Abstract
The identification of the molecular mechanisms controlling cardiomyocyte proliferation during the embryonic, fetal, and early neonatal life appears of paramount interest in regard to exploiting this information to promote cardiac regeneration. Here, we show that the proliferative potential of neonatal rat cardiomyocytes is powerfully stimulated by the sustained activation of the Notch pathway. We found that Notch1 is expressed in proliferating ventricular immature cardiac myocytes (ICMs) both in vitro and in vivo, and that the number of Notch1-positive cells in the heart declines with age. Notch1 expression in ICMs paralleled the expression of its Jagged1 ligand on non-myocyte supporting cells. The inhibition of Notch signaling in ICMs blocked their proliferation and induced apoptosis; in contrast, its activation by Jagged1 or by the constitutive expression of its activated form using an adeno-associated virus markedly stimulated proliferative signaling and promoted ICM expansion. Maintenance or reactivation of Notch signaling in cardiac myocytes might represent an interesting target for innovative regenerative therapy.
Introduction
The Notch signaling pathway plays a key role at multiple steps of morphogenesis during embryonic development as well as in a wide variety of processes during adult life. In particular, Notch activation appears to finely tune the balance between proliferation and differentiation of stem and progenitor cells in several different settings, including hematopoietic and nervous systems, skin, gut, and heart (for reviews see Bray, 2006; Chiba, 2006; Hurlbut et al., 2007; Nemir and Pedrazzini, 2008; Niessen and Karsan, 2008). Physiological activation of Notch signaling requires cell–cell contact and occurs through binding of the Notch receptor to one of its ligands (Delta and Jagged in vertebrates and Serrate in invertebrates) followed by the proteolytic release of the intracellular domain (ICD) of Notch (Notch-ICD) and its translocation into the nucleus (De Strooper et al., 1999). Once inside the nucleus, Notch-ICD interacts with transcription regulators of the CSL family (CBF1, Su(H), and Lag-1), triggering the activation of genes of the hairy and enhancer of split (HES) family (Jarriault et al., 1995; Iso et al., 2003). Experimental evidence obtained in Xenopus laevis, zebrafish, chicken, and mammals has demonstrated that Notch signaling controls mesodermal commitment, including cardiogenic differentiation, during development (Rones et al., 2000; Loomes et al., 2002; Beis et al., 2005; Rutenberg et al., 2006; Del Monte et al., 2007; Grego-Bessa et al., 2007). Of interest, mice lacking the genes encoding either Notch receptors or their ligands show early embryonic lethality, which is mainly the consequence of cardiovascular abnormalities. This observation supports the notion that the Notch pathway essentially participates in the process of cardiac and vascular development (Gridley, 1997; Gridley, 2003; Shawber and Kitajewski, 2004; Del Monte et al., 2007; Grego-Bessa et al., 2007). Several other observations also favor this conclusion. In humans, the Alagille syndrome, a congenital disorder associated with heart abnormalities, has been linked to haploinsufficiency of Jagged1 (Li et al., 1997; Oda et al., 1997; McElhinney et al., 2002). In mice, combined haploinsufficiency of Jagged1 and Notch2 results in cardiac defects reminiscent of this syndrome (McCright et al., 2002). The Notch signaling pathway also appears to be involved in cardiac valve development (Timmerman et al., 2004): in humans, heterozygous mutations disrupt normal development of the aortic and, occasionally, mitral valves (Garg et al., 2005). Although these models demonstrate the importance of Notch in heart development, little is known about the cellular and molecular mechanisms of Notch function in this district.
The emerging concept that the heart is not a postmitotic organ but rather a dynamic tissue maintained by a balance between cell death and cardiomyocyte differentiation from precursor cells implies the existence of a pool of resident stem cells able to trigger heart repair and the maintenance of tissue homeostasis (Kajstura et al., 2000; Hierlihy et al., 2002; Beltrami et al., 2003; Oh et al., 2003; Matsuura et al., 2004; Messina et al., 2004; Laugwitz et al., 2005). The understanding of the molecular and cellular mechanisms controlling cardiac precursor cell proliferation and differentiation appears of paramount interest with regard to exploiting this information to promote cardiac regeneration in adults.
In this paper, we show that the proliferative potential of immature cardiac myocytes (ICMs) is powerfully stimulated by the sustained activation of the Notch pathway. We found that Notch1 was highly expressed in early neonatal proliferating cardiomyocytes and that its activation protected these cells from apoptosis; the ligand Jagged1 was instead mainly detectable in non-myocyte cardiac stromal cells. The levels of expression of Notch1 were found to decline together with the loss of the cardiomyocyte proliferative potential, both in vitro and in vivo; consistent with this observation, the sustained activation of Notch1 determined a remarkable expansion of cardiac cells in vitro. Similar to the role of the Notch pathway in preserving progenitor cells in other tissues, these results indicate that immature, still proliferating cardiomyocytes rely on Notch1 activity for cell renewal, thus raising the exciting the possibility that the forced activation of this pathway might lead to the expansion of the pool of poorly differentiated, committed cardiac cells.
Results
The number of Notch1-positive, BrdU-positive cardiomyocytes in vivo declines with age
We started by analyzing the number of cycling cells in neonatal and adult rat hearts. For this purpose, animals at birth and at 9 mo of age (adults) were treated with BrdU, and after 1 wk, the presence of cells that had incorporated the nucleotide analogue was assessed by immunofluorescence. In agreement with published observations (Chen et al., 2007; Kang and Koh, 1997), >30% of nuclei of the α-actinin–positive neonatal cardiomyocytes scored positive for BrdU incorporation, whereas <3% of cells had incorporated BrdU in the adult hearts (Fig. 1, A and B, for representative images at different magnifications; and Fig. 1 C for quantification). To confirm the cardiac lineage specificity of the proliferating cells, double labeling with the cardiomyocyte-specific nuclear marker GATA-4 and BrdU was performed on heart sections of neonatal rats. In agreement with the α-actinin stainings, the nuclei of several proliferating cells scored positive for both GATA-4 and BrdU (Fig. 1 D). To further confirm these findings, heart sections from neonatal and adult animals were stained with an antibody against cyclin A, a G1/S marker that is only expressed in proliferating cells. A large number (∼30%) of nuclei in the neonatal hearts scored positive for this antibody. In sharp contrast, many fewer cells (<3%) were detected in adult hearts (Fig. 1 E for representative images and Fig. 1 F for quantification). Of interest, we noticed that the neonatal hearts also contained a large number of α-actinin–positive cells that were positive for the expression of the Notch1 receptor (Fig. 1 G). When these neonatal hearts were stained for both BrdU incorporation and Notch1 expression, several of the BrdU-positive cells were found to express Notch1 (Fig. 1 H) and, in particular, its activated intracellular form detected by an antibody against Val1744 (Fig. 1 I). Upon quantification, ∼19% of nuclei in the neonatal hearts were found to be double-positive for both BrdU and Notch1 (Fig. 1 C). In both neonatal and adult hearts, Jagged1 was not detectable in mature cardiomyocytes, whereas its expression was confined to patchy areas of α-actinin–negative cells and the endothelial surface lining cardiac muscle fibers (Fig. 1 L and not depicted).
Figure 1.
The number of Notch1-positive, BrdU-positive cardiomyocytes in vivo declines with age. (A) Low (top)- and high (bottom)-magnification representative images of heart sections from newborn rats, immunostained for BrdU (red) and α-actinin (green). Nuclei were marked by DAPI (blue). Arrows indicate proliferating myocardial cells. (B) High-magnification representative images of α-actinin–positive cardiac myocytes (green) from newborn rat hearts showing positivity to BrdU immunostaining (red). Nuclei were marked by DAPI. (C) Quantification of BrdU-positive and BrdU/Notch1-double-positive myocardial cells in newborn and adult rat hearts. Results indicate mean ± SEM, n = 6. (D) High magnification representative images of heart sections from neonatal rats immunostained for GATA-4 (red) and BrdU (green). Nuclei were marked by DAPI (blue). Arrows indicate proliferating myocardial cells. (E) Low (top)- and high (bottom)-magnification representative images of heart sections from newborn rats, immunostained for cyclin A (red) and α-actinin (green). Nuclei were stained by DAPI (blue). Arrows indicate proliferating myocardial cells. (F) Quantification of cyclin A–positive myocardial cells in newborn and adult rats. Results indicate mean ± SEM, n = 6. (G) Low (top)- and high (top)-magnification representative images of heart sections from newborn rats showing strong immunoreactivity for Notch1 (red) of α-actinin (green)-positive cardiomyocytes. Arrows indicate Notch1-positive myocardial cells. (H and I) High-magnification images of representative cells from a newborn heart section showing positivity of BrdU incorporation (green) and expression of the Notch1 cell membrane receptor (H) and its activated intracellular form (I, red). Nuclei were stained with DAPI. (L) Representative images of both newborn (top) and adult (bottom) rat heart sections showing a patchy staining for Jagged1 (red) in stromal areas of myocardial tissue, the latter positive for α-actinin staining (green). Nuclei were stained with DAPI. In A, E, and G–L, the largest figure in each panel shows fluorescence in all three channels; the two smaller panels show the red/green (top) and red/blue (bottom) channels. Bars: (A, top) 200 μm; (A, bottom) 10 μm; (B and D) 10 μm; (E, top) 100 μm; (E, bottom) 10 μm; (G, top) 200 μm; (G, bottom) μm; (H and I) 10 μm. (L) 100 μm.
