Abstract
Silencing mediator for retinoic acid and thyroid hormone receptor (SMRT) is a transcriptional corepressor that participates in diverse signaling pathways and human diseases. However, regulation of SMRT stability remains largely unexplored. We show that the peptidyl-prolyl isomerase Pin1 interacts with SMRT both in vitro and in mammalian cells. This interaction requires the WW domain of Pin1 and SMRT phosphorylation. Pin1 regulates SMRT protein stability, thereby affecting SMRT-dependent transcriptional repression. SMRT phosphorylation at multiple sites is required for Pin1 interaction, and these sites can be phosphorylated by Cdk2, which interacts with SMRT. Cdk2-mediated phosphorylation of SMRT is required for Pin1 binding and decreases SMRT stability, whereas mutation of these phosphorylation sites abrogates Pin1 binding and stabilizes SMRT. Finally, decreases in SMRT stability occur in response to the activation of Her2/Neu/ErbB2, and this receptor functions upstream of both Pin1 and Cdk2 in the signaling cascade that regulates SMRT stability and cellular response to tamoxifen.
Introduction
Silencing mediator for retinoic acid and thyroid hormone receptor (SMRT) and nuclear receptor corepressor (N-CoR) are two closely related transcriptional corepressors that were isolated in a search for factors that mediate transcriptional repression by nuclear hormone receptors (Chen and Evans, 1995; Horlein et al., 1995; Sande and Privalsky, 1996; Seol et al., 1996; Ordentlich et al., 1999; Park et al., 1999). The repression activities of SMRT and N-CoR are manifested through association with class I and II histone deacetylases (HDACs; Alland et al., 1997; Nagy et al., 1997; Huang et al., 2000; Kao et al., 2000; Fischle et al., 2002). Both SMRT and N-CoR form stable complexes with and serve as activating cofactors for HDAC3 (Guenther et al., 2001; Fischle et al., 2002; Guenther et al., 2002). In addition to nuclear hormone receptors, SMRT and N-CoR also participate in diverse signaling pathways through interactions with a variety of transcription factors (Kao et al., 1998; Tsai et al., 2004; Goodson et al., 2005) and are required for normal mammalian development (Jepsen et al., 2000, 2007). Corepressors have been shown to be involved in several human diseases, most notably breast cancers and acute promyelocytic leukemias (Khan et al., 2004; Privalsky, 2004). The regulation of N-CoR stability has been implicated in several normal and aberrant cellular pathways (Zhang et al., 1998; Khan et al., 2004; Perissi et al., 2004); however, the mechanism of SMRT stability regulation has not been clearly defined.
SMRT contains at least three different types of functional domains. Near the N terminus are two Swi/Ada/N-CoR/TFIID motifs in addition to two receptor interaction domains near the C terminus (Privalsky, 2004). SMRT also contains at least four independent repression domains (RDs; I–IV). Because diverse proteins are recruited to these RDs, we sought to identify novel regulators of SMRT by using RDs III and IV as bait in a yeast two-hybrid screen. We identified the peptidyl-prolyl cis-trans isomerase, Pin1, as a SMRT-interacting protein.
Pin1 is comprised of an N-terminal protein-binding WW domain and a C-terminal peptidyl-prolyl isomerase (PPIase) domain (Lu et al., 1996; Yeh and Means, 2007). The WW domain of Pin1 binds preferentially to phospho-Ser-Pro– (pS-P) or phospho-Thr-Pro (pT-P)–containing peptide sequences (Ranganathan et al., 1997; Yaffe et al., 1997), and the enzyme domain also preferentially isomerizes the prolyl bond after pT-P or pS-P. Pin1 is frequently localized to nuclei and serves as a regulatory protein for a variety of proteins associated with transcription, including cyclin E1, c-Myc, p53, SRC-3, and the retinoic acid receptor (Zacchi et al., 2002; Zheng et al., 2002; Yeh et al., 2004, 2006; Brondani et al., 2005; Yi et al., 2005; van Drogen et al., 2006). In the case of all of these transcription factors, the binding of Pin1 to a phosphorylated motif regulates the stability of its target protein. In this study, we characterize the interaction between SMRT and Pin1. We find that Pin1 binds to phosphorylated SMRT, identify the relevant protein kinase as Cdk2, and show that Cdk2 and Pin1 facilitate the degradation of SMRT. We also demonstrate that Cdk2 and Pin1 are required for ErbB2-mediated degradation of SMRT protein. Together, our data reveal a novel mechanism by which SMRT is regulated in cells.
Results
SMRT interacts with Pin1 in a phosphorylation-dependent manner
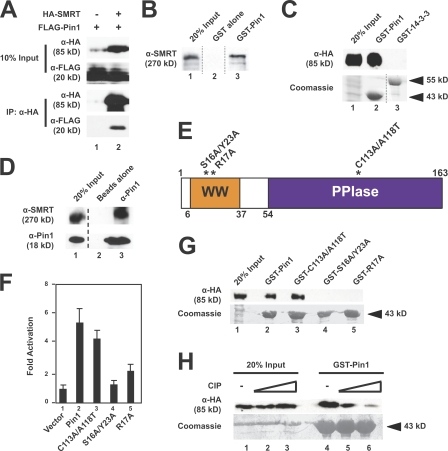
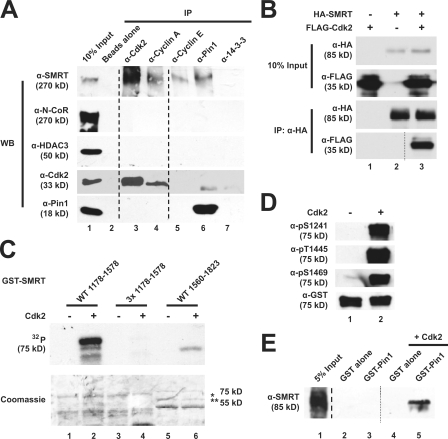
In a search for proteins that may regulate SMRT activity, yeast two-hybrid screens were performed using pGal4-SMRT (including RDs III and IV) as bait against a library derived from 17-d-old mouse embryos. Among the interacting clones, mouse Pin1 was isolated six times, with the longest insert encoding full-length Pin1. To test whether exogenous SMRT and Pin1 interact in mammalian cells, we used extracts of transfected mammalian cells for coimmunoprecipitations (Fig. 1 A). Using α-HA antibodies, we were able to coimmunoprecipitate FLAG-Pin1 with HA-SMRT (1,178–1,823; Fig. 1 A, lane 2), indicating that these proteins interact. This interaction was confirmed by showing that endogenous SMRT was able to bind GST–Pin1 (Fig. 1 B, lane 3) but not GST alone (Fig. 1 B, lane 2). Because both Pin1 and 14-3-3 proteins interact with substrates in a phosphorylation-dependent manner (Yaffe and Elia, 2001), we tested whether 14-3-3 proteins were also able to interact with SMRT (Fig. 1 C). GST–Pin1 (Fig. 1 C, lane 2) but not GST–14-3-3ε (Fig. 1 C, lane 3) was able to pull down HA-SMRT (1,178–1,823). Finally, Pin1 was also able to coimmunoprecipitate endogenous SMRT (Fig. 1 D, lane 3), whereas protein A beads alone were unable to do so (Fig. 1 D, lane 2). These data confirm Pin1 to be a SMRT-interacting protein both in vitro and in mammalian cells and verify the interaction originally observed in the yeast two-hybrid screen.
Figure 1.
Pin1 is a SMRT-associating protein. (A) Transfected SMRT and Pin1 interact in mammalian cells. HeLa cells were transfected with either FLAG-Pin1 alone or with both FLAG-Pin1 and HA-SMRT (1,178–1,823). WCEs expressing these proteins were subjected to coimmunoprecipitation with α-HA–conjugated agarose beads followed by immunoblotting with the indicated antibodies. (B) Pin1 interacts with SMRT in vitro. Purified, immobilized GST–Pin1 fusion protein was incubated with HeLa nuclear extracts, and bead fractions were subjected to immunoblotting with α-SMRT antibodies. (C) SMRT interacts with Pin1 but not 14-3-3ε. GST pull downs were performed using purified GST–Pin1, GST–14-3-3ε, and HeLa WCEs expressing HA-SMRT (1,178–1,823). Top, immunoblotting with α-HA; bottom, Coomassie staining. (D) Endogenous SMRT and Pin1 interact in mammalian cells. HeLa nuclear extracts were subjected to coimmunoprecipitation with Pin1 antibodies and immunoblotting with the indicated antibodies. Unrelated lanes were removed. (E) Schematic representation of human Pin1. Point mutations used in this study are indicated by *. Pin1 contains both a WW domain and a PPIase domain. Amino acids are indicated by numbers. (F) The Pin1 WW domain is critical for interaction with SMRT in yeast. Y190 cells expressing GAL4 DBD SMRT (1,178–1,823) and the indicated GAL4 activation domain Pin1 constructs were subjected to β-galactosidase assays as described in Materials and methods. Error bars represent ±SD. (G) The Pin1 WW domain is essential for SMRT association in vitro. GST pull downs were performed using the indicated GST–Pin1 proteins and HeLa WCEs expressing HA-SMRT (1,178–1,823). Top, immunoblotting with α-HA; bottom, Coomassie staining. (H) Phosphatase treatment disrupts the SMRT–Pin1 interaction in vitro. HeLa WCEs expressing HA-SMRT (1,178–1,823) were treated with increasing amounts of calf intestinal phosphatase followed by GST pull downs. Top, immunoblotting with α-HA; bottom, Coomassie staining.
