the manufacturer's instructions. A 620-bp 5′ cDNA fragment digested with BamHI and SmaI was 32P-labeled using the Rediprime II random prime labeling system (GE Healthcare), as recommended by the manufacturer. Membranes were exposed using an imaging analyzer (BAS-2500 MAC; Fujifilm).
Antibodies
The following primary antibodies were used: monoclonal mouse anti-keratin 8 (Ks 8.7; Progen Pharmaceuticals), monoclonal mouse anti-keratin 18 (CY-90; Sigma-Aldrich), polyclonal mouse anti-pan keratin (Sigma-Aldrich), polyclonal guinea pig anti-K8/18 (Progen Pharmaceuticals), polyclonal guinea pig anti–desmoplakin 1 (Progen Pharmaceuticals), monoclonal mouse anti–desmoplakin 1 and 2 (Progen Pharmaceuticals), monoclonal mouse anti–ZO-1 (1; BD Biosciences), monoclonal rat anti–ZO-1 (BM173; Acris Antibodies, GmbH), monoclonal rat anti–E-cadherin (ECCD-2; EMD), monoclonal mouse anti-neurofilaments, monoclonal rat anti–platelet/endothelial cell adhesion molecule (anti-PECAM; CD31; BD Biosciences), monoclonal mouse anti–α-tubulin (B-5-1-2; Sigma-Aldrich), monoclonal mouse anti–claudin-2 (12H12; Invitrogen), monoclonal mouse anti–desmocollin-2/3 (7G6; Invitrogen), monoclonal mouse anti–desmoglein 2 (10G11; Progen Pharmaceuticals), monoclonal mouse anti–nectin-1 (CK8; Invitrogen), monoclonal mouse anti–β-catenin (14; BD Biosciences), polyclonal rabbit anti-ezrin (Millipore), rabbit anti-Par3 polyclonal antibody (provided by S. Ohno, Yokohama City University, Yokohama, Kanagawa, Japan; Millipore), monoclonal mouse anti-occludin (OC-3F10; Invitrogen), monoclonal rat anti–nectin-2 (502–57; HyCult Biotechnology), polyclonal rabbit anti-GFP (Santa Cruz Biotechnology, Inc.), polyclonal rabbit anti-PKCζ (Santa Cruz Biotechnology, Inc.), and polyclonal rabbit anti–glyceraldehyde 3-phosphate dehydrogenase (anti-GAPDH) conjugated to HRP (Abcam). Monoclonal mouse anti-afadin antibody was a gift from S. Tsukita (Osaka University, Osaka, Japan). Species-specific secondary antibodies were conjugated to Alexa Fluor 488, 555, 594, and 680 (Invitrogen) or Cy3 (GE Healthcare). F-actin was stained with Alexa Fluor 488 phalloidin (Invitrogen).
Scanning electron microscopy
Cells were cultured on coverslips. They were fixed with 2.5% glutaraldehyde and 2% formaldehyde in 0.1 M cacodylate buffer, pH 7.4, for 2 h at room temperature, followed by postfixation with the same buffer containing 1% OsO4 on ice for 1 h. Samples were then washed with distilled water, dehydrated through a graded series of ethanols, transferred into isoamyl acetate, and dried in a critical point drier (JCPD-5; JEOL, Ltd.) after substitution with liquid CO2. Dried samples were coated with gold using a gold sputter coater (JFC-1600; JEOL, Ltd.) and were examined under a scanning electron microscope (JSM-5610LV; JEOL, Ltd.).
Immunoelectron microscopy
Cells were fixed with 2% formaldehyde in PBS buffer for 15 min and permeabilized with 0.1% saponin in PBS for 15 min, followed by treatment with anti-Albatross antibodies for 1 h. After washing with PBS, samples were incubated with colloidal gold (1.4 nm)- and Alexa Fluor 488–conjugated secondary antibodies (Alexa Fluor 488–FluoroNanogold–anti–rabbit Fab′; Nanoprobes) for 1 h. Fluorescence images of the sample were obtained for confirmation by fluorescence microscopy. Samples were then fixed with 1% glutaraldehyde in 0.1 M Hepes, pH 7.5, for 10 min. Gold particles in the sample were intensified by use of an HQ sliver kit (Nanoprobes) according to the manufacturer's instructions. After postfixation in 0.1% OsO4 on ice for 90 min, the samples were stained with 0.5% aqueous uranyl acetate for 2 h at room temperature, dehydrated through graded concentrations of ethanol, and embedded in Polybed 812 (PolyScience). Coverslips were removed by immersing the sample in hydrogen fluoride on ice for 60 min. The sample blocks were further embedded in Polybed 812. Ultrathin sections were cut, double-stained with uranyl acetate and lead citrate, and viewed under a transmission electron microscope (JEM 1010; JEOL, Ltd.).