These observations raised the intriguing possibility that the Notch1-positive, BrdU-positive cells might represent immature cardiomyocytes that were still in proliferation in the neonatal hearts.
Notch-1 expression is down-regulated during cardiomyocytes differentiation
To further investigate the role of Notch in the regulation of cardiomyocytes proliferation, the expression of the different Notch receptors and ligands was analyzed in primary cultures of cells extracted from neonatal rat myocardium. To specifically distinguish between cardiac myocytes and cardiac stromal cells, we obtained cultures highly enriched in α-actinin–positive cardiomyocytes by three subsequent sub-plating steps (>90% purity; see Materials and methods) along with cultures of α-actinin–negative stromal cells (<1% α-actinin positivity). Both cultures were maintained for 7–14 d. In the cardiac myocytes cultures, after 24 h, cells were small, had a rounded shape, and expressed poorly organized α-actinin; we refer to these myocytes as ICMs (Fig. 2 A, day 1). During the first week of culture, ICMs progressed along their differentiation by increasing in size and showed progressively more organized sarcomers; from day 3, mature cardiomyocytes started beating. At all time points, including day 1, the cells were positive for α-actinin and Nkx2.5 (Fig. 2 B), which is indicative of their committed cardiomyocyte identity. About 5% of the day-1 immature ICMs scored positive for BrdU incorporation; this percentage decreased to ∼2% and ∼1.4% at days 3 and 7 after plating (see Fig. 2 C for representative images and Fig. 5 C for quantification). Consistent with the BrdU incorporation results, when the distribution of cardiomyocytes in the different phases of the cell cycle was analyzed by flow cytometry after propidium iodide (PI) staining, the percentage of cells in the G1 and S phases was found to progressively decrease along with cardiomyocyte differentiation in favor of a G2/M-specific profile (Fig. 2 D). This result is in agreement with analogous findings presented in the accompanying manuscript by Campa et al. (see p. 129 of this issue). At days 1, 3, and 7 after plating, the levels of expression of individual genes in the Notch pathway were assessed by transcript-specific RT-PCR in both purified cardiomyocyte and stromal cell cultures (Fig. 2 E). At day 1, ICMs expressed high levels of Notch1, whereas Notch2 was barely detectable; the levels of Notch1 appeared to progressively decrease along cardiomyocyte differentiation and became almost undetectable at day 7. In a reciprocal fashion, the levels of Jagged1 in cardiomyocytes progressively increased with time; Jagged2 and Delta1 were not detected at any time in these cells. In supporting stromal cells, no expression of Notch1 was detectable at day 1, whereas its levels progressively increased at days 3 and 7. In contrast, at all time points, these cells expressed high levels of Jagged1, Notch2, and its specific downstream effector Hey1 (Rutenberg et al., 2006). No significant differences in the expression of the putative target genes of the Notch pathway, such as Hes1-5, Hey1, and Hey2, were observed in immature and fully differentiated cardiomyocytes, as well as in stromal cells, which indicates that regulation of these target genes is also dependent on pathways different from Notch (also see the following paragraph).
Figure 2.
Time course of expression of Notch signaling components during cardiomyocyte differentiation. (A) Representative images of purified neonatal cardiomyocytes stained for α-actinin (green) at different time points in culture. After 24 h from plating (day 1), ICMs are detected as small, round cells expressing poorly organized α-actinin; at day 3, they appear larger and show more organized sarcomeres; at day 7, they are differentiated, beating cells. Nuclei were stained with DAPI. (B) Representative images of Nkx2.5-positive cardiomyocytes during their in vitro differentiation. Green, α-actinin; red, Nkx2.5; blue, DNA (DAPI). (C) BrdU incorporation along cardiomyocyte differentiation. ICMs (day 1) are detected as small, round-shaped, BrdU-positive cells. The number of BrdU-positive cardiomyocytes progressively decreases along differentiation. Green, α-actinin; red, BrdU; blue, DNA (DAPI). The panels show representative images; for quantification, see Fig. 5 C. (D) Cell cycle distribution of cardiomyocytes at different times after plating. (top) Flow cytometry cell cycle profiles after DNA staining using PI and analysis of 10,000 cell counts; (bottom) histogram shows cell cycle distribution. Results are representative of at least three independent experiments. (E) RT-PCR analysis of the expression of individual genes in the Notch pathway in purified cardiomyocytes (CM) and stromal cells (S) at the indicated days of culture. RNA from total embryo extracts was used as a positive control (+). (F) Western blot analysis of Notch1 and Jagged1 proteins in purified cardiomyocytes (CM) and stromal cells (S) at the indicated time points. Notch1 proteins were detected in ICMs at day 1 of culture, progressively decreasing along cardiomyocyte differentiation at days 3 and 7. Specific signals became apparent in stromal cells only at day 7. Jagged1 was not detectable in ICMs (day 1), though it progressively increased at days 3 and 7. In stromal cells, Jagged1 was expressed at all time points. (G) Western blot analysis of activated Notch1 (p110-ICD) in cardiomyocytes. (H) Cellular localization of Notch1 and Jagged1 proteins during cardiomyocyte differentiation. Cardiomyocytes at days 1, 3, and 7 of culture were immunostained using antibodies against Notch1, Val1744-ICD, Jagged-1 (red), and α-actinin (green); nuclei were marked with DAPI (blue). The largest figure in each panel shows fluorescence in all three channels; the two smaller figures show the blue/green (top) and red/blue (bottom) channels. Bars: (A) 50 μm; (B, C, and H) 10 μm.
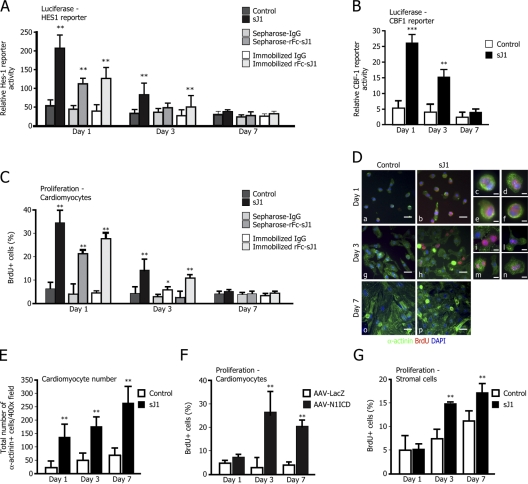
Figure 5.