Pin1 is a 163–amino acid protein that contains two known functional domains, a WW domain (amino acids 6–37) and a PPIase domain (amino acids 54–163; Fig. 1 E). Pin1 associates with its interacting partners through the WW domain, a protein–protein interaction module that has been shown to bind specifically to pS-P or pT-P dipeptide motifs (Sudol et al., 2001; Yaffe and Elia, 2001). Mutations of Pin1 such as S16A/Y23A and R17A are unable to bind pS/pT-P motifs and thus abrogate this binding (Shen et al., 1998; Lu et al., 1999). We generated these mutants of Pin1 as well as a C113A/A118T mutant that has decreased PPIase activity (Shen et al., 1998) by site-directed mutagenesis, and yeast two-hybrid assays were used to determine whether the WW domain or isomerase activity is critical for SMRT interaction (Fig. 1 F). SMRT (1,178–1,823) fused to the Gal4 DNA-binding domain (DBD) was cotransformed with wild-type (WT) Pin1 or one of these mutated forms of Pin1, fused to the Gal4 activation domain into yeast cells, and tested for β-galactosidase activity. C113A/A118T retained SMRT binding activity (Fig. 1 F, lane 3), suggesting that enzyme activity was not required for binding, whereas both WW domain mutants (Fig. 1 F, lanes 4 and 5) failed to bind SMRT as indicated by loss of β-galactosidase activation. GST pull-down assays confirmed the yeast two-hybrid data by demonstrating that both WT Pin1 and the C113A/A118T enzymatic mutant bound HA-SMRT (1,178–1,823; Fig. 1 G, lanes 2 and 3), but the S16A/Y23A and R17A WW mutants were unable to bind HA-SMRT (1,178–1,823; Fig. 1 G, lanes 4 and 5). Together, these data indicate that the integrity of the WW domain but not PPIase activity is essential for the Pin1–SMRT interaction both in vitro and in yeast two-hybrid assays.
The importance of the WW domain of Pin1 for SMRT interaction suggested that phosphorylation of SMRT might be involved in this interaction. As an initial test of this hypothesis, extracts of HeLa cells expressing HA-SMRT (1,178–1,823) were incubated with increasing concentrations of calf intestinal phosphatase before incubation with GST–Pin1. Fig. 1 H shows that phosphatase treatment of extracts dramatically reduced the association of SMRT with Pin1 in vitro (lanes 5 and 6). These data indicate that phosphorylation of SMRT is very likely to play a critical role in Pin1 binding.
Pin1 affects SMRT stability
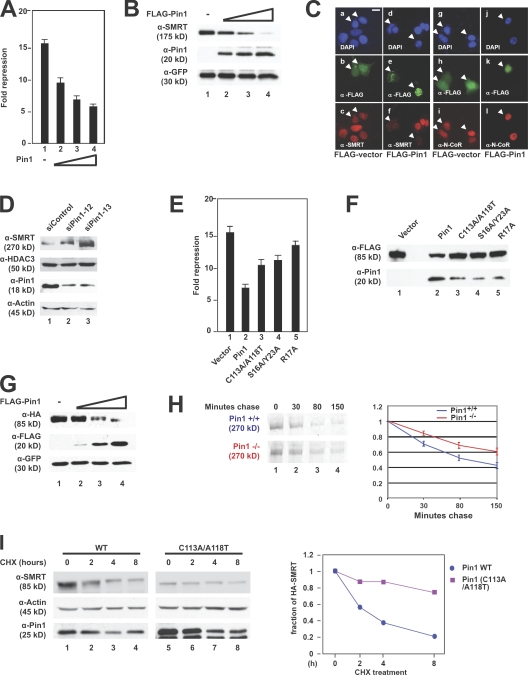
To examine the functional significance of the SMRT and Pin1 association, we tested whether ectopic expression of Pin1 affected the transcriptional repressor activity of SMRT. Transient transfections showed that Gal4-SMRT (1,050–1,823) potently repressed basal transcription of a Gal4 reporter (Fig. 2 A, lane 1), but coexpression of Pin1 reduced SMRT repression activity in a dose-dependent manner (Fig. 2 A, lanes 2–4). Because Pin1 compromised SMRT repressor activity, we hypothesized that Pin1 might modulate SMRT-mediated repression by affecting its steady-state levels, as has been shown for other nuclear targets of Pin1. To test this possibility, SMRT (1,012–2,507) and Pin1 were coexpressed in CV-1 cells. As shown in Fig. 2 B, expression of Pin1 had a negative, dose-dependent effect on SMRT protein levels. We also evaluated the effect of Pin1 on the expression of endogenous SMRT by immunofluorescence microscopy. SMRT levels were significantly decreased in cells transfected with FLAG-Pin1 (Fig. 2 C, compare c with f), whereas the levels of the closely related corepressor N-CoR did not change (Fig. 2 C, i and l). We confirmed these observations using siRNA targeting Pin1. Fig. 2 D shows that when endogenous Pin1 levels were decreased by siRNA, endogenous SMRT protein levels increased (lanes 2 and 3), whereas HDAC3 levels showed negligible differences. Together, these data suggest that Pin1 regulates SMRT abundance.
Figure 2.
Pin1 down-regulates SMRT protein levels. (A) Overexpression of Pin1 relieves SMRT repression. CV-1 cells were transiently transfected with Gal4-SMRT (1,050–1,823), increasing amounts of Pin1, and a thymidine kinase–luciferase reporter. Luciferase activity was measured 36–48 h after transfection; all experiments were performed in triplicate. Error bars represent ±SD. (B) Overexpression of Pin1 has a negative effect on SMRT steady-state levels. SMRT (1,012–2,507), FLAG-Pin1, and GFP were cotransfected into CV-1 cells. WCEs were subjected to immunoblotting with the indicated antibodies. (C) Ectopic expression of Pin1 affects endogenous SMRT levels in MCF-7 cells. Cells were transfected with FLAG-Pin1 or FLAG-vector and immunostained with the indicated antibodies 24 h after transfection. (top) DAPI staining; (middle) α-FLAG staining; (bottom) SMRT or N-CoR staining. Arrowheads indicate transfected cells. Bar, 5 μm. (D) siRNA-mediated knockdown of Pin1 increases SMRT protein levels. HeLa cells were transfected with either a control oligonucleotide or one of two Pin1-targeting siRNAs. Cells were harvested 72 h later, and WCEs were subjected to immunoblotting with the indicated antibodies. (E) Pin1 mutants lose the ability to relieve SMRT repression. CV-1 cells were transiently transfected with Gal4-SMRT (1,050–1,823), Pin1 WT or mutants, and a thymidine kinase–luciferase reporter. Luciferase activity was measured 36–48 h later; all experiments were performed in triplicate. Error bars represent ±SD. (F) Pin1 mutants lose the ability to down-regulate SMRT levels. CV-1 cells were cotransfected with FLAG-SMRT (1,178–1,823) and HA-tagged Pin1 WT or mutants. WCEs were subjected to immunoblotting with the indicated antibodies. (G) Exogenous overexpression of Pin1 in Pin1−/− MEFs decreases SMRT levels. HA-SMRT (1,178–1,823), FLAG-Pin1, and GFP were cotransfected into Pin1−/− MEFs, and immunoblotting was performed using the indicated antibodies. (H) SMRT half-life is increased in Pin1−/− MEFs. Pin1+/+ and Pin1−/− MEFs were pulse labeled with [35S]methionine and [35S]cysteine and chased with cold growth medium as described in Materials and methods. WCEs were prepared and subjected to immunoprecipitation, and protein levels were detected by autoradiography. Left, 35S autoradiography; right, quantification of immunoprecipitated 35S-labeled SMRT. Error bars represent ±SD. (I) SMRT destabilization requires Pin1 isomerase activity. HeLa cells stably expressing shRNA against Pin1 were transfected with SMRT (1,178–1,823) and either WT Pin1 or Pin1 (C113A/A118T) and treated with 100 μg/ml CHX for the indicated times, and immunoblots were performed using the indicated antibodies. Left, immunoblot; right, quantification of SMRT levels normalized to actin levels.
To determine whether SMRT interaction or the prolyl isomerase activity of Pin1 is critical for modulating the transcriptional repression activity of SMRT, the Pin1 mutants described in Fig. 1 E were used. We found that all three Pin1 mutants were defective in their ability to relieve Gal4-SMRT (1,050–1,823) activity to varying degrees (Fig. 2 E, lanes 3–5), indicating that both binding (from the WW domain) and enzymatic activity (from the PPIase domain) are required for Pin1 to affect SMRT-mediated repression. Because ectopic expression of Pin1 affected SMRT protein levels, we next examined the effect of the Pin1 mutants on SMRT protein levels. To test this, FLAG-SMRT (1,178–1,823) was cotransfected into cells with or without WT or mutant forms of HA-Pin1. As seen in Fig. 2 F, WT HA-Pin1 decreased SMRT steady-state levels (lane 2) to a much greater extent than did any of the three mutants (lanes 3–5). Together, these data indicate that both domains of Pin1 participate in the regulation of SMRT accumulation. Additionally, it is likely that a Pin1-dependent decrease in SMRT protein levels is responsible for the loss of repression activity (Fig. 2, A and E).
To further examine the effect of ectopic Pin1 on SMRT, we used Pin1−/− mouse embryonic fibroblast (MEF) cells (Yeh et al., 2006). Consistent with previous data, ectopic expression of FLAG-Pin1 in Pin1−/− MEFs dramatically decreased HA-SMRT (1,178–1,823) protein levels in a dose-dependent manner (Fig. 2 G), indicating that this mechanism is active in MEFs. We then determined whether the presence of endogenous Pin1 altered SMRT stability by performing metabolic pulse-chase labeling assays to measure the half-life of SMRT in Pin1+/+ and Pin1−/− MEFs. MEF cells were pulse labeled with [35S]methionine and [35S]cysteine followed by a chase with unlabeled amino acids. Cell extracts were subjected to immunoprecipitation with SMRT antibodies and autoradiography. Fig. 2 H shows that SMRT was more stable in Pin1−/− MEFs than in Pin+/+ MEFs. To confirm that the effect on SMRT half-life was because of Pin1, we transfected HeLa cells stably expressing short hairpin RNA against Pin1 (Reineke et al., 2008) with either WT or mutant Pin1 (C113A/A118T). Fig. 2 I shows that WT Pin1 decreased SMRT stability, whereas the mutant had little effect. From these data, we conclude that Pin1 is a SMRT-interacting protein that regulates SMRT protein levels by modulating SMRT half-life.