Generation of knockdown cells
Albatross knockdown cell lines were produced by RNA interference with two target sequences of human Albatross mRNA, 5′-GGTGCTAGAAACATCGTAC-3′ and 5′-GGGAGTTCAAGCTAGACAA-3′, independently. To establish these cell lines, shRNA of these sequences (for line 1050 and 1160, respectively) were introduced into cells with the pSI-CMSCVpuro-H1R retroviral vector containing the H1 promoter for shRNA and a puromycin resistance gene (Yugawa et al., 2007). They were then selected in 20 μg/ml puromycin-containing medium for A549 cells. Subsequently, these cells were maintained in medium containing 2 μg/ml puromycin. As a negative control, we used the target sequence for EGFP. In the experiments, all Albatross knockdown phenotypes were confirmed, both for 1050 and 1160 cell lines. Par3 knockdown cell lines were also produced as described above, with two previously reported target sequences of human Par3 mRNA, 5′-GGCATGGAGACCTTGGAAG-3′ and 5′-GACACAGAAGAAAGTTCAA-3′ for lines 1555 and 1556, respectively (Nishimura et al., 2005; Ooshio et al., 2007).
Cell aggregation assays
Cell aggregation assays were accomplished according to the method described previously (Takeichi, 1977), with slight modifications. Cells were trypsinized to produce single cells. Then they were centrifuged and resuspended in Hepes-buffered saline (25 mM Hepes-NaOH, pH 7.2, 137 mM NaCl, 5 mM KCl, 0.7 mM Na2HPO4, and 6 mM dextrose) in the presence of 1 mM CaCl2 at a density of 106 cells/ml. Aliquots (0.5 ml) of suspension were placed in 12-well plates (BD Biosciences) precoated with BSA. After 30-min shaking on a gyratory shaker at 37°C at 80 rpm to allow cell aggregation, the free cells were enumerated with a hemocytometer.
Rescue experiments
cDNAs encoding GFP or GFP-inserted mouse Albatross at 624 amino acids were cloned into expression vectors driven by the herpes simplex virus thymidine kinase promoter with a neomycin resistance gene. They were transfected into cloned Albatross knockdown cells from A1160 with Lipofectamine LTX (Invitrogen) according to the manufacturer's instructions, and each pooled stable line was developed with incubation in medium containing 1 mg/ml neomycin for 5 d.
Keratin-introduced SW13 system
cDNAs encoding full-length human K8/18 were cloned into the mammalian expression vector pIRESpuro3 or one modified with a CAG promoter (Niwa et al., 1991) for stable expression. For transient expression, the pDR2 (Clontech Laboratories, Inc.) vector was used. Transfection into Epstein-Barr virus nuclear antigen–expressing SW13 cells was performed as described above. Two pooled stable lines were developed independently with incubation in medium containing 0.5 μg/ml puromycin for 5 d. For Albatross siRNA experiments, siRNA with target sequences for lines 1050 and 1160, HP-validated siRNA SI03649030 and SI03649058 for gene FBF1 (QIAGEN) or a negative control (QIAGEN) were transfected into cells with Lipofectamine RNAiMAX (Invitrogen) according to the manufacturer's instructions, with slight modifications. In brief, forward transfection was performed for cells at 48 h after reverse transfection, followed by fixation.
Online supplemental material
Fig. S1 shows cloning of human Albatross. Fig. S2 shows that in epithelial tissues, Albatross is not only localized on keratin filaments but also in the vicinity of AJCs. Fig. S3 shows that Albatross regulates AJC formation and associates with Par3. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200803133/DC1.
Supplementary Material
Acknowledgments
We are grateful to Dr. S. Tsukita and Dr. S. Ohno for their offer of valuable samples and suggestions, and we greatly appreciate the kind comments and advice of Dr. T. M. Magin (Bonn University, Germany). We thank Dr. K. Nagata (Aichi Cancer Center Research Institute) for his support and Dr. Malcolm Moore for critical comments on the language in the manuscript.