sJ1 and Notch-ICD transduction enhance cardiomyocyte proliferation. (A) Treatment with sJ1 activates HES1 promoter transcription. Neonatal cardiomyocytes were assayed for HES1 reporter transactivation upon stimulation with conditioned medium from NIH-3T3 cells secreting sJ1 or rFc-sJ1 either preclustered on Sepharose beads or immobilized on plates. The respective controls were the supernatant from mock-transfected NIH-3T3 cells (Control), Sepharose-IgG, and immobilized IgG. The transactivation effect was evaluated at the indicated time points after reporter transfection. The histogram represents mean values ± SEM, n = 6. **, P < 0.001. (B) Treatment with sJ1 markedly enhances expression of the CBF1-luciferase reporter. Control or sJ1-conditioned medium (sJ1) was added to cardiomyocytes for 24 h after reporter transfection; the transactivation effect was evaluated at the indicated time points. The histogram represents mean values ± SEM, n = 6; **, P < 0.001. (C) sJ1 stimulation increases BrdU incorporation in cardiac myocytes. Neonatal cardiomyocytes were cultured in the presence of either sJ1- or control-conditioned medium, or either hIgG or rFc-sJ1 ligand preclustered onto Sepharose beads, and pulsed for BrdU incorporation. In a mirror experiment, neonatal cardiomyocytes were seeded onto rFc-sJ1 (or normal h-IgG as a control) precoated slides. After BrdU immunostaining, the total number of BrdU+ cells per well was scored. The histograms show mean ± SEM of the percentage of BrdU+ nuclei from at least three fields of ∼200 cells per field. Results are representative of at least three independent experiments. **, P < 0.001. (D) Low (panels a, b, g, h, o, and p)- and high (panels c–f and i–n)-magnification representative images of BrdU+ cardiomyocytes at different time points, after treatment with control- or sJ1-conditioned medium. Green, α-actinin; red, BrdU; blue, DNA (DAPI). Bars: (low magnification) 50 μm; (high magnification) 10 μm. The examples shown are representative of more than five experiments with similar outcomes. (E) sJ1 remarkably stimulates cardiomyocyte proliferation. Cardiac myocytes were cultured in the presence of sJ1 (sJ1)- or control-conditioned media. After specific immunostaining, the total number of α-actinin–positive cells per well was scored. The histogram shows mean ± SEM of at least 8 wells per time point from three independent experiments. **, P < 0.001. (F) AAV8-N1ICD strongly stimulates BrdU incorporation in cardiomyocytes. Neonatal cardiomyocytes were infected either with AAV8-LacZ or AAV8-N1ICD and pulsed for BrdU incorporation at different time points. After BrdU immunostaining, the total number of BrdU+ cells per well was scored. The histograms show mean ± SEM of the percentage of BrdU+ nuclei from at least three fields of ∼200 cells per field. Results are representative of at least three independent experiments. **, P < 0.001. (G) SJ1 stimulates BrdU incorporation in stromal cells. Stromal cells were cultured in the presence of sJ1 (sJ1)- or control-conditioned medium and pulsed for BrdU incorporation. After BrdU immunostaining, the total number of BrdU+ cells per well was scored. The histograms show mean ± SEM of the percentage of BrdU+ nuclei from at least three fields of ∼200 cells per field. **, P < 0.001.
The expression profiles of the Notch1 and Jagged1 proteins in differentiating cardiomyocytes and in the stromal cells were further analyzed by Western blotting. A polyclonal antiserum directed against the C terminus of Notch1 detected both the 300-kD, full-length protein and the p120 cleaved transmembrane Notch1 subunit in ICMs at day 1 of culture, whereas positivity progressively decreased at days 3 and 7 (Fig. 2 F). No specific signals were detected in stromal cells at days 1 and 3, although these became apparent at day 7. Consistent with the RT-PCR findings, the Jagged1 protein was not expressed in ICMs (day 1), though it progressively increased at days 3 and 7. In stromal cells, the levels of the protein remained relatively high at all time points (Fig. 2 F). Finally, the levels of activated Notch1 (ICD) paralleled the expression pattern of the full-length protein (Figs. 2 G and Fig. S1 A, available at http://www.jcb.org/cgi/content/full/jcb.200806091/DC1).
The cellular localization of Notch1 and Jagged1 proteins during cardiomyocytes differentiation was further studied by immunofluorescence (Fig. 2 H). Staining with an antibody that recognizes the C terminus of Notch confirmed the expression of the protein in ICMs (day 1) and its progressive disappearance afterward. Consistent with these findings, very strong immunoreactivity for activated Notch1-ICD (an antibody against Val1744) was found in small, round-shaped ICMs at day 1 (Figs. 2 H and Fig. S1 B), whereas differentiated cardiomyocytes at day 7 were almost completely negative. In agreement with the RT-PCR and Western blot analyses, and in contrast to cardiomyocytes, stromal cells were positive for both anti-Notch1 antibodies at days 3 and 7. The Notch ligand Jagged1 was not detectable in ICMs at day 1, whereas an increasingly stronger signal was detected at later times, as the cardiomyocyte acquired a fully differentiated and beating phenotype. Stromal cells showed a strong immunoreactivity for Jagged1 at all time points.
To further characterize the in vitro differentiation of neonatal ICMs, we analyzed the pattern of expression of Numb, an inhibitor of Notch signaling that interacts with the intracellular portion of Notch and prevents its nuclear translocation (Guo et al., 1996; Wakamatsu et al., 1999). At day 1, Numb expression was almost undetectable in ICMs, although it gradually increased at day 3 and 7, as detected by both transcript-specific PCR and Western blotting (Fig. 3, A and B, respectively).
Figure 3.
Numb expression is developmentally regulated in cardiomyocytes. (A and B) RT-PCR (A) and Western blot (B) analyses for Numb expression in cultured cardiomyocytes (CM). (C) Immunofluorescence staining for Numb along cardiomyocyte differentiation. Green, α-actinin; red, Numb; blue, DNA (DAPI). (D) Asymmetrical Numb localization in adjacent ICMs. The largest figure in each panel shows fluorescence in all three channels; the two smaller figures show the blue/green (top) and red/blue (bottom) channels. Green, α-actinin; red, Numb; blue, DNA. Bars, 10 μm.
The levels of Numb along cardiomyocyte differentiation were further analyzed by immunofluorescence staining using specific antibodies. The protein was expressed at very low levels at day 1, though its levels progressively increased at days 3 and 7 (Fig. 3 C). Interestingly, asymmetrical intracellular localization of Numb was detected in dividing cardiomyocytes at day 1 (Fig. 3 D), which suggests that Numb-mediated Notch down-regulation might participate in the loss of replicative potential and differentiation of ICMs.
Together, these results show that the loss of proliferative potential of isolated cardiac myocytes parallels the down-regulation of expression of the Notch1 receptor and the progressive expression of Jagged1. In contrast, Jagged1 is constantly expressed in supporting stromal cells, whereas the levels of Notch1 increase over time and parallel their increased proliferation rate.
Inhibition of Notch signaling blocks cardiomyocyte proliferation and induces apoptosis
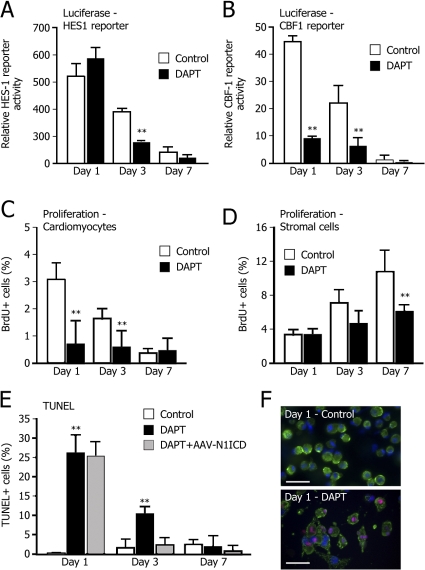
To further investigate the role of Notch1 activation in primary cardiomyocytes, cells at different days of culture were transfected with a luciferase construct under the control of two different Notch1-responsive promoters: the natural HES1 promoter (Jarriault et al., 1995) and an artificial promoter containing four repeats of the CBF1 binding sites (HES1 and CBF1 reporters, respectively; Hsieh et al., 1996). Parallel to the Notch1 transcript and protein levels, activation of the natural HES1 reporter was high at day 1, although it progressively decreased at later times (an approximately fivefold decrease in promoter activation between day 1 and 7; Fig. 4 A). Interestingly, this promoter turned out to be inhibited by the γ-secretase inhibitor only in differentiated cardiomyocytes at days 3 and 7, whereas it was totally unaffected by N-(N-[3,5-difluorophenacetyl]-L-alanyl)-S-phenylglycine t-butyl ester (DAPT) treatment in ICMs (Fig. 4 A). This observation prompts us to speculate that, in immature cardiomyocytes, the HES1 promoter might be a direct target of CREB activation, as demonstrated in other cell types (not further investigated here; Herzig et al., 2003). The CBF1 reporter showed an identical response at the various time points (>40 decrease in promoter activation between day 1 and day 7; Fig. 4 B); cell treatment with DAPT markedly suppressed promoter activation (a more than fivefold decrease at day 1), which is consistent with the conclusion that CBF1 reporter activation in cultured cardiomyocytes was strictly dependent on Notch1 activity.
Figure 4.