Mapping Pin1 interaction sites
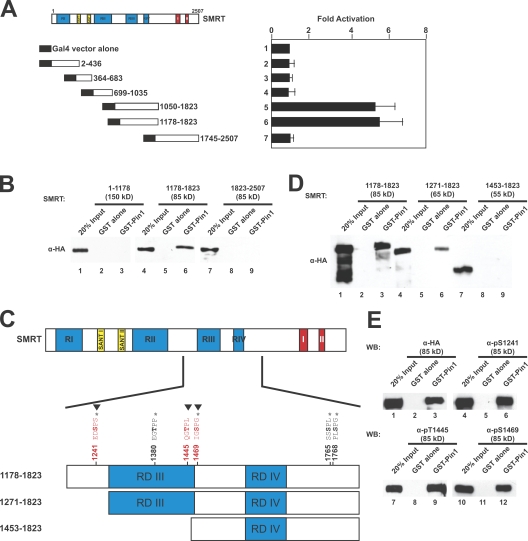
To elucidate the mechanism by which Pin1 modulates SMRT stability, we first mapped the pS/T-P motifs in SMRT responsible for Pin1 binding. SMRT deletion constructs fused to the yeast Gal4 DBD (Fig. 3 A, left) were used in yeast two-hybrid assays. As shown in Fig. 3 A, we found that Pin1 does not interact with the N- or C-terminal regions of SMRT; rather, Pin1 specifically interacts with amino acids 1,178–1,823 of SMRT (containing RDs III and IV). These results were confirmed by GST pull downs (Fig. 3 B) showing that Pin1 interacts specifically with the SMRT fragment corresponding to RDs III and IV (Fig. 3 B, lane 6) but neither the N- nor C-terminal fragments (Fig. 3 B, lanes 3 and 9).
Figure 3.
Mapping the Pin1 interaction domain in SMRT. (A) Yeast two-hybrid mapping of the Pin1 interaction domain. Left, diagram of Gal4-DBD-SMRT fragments used; right, normalized β-galactosidase activity. Amino acids are indicated by numbers. Error bars represent ±SD. (B) GST pull down mapping of Pin1-interacting regions in SMRT. HeLa WCEs expressing the indicated HA-SMRT constructs were subjected to pull downs using GST–Pin1 and immunoblotted with anti-HA antibodies. (C) Schematic diagram of SMRT showing in vivo phosphorylation sites as mapped by mass spectrometry (*), Cdk consensus motifs (▾), and deletion constructs used in Fig. 3 D. Amino acids are indicated by numbers. (D) GST pull downs of N-terminal deletions of HA-SMRT. HeLa WCEs expressing the indicated HA-SMRT constructs were subjected to pull downs using GST–Pin1 and immunoblotted with anti-HA antibodies. (E) Phosphorylated SMRT interacts with Pin1 in vitro. HeLa WCEs expressing HA-SMRT (1,178–1,823) were subjected to pull downs using GST–Pin1 and immunoblotting with the indicated antibodies. WB, Western blot.
Because the interaction between SMRT and Pin1 is dependent on the phosphorylation status of SMRT (Fig. 1 H), we used mass spectrometry to identify in vivo phosphorylation sites in FLAG-SMRT (1,178–1,823). Four phosphopeptide sequences were identified, indicating that S1241, T1373, T1469, S1762, and S1765 were phosphorylated (Fig. 3 C, *). We used N-terminal deletion constructs (1,281–1,823 and 1,463–1,823; Fig. 3 C) expressed in HeLa cells to determine whether any of the identified phosphorylation sites were critical for interaction with GST–Pin1. Fig. 3 D shows that deletion past the first phosphorylation site (S1241) resulted in some loss of GST–Pin1 binding (lane 6), whereas further deletion resulted in complete loss (lane 9). Among the phosphorylated sites, S1241 and S1469 are both consensus Cdk target sites of the motif S/T-P-X-R/K. An additional possible Cdk site at T1445 was not phosphorylated based on the mass spectrometry analysis (Fig. 3 C, Cdk sites indicated by ▾). Because loss of two potential Cdk sites abrogated GST–Pin1 interaction (S1241 and T1445; Fig. 3 D), we next investigated whether Pin1 could interact with SMRT phosphorylated at those three Cdk sites. We performed pull downs with GST–Pin1 and immunoblotted using phosphospecific antibodies for the three Cdk sites (Fig. 3 E). We found that GST–Pin1 interacted with HA-SMRT (1,178–1,823) that was phosphorylated at these three Cdk sites (Fig. 3 E, lanes 6, 9, and 12). Collectively with the phosphatase data (Fig. 1 H), these results reveal that phosphorylation of SMRT is critical for its interaction with Pin1.
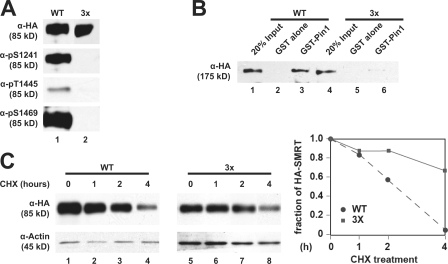
Because deletion of Cdk sites in SMRT leads to loss of Pin1 binding, we next investigated the importance of these sites by generating a Ser→Ala and Thr→Ala triple mutant SMRT (SMRT 3×; Fig. 3 C, ▾). The phosphospecific antibodies were unable to detect this mutant (Fig. 4 A, lane 2), confirming the specificity of the antibodies. SMRT 3× (1,012–2,507) was much less able to interact with GST–Pin1 (Fig. 4 B, lane 6) than the WT, indicating that intact Cdk consensus sites are critical for interaction with Pin1. In addition, we also generated double and single Cdk mutants in SMRT and found that single mutations had no significant effect on Pin1 binding, whereas the double mutants showed only slight decreases in Pin1 binding (unpublished data). SMRT 3× (1,178–1,823) was also more stable than the WT protein, as shown by cycloheximide (CHX) treatment in HeLa cells (Fig. 4 C). These data indicate that consensus Cdk sites in SMRT are phosphorylated and critical for the SMRT–Pin1 interaction.
Figure 4.
Mutations of Cdk sites in SMRT disrupt Pin1 interaction. (A) Phosphospecific antibodies recognize WT but not mutant SMRT. HeLa cells were transfected with either WT HA-SMRT (1,178–1,823) or a 3× mutant (S1241A/T1445A/S1469A), and WCEs were prepared for immunoblotting with the indicated antibodies. (B) Mutation of Cdk sites in SMRT disrupts Pin1 interaction in vitro. HeLa cells were transfected with HA-SMRT WT (1,012–2,507) or the 3× mutant, and WCEs were subjected to pull downs with GST–Pin1 and immunoblotted with α-HA antibodies. (C) Mutation of Cdk sites stabilizes SMRT. HeLa cells were transfected with HA-SMRT (1,178–1,823) or the 3× mutant and treated with 100 μg/ml CHX for the indicated times. WCEs were immunoblotted with the indicated antibodies. Left, immunoblot; right, quantification of SMRT levels normalized to actin levels.
SMRT is a Cdk2 target
Because mutation of putative Cdk sites in SMRT abrogated both Pin1 binding and its effect on SMRT stability, we hypothesized that phosphorylation by Cdk–cyclin complexes might be responsible for generating Pin1-binding sites. Fig. 5 A shows that endogenous Cdk2 and its activating cyclins A and E were capable of coimmunoprecipitating endogenous SMRT from HeLa nuclear extracts (lanes 3–5). However, cyclin E did not efficiently immunoprecipitate Cdk2. Interestingly, neither N-CoR nor HDAC3 was detected in these immunocomplexes. To confirm these observations, we coexpressed HA-SMRT (1,178–1,823) and Cdk2 in mammalian cells. As expected, Cdk2 coimmunoprecipitated with SMRT (Fig. 5 B, lane 3). Furthermore, Cdk2 phosphorylated WT GST-SMRT (1,178–1,578; Fig. 5 C, lane 2) but not the GST-3× mutant (1,178–1,578; Fig. 5 C, lane 4) or a GST–SMRT (1,560–1,823) fragment that does not contain any consensus Cdk phosphorylation sites (Fig. 5 C, lane 6), indicating that S1241, T1445, and S1469 are required for Cdk2-dependent phosphorylation in vitro. Additionally, Cdk2 specifically phosphorylated all three of the predicted Cdk sites in vitro (Fig. 5 D, lane 2). Addition of active Cdk2 to purified His6-SMRT (1,178–1,823) facilitated Pin1 binding in vitro (Fig. 5 E, lane 5). Pin1 binding was not observed in the absence of kinase (Fig. 5 E, lane 3), indicating that phosphorylation is required for the interaction. These data indicate that Cdk2 can bind and phosphorylate SMRT and is likely to be a kinase that facilitates Pin1 binding by generating pS/T-P sites.
Figure 5.
Cdk2 interacts with and phosphorylates SMRT to generate Pin1-binding sites. (A) Endogenous SMRT coimmunoprecipitates with Cdk2 in HeLa nuclear extracts. Coimmunoprecipitations were performed as described in Fig. 1 D using the indicated antibodies (top) followed by immunoblotting with the indicated antibodies (left). Unrelated lanes were removed. (B) Exogenous Cdk2 and SMRT interact. HA-SMRT (1,178–1,823) and FLAG-Cdk2 were cotransfected into HeLa cells. Coimmunoprecipitations were performed as in Fig. 1 A and immunoblotted with the indicated antibodies. (C) Cdk2 phosphorylates SMRT in vitro. Purified GST-SMRT proteins (WT 1,178–1,578, 3× mutant 1,178–1,578, and WT 1,560–1,823) were incubated with purified Cdk2 and [32P]ATP. Samples were resolved by SDS-PAGE and transferred to a polyvinylidene difluoride membrane before autoradiography. Top, autoradiography; bottom, Coomassie staining; *, GST-SMRT (1,178–1,578); **, GST-SMRT (1,560–1,823). (D) Cdk2 phosphorylates S1241, T1445, and S1469 of SMRT. In vitro kinase assays were performed as aforementioned using GST-SMRT (1,178–1,588), Cdk2, and unlabeled ATP. Immunoblotting was performed with the indicated antibodies. (E) Cdk2 phosphorylation of SMRT generates Pin1-binding sites. His6-SMRT (1,178–1,823) was subjected to in vitro kinase assays as described in D. After kinase reactions, samples were subjected to GST pull downs with GST–Pin1. Immunoblotting was performed with the indicated antibodies. Unrelated lanes were removed.