This work was supported by Grants-in-Aid for Scientific Research on Priority Areas (19057016), Exploratory Research (20657039), and Scientific Research (B; 20370082) from the Ministry of Education, Culture, Sports, Science and Technology of Japan; by a Grant-in-Aid for the Third Term Comprehensive 10-Year Strategy for Cancer Control from the Ministry of Health and Welfare, Japan; by the Naito Foundation; and by the Uehara Memorial Foundation.
M. Sugimoto and A. Inoko contributed equally to this paper.
Abbreviations used in this paper: AJ, adherens junctions; AJC, apical junctional complex; BC, bile canaliculi; DS, desmosomes; GAPDH, glyceraldehyde 3-phosphate dehydrogenase; IF, intermediate filament; K8/18, keratin 8 and 18; MBP, maltose-binding protein; TJ, tight junctions; TPHD, trichohyalin and plectin homology domain.
References
- Bornslaeger, E.A., C.M. Corcoran, T.S. Stappenbeck, and K.J. Green. 1996. Breaking the connection: displacement of the desmosomal plaque protein desmoplakin from cell-cell interfaces disrupts anchorage of intermediate filament bundles and alters intercellular junction assembly. J. Cell Biol. 134:985–1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caulin, C., C.F. Ware, T.M. Magin, and R.G. Oshima. 2000. Keratin-dependent, epithelial resistance to tumor necrosis factor-induced apoptosis. J. Cell Biol. 149:17–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, X., and I.G. Macara. 2005. Par-3 controls tight junction assembly through the Rac exchange factor Tiam1. Nat. Cell Biol. 7:262–269. [DOI] [PubMed] [Google Scholar]
- Coulombe, P.A., and P. Wong. 2004. Cytoplasmic intermediate filaments revealed as dynamic and multipurpose scaffolds. Nat. Cell Biol. 6:699–706. [DOI] [PubMed] [Google Scholar]
- Fuchs, E., and D.W. Cleveland. 1998. A structural scaffolding of intermediate filaments in health and disease. Science. 279:514–519. [DOI] [PubMed] [Google Scholar]
- Gilbert, S., A. Loranger, N. Daigle, and N. Marceau. 2001. Simple epithelium keratins 8 and 18 provide resistance to Fas-mediated apoptosis. The protection occurs through a receptor-targeting modulation. J. Cell Biol. 154:763–773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirose, T., M. Karasawa, Y. Sugitani, M. Fujisawa, K. Akimoto, S. Ohno, and T. Noda. 2006. PAR3 is essential for cyst-mediated epicardial development by establishing apical cortical domains. Development. 133:1389–1398. [DOI] [PubMed] [Google Scholar]
- Inada, H., I. Izawa, M. Nishizawa, E. Fujita, T. Kiyono, T. Takahashi, T. Momoi, and M. Inagaki. 2001. Keratin attenuates tumor necrosis factor-induced cytotoxicity through association with TRADD. J. Cell Biol. 155:415–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izawa, I., and M. Inagaki. 2006. Regulatory mechanisms and functions of intermediate filaments: a study using site- and phosphorylation state-specific antibodies. Cancer Sci. 97:167–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izawa, I., M. Nishizawa, K. Ohtakara, K. Ohtsuka, H. Inada, and M. Inagaki. 2000. Identification of Mrj, a DnaJ/Hsp40 family protein, as a keratin 8/18 filament regulatory protein. J. Biol. Chem. 275:34521–34527. [DOI] [PubMed] [Google Scholar]
- James, P., J. Halladay, and E.A. Craig. 1996. Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics. 144:1425–1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, J.C., and K.J. Green. 1991. Intermediate filament-plasma membrane interactions. Curr. Opin. Cell Biol. 3:127–132. [DOI] [PubMed] [Google Scholar]
- Kim, S., P. Wong, and P.A. Coulombe. 2006. A keratin cytoskeletal protein regulates protein synthesis and epithelial cell growth. Nature. 441:362–365. [DOI] [PubMed] [Google Scholar]