Inhibition of Notch signaling blocks cardiomyocyte proliferation and induces apoptosis. (A and B) DAPT treatment markedly suppresses both HES1- and CBF1-luciferase reporter activation. DMSO-containing buffer (Control) or DAPT in DMSO buffer (DAPT) was added to cardiac myocytes for 24 h after reporter gene transfection; the transactivation effect was evaluated at the indicated time points. The histogram shows mean values ± SEM, n = 6. **, P < 0.001. (C and D) DAPT blocks proliferation in cardiomyocytes and stromal cells. Purified cardiac myocytes and, in a mirror experiment, stromal cells were cultured in the presence of DAPT or DMSO-containing buffer (Control). After pulse-labeling with BrdU and specific immunostaining, the total number of BrdU+ cells per well was evaluated. The histograms show mean ± SEM of the percentage of BrdU+ nuclei from at least three views of ∼200 cells per view. Results are representative of at least three independent experiments. **, P < 0.001. (E) DAPT treatment markedly increases cardiomyocyte apoptosis through a specific Notch-dependent pathway. A TUNEL assay was performed on cardiomyocytes in the presence of DAPT or DMSO buffer (Control). Transduction with AAV8-N1ICD protected cardiac myocytes from apoptosis at days 3 and 7, although it was ineffective at day 1, which is consistent with the delayed kinetics of AAV transduction. The histograms show mean ± SEM of the percentage of BrdU+ nuclei from at least three views of ∼200 cells per view. Results are representative of at least three independent experiments. **, P < 0.001. (F) Representative images of TUNEL-positive cardiomyocytes (day 1 of culture) upon DAPT or DMSO buffer (Control) treatment. Green, α-actinin; red, TUNEL-positive nuclei; blue, DNA (DAPI). Bar, 50 μm.
To assess whether the block of Notch activation by γ-secretase inhibition might impair cardiomyocyte proliferation, the levels of BrdU incorporation at days 1, 3, and 7 of culture were analyzed. Treatment with DAPT determined a marked decrease in the number of BrdU-positive cardiomyocytes at days 1 and 3, respectively (Fig. 4 C).
When the same analysis was performed in cultures of stromal cells, DAPT was also found to significantly decrease the proliferative potential of these cells at day 7 only (Fig. 4 D), which is consistent with a role of Notch1 activation in sustaining also the proliferation of these cells at later time points. Cardiomyocyte treatment with DAPT was found to markedly increase the number of apoptotic cells at day 1 (from 3.6 to 26%), as assessed by the TUNEL assay (Fig. 4 E for quantification and Fig. 4 F for representative images). Of interest, the induction of apoptosis upon γ-secretase inhibition was progressively less profound at the later time points (9.3 and 3.6% of apoptotic cells at days 3 and 7, respectively, compared with 3.9 and 3.3% normally detected at the same time points in the absence of any treatment).
Gene transfer in primary cardiac myocytes can be efficiently achieved using viral vectors based on the adeno-associated virus serotype 8 (AAV8), as shown in Fig. S2 (available at http://www.jcb.org/cgi/content/full/jcb.200806091/DC1). The constitutive expression of the activated Notch1 ICD (N1-ICD), driven by AAV8, provided significant protection to cardiac myocytes from apoptosis (∼4 and ∼1.5% of apoptotic cells at days 3 and 7 after transduction, respectively, as compared with 9.22 and 3.3% apoptotic cells detected at the same time points in the presence of DAPT; Fig. 4 E). Of notice, the proapoptotic effect of DAPT was not rescued by AAV8-N1ICD transduction at day 1, a result that is consistent with the kinetics of transgene expression after AAV8 transduction, which reaches its maximum at 72 h after infection (Fig. S2). This finding is consistent with the biology of AAV transduction in a variety of other cell types in vitro and in vivo (Palomeque et al., 2007; Zincarelli et al., 2008).
Together, these results strongly indicate that Notch1 activation at the early time points not only triggers cell proliferation but is also required to maintain cell viability and prevent apoptosis.
Soluble Jagged1 enhances cardiomyocyte proliferation
The observation that the inhibition of the Notch pathway impairs cardiomyocytes proliferation suggested that, on the contrary, its stimulation might lead to their increased proliferation. To directly assess this possibility, we cultured freshly isolated neonatal cardiomyocytes in the presence of different sources of its Jagged1 ligand, including conditioned medium from NIH-3T3 cells secreting a soluble form of Jagged1 (sJ1; Wong et al., 2000) and recombinant fragment crystallizable (Fc)-fused soluble Jagged1 (rFc-sJ1) either preclustered on Sepharose beads (Vas et al., 2004) or immobilized on plates (Koyanagi et al., 2007). Immature cardiomyocytes at day 1, cultured in the presence of any of the three sources of sJ1, showed a marked up-regulation of the HES1 promoter (Fig. 5 A). This transactivation effect was still evident at day 3, although it became almost undetectable in differentiated cardiomyocytes at day 7. A conditioned medium containing the Delta1 ligand had no apparent effect when used under the same conditions (unpublished data). Of interest, the conditioned medium containing sJ1 was even more potent than the two forms of immobilized rFc-sJ1, which is consistent with the possibility that sJ1 might spontaneously cluster on the surface of the target cells by virtue of its interactions with various proteins of the extracellular matrix and cell surface, as already described for other soluble growth factors (for reviews see Taipale and Keski-Oja, 1997; Cool and Nurcombe, 2006; Guerrini et al., 2007). In a consistent manner, sJ1 was also able to activate transcription of the CBF1 promoter in cardiomyocytes at days 1 and 3 (but not at day 7) of culture (Fig. 5 B). The sensitivity to sJ1 stimulation correlated with the presence of the active Notch1 receptor in cardiac myocytes. Upon sJ1 stimulation, day-1 and, to a lesser extent, day-3 cardiomyocyte extracts showed markedly increased levels of N1-ICD, whereas DAPT treatment completely abolished the signal in sJ1-stimulated cardiac myocytes at any time point; no signal was detected using the Val 1744 antiserum in day-7 cardiac myocytes under any condition (Fig. S1 A).
Consistent with the transactivation results, the treatment of neonatal cardiomyocytes with either sJ1 or the two different types of immobilized rFc-sJ1 proteins determined a marked increase in the extent of BrdU incorporation. The number of BrdU-positive cells was ∼34% in the sJ1-treated sample versus 7% in the absence of treatment at day 1, and 11% versus 4.4% at day 3; BrdU incorporation was consistently increased upon treatment with immobilized rFc-sJ1 (quantification and representative images are shown in Fig. 5 (C and D, respectively). At day 7 of culture, when Notch1 was no longer expressed in cardiomyocytes, sJ1 addition had no apparent effect on BrdU incorporation. Remarkably, the activation of the Notch1 pathway by sJ1 lead to a significant increase in the total number of cardiomyocytes in culture, which, at day 7 of stimulation, was almost fivefold compared to the control (Fig. 5 E). The effect of Notch1 stimulation on cardiomyocyte proliferation was further strengthened by the observation that the forced expression of N1-ICD in cardiomyocytes by AAV8-N1ICD transduction stimulated BrdU incorporation in day-3 and, remarkably, also in day-7 cardiomyocytes (21.1% vs. 2.8% in AAV8-N1ICD vs. AAV-LacZ transduced cardiomyocytes; Fig. 5 F).
Finally, in contrast to cardiomyocytes, sJ1 did not affect BrdU incorporation in stromal cells at day 1, in keeping with the absence of the Notch1 receptor in these cells at this time point (Fig. 5 G). At days 3 and 7, when Notch1 became expressed, the number of cells incorporating BrdU started to increase, although at a level remarkably lower than in early cardiomyocytes (17.2% vs. 11% BrdU-positive stromal cells at day 7 in the presence or absence of sJ1, respectively).
Notch1 activation parallels cell cycle protein expression in immature cardiomyocytes
The levels of several proteins involved in cell cycle regulation were analyzed by immunoblotting in cardiomyocytes at different time points in culture and upon sJ1 stimulation. The arrest in proliferation and decrease in BrdU incorporation over time were found to correlate with the progressive increase of the cyclin-dependent kinase inhibitors (CKIs) p21Cip1 and p27Kip1, and of the Cdt1 inhibitor geminin (Fig. 6 A; Wohlschlegel et al., 2000). Consistent with the increase of these factors, the levels of cyclin D1, cyclin A, and cyclin E were significantly lower in cardiomyocytes at days 7 and 14 of culture. These observations reinforce similar findings of Campa et al. (2008) on the specific role of cyclin D1 regulation in the cardiomyocyte cell cycle. Remarkably, sJ1 stimulation reversed cell cycle exit at days 1 and 3, whereas it had no effect at later stages. Cardiomyocyte proliferation correlated with the presence of high levels of phosphorylated p38 MAPK; conversely, differentiated cells expressed Y705-phosphorylated p-STAT3, a known marker of terminal differentiation in other tissues (Androutsellis-Theotokis et al., 2006). The sensitivity to sJ1 stimulation was strictly dependent on the presence of the Notch1 receptor on cardiomyocytes. At days 1 and 3, both the Notch1 precursor p300 and the cleaved p120 and p110 forms were present, and sJ1 stimulation further increased their levels while inhibiting its own expression (evident at day 3 in Fig. 6 A). At days 7 and 14, p300 was undetectable, and its cleaved forms progressively disappeared. In contrast, expression of Jagged1 became detectable and was negatively regulated by sJ1 stimulation in a negative feedback loop. Collectively, these results support the conclusion that Notch activation powerfully stimulates immature cardiac myocyte proliferation and expansion during the early times of culture, whereas it progressively becomes ineffective along the differentiation process and is incapable on its own to promote reentry of differentiated cardiomyocytes into the cell cycle.