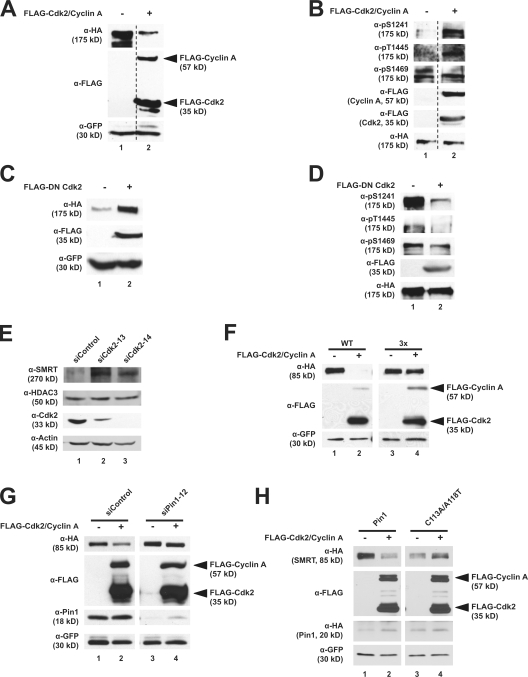
If Cdk2 generates Pin1-binding sites in vivo, we hypothesized that Cdk2 may have an effect similar to that of Pin1 on SMRT stability. To test this hypothesis, we coexpressed Cdk2 and cyclin A in HeLa cells to determine the effects on SMRT steady-state levels. As seen in Fig. 6 A, Cdk2 and cyclin A coexpression decreased HA-SMRT (1,012–2,507) protein levels (lane 2). Additionally, overexpression of Cdk2 and cyclin A increased HA-SMRT (1,012–2,507) phosphorylation at two of the three putative Cdk sites (Fig. 6 B, lane 2) when these extracts were normalized for HA-SMRT (1,012–2,507) levels. Consistent with these results, expression of a dominant-negative (DN) mutant form of Cdk2 (D145N) led to the accumulation of HA-SMRT (1,012–2,507) protein (Fig. 6 C) and a marked decrease in HA-SMRT (1,012–2,507) phosphorylation at the predicted Cdk sites as visualized by immunoblotting with phosphospecific antibodies (Fig. 6 D). We further confirmed these observations using siRNA targeting Cdk2. Fig. 6 E shows that endogenous SMRT levels increase when Cdk2 levels are knocked down. Furthermore, Cdk2 overexpression significantly decreased steady-state protein levels of WT HA-SMRT (1,178–1,823) but not HA-SMRT 3× (1,178–1,823), indicating that Cdk phosphorylation sites are critical for this regulation (Fig. 6 F, compare lanes 1 and 2 with lanes 3 and 4). Together, these data indicate that Cdk2 is a critical regulator of SMRT stability.
Figure 6.
Cdk2 stimulates SMRT phosphorylation in HeLa cells. (A) Coexpression of Cdk2 and cyclin A decreases SMRT steady-state levels. HeLa cells were cotransfected with HA-SMRT (1,012–2,507), GFP, FLAG-Cdk2, and FLAG–cyclin A as indicated. Unrelated lanes were removed. (B) Coexpression of Cdk2 and cyclin A increases SMRT phosphorylation. HeLa cells were transfected as in A and treated with 20 nM calyculin A for 1 h before harvest. Unrelated lanes were removed. (C) DN Cdk2 increases SMRT steady-state levels. HeLa cells were transfected with HA-SMRT (1,012–2,507), GFP, and FLAG-DN-Cdk2 (D145N), and WCEs were immunoblotted with the indicated antibodies. (D) DN Cdk2 decreases SMRT phosphorylation. HeLa cells were transfected as in C and treated with 20 nM calyculin A 1 h before harvest. WCEs were subjected to immunoblotting with indicated antibodies. (E) siRNA-mediated knockdown of Cdk2 increases SMRT protein levels. Transfections were performed as in Fig. 2 D. (F) Cdk2 overexpression does not affect mutant SMRT protein levels. HeLa cells were cotransfected with HA-SMRT (1,188–1,833) 3× mutant, GFP, FLAG-Cdk2, and FLAG–cyclin A as indicated. (G) Endogenous Pin1 is required for Cdk2-mediated SMRT degradation. HeLa cells were cotransfected with HA-SMRT (1,188–1,833), GFP, FLAG-Cdk2, FLAG–cyclin A, and either siRNA-targeting Pin1 or control oligonucleotides as indicated. (H) Mutant Pin1 can block Cdk2-mediated SMRT degradation. HeLa cells were cotransfected with HA-SMRT (1,188–1,833), GFP, FLAG-Cdk2, FLAG–cyclin A, and either WT HA-Pin1 or the C113A/A118/T mutant as indicated. (A–H) WCEs were subjected to immunoblotting with the indicated antibodies.
To examine whether Cdk2-mediated SMRT destabilization is dependent on Pin1, we used siRNA targeting Pin1 to deplete endogenous Pin1 from cells. Fig. 6 G shows that Pin1 knockdown blocked the ability of Cdk2–cyclin A to decrease steady-state levels of HA-SMRT (1,178–1,823; compare lanes 1 and 2 with lanes 3 and 4). Furthermore, a Pin1 mutant with decreased isomerization activity, C113A/A118T, also blocked the destabilization effects of Cdk2 on HA-SMRT (1,178–1,823; Fig. 6 H). These data indicate that endogenous Pin1 contributes to HA-SMRT (1,178–1,823) degradation through a Cdk2-dependent pathway.
ErbB2 destabilizes SMRT protein level
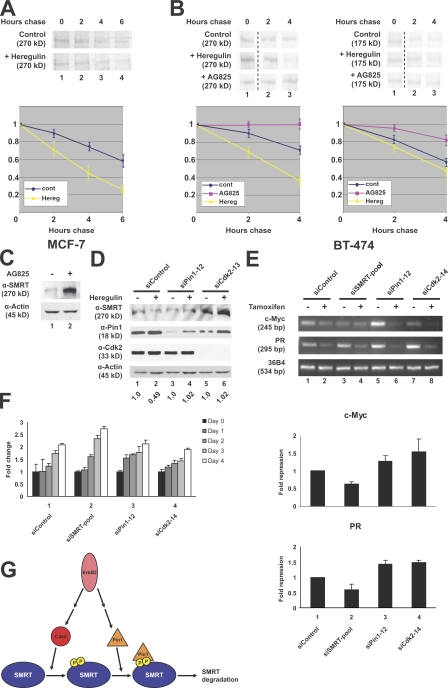
Because ErbB2/Her2/Neu has been shown to increase Cdk activity and up-regulate Pin1 expression in certain breast cancers (Ryo et al., 2002; Timms et al., 2002), we next investigated whether ErbB2 activity could modulate SMRT half-life. Using heregulin, a small protein activator of ErbB2, and AG825, a chemical inhibitor of ErbB2, we analyzed SMRT half-life. MCF-7 cells show a decrease in SMRT half-life when treated with heregulin as compared with the vehicle control (Fig. 7 A). Furthermore, in BT-474 cells that overexpress ErbB2, both transfected HA-SMRT (1,012–2,507; Fig. 7 B, right) and endogenous SMRT (Fig. 7 B, left) half-life decreased upon heregulin treatment. Conversely, upon treatment with the ErbB2 inhibitor AG825, both SMRT half-life (Fig. 7 B) and SMRT protein levels increased (Fig. 7 C). Finally, to test whether ErbB2 acts upstream of Cdk2 and Pin1, we used siRNA targeting either Pin1 or Cdk2 to evaluate SMRT protein levels. BT-474 breast cancer cells were transfected with siRNAs followed by treatment with heregulin. Fig. 7 D shows that heregulin decreased SMRT protein levels by half (lanes 1 and 2), whereas siRNA targeting either Pin1 or Cdk2 blocked this effect (lanes 3–6). Collectively, these results indicate that ErbB2 signaling upstream of Cdk2 and Pin1 is a potential regulatory cascade involved in regulating the stability of SMRT.
Figure 7.
ErbB2 attenuates SMRT levels in breast cancer cells. (A) SMRT half-life in MCF-7 cells. Cells were pulse labeled with [35S]methionine and [35S]cysteine and chased with cold growth medium. Additionally, cells were treated with 100 ng/ml heregulin or a vehicle control for 30 min before labeling. Top, 35S autoradiography; bottom, quantification of immunoprecipitated 35S-labeled SMRT. (B) SMRT half-life in BT-474 cells. Experiments were performed according to the description in A with the addition of 0.1μM AG825 treatment. Unrelated lanes were removed. Left, endogenous SMRT; right, transfected HA-SMRT (1,012–2,507); top, 35S autoradiography; bottom, quantification of immunoprecipitated 35S-labeled SMRT. (C) AG825 treatment increases SMRT protein levels. BT-474 cells were treated with 0.1 μM AG825 or vehicle control for 48 h. WCEs were immunoblotted with the indicated antibodies. (D) ErbB2 controls SMRT levels through a Pin1- and Cdk2-dependent pathway. BT-474 cells were transfected with siRNAs for 24 h followed by treatment with 100 ng/ml heregulin for 48 h. WCEs were subjected to immunoblotting with the indicated antibodies. Quantification of SMRT levels normalized to actin is shown below the blot; quantification was performed in Photoshop. (E) SMRT is required for ERα-dependent repression in response to tamoxifen. BT-474 cells were transfected with siRNAs for 48 h followed by treatment with 100 nM 4-hydroxytamoxifen for 72 h. Total RNA was harvested followed by cDNA generation. RT-PCR was performed using primers targeting the indicated genes. Top, ethidium bromide staining; bottom, quantification of three sets of experiments for each gene. (F) SMRT is required for tamoxifen-dependent inhibition of cell proliferation. BT-474 cells were transfected with siRNAs as in E. 48 h after transfection, cells were counted, split evenly onto plates, and treated with 100 nM 4-hydroxytamoxifen for the indicated times. Proliferation was measured every 24 h. Error bars represent ±SD. (G) Model of SMRT regulation by Pin1 and Cdk2. SMRT is phosphorylated by Cdk2–cyclin A complexes. These phosphorylation sites serve as binding sites for Pin1, which subsequently targets SMRT for degradation. Additionally, Pin1 may act in other pathways to affect tamoxifen resistance, as knockdown of Pin1 in BT-474 cells did not significantly up-regulate SMRT protein levels.