- Kiyono, T., S.A. Foster, J.I. Koop, J.K. McDougall, D.A. Galloway, and A.J. Klingelhutz. 1998. Both Rb/p16INK4a inactivation and telomerase activity are required to immortalize human epithelial cells. Nature. 396:84–88. [DOI] [PubMed] [Google Scholar]
- Mertens, A.E., T.P. Rygiel, C. Olivo, R. van der Kammen, and J.G. Collard. 2005. The Rac activator Tiam1 controls tight junction biogenesis in keratinocytes through binding to and activation of the Par polarity complex. J. Cell Biol. 170:1029–1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mertens, A.E., D.M. Pegtel, and J.G. Collard. 2006. Tiam1 takes PARt in cell polarity. Trends Cell Biol. 16:308–316. [DOI] [PubMed] [Google Scholar]
- Nelson, W.J. 2003. Adaptation of core mechanisms to generate cell polarity. Nature. 422:766–774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura, T., T. Yamaguchi, K. Kato, M. Yoshizawa, Y. Nabeshima, S. Ohno, M. Hoshino, and K. Kaibuchi. 2005. PAR-6-PAR-3 mediates Cdc42-induced Rac activation through the Rac GEFs STEF/Tiam1. Nat. Cell Biol. 7:270–277. [DOI] [PubMed] [Google Scholar]
- Nishizawa, M., I. Izawa, A. Inoko, Y. Hayashi, K. Nagata, T. Yokoyama, J. Usukura, and M. Inagaki. 2005. Identification of trichoplein, a novel keratin filament-binding protein. J. Cell Sci. 118:1081–1090. [DOI] [PubMed] [Google Scholar]
- Niwa, H., K. Yamamura, and J. Miyazaki. 1991. Efficient selection for high-expression transfectants with a novel eukaryotic vector. Gene. 108:193–199. [DOI] [PubMed] [Google Scholar]
- Ooshio, T., N. Fujita, A. Yamada, T. Sato, Y. Kitagawa, R. Okamoto, S. Nakata, A. Miki, K. Irie, and Y. Takai. 2007. Cooperative roles of Par-3 and afadin in the formation of adherens and tight junctions. J. Cell Sci. 120:2352–2365. [DOI] [PubMed] [Google Scholar]
- Perez-Moreno, M., C. Jamora, and E. Fuchs. 2003. Sticky business: orchestrating cellular signals at adherens junctions. Cell. 112:535–548. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Boulan, E., and W.J. Nelson. 1989. Morphogenesis of the polarized epithelial cell phenotype. Science. 245:718–725. [DOI] [PubMed] [Google Scholar]
- Schmidt, T., H. Karsunky, B. Frass, W. Baum, A. Denzel, and T. Moroy. 2000. A novel protein (Fbf-1) that binds to CD95/APO-1/FAS and shows sequence similarity to trichohyalin and plectin. Biochim. Biophys. Acta. 1493:249–254. [DOI] [PubMed] [Google Scholar]
- Takeichi, M. 1977. Functional correlation between cell adhesive properties and some cell surface proteins. J. Cell Biol. 75:464–474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukita, S., and S. Tsukita. 1989. Isolation of cell-to-cell adherens junctions from rat liver. J. Cell Biol. 108:31–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukita, S., M. Furuse, and M. Itoh. 2001. Multifunctional strands in tight junctions. Nat. Rev. Mol. Cell Biol. 2:285–293. [DOI] [PubMed] [Google Scholar]
- Umeda, K., J. Ikenouchi, S. Katahira-Tayama, K. Furuse, H. Sasaki, M. Nakayama, T. Matsui, S. Tsukita, M. Furuse, and S. Tsukita. 2006. ZO-1 and ZO-2 independently determine where claudins are polymerized in tight-junction strand formation. Cell. 126:741–754. [DOI] [PubMed] [Google Scholar]
- Van Criekinge, W., M. van Gurp, E. Decoster, P. Schotte, M. Van de Craen, W. Fiers, P. Vandenabeele, and R. Beyaert. 1998. Use of the yeast three-hybrid system as a tool to study caspases. Anal. Biochem. 263:62–66. [DOI] [PubMed] [Google Scholar]
- Yugawa, T., K. Handa, M. Narisawa-Saito, S. Ohno, M. Fujita, and T. Kiyono. 2007. Regulation of Notch1 gene expression by p53 in epithelial cells. Mol. Cell. Biol. 27:3732–3742. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.