Figure 6.
Notch1 activation parallels cardiomyocyte proliferation. (A) sJ1 treatment activates cell cycle–related signaling. Cardiomyocytes were cultured in the presence of control (−)- or sJ1-conditioned medium (+) before immunoblotting for cell cycle–specific effectors. (B) Model of Notch1-dependent regulation of ICM proliferation and differentiation. See Discussion for details. Bars, 10 μm.
Discussion
In this study, we provide evidence that activation of the Notch1 pathway maintains proliferation and permits expansion of ex vivo cultured ICMs. We found that Notch1 is highly expressed by immature, still dividing, cardiac myocytes, whereas Jagged1 is mainly expressed on supporting stromal cells. The loss of proliferative potential by isolated myocytes parallels the down-regulation of the Notch1 receptor and the progressive expression of Jagged1. On the contrary, we show that the sustained, constitutive activation of Notch1 signaling directly correlates with an increase in ICM proliferation.
Our results indicate that the multidirectional Notch receptor/ligand signaling paradigm also extends to cardiac cells. According to this paradigm, it can be envisaged that the stromal microenvironment, expressing Jagged1, triggers an inductive signal to ICMs to expand their number, keeping them in a highly proliferative state. This is supported by our observation that activation of Notch signaling by both exogenous soluble ligand stimulation and intracellular AAV-mediated constitutive expression of N1-ICD enhance ICM proliferation, delay newly formed cardiomyocytes from rapid differentiation into mature beating cells (normally within 24 h from plating), and extend their lifespan in culture. However, Notch receptor activation does not revert newly formed cardiomyocytes back into precursor cells but keeps the ICM proliferating window open for a longer time, stimulating their increase in number.
The early embryonic lethality caused by cardiovascular defects of mice lacking genes of the Notch signaling cascade strongly supports a key function for Notch in the process of cardiac and vascular development (Loomes et al., 1999; Rones et al., 2000; Loomes et al., 2002; for reviews see High and Epstein, 2008; Niessen and Karsan, 2008). Indeed, Notch signaling has been found to exert a suppressive role during myocardial specification in several different species (Park et al., 1998; Rones et al., 2000; Han and Bodmer, 2003; Chau et al., 2006). Consistent with these findings, several in vitro studies have also implied a suppressive role of Notch on mesodermal commitment and cardiac differentiation. RBP-J–deficient embryonic stem cells show enhanced mesodermal and cardiogenic differentiation, which is likely attributable to the lack of Notch signaling (Schroeder et al., 2003; Nemir et al., 2006; Schroeder et al., 2006). In addition, constitutive activation of Notch in mesodermal progenitors blocks cardiomyocyte production in vitro (Schroeder et al., 2006), and forced Notch expression in mesodermal cardiac progenitor cells expressing Mesp1 during development prevents cardiogenesis and results in cardiac abnormalities in transgenic mice (Watanabe et al., 2006).
The contradiction between the findings postulating a suppressive role of Notch during early cardiac development and our results, which clearly show that Notch activation is essential to promote ICM proliferation, is only apparent. In fact, Notch signaling is known to be exquisitely sensitive to dosage, developmental timing, and cellular context (Artavanis-Tsakonas et al., 1999). It is therefore not surprising that, after mesodermal specification and cardiac commitment, the reactivation of Notch signaling is required to sustain the proliferation of the pool of ICMs still able to undergo mitosis.
Our results demonstrate the existence, both in vitro and in vivo in neonatal animals, of a pool of N1-ICD–positive cells that express early cardiac markers (GATA4 and Nkx2.5) and incorporate BrdU. Loss of the proliferative potential of these cells parallels the progressive disappearance of Notch1, which is consistent with the conclusion that Notch1 might primarily act by preventing terminal differentiation in this expanding proliferating compartment. On the basis of the knowledge currently available on Notch function and our results, we would like to propose the following model (Fig. 6 B). Although Notch1 is essential to sustain ICM proliferation, its levels progressively decrease in vitro parallel to cardiomyocyte differentiation. This determines a progressive withdrawal of differentiated cardiomyocytes from the cell cycle. By a positive feedback regulation, the Jagged1 ligand starts to be expressed on the surface of these cells. At this point, by a lateral induction mechanism, cardiac myocytes themselves behave as signal-sending cells to their stromal counterparts, the proliferation of which is also sustained at later stages of culture by activation of de novo expressed Notch1 itself. This model is also consistent with the observation that the levels of expression of the Notch antagonist Numb increase as long as the ICM proliferation diminishes, thus inversely correlating with the levels of Notch1. An asymmetrical localization of Numb has been described during embryogenesis, where it is involved in cell fate determination in different tissues (Rhyu et al., 1994; Carmena et al., 1998; Park et al., 1998; Artavanis-Tsakonas et al., 1999; Wakamatsu et al., 1999) and in satellite cell activation during skeletal muscle regeneration (Conboy and Rando, 2002). Moreover, high levels of Notch signaling lead to a reduction of Numb protein, revealing a reciprocal negative regulation between Notch and Numb (Chapman et al., 2006). Our results indicate that this regulation also occurs in mammalian cardiomyogenesis. Whether Numb expression might also play a role in vivo still remains to be determined. In this respect, however, it is intriguing to notice that Numb-positive progenitor cells have been found in the adult mouse heart, lying in defined interstitial structures reminiscent of stem cell niches (Urbanek et al., 2006).
ICM cells showed shortening of the G1 phase, showed low or undetectable levels of the p27Kip1 and p21Cip1 CKIs and of the Cdt1 inhibitor geminin, and expressed cyclins D1, A, and E, an overall profile that is characteristic of cells with a high replicative turnover. In addition, cell treatment with sJ1 further enhanced these features. In contrast, fully differentiated cardiomyocytes expressed abundant levels of CKIs, whereas the G1/S cyclins became almost undetectable. These features are perfectly in agreement with the observations presented in the accompanying paper by Campa et al. (2008), which shows that the forced activation of Notch2 by ICD gene transfer promotes reentry of cardiomyocytes into the cell cycle but does not bypass a checkpoint that blocks these cells in the G2/M phase.
Our experiments raise the possibility that the Notch pathway might also mediate the expansion of the pool of ICMs through protection from apoptotic cell death. Indeed, we observed that γ-secretase–mediated block of Notch signaling in ex vivo cultured ICMs leads to massive cell apoptosis and that this effect is prevented by N1-ICD transduction. In this respect, it is worth noting that the role of Notch signaling in the regulation of apoptosis in other mammalian systems is not univocal (Mason et al., 2000; Lutolf et al., 2002; Noseda et al., 2004) and might thus be dependent on the differentiation and proliferation state of the cells.
Collectively, in this work, we provide the first evidence that activation of the Notch pathway mediates the expansion of a pool of immature still-dividing cardiac myocytes by promoting their proliferation and protecting them from apoptosis. Our results also show that the in vitro treatment of neonatal cardiomyocyte cultures with an sJ1 agonist, although highly successful in expanding ICM proliferation at earlier stages, becomes totally ineffectual when Notch1 is no longer expressed. These observations are contrary to the possibility that treatment with sJ1 might reverse differentiation but prompt its use for the in vitro expansion of the pool of immature, still-proliferating cardiomyocytes. Thus, sJ1 might turn out to be a powerful growth factor able to expand the dividing cardiac precursor cell population when starting with various sources of cardiac stem cells.
Materials and methods
Constructs, cell lines and antibodies
The insertless pMEXneo and sJ1 constructs, together with stable NIH-3T3 transfectants, were provided by I. Prudovsky (Center for Molecular Medicine, Maine Medical Center Research Institute, Scarborough, ME) and cultured as described previously (Wong et al., 2000); the pCS2-ICV-6MT plasmid was provided by R. Kopan (Molecular Biology and Pharmacology, Washington University, St. Louis, MO); the Delta1Fc construct (Wang et al., 1998) was provided by G. Weinmaster (David Geffen School of Medicine at University of California, Los Angeles, Los Angeles, CA); wild-type (4xwtCBF1Luc) and mutant (4xmtCBF1luc) reporters were provided by S.D. Hayward (John Hopkins University School of Medicine, Baltimore, MD); and the pHes1-Luc reporter construct was provided by R. Kageyama (Institute for Virus Research, Kyoto University, Kyoto, Japan).