To further characterize the role of the Cdk2- and Pin1-dependent SMRT degradation pathway in tamoxifen resistance, we treated BT-474 cells with siRNA targeting either SMRT, Pin1, or Cdk2 to examine gene expression. After siRNA treatment, cells were treated with or without 4-hydroxytamoxifen for 72 h followed by RNA extraction and RT-PCR. We examined the expression of known ERα gene targets such as c-Myc and progesterone receptor (PR). As shown in Fig. 7 E, tamoxifen treatment repressed both c-Myc and PR expression but not the control gene 36B4 (lanes 1 and 2), whereas knockdown of SMRT compromised tamoxifen-mediated repression of both c-Myc and PR expression (lanes 3 and 4), and knockdown of Pin1 or Cdk2 increased repression of both genes (lanes 5–8). Interestingly, knockdown of Pin1 led to increases in c-Myc and PR transcription, whereas Cdk2 knockdown increased PR transcript levels. These data indicate that SMRT is critical for ERα-dependent gene repression in response to tamoxifen. Because tamoxifen suppresses cell proliferation of BT-474 cells, we also examined whether knockdown of SMRT, Pin1, or Cdk2 affected proliferation rates. Fig. 7 F shows that knockdown of SMRT increased cell proliferation when compared with siControl, whereas Cdk2 showed a slight decrease in cell proliferation compared with the control. Together, these data indicate that control of SMRT stability likely has multiple cellular effects, including gene expression and cell proliferation.
Discussion
Here we show that the corepressor SMRT interacts with both the PPIase Pin1 and the cell cycle–dependent kinase Cdk2, two proteins that are downstream effectors of Her2/Neu/ErbB2 (Neve et al., 2000; Ryo et al., 2002). Pin1 requires an intact WW domain and phosphorylation of SMRT for this interaction. Cdk2 phosphorylates SMRT at consensus Cdk motifs to generate Pin1-binding sites and consequently targets SMRT for degradation, the latter also requiring the PPIase activity of Pin1 (Fig. 7 G). Consistent with these observations, activation of Her2/Neu/ErbB2 promotes SMRT degradation, whereas inhibition of Her2/Neu/ErbB2 stabilizes SMRT. These data suggest a concerted regulation of a transcriptional corepressor initiated by extracellular stimuli functioning to activate a transmembrane tyrosine kinase receptor.
In tamoxifen-responsive breast cancer cells, tamoxifen-bound ERα recruits corepressor complexes to inhibit ERα-target gene expression (Smith et al., 1997; Fleming et al., 2004). Decreased levels of corepressors correlate with acquired tamoxifen resistance (Lavinsky et al., 1998), whereas high expression of coactivators favors an agonist effect for tamoxifen (Shang and Brown, 2002; Shou et al., 2004). Overexpression of SMRT or N-CoR promotes the antagonist activity of tamoxifen in transient transfection assays (Jackson et al., 1997). The antiproliferative activity of tamoxifen therefore depends on the relative abundance of corepressors and coactivators (Graham et al., 2000a,b; Fujita et al., 2003). Indeed, both SMRT and N-CoR are required for the antiproliferative effects of tamoxifen in MCF-7 cells (Keeton and Brown, 2005). Additionally, certain coactivators such as AIB1/SRC-3 have been shown to be up-regulated in ErbB2-positive breast tumors, which would further disrupt the corepressor/coactivator ratio (Osborne et al., 2003), and it is interesting that SRC-3 turnover is also regulated by Pin1 (Yi et al., 2005) and reversible phosphorylation (Wu et al., 2007). Here we show that SMRT protein levels can be modulated by the oncogene Her2/Neu/ErbB2 to alter the ratio of corepressors to coactivators, thereby elucidating part of the potential mechanism underlying tamoxifen resistance and aberrant transcriptional regulation in breast cancers.
Our studies clearly demonstrate that SMRT has distinct functions from N-CoR, as knockdown of SMRT is sufficient to desensitize cells to tamoxifen-mediated inhibition of PR and c-Myc expression and cell growth. Furthermore, overexpression of Pin1 in MCF-7 cells decreases SMRT protein levels but not N-CoR. Indeed, recent studies show that these two corepressors can be targets of different cellular pathways (Jepsen et al., 2000, 2007; Jonas and Privalsky, 2004; Yu et al., 2006), including growth factor signaling, DNA damage response, and normal mammalian development, and likely regulate some nonoverlapping genes. As mentioned previously, knockdown of both SMRT and N-CoR is required to overcome the antiproliferative effects of tamoxifen in MCF-7 cells (Keeton and Brown, 2005). Furthermore, N-CoR protein levels are decreased in tamoxifen-resistant tumors from a mouse model (Lavinsky et al., 1998). This indicates that a global decrease in corepressor expression is required for breast cancer progression and that there is an analogous pathway leading to the accelerated degradation of N-CoR in ErbB2-positive/tamoxifen-resistant breast cancers. It is probable that ErbB2-positive/tamoxifen-resistant breast cancers exploit a normal cellular pathway leading to the degradation of SMRT.
Pin1 has been shown to collaborate with several kinases to modulate protein stability, including MAPK, GSK3β, Cdk1, and Cdk2 (Ryo et al., 2001; Yeh et al., 2004, 2006; Pastorino et al., 2006). Our observation that Cdk2 phosphorylates SMRT to regulate its abundance supports the idea that Pin1 exerts diverse functions in different signaling pathways. We hypothesize that compartmental and temporal regulation of the association between Pin1 and its targets fine tunes Pin1 activity. For example, several Pin1-mediated degradation pathways are likely to be cell cycle dependent, as multiple cell cycle regulatory proteins are involved (Yeh and Means, 2007). It will be highly informative to explore whether SMRT protein levels are regulated in a cell cycle–dependent manner, as has been suggested previously (Park et al., 1999). Like SMRT, N-CoR also contains seven consensus Cdk phosphorylation sites. However, of the three sites identified here, only serine 1469 (S1469) is not conserved between SMRT and N-CoR. Intriguingly, S1469 is the phosphorylation site that does not change in response to Cdk2 overexpression. Therefore, it is likely that there are other kinases that may play a role in this complex regulatory pathway.
Pin1 is thought to be an important regulator of tumorigenesis and is overexpressed or underexpressed in different tumors (Bao et al., 2004; Mantovani et al., 2007; Yeh and Means, 2007); thus, it can potentially act as either an oncogene or a tumor-suppressor gene, depending on cellular context. ErbB2 activation stimulates Pin1 transcription, and in turn Pin1 stimulates the transformative properties of ErbB2-positive cells (Ryo et al., 2002). Interestingly, although activation of ErbB2 decreases SMRT protein levels in BT-474 cells, Pin1 knockdown does not affect SMRT levels (Fig. 7 D). However, knockdown of Pin1 sensitizes cells to tamoxifen-mediated repression of c-Myc and PR expression without affecting BT-474 cell proliferation (Fig. 7, E and F). These data suggest that Pin1 may affect tamoxifen sensitivity through other pathways independent of controlling SMRT protein levels.
Pin1 is capable of promoting both stabilization and degradation of several target proteins (Yeh and Means, 2007); however, in many cases, the mechanisms underlying these opposing activities remain largely unexplored. In the case of c-Myc, Pin1 promotes a conformational change that allows rapid dephosphorylation of a specific threonine residue that is required for the binding of a ubiquitin E3 ligase (Yeh et al., 2004). In other cases, the conformational switch to certain prolyl isomers (cis or trans) also makes the target protein better substrates for particular E3 ubiquitin ligases (and therefore better substrates for degradation) or protein phosphatases (Yi et al., 2005; van Drogen et al., 2006; Yeh et al., 2006; Wu et al., 2007; Yeh and Means, 2007). Proteins such as c-Myc, cyclin E, and SRC-3 use a common Pin1-interacting motif to facilitate turnover by the E3 ligase Fbw7. As this motif does not exist in SMRT, it is likely that a different E3 ligase may be involved. Thus, identification of components of the degradation machinery responsible for Pin1- and Cdk2-dependent SMRT degradation warrants further investigation and may reveal potential new therapeutic targets for treatment of certain breast cancers.
Phosphorylation of SMRT by Cdk2 may participate in some integral aspect of cell cycle regulation. Certain genes required for cell cycle progression may require removal or degradation of SMRT from their promoters in order for cells to proliferate. Another interesting hypothesis is that SMRT may play roles other than that of a corepressor as recent studies have indicated, including involvement in DNA damage repair pathways and cell cycle regulation (Li et al., 2006; Yu et al., 2006). Additionally, it will be important to explore other potential regulators of SMRT stability and activity that may play a role in human development and disease generation or progression. Our observation that cyclin E coimmunoprecipitated SMRT but not Cdk2 is intriguing. This could be caused by the inaccessibility of the epitope by cyclin E antibodies. Alternatively, but not exclusively, these data suggest that the association of cyclin E with SMRT is independent of Cdk2. It will be interesting to further investigate the functional significance of the association between SMRT and cyclin E.
The ability of Pin1 to control SMRT protein levels appears to be cell type dependent, as Pin1 knockdown significantly increases SMRT protein steady-state levels in HeLa but not in BT-474 cells (Fig. 2 D and Fig. 7 D). In contrast, knockdown of Cdk2 increases SMRT protein levels in both HeLa and BT-474 cells (Fig. 6 E and Fig. 7 D). These observations suggest that Cdk2 may use Pin1-dependent or -independent pathways to promote SMRT degradation. ErbB2 activation stimulates Cdk2 activity, and knockdown of Cdk2 significantly increases steady-state levels of SMRT proteins and blocks ErbB2-mediated SMRT degradation, indicating that Cdk2 is an integral component in the ErbB2-dependent SMRT degradation pathway. Notably, knockdown of Cdk2 abolishes ErbB2-activated gene expression (Fig. 7 E) and cell proliferation (Fig. 7 F). Our data favor an oncogenic role for Cdk2 in SMRT destabilization and suggest that decreases in SMRT protein levels correlate with relief in gene repression and increases in proliferation, both indicators of tamoxifen resistance, whereas increases in SMRT levels correlate with tamoxifen sensitivity.
Materials and methods
Yeast methods
Yeast two-hybrid screens and assays were performed using standard methods described previously (Kao et al., 2000). A yeast two-hybrid library from mouse 17-d-old embryos (Stratagene) and pGBT9-hSMRT (a region encompassing amino acids 1,060–1,823 of human SMRT) were cotransformed into the yeast strain Y190. Approximately 5 × 106 yeast transformants were screened and selected on yeast minimal medium Leu-Trp-His plates containing 40 mM 3-aminotriazole (Sigma-Aldrich). After 7 d, colonies were picked, and the interactions were confirmed by β-galactosidase assays. Plasmids were recovered from the yeast and retransformed into yeast along with the bait construct. Positive clones were subjected to sequencing. Liquid β-galactosidase assays were performed as described by the manufacturer (Clontech Laboratories, Inc.), and the data represent the mean of duplicate reactions of two colonies.
Plasmid construction
pGBT9-SMRT yeast two-hybrid plasmids have been previously described (Kao et al., 2000). Pin1, Cdk2, and cyclin A expression vectors were generated by PCR from a HeLa library and subcloned into the CMX-1F or -1H vector (Gao et al., 2006). HA full-length SMRT and HA-SMRT (1–1,178) were derived from previously described plasmids (Park et al., 1999); HA-SMRT (1,012–2,507), HA-SMRT (1,823–2,507), and HA- and FLAG-SMRT (1,178–1,823) were derived from previously described plasmids (Chen and Evans, 1995). GST–Pin1, GST-SMRT (1,178–1,578), and GST-SMRT (1,560–1,823) were generated from HA-Pin1 and HA-SMRT (1,178–1,823), respectively, and cloned into the pGEX vector (GE Healthcare). GST–14-3-3ε, reporter constructs, and β-galactosidase expression vectors have been previously described (Kao et al., 2001). Site-directed mutagenesis was performed using the QuikChange kit (Stratagene).
Cell culture and transfection
HeLa, MCF-7, and CV-1 cells were grown in standard DME (Sigma-Aldrich) supplemented with 10% FBS, 50 U/ml penicillin G, and 50 μg/ml streptomycin sulfate at 37°C in 5% CO2. MEF cells were grown in DME (Invitrogen) supplemented with 10% heat-inactivated FBS. BT-474 cells were grown in DME (American Type Culture Collection) supplemented with 10% FBS. Transfections were performed using either Lipofectamine (CV-1) or Lipofectamine 2000 (HeLa, MCF-7, MEFs, and BT-474; Invitrogen) and harvested 48 h after transfection. Transfections with siRNA (Thermo Fisher Scientific) were performed using Lipofectamine 2000, and cells were harvested 72 h after transfection. Cells were treated with 20 nM calyculin A (BIOMOL International, L.P.) to inhibit phosphatase activity for 1 h before harvest where noted. Cells were treated with 100 μg/ml CHX (Sigma-Aldrich) for the indicated times before harvest where noted.
In vitro protein–protein interaction assays
GST fusion proteins, GST, GST–Pin1 (WT or mutant), and GST–14-3-3ε were expressed in an Escherichia coli DH5α strain and affinity purified on glutathione–Sepharose 4B beads. In vitro pull-down assays were performed by incubating GST–Pin1 with nuclear extracts (Dignam et al., 1983) or whole cell lysates from HeLa cells according to our published protocol (Kao et al., 2000) for 1 h at 4°C. After extensive washes, SDS-PAGE sample buffer was added to the beads, boiled, and separated by SDS-PAGE. For phosphatase treatments, extracts prepared from HA-SMRT– expressing cells were treated with increasing concentrations of calf intestinal phosphatase (Roche) for 30 min at 30°C before GST pull-down assays. His6-SMRT (1,178–1,823) was purified from E. coli BL-21 cells using nickel–nitrilotriacetic acid agarose beads (QIAGEN).
Coimmunoprecipitations
Transiently transfected HeLa whole cell extracts (WCEs) were prepared according to our published protocol (Kao et al., 2000). Immunoprecipitations for transfected samples were performed using anti-Flag or anti-HA M2 beads (Sigma-Aldrich). Endogenous coimmunoprecipitations were performed using the indicated antibodies (anti-Cdk2, cyclin A, cyclin E, and 14-3-3 [Santa Cruz Biotechnology, Inc.]; anti-Pin1 [Millipore]) with HeLa nuclear extracts. Extracts were incubated with antibodies and protein A beads (RepliGen Corp.) for 3 h at 4°C. Immunopellets were washed extensively and subjected to SDS-PAGE followed by immunoblot analyses with the antibodies anti-Flag (Sigma-Aldrich), anti-HA (Roche), and anti-Pin1 (Millipore) and anti-Cdk2, anti–cyclin A, anti–14-3-3, and anti-HDAC3 (Santa Cruz Biotechnology, Inc.). SMRT and N-CoR antibodies were purified in our laboratory; phosphospecific SMRT antibodies were generated and purified by Affinity BioReagents.
Transient transfection reporter assays
CV-1 cells were cotransfected with 16.6–66.6 ng pCMX-Gal4 and pCMX-Gal4-SMRT (1,060–1,823) constructs (Nagy et al., 1997), 100 ng pMH100–thymidine kinase–Luc, and 100 ng pCMX-LacZ in DME growth medium using Lipofectamine. The amount of DNA in each transfection was kept constant by the addition of parachlorometaxylenol. Cells were harvested and assayed for luciferase activity 36–48 h after transfection. The luciferase activity was normalized to the β-galactosidase activity. Each transfection was performed in triplicate and repeated at least two times.
In vitro kinase assays
GST-SMRT (1,178–1,578 and 1,560–1,823) constructs were purified from E. coli and incubated with purified Cdk2 (New England Biolabs, Inc.) according to the manufacturer's protocol. In brief, equal amounts of purified GST-tagged proteins were incubated with 100 U of purified Cdk2–cyclin A complexes in the associated buffer (NEB) for 30 min at 30°C. For autoradiography, [32P]ATP (PerkinElmer) was used. Reactions were stopped with SDS-PAGE sample buffer, separated by SDS-PAGE, transferred to polyvinylidene difluoride membrane, and subjected to autoradiography, immunoblotting (anti-GST; Santa Cruz Biotechnology, Inc.), or Coomassie staining.
Mass spectrometry
HeLa WCEs transfected with FLAG-SMRT (1,178–1,823) and treated with 20 nM calyculin A were subjected to coimmunoprecipitations using anti-FLAG affinity beads (Sigma-Aldrich) and separated by SDS-PAGE. The gel was Coomassie stained, and protein bands were cut out, washed and destained (50% ethanol and 5% acetic acid), reduced and alkylated (DTT and iodoacetamide), dehydrated (acetonitrile), and dried in a speed vac. Samples were digested using trypsin (20 ng/μL in 50 mM ammonium bicarbonate), and peptides were analyzed by liquid chromatography-mass spectrometry (linear ion trap; Finnigan LTQ).
Pulse-chase labeling
Cells were starved 10 min before labeling with DME lacking cysteine and methionine. Cells were then pulse labeled for 30 min with DME containing [35S]Met/Cys (PerkinElmer), washed twice, and chased in DME with unlabeled Met/Cys for the indicated times. Before labeling, cells were treated with either 100 ng/ml heregulin (R&D Systems) or 0.1 μM AG825 (EMD) for 30 min. Cells were lysed, and WCEs were subjected to immunoprecipitation overnight with either anti-SMRT (our own) or anti-HA antibodies (Sigma-Aldrich), separated by SDS-PAGE, and analyzed by autoradiography. Bands were quantified using an imaging system (VersaDoc; Bio-Rad Laboratories).
Immunofluorescent microscopy
Immunofluorescent staining was performed at room temperature as described previously (Reineke et al., 2008) using the antibodies anti-SMRT (ABR) and anti-FLAG (Santa Cruz Biotechnology, Inc.). N-CoR antibodies were purified in our laboratory. Secondary antibodies (conjugated to Alexa Fluor 488 or 594) were purchased from Invitrogen. Coverslips were mounted using mounting medium with DAPI (Vectashield; Vector Laboratories). Cells were analyzed using a fluorescent microscope (DMLB; Leica) using a 40× lens (Leica) with a numerical aperture of 506744. Images were captured with a camera (7.2 Color Mosaic; Diagnostic Instruments, Inc.) and acquired using SPOT Advanced (Diagnostic Instruments, Inc.).
RT-PCR
BT-474 cells were transfected with siRNA as described in Cell culture and transfection. 48 h after transfection, cells were treated with 100 nM 4-hydroxytamoxifen (Sigma-Aldrich) or ethanol vector control. 72 h after treatment, total RNA was harvested using PrepEase RNA Spin kits (USB). cDNA was generated using Superscript II reverse transcription and oligo-dT primers (Invitrogen). PCR reactions were performed using the following primers: 36B4 forward 5′-TGTTTCATTGTGGGAGCAGAC-3′, 36B4 reverse 5′-AAGCACTTCAGGGTTCTAGAT-3′, c-Myc forward 5′-ATGAAAAGGCCCCCAAGGTAGTTAT-3′, c-Myc reverse 5′-GCATTTGATCATGCATTTGAAACAA-3′, PR forward 5′-CCATGTGGCAGATCCCACAGGAGTT-3′, and PR reverse 5′-TGGAAATTCAACACTCAGTGCC-3′. Quantification was performed in Photoshop (Adobe).
Cell proliferation
BT-474 cells were transfected with siRNA as described for RT-PCR. 48 h after transfection, cells were counted, and 1,500 cells were plated into 96-well plates. Cells were treated with either 100 nM 4-hydroxytamoxifen or ethanol vehicle control. The day 0 time point was taken ∼12 h after plating. Cell proliferation was measured every 24 h using the CyQUANT NF Cell Proliferation Assay kit (Invitrogen).