Anti-Notch-1 (C-20), anti-Jagged1 (H-114), anti-Delta (F15), anti–cyclin-A (C19), anti–cyclin-D1 (C20), anti–cyclin-E (C19), anti-p27Kip1 (C-19), and anti–GATA-4 antibodies were obtained from Santa Cruz Biotechnology, Inc.; anti–cleaved Notch-1 (Val1744), anti–phospho-p38 MAPK, and anti–phospho-STAT3 (Tyr705) were obtained from Cell Signaling Technology; antisarcomeric α-actinin (EA-53) and anti-p21Cip1 antibodies were obtained from AbCam; anti-Numb antibodies were obtained from Millipore; and FITC-coupled anti-BrdU antibodies were obtained from BD Biosciences. Anti-geminin antibodies were a gift from R. Laskey (Hutchinson/Medical Research Council Research Center, Cambridge, UK).
BrdU was obtained from Boehringer-Mannheim/Roche. Hoechst 33258, PI, RNase A, and all other chemicals were purchased from Sigma-Aldrich.
Production, purification, and characterization of rAAV vectors
rAAV vectors were prepared by the AAV Vector Unit at the International Centre for Genetic Engineering and Biotechnology Trieste (http://www.icgeb.org/RESEARCH/TS/COREFACILITIES/AVU.htm), as described previously (Arsic et al., 2004). In brief, infectious AAV8 vector particles were generated in HEK293 cells by cotransfecting each vector plasmid (pAAV-N1ICD and pAAV-LacZ) together with the packaging plasmid (pAAV8-2; Gao et al., 2002) and helper plasmid (pHELPER; Stratagene), expressing AAV and adenovirus helper functions, respectively. Viral stocks were obtained by CsCl2 gradient centrifugation; rAAV titers, determined by measuring the copy number of viral genomes in pooled, dialyzed gradient fractions, as described previously (Zentilin et al., 2001), were in the range of ∼1012 to ∼1013 genome copies per milliliter.
Animals
Animal care and treatment were conducted in conformity with institutional guidelines in compliance with national and international laws and policies (European Economic Community Council Directive 86/609, OJL 358, December 12, 1987). Wistar rats were purchased from Charles River Laboratories Italia Srl and maintained under controlled environmental conditions. When indicated, animals were treated for 2 d with 20 mg/kg of BrdU before killing.
Culture of neonatal rat cardiomyocytes
Neonatal rat ventricular myocytes were plated at low density in DME/4.5 g glucose containing 5% bovine calf serum and 2 mg/ml vitamin B12 and cultured as described previously (Deng et al., 2000). In brief, ventricles from 2–3 litters of neonatal Wistar rats were separated from the atria using scissors and then dissociated in CBFHH (calcium and bicarbonate-free Hanks with Hepes) buffer containing 2 mg/ml of trypsin and 20 μg/ml of DNase II. Cells were preplated for 2 h into 100-mm dishes in DME containing 5% defined bovine calf serum (HyClone); stromal cells were then separated from unattached myocytes that were plated at a density of 500 cells/mm2 into 60-mm dishes (for protein signaling, cytochemistry, RNA, and protein analysis; Falcon), multi-well plates (for biochemical assays; Costar) or multiwell slides coated with 0.2% gelatin (for immunofluorescence staining). For RT-PCR and Western blot analyses, three subsequent 1-h-long preplating steps were performed in order to strongly enrich the cell preparations in cardiomyocytes (∼90% purity). For the experiments involving cardiomyocyte transduction using AAV vectors, neonatal rat cardiomyocytes, immediately after plating onto 4-well slides coated with 0.2% gelatin at a density of 5 × 104 cells per well, were infected with 2 × 1010 genome copies per well. After 12 h, the culture medium was changed and cells were subjected to the different treatments and subsequent analyses.
RT-PCR analysis
Total mRNA was purified from cultured cardiomyocytes at 1, 3, or 7 d after plating. mRNA was reverse-transcribed using Superscript II (Invitrogen) with random hexamers (10 μM) as a primer in a 35-μl reaction containing 1× Superscript II RT buffer (Invitrogen), 10 μM dATP, dTTP, dCTP, and dGTP, and 20 U RNasin (Invitrogen). After 2 h at 42°C, the reaction was terminated by adding 365 μl of H2O and boiling for 2 min. For PCR amplification, specific primer pairs (0.5 μM each) were incubated with 50 ng cDNA and 1 U Herculase enhanced DNA polymerase (Stratagene) in a 20-μl reaction mixture that included 1× buffer and 100 μM dATP, dCTP, dGTP, and dTTP. Typical cycle parameters were 1 min at 92°C, 1 min at 59°C, and 3 min at 72°C for 20 cycles, followed by a cycle at 72°C for 7 min. The whole reaction was then fractionated on 1.2% agarose gel and the PCR product was visualized by ethidium bromide staining.
The primers for RT-PCR analysis were as follows: Notch1 forward primer, 5′-GCAGCCACAGAACTTACAAATCCAG-3′, and reverse primer, 5′-TAAATGCCTCTGGAATGTGGGTGAT-3′ (445-bp amplified fragment); Notch2 forward primer, 5′-GCTGTCCTCTTCATGCTGCA-3′, and reverse primer, 5′-AGCAGAAGTCAAGACAGTC-3′ (505 bp); Notch3 forward primer, 5′-TTCCAGATTCTCATCAGGAA-3′, and reverse primer, 5′-TCCTGGTGGGCAGAGCAATG-3′ (480 bp); Notch4 forward primer, 5′-CCTCAAGCAGGGACCGGGTC-3′, and reverse primer, 5′-AGGCAGAAGTGGATGAGGAC-3′ (280-bp amplified fragment); Jagged1 forward primer, 5′-CCACAAAGGGCACGGCGAGTG-3′, and reverse primer, 5′-TGCATGGGGTTTTTGATTTGG-3′ (668 bp); Jagged2 forward primer, 5′-ACATGCTATGACAGCGGCGA-3′, and reverse primer, 5′-CCAGGAACTCCCATGTGGGA-3′ (480 bp); Delta1 forward primer, 5′-CGACCTCGCAACAGAAAAC-3′, and reverse primer, 5′-ATGGAGACAGCCTGGGTATC-3′ (397 bp); HES1 forward primer, 5′-GGCAGGCGCACCCCGCCTTG-3′, and reverse primer, 5′-GCAGCCAGGCTGGAGAGGCT-3′ (170 bp); Hes2 forward primer, 5′-CTTCTGTTTTCGCTCTGCCTT-3′, and reverse primer, 5′-TCGAGGTAGCTATCCAAAGAC-3′ (352 bp); Hes3 forward primer, 5′-TCAGCATCTTTATCAGGCCTC-3′, and reverse primer, 5′-CTAGCCAAAGTCCTTGCAATG-3′ (302 bp); HES5 forward primer, 5′-ACCGCATCAACAGCAGCATT-3′, and reverse primer, 5′-AGGCTTTGCTGTGCTTCAGGT-3′ (135 bp); HEY1 forward primer, 5′-ACCCAATGGACTCCACACATC-3′, and reverse primer, 5′-TGTTCCACTGCTTGTCTGCTG-3′ (352 bp); HEY2 forward primer 5′-GAGATCTTGCAGATGACTGTG-3′, and reverse primer, 5′-TGTCGAAGGACTCAGTAGATG-3′ (178 bp); Numb forward primer, 5′-AAACCAGTGACATTAGTGGC-3′, and reverse primer, 5′-TGAGGCCTGCGGCTGCTGGG-3′ (326 bp); and GAPDH forward primer, 5′-ATTGTCAGCAATGCATCCTGCA-3′, and reverse primer, 5′-AGACAACCTGGTCCTCAGTGTA-3′ (320 bp). In all experiments, RT-PCR was performed on total embryo RNA as a positive control (+).
Western blotting
For protein analysis, samples were lysed in RIPA buffer (50 mM Tris-HCl, 150 mM NaCl, 5 mM EDTA, 5 mM EGTA, 0.5% NaDoc, 1% Triton X-100, and 0.1% SDS) containing 90 μg/ml PMSF, 1 mM NaVO4, 20 μg/ml aprotinin, and 20 μg/ml leupeptin (Sigma-Aldrich). Protein concentration was determined by the Bradford method (Bio-Rad Laboratories); equal amounts of protein were resolved on SDS-PAGE minigels and transferred to nitrocellulose membranes (GE Healthcare). Immunoblots were blocked in 5% nonfat dry milk in TBST (50 mM Tris-HCl, pH 7.4, 200 mM NaCl, and 0.1% Tween 20). Membranes were incubated with primary antibodies overnight and washed in Tris-buffered saline, 0.1% Tween 20. Secondary antibodies were diluted in blocking buffer and incubated with the membranes for 45 min at room temperature. Proteins were detected by enhanced chemiluminescence (GE Healthcare).