Acknowledgments
We thank R. Evans and D. Chen for plasmids and reagents, N. Ogba and M. Montano for primers, Y.C. Yang for use of microscope facilities, and M. Kinter for peptide sequencing assistance. We also thank E. Reineke, D. Samols, and E. Stavnezer for critically reading this manuscript.
This work was supported by the American Cancer Society (grant RSG-04-052-01-GMC) and the Ohio Cancer Research Associates (grant to H.-Y. Kao) and by the National Institutes of Health (grant CA082845 to A.R. Means and grant T32GM08056 to K.J. Stanya). H.-Y. Kao is a recipient of the James T. Pardee–Carl A. Gerstacker Assistant Professor of Cancer Research Faculty Chair in Cancer Research at Case Western Reserve University Cancer Center.
Abbreviations used in this paper: CHX, cycloheximide; DBD, DNA-binding domain; DN, dominant negative; HDAC, histone deacetylase; MEF, mouse embryonic fibroblast; N-CoR, nuclear receptor corepressor; PPIase, peptidyl-prolyl isomerase; PR, progesterone receptor; pS-P, phospho-Ser-Pro; pT-P, phospho-Thr-Pro; RD, repression domain; SMRT, silencing mediator for retinoic acid and thyroid hormone receptor; WCE, whole cell extract; WT, wild type.
References
- Alland, L., R. Muhle, H. Hou Jr., J. Potes, L. Chin, N. Schreiber-Agus, and R.A. DePinho. 1997. Role for N-CoR and histone deacetylase in Sin3-mediated transcriptional repression. Nature. 387:49–55. [DOI] [PubMed] [Google Scholar]
- Bao, L., A. Kimzey, G. Sauter, J.M. Sowadski, K.P. Lu, and D.G. Wang. 2004. Prevalent overexpression of prolyl isomerase Pin1 in human cancers. Am. J. Pathol. 164:1727–1737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brondani, V., Q. Schefer, F. Hamy, and T. Klimkait. 2005. The peptidyl-prolyl isomerase Pin1 regulates phospho-Ser77 retinoic acid receptor alpha stability. Biochem. Biophys. Res. Commun. 328:6–13. [DOI] [PubMed] [Google Scholar]
- Chen, J.D., and R.M. Evans. 1995. A transcriptional co-repressor that interacts with nuclear hormone receptors. Nature. 377:454–457. [DOI] [PubMed] [Google Scholar]
- Dignam, J.D., R.M. Lebovitz, and R.G. Roeder. 1983. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 11:1475–1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischle, W., F. Dequiedt, M.J. Hendzel, M.G. Guenther, M.A. Lazar, W. Voelter, and E. Verdin. 2002. Enzymatic activity associated with class II HDACs is dependent on a multiprotein complex containing HDAC3 and SMRT/N-CoR. Mol. Cell. 9:45–57. [DOI] [PubMed] [Google Scholar]
- Fleming, F.J., A.D. Hill, E.W. McDermott, N.J. O'Higgins, and L.S. Young. 2004. Differential recruitment of coregulator proteins steroid receptor coactivator-1 and silencing mediator for retinoid and thyroid receptors to the estrogen receptor-estrogen response element by beta-estradiol and 4-hydroxytamoxifen in human breast cancer. J. Clin. Endocrinol. Metab. 89:375–383. [DOI] [PubMed] [Google Scholar]
- Fujita, T., Y. Kobayashi, O. Wada, Y. Tateishi, L. Kitada, Y. Yamamoto, H. Takashima, A. Murayama, T. Yano, T. Baba, et al. 2003. Full activation of estrogen receptor alpha activation function-1 induces proliferation of breast cancer cells. J. Biol. Chem. 278:26704–26714. [DOI] [PubMed] [Google Scholar]
- Gao, C., X. Li, M. Lam, Y. Liu, S. Chakraborty, and H.Y. Kao. 2006. CRM1 mediates nuclear export of HDAC7 independently of HDAC7 phosphorylation and association with 14-3-3s. FEBS Lett. 580:5096–5104. [DOI] [PubMed] [Google Scholar]
- Goodson, M., B.A. Jonas, and M.A. Privalsky. 2005. Corepressors: custom tailoring and alterations while you wait. Nucl. Recept. Signal. 3:e003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham, J.D., D.L. Bain, J.K. Richer, T.A. Jackson, L. Tung, and K.B. Horwitz. 2000. a. Nuclear receptor conformation, coregulators, and tamoxifen-resistant breast cancer. Steroids. 65:579–584. [DOI] [PubMed] [Google Scholar]
- Graham, J.D., D.L. Bain, J.K. Richer, T.A. Jackson, L. Tung, and K.B. Horwitz. 2000. b. Thoughts on tamoxifen resistant breast cancer. Are coregulators the answer or just a red herring? J. Steroid Biochem. Mol. Biol. 74:255–259. [DOI] [PubMed] [Google Scholar]
- Guenther, M.G., O. Barak, and M.A. Lazar. 2001. The SMRT and N-CoR corepressors are activating cofactors for histone deacetylase 3. Mol. Cell. Biol. 21:6091–6101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guenther, M.G., J. Yu, G.D. Kao, T.J. Yen, and M.A. Lazar. 2002. Assembly of the SMRT-histone deacetylase 3 repression complex requires the TCP-1 ring complex. Genes Dev. 16:3130–3135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horlein, A.J., A.M. Naar, T. Heinzel, J. Torchia, B. Gloss, R. Kurokawa, A. Ryan, Y. Kamei, M. Soderstrom, C.K. Glass, et al. 1995. Ligand-independent repression by the thyroid hormone receptor mediated by a nuclear receptor co-repressor. Nature. 377:397–404. [DOI] [PubMed] [Google Scholar]
- Huang, E.Y., J. Zhang, E.A. Miska, M.G. Guenther, T. Kouzarides, and M.A. Lazar. 2000. Nuclear receptor corepressors partner with class II histone deacetylases in a Sin3-independent repression pathway. Genes Dev. 14:45–54. [PMC free article] [PubMed] [Google Scholar]
- Jackson, T.A., J.K. Richer, D.L. Bain, G.S. Takimoto, L. Tung, and K.B. Horwitz. 1997. The partial agonist activity of antagonist-occupied steroid receptors is controlled by a novel hinge domain-binding coactivator L7/SPA and the corepressors N-CoR or SMRT. Mol. Endocrinol. 11:693–705. [DOI] [PubMed] [Google Scholar]
- Jepsen, K., O. Hermanson, T.M. Onami, A.S. Gleiberman, V. Lunyak, R.J. McEvilly, R. Kurokawa, V. Kumar, F. Liu, E. Seto, et al. 2000. Combinatorial roles of the nuclear receptor corepressor in transcription and development. Cell. 102:753–763. [DOI] [PubMed] [Google Scholar]
- Jepsen, K., D. Solum, T. Zhou, R.J. McEvilly, H.J. Kim, C.K. Glass, O. Hermanson, and M.G. Rosenfeld. 2007. SMRT-mediated repression of an H3K27 demethylase in progression from neural stem cell to neuron. Nature. 450:415–419. [DOI] [PubMed] [Google Scholar]
- Jonas, B.A., and M.L. Privalsky. 2004. SMRT and N-CoR corepressors are regulated by distinct kinase signaling pathways. J. Biol. Chem. 279:54676–54686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kao, H.Y., P. Ordentlich, N. Koyano-Nakagawa, Z. Tang, M. Downes, C.R. Kintner, R.M. Evans, and T. Kadesch. 1998. A histone deacetylase corepressor complex regulates the Notch signal transduction pathway. Genes Dev. 12:2269–2277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kao, H.Y., M. Downes, P. Ordentlich, and R.M. Evans. 2000. Isolation of a novel histone deacetylase reveals that class I and class II deacetylases promote SMRT-mediated repression. Genes Dev. 14:55–66. [PMC free article] [PubMed] [Google Scholar]
- Kao, H.Y., A. Verdel, C.C. Tsai, C. Simon, H. Juguilon, and S. Khochbin. 2001. Mechanism for nucleocytoplasmic shuttling of histone deacetylase 7. J. Biol. Chem. 276:47496–47507. [DOI] [PubMed] [Google Scholar]
- Keeton, E.K., and M. Brown. 2005. Cell cycle progression stimulated by tamoxifen-bound estrogen receptor-alpha and promoter-specific effects in breast cancer cells deficient in N-CoR and SMRT. Mol. Endocrinol. 19:1543–1554. [DOI] [PubMed] [Google Scholar]
- Khan, M.M., T. Nomura, T. Chiba, K. Tanaka, H. Yoshida, K. Mori, and S. Ishii. 2004. The fusion oncoprotein PML-RARalpha induces endoplasmic reticulum (ER)-associated degradation of N-CoR and ER stress. J. Biol. Chem. 279:11814–11824. [DOI] [PubMed] [Google Scholar]
- Lavinsky, R.M., K. Jepsen, T. Heinzel, J. Torchia, T.M. Mullen, R. Schiff, A.L. Del-Rio, M. Ricote, S. Ngo, J. Gemsch, et al. 1998. Diverse signaling pathways modulate nuclear receptor recruitment of N-CoR and SMRT complexes. Proc. Natl. Acad. Sci. USA. 95:2920–2925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Y., G.D. Kao, B.A. Garcia, J. Shabanowitz, D.F. Hunt, J. Qin, C. Phelan, and M.A. Lazar. 2006. A novel histone deacetylase pathway regulates mitosis by modulating Aurora B kinase activity. Genes Dev. 20:2566–2579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu, K.P., S.D. Hanes, and T. Hunter. 1996. A human peptidyl-prolyl isomerase essential for regulation of mitosis. Nature. 380:544–547. [DOI] [PubMed] [Google Scholar]
- Lu, P.J., X.Z. Zhou, M. Shen, and K.P. Lu. 1999. Function of WW domains as phosphoserine- or phosphothreonine-binding modules. Science. 283:1325–1328. [DOI] [PubMed] [Google Scholar]