Luciferase assays
To detect Notch1 activity under the different experimental conditions, neonatal cardiomyocytes were seeded onto 0.2% gelatin-coated 24-well culture plates (5 × 104 cells per well) and cotransfected at the stated time points with 0.2 μg of reporter plasmid and 0.01 μg pRL-Renilla (which was used as control) by using FuGENE6 transfection reagent (Roche). The pHes1-Luc reporter construct, wild-type (4xwtCBF1Luc), and mutant (4xmtCBF1luc) CBF1 reporters (Hsieh et al., 1996) were cotransfected with the control plasmid pRL-CMV (Promega). In some cases, a plasmid expressing Notch1-ICD was included as a positive control. After incubation for 6 h, the medium was removed and the cells were fed in serum-free medium with either sJ1 or pMexNeo concentrated supernatants and either hIgG or recombinant Fc-sJ1 (R&D Systems) preclustered onto Sepharose beads (Vas et al., 2004). In a parallel experiment, neonatal cardiomyocytes were seeded onto 24-well culture plates (5 × 104 cells per well) precoated with recombinant Fc-sJ1 or normal h-IgG as a control (5 μg/ml in 0.05 M carbonate-bicarbonate buffer, pH 9.6). 24 h after treatment, the cell monolayers were washed with PBS, harvested by scraping, and resuspended in PLB buffer (Promega). Firefly and Renilla luciferase activities were assayed with the Dual-luciferase assay kit (Promega). Firefly luciferase activity was corrected for the transfection efficiency by using the control Renilla luciferase activity in each sample.
Immunocytochemical and immunohistochemical staining
The primary antibodies used were as follows: rabbit polyclonal anti-cleaved N1 (Val1744), 1:100 (Cell Signaling Technology); both rabbit and goat polyclonal anti-Notch1 (C-20), 1:100 (Santa Cruz Biotechnology, Inc.); rabbit polyclonal anti-Jagged1 (H-114), 1:100 (Santa Cruz Biotechnology, Inc.); mouse anti–α-sarcomeric actinin (EA-53), 1:100 (AbCam); rat monoclonal (BU1/75) anti-BrdU, 1:100 (AbCam); FastImmune anti-BrdU monoclonal antibodies with Dnase (Becton Dickinson); and mouse monoclonal anti-GATA4 (G-4), 1:100 (Santa Cruz Biotechnology, Inc.). The secondary antibodies used were as follows: goat anti–mouse conjugated to Alexa Fluor 488; goat anti–rabbit conjugated to Alexa Fluor 594; and donkey anti–rat conjugated to Alexa Fluor 488, all at 1:1,000 (Invitrogen). Cells were fixed in PBS containing 3% PFA and 2% sucrose for 10 min; aldehydes were quenched with 0.1 M glycine in PBS for 15 min at room temperature. Cells were permeabilized with 0.1% Triton X-100 for 5 min, blocked in 2% BSA and 0.05% sodium azide in PBS (blocking buffer) for 1 h, and incubated with primary antibodies from 2 h to overnight. Alexa Fluor 488– or Alexa Fluor 594–conjugated secondary antibodies (Invitrogen) were incubated for 30 min to 1 h at room temperature. All washes were in PBS 0.2% Tween 20. Cell nuclei were stained with DAPI, and samples were mounted in Vectashield (Vector Laboratories) with ProLong Gold (Invitrogen). For histological analysis, frozen sections (5-μm-thick) were fixed in cold acetone (−20°C) and blocked for 30 min with 5% goat or horse serum in PBS, depending on the secondary antibody. Primary antibodies were used diluted 1:100 in blocking buffer (2% BSA and 0.05% sodium azide in PBS). Secondary antibodies used were Alexa Fluor 488– or Alexa Fluor 594–conjugated antibodies (Invitrogen). Nuclei were counterstained with DAPI. Images were acquired at room temperature with a DMLB upright fluorescence microscope (Leica) equipped with a charge-coupled device camera (CoolSNAP CF; Roper Scientific) using MetaView 4.6 quantitative analysis software (MDS Analytical Technologies). For image acquisition, the following objectives were used: HCX PL Fluotar 100×/1.30 NA, HCX PL apocromatic 63×/1.32-0.6 NA, HCX PL Fluotar 40×/0.75 NA, HCX PL N-Plan 20×/0.40 NA, and HCX PL N-Plan 10×/0.25 NA (all from Leica). Within each experiment, instrument settings were kept constant. For all image quantification, data collected from three independent experiments were used to calculate the mean, SEM, and p-value using the Student's t test for unpaired samples.
To stain for AAV8-LacZ expression, transduced neonatal rat cardiomyocytes were fixed in PBS containing 2% formaldehyde and 0.2% glutaraldehyde at 4°C for 5 min after 24, 48, and 72 h of culture. After washing with PBS three times, sections were stained with 1 mg/ml X-gal in staining solution (40 mM sodium phosphate [dibasic], 40 mM citric acid, 150 mM NaCl, 2 mM MgCl2, 5 mM potassium ferricyanide, and 5 mM potassium ferricyanide) at 37°C overnight. Cyanide salts and Xgal (5-bromo-4-chloro-3-indolyl-β-d-galactoside; Thermo Fisher Scientific) were added from freshly made stocks in PBS and dimethylformamide, respectively. Cells were then washed three times with PBS for 5 min each at room temperature before microscopic examination and photography.
Treatment with sJ1
Conditioned media were prepared from subconfluent pCDNA3.1-SJ1 and pMexNeo NIH-3T3 stable transfectants grown for 3 d in 15-cm dishes in the presence of 2% FCS DME. Media were centrifuged at low speed to remove cellular debris and concentrated 20-fold with a Centriplus-30 filter (Amicon; Millipore) at 4°C. For the different assays, rat cardiomyocytes were fed in serum-free medium supplemented with 10× diluted sJ1 or pMexNeo concentrated supernatants, then subjected to analysis at the indicated time points.
Treatment with γ-secretase inhibitors
Neonatal rat cardiomyocytes (2.0 × 106) were plated either on 0.2% gelatin-coated microscope slices (for immunofluorescence analysis) or on multiwell plates (for biochemical assays; Costar). To check γ-secretase inhibitors effect on BrdU incorporation, 10–35 μM DAPT was added to cell cultures and an equal amount of DMSO as a control. After 22 h, cardiomyocytes were pulse-labeled with Brdu, and specific staining was performed as described in “BrdU pulse labeling and detection.” To check DAPT-induced apoptosis, 10–35 μM DAPT and an equal amount of DMSO as a control were added to neonatal rat cardiomyocytes plated onto 0.2% gelatin-coated glass chamber slides. After 22 h of culture, cardiomyocytes were fixed and a TUNEL assay was performed as described in “TUNEL staining.” To check the γ-secretase inhibitor's effect on cleaved Notch-1 intracellular detection, 10–35 μM DAPT was added to cell cultures for 22 h; an equal amount of DMSO was added as a control. For luciferase assays, 10–35 μM DAPT was added to cell cultures 6 h after the reporter's transfection and luciferase activity was checked 24 h later, as described in “Luciferase assays.”
PI staining
For PI staining, cells were washed in 1× PBS at room temperature, fixed in 1 ml of cold 70% ethanol, and centrifuged (1,500 g for 10 min). Cell pellets were resuspended in 0.5 ml of 1× PBS containing 0.2 mg/ml RNase-A (Roche) and incubated at 37°C for 30 min, after which 0.5 ml of 80 mg/ml PI (Sigma-Aldrich) was added. Cells were incubated with PI for at least 10 min at 4°C before analysis. PI incorporation was evaluated by flow cytometry using a FACSCalibur flow cytometer (Becton Dickinson) and BD CellQuest Pro software (Becton Dickinson).
BrdU pulse labeling and detection
After pulse-labeling of cardiomyocyte with BrdU (10 μM for 40 min), BrdU staining was performed according to the manufacturer's instructions (BD Biosciences). In brief, cultures were fixed in 3% PFA and 2% sucrose in PBS, pH 7.6; washed three times for 10 min with PBS 1% Triton X-100; and incubated with 1 M HCl for 10 min on ice, 2 M HCl for 10 min at room temperature, and with 2 M HCl for 20 min at 37°C. After DNA denaturation, cells were incubated with 0.1 M sodium-borate buffer, pH 8.4, for 12 min at room temperature, then washed three times with PBS 1% Triton X-100, blocked with 2% BSA in PBS 1 h at room temperature, and incubated with anti-BrdU antibodies O/N at 4°C. All subsequent steps were as described in “Immunocytochemical and immunohistochemical staining.” Each slide was then analyzed by counting, in a blind fashion, the number of BrdU-positive and BrdU-negative cells over 10 high-magnification fields for each condition. BrdU detection on some hystological sections was performed using the FastImmune anti-BrdU monoclonal antibodies with Dnase I, according to the manufacturer's instructions (Becton Dickinson).