- Mantovani, F., F. Tocco, J. Girardini, P. Smith, M. Gasco, X. Lu, T. Crook, and G. Del Sal. 2007. The prolyl isomerase Pin1 orchestrates p53 acetylation and dissociation from the apoptosis inhibitor iASPP. Nat. Struct. Mol. Biol. 14:912–920. [DOI] [PubMed] [Google Scholar]
- Nagy, L., H.Y. Kao, D. Chakravarti, R.J. Lin, C.A. Hassig, D.E. Ayer, S.L. Schreiber, and R.M. Evans. 1997. Nuclear receptor repression mediated by a complex containing SMRT, mSin3A, and histone deacetylase. Cell. 89:373–380. [DOI] [PubMed] [Google Scholar]
- Neve, R.M., H. Sutterluty, N. Pullen, H.A. Lane, J.M. Daly, W. Krek, and N.E. Hynes. 2000. Effects of oncogenic ErbB2 on G1 cell cycle regulators in breast tumour cells. Oncogene. 19:1647–1656. [DOI] [PubMed] [Google Scholar]
- Ordentlich, P., M. Downes, W. Xie, A. Genin, N.B. Spinner, and R.M. Evans. 1999. Unique forms of human and mouse nuclear receptor corepressor SMRT. Proc. Natl. Acad. Sci. USA. 96:2639–2644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osborne, C.K., V. Bardou, T.A. Hopp, G.C. Chamness, S.G. Hilsenbeck, S.A. Fuqua, J. Wong, D.C. Allred, G.M. Clark, and R. Schiff. 2003. Role of the estrogen receptor coactivator AIB1 (SRC-3) and HER-2/neu in tamoxifen resistance in breast cancer. J. Natl. Cancer Inst. 95:353–361. [DOI] [PubMed] [Google Scholar]
- Park, E.J., D.J. Schroen, M. Yang, H. Li, L. Li, and J.D. Chen. 1999. SMRTe, a silencing mediator for retinoid and thyroid hormone receptors-extended isoform that is more related to the nuclear receptor corepressor. Proc. Natl. Acad. Sci. USA. 96:3519–3524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pastorino, L., A. Sun, P.J. Lu, X.Z. Zhou, M. Balastik, G. Finn, G. Wulf, J. Lim, S.H. Li, X. Li, et al. 2006. The prolyl isomerase Pin1 regulates amyloid precursor protein processing and amyloid-beta production. Nature. 440:528–534. [DOI] [PubMed] [Google Scholar]
- Perissi, V., A. Aggarwal, C.K. Glass, D.W. Rose, and M.G. Rosenfeld. 2004. A corepressor/coactivator exchange complex required for transcriptional activation by nuclear receptors and other regulated transcription factors. Cell. 116:511–526. [DOI] [PubMed] [Google Scholar]
- Privalsky, M.L. 2004. The role of corepressors in transcriptional regulation by nuclear hormone receptors. Annu. Rev. Physiol. 66:315–360. [DOI] [PubMed] [Google Scholar]
- Ranganathan, R., K.P. Lu, T. Hunter, and J.P. Noel. 1997. Structural and functional analysis of the mitotic rotamase Pin1 suggests substrate recognition is phosphorylation dependent. Cell. 89:875–886. [DOI] [PubMed] [Google Scholar]
- Reineke, E.L., M. Lam, Q. Liu, Y. Liu, K.J. Stanya, K.S. Chang, A.R. Means, and H.Y. Kao. 2008. Degradation of the tumor suppressor PML by Pin1 contributes to the cancer phenotype of breast cancer MDA-MB-231 cells. Mol. Cell. Biol. 28:997–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryo, A., M. Nakamura, G. Wulf, Y.C. Liou, and K.P. Lu. 2001. Pin1 regulates turnover and subcellular localization of beta-catenin by inhibiting its interaction with APC. Nat. Cell Biol. 3:793–801. [DOI] [PubMed] [Google Scholar]
- Ryo, A., Y.C. Liou, G. Wulf, M. Nakamura, S.W. Lee, and K.P. Lu. 2002. PIN1 is an E2F target gene essential for Neu/Ras-induced transformation of mammary epithelial cells. Mol. Cell. Biol. 22:5281–5295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sande, S., and M.L. Privalsky. 1996. Identification of TRACs (T3 receptor-associating cofactors), a family of cofactors that associate with, and modulate the activity of, nuclear hormone receptors. Mol. Endocrinol. 10:813–825. [DOI] [PubMed] [Google Scholar]
- Seol, W., M.J. Mahon, Y.K. Lee, and D.D. Moore. 1996. Two receptor interacting domains in the nuclear hormone receptor corepressor RIP13/N-CoR. Mol. Endocrinol. 10:1646–1655. [DOI] [PubMed] [Google Scholar]
- Shang, Y., and M. Brown. 2002. Molecular determinants for the tissue specificity of SERMs. Science. 295:2465–2468. [DOI] [PubMed] [Google Scholar]
- Shen, M., P.T. Stukenberg, M.W. Kirschner, and K.P. Lu. 1998. The essential mitotic peptidyl-prolyl isomerase Pin1 binds and regulates mitosis-specific phosphoproteins. Genes Dev. 12:706–720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shou, J., S. Massarweh, C.K. Osborne, A.E. Wakeling, S. Ali, H. Weiss, and R. Schiff. 2004. Mechanisms of tamoxifen resistance: increased estrogen receptor-HER2/neu cross-talk in ER/HER2-positive breast cancer. J. Natl. Cancer Inst. 96:926–935. [DOI] [PubMed] [Google Scholar]
- Smith, C.L., Z. Nawaz, and B.W. O'Malley. 1997. Coactivator and corepressor regulation of the agonist/antagonist activity of the mixed antiestrogen, 4-hydroxytamoxifen. Mol. Endocrinol. 11:657–666. [DOI] [PubMed] [Google Scholar]
- Sudol, M., K. Sliwa, and T. Russo. 2001. Functions of WW domains in the nucleus. FEBS Lett. 490:190–195. [DOI] [PubMed] [Google Scholar]
- Timms, J.F., S.L. White, M.J. O'Hare, and M.D. Waterfield. 2002. Effects of ErbB-2 overexpression on mitogenic signalling and cell cycle progression in human breast luminal epithelial cells. Oncogene. 21:6573–6586. [DOI] [PubMed] [Google Scholar]
- Tsai, C.C., H.Y. Kao, A. Mitzutani, E. Banayo, H. Rajan, M. McKeown, and R.M. Evans. 2004. Ataxin 1, a SCA1 neurodegenerative disorder protein, is functionally linked to the silencing mediator of retinoid and thyroid hormone receptors. Proc. Natl. Acad. Sci. USA. 101:4047–4052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Drogen, F., O. Sangfelt, A. Malyukova, L. Matskova, E. Yeh, A.R. Means, and S.I. Reed. 2006. Ubiquitylation of cyclin E requires the sequential function of SCF complexes containing distinct hCdc4 isoforms. Mol. Cell. 23:37–48. [DOI] [PubMed] [Google Scholar]
- Wu, R.C., Q. Feng, D.M. Lonard, and B.W. O'Malley. 2007. SRC-3 coactivator functional lifetime is regulated by a phospho-dependent ubiquitin time clock. Cell. 129:1125–1140. [DOI] [PubMed] [Google Scholar]
- Yaffe, M.B., and A.E. Elia. 2001. Phosphoserine/threonine-binding domains. Curr. Opin. Cell Biol. 13:131–138. [DOI] [PubMed] [Google Scholar]
- Yaffe, M.B., M. Schutkowski, M. Shen, X.Z. Zhou, P.T. Stukenberg, J.U. Rahfeld, J. Xu, J. Kuang, M.W. Kirschner, G. Fischer, et al. 1997. Sequence-specific and phosphorylation-dependent proline isomerization: a potential mitotic regulatory mechanism. Science. 278:1957–1960. [DOI] [PubMed] [Google Scholar]
- Yeh, E.S., and A.R. Means. 2007. PIN1, the cell cycle and cancer. Nat. Rev. Cancer. 7:381–388. [DOI] [PubMed] [Google Scholar]
- Yeh, E., M. Cunningham, H. Arnold, D. Chasse, T. Monteith, G. Ivaldi, W.C. Hahn, P.T. Stukenberg, S. Shenolikar, T. Uchida, et al. 2004. A signalling pathway controlling c-Myc degradation that impacts oncogenic transformation of human cells. Nat. Cell Biol. 6:308–318. [DOI] [PubMed] [Google Scholar]
- Yeh, E.S., B.O. Lew, and A.R. Means. 2006. The loss of PIN1 deregulates cyclin E and sensitizes mouse embryo fibroblasts to genomic instability. J. Biol. Chem. 281:241–251. [DOI] [PubMed] [Google Scholar]
- Yi, P., R.C. Wu, J. Sandquist, J. Wong, S.Y. Tsai, M.J. Tsai, A.R. Means, and B.W. O'Malley. 2005. Peptidyl-prolyl isomerase 1 (Pin1) serves as a coactivator of steroid receptor by regulating the activity of phosphorylated steroid receptor coactivator 3 (SRC-3/AIB1). Mol. Cell. Biol. 25:9687–9699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, J., C. Palmer, T. Alenghat, Y. Li, G. Kao, and M.A. Lazar. 2006. The corepressor silencing mediator for retinoid and thyroid hormone receptor facilitates cellular recovery from DNA double-strand breaks. Cancer Res. 66:9316–9322. [DOI] [PubMed] [Google Scholar]
- Zacchi, P., M. Gostissa, T. Uchida, C. Salvagno, F. Avolio, S. Volinia, Z. Ronai, G. Blandino, C. Schneider, and G. Del Sal. 2002. The prolyl isomerase Pin1 reveals a mechanism to control p53 functions after genotoxic insults. Nature. 419:853–857. [DOI] [PubMed] [Google Scholar]
- Zhang, J., M.G. Guenther, R.W. Carthew, and M.A. Lazar. 1998. Proteasomal regulation of nuclear receptor corepressor-mediated repression. Genes Dev. 12:1775–1780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng, H., H. You, X.Z. Zhou, S.A. Murray, T. Uchida, G. Wulf, L. Gu, X. Tang, K.P. Lu, and Z.X. Xiao. 2002. The prolyl isomerase Pin1 is a regulator of p53 in genotoxic response. Nature. 419:849–853. [DOI] [PubMed] [Google Scholar]