TUNEL staining
Freshly isolated cardiomyocytes were cultured on glass chamber slides coated with 0.2% gelatin. At stated time points, 10–35 μM DAPT was added to cell cultures; 24 h later, slides were fixed as described in “Immunocytochemical and immunohistochemical staining” and then stained for DNA fragmentation characteristic of apoptosis by the TUNEL reaction, as described previously (Iredale et al., 1998), with the modifications described to reduce false positivity (Stahelin et al., 1998). Each slide was then analyzed by counting, in a blind fashion, the number of TUNEL-positive and TUNEL-negative cells over 10 high-magnification fields for each condition.
Online supplemental material
Fig. S1 shows the detection of activated Notch1 upon either γ-secretase or sJ1 treatments during cardiomyocyte differentiation. Fig. S2 shows the time course of AAV transduction of in vitro cultured cardiomyocytes. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200806091/DC1.
Supplementary Material
Acknowledgments
The authors are grateful to C. Long for his precious suggestions and helpful discussion; to Giulia Felician, Marina Dapas, and Michela Zotti for their outstanding technical support; to Mauro Sturnega for his invaluable help in animal experimentation; and to Suzanne Kerbavcic for excellent editorial assistance.
This work was supported by a grant from the “Fondazione CR Trieste” of Trieste, Italy.
Abbreviations used in this paper: AAV, adeno-associated virus; CKI, cyclin-dependent kinase inhibitor; DAPT, N-(N-[3,5-difluorophenacetyl]-L-alanyl)-S-phenylglycine t-butyl ester; Fc, fragment crystallizable; HES, hairy and enhancer of split; ICD, intracellular domain; ICM, immature cardiac myocyte; PI, propidium iodide; rFc-sJ1, recombinant Fc-fused soluble Jagged1; sJ1, soluble Jagged1.
References
- Androutsellis-Theotokis, A., R.R. Leker, F. Soldner, D.J. Hoeppner, R. Ravin, S.W. Poser, M.A. Rueger, S.K. Bae, R. Kittappa, and R.D. McKay. 2006. Notch signaling regulates stem cell numbers in vitro and in vivo. Nature. 442:823–826. [DOI] [PubMed] [Google Scholar]
- Arsic, N., S. Zacchigna, L. Zentilin, G. Ramirez-Correa, L. Pattarini, A. Salvi, G. Sinagra, and M. Giacca. 2004. Vascular endothelial growth factor stimulates skeletal muscle regeneration in vivo. Mol. Ther. 10:844–854. [DOI] [PubMed] [Google Scholar]
- Artavanis-Tsakonas, S., M.D. Rand, and R.J. Lake. 1999. Notch signaling: cell fate control and signal integration in development. Science. 284:770–776. [DOI] [PubMed] [Google Scholar]
- Beis, D., T. Bartman, S.W. Jin, I.C. Scott, L.A. D'Amico, E.A. Ober, H. Verkade, J. Frantsve, H.A. Field, A. Wehman, et al. 2005. Genetic and cellular analyses of zebrafish atrioventricular cushion and valve development. Development. 132:4193–4204. [DOI] [PubMed] [Google Scholar]
- Beltrami, A.P., L. Barlucchi, D. Torella, M. Baker, F. Limana, S. Chimenti, H. Kasahara, M. Rota, E. Musso, K. Urbanek, et al. 2003. Adult cardiac stem cells are multipotent and support myocardial regeneration. Cell. 114:763–776. [DOI] [PubMed] [Google Scholar]
- Bray, S.J. 2006. Notch signalling: a simple pathway becomes complex. Nat. Rev. Mol. Cell Biol. 7:678–689. [DOI] [PubMed] [Google Scholar]
- Campa, V.C.S.C.E.C.B.M., R. Gutiérrez-Lanza, F. Cerignoli, R. Díaz-Trelles, B. Nelson, T. Tsuji, M. Barcova, W. Jiang, and M. Mercola. 2008. Notch activates cell cycle reentry and progression in quiescent cardiomyocytes. J. Cell Biol. 182:129–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmena, A., S. Gisselbrecht, J. Harrison, F. Jimenez, and A.M. Michelson. 1998. Combinatorial signaling codes for the progressive determination of cell fates in the Drosophila embryonic mesoderm. Genes Dev. 12:3910–3922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman, G., L. Liu, C. Sahlgren, C. Dahlqvist, and U. Lendahl. 2006. High levels of Notch signaling down-regulate Numb and Numblike. J. Cell Biol. 175:535–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chau, M.D., R. Tuft, K. Fogarty, and Z.Z. Bao. 2006. Notch signaling plays a key role in cardiac cell differentiation. Mech. Dev. 123:626–640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, X., R.M. Wilson, H. Kubo, R.M. Berretta, D.M. Harris, X. Zhang, N. Jaleel, S.M. Macdonnell, C. Bearzi, J. Tillmanns, et al. 2007. Adolescent feline heart contains a population of small proliferative ventricular myocytes with immature physiological properties. Circ. Res. 100:536–544. [DOI] [PubMed] [Google Scholar]
- Chiba, S. 2006. Notch signaling in stem cell systems. Stem Cells. 24:2437–2447. [DOI] [PubMed] [Google Scholar]
- Conboy, I.M., and T.A. Rando. 2002. The regulation of Notch signaling controls satellite cell activation and cell fate determination in postnatal myogenesis. Dev. Cell. 3:397–409. [DOI] [PubMed] [Google Scholar]
- Cool, S.M., and V. Nurcombe. 2006. Heparan sulfate regulation of progenitor cell fate. J. Cell. Biochem. 99:1040–1051. [DOI] [PubMed] [Google Scholar]
- Del Monte, G., J. Grego-Bessa, A. Gonzalez-Rajal, V. Bolos, and J.L. De La Pompa. 2007. Monitoring Notch1 activity in development: Evidence for a feedback regulatory loop. Dev. Dyn. 236:2594–2614. [DOI] [PubMed] [Google Scholar]
- Deng, X.F., D.G. Rokosh, and P.C. Simpson. 2000. Autonomous and growth factor induced hypertrophy in cultured neonatal mouse cardiac myocytes. Comparison with rat. Circ. Res. 87:781–788. [DOI] [PubMed] [Google Scholar]
- De Strooper, B., W. Annaert, P. Cupers, P. Saftig, K. Craessaerts, J.S. Mumm, E.H. Schroeter, V. Schrijvers, M.S. Wolfe, W.J. Ray, et al. 1999. A presenilin-1-dependent gamma-secretase-like protease mediates release of Notch intracellular domain. Nature. 398:518–522. [DOI] [PubMed] [Google Scholar]
- Gao, G.P., M.R. Alvira, L. Wang, R. Calcedo, J. Johnston, and J.M. Wilson. 2002. Novel adeno-associated viruses from rhesus monkeys as vectors for human gene therapy. Proc. Natl. Acad. Sci. USA. 99:11854–11859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garg, V., A.N. Muth, J.F. Ransom, M.K. Schluterman, R. Barnes, I.N. King, P.D. Grossfeld, and D. Srivastava. 2005. Mutations in NOTCH1 cause aortic valve disease. Nature. 437:270–274. [DOI] [PubMed] [Google Scholar]
- Grego-Bessa, J., L. Luna-Zurita, G. del Monte, V. Bolos, P. Melgar, A. Arandilla, A.N. Garratt, H. Zang, Y.S. Mukouyama, H. Chen, et al. 2007. Notch signaling is essential for ventricular chamber development. Dev. Cell. 12:415–429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gridley, T. 1997. Notch signaling in vertebrate development and disease. Mol. Cell. Neurosci. 9:103–108. [DOI] [PubMed] [Google Scholar]
- Gridley, T. 2003. Notch signaling and inherited disease syndromes. Hum Mol Genet. 12 Spec No 1:R9–13. [DOI] [PubMed] [Google Scholar]
- Guerrini, M., M. Hricovini, and G. Torri. 2007. Interaction of heparins with fibroblast growth factors: conformational aspects. Curr. Pharm. Des. 13:2045–2056. [DOI] [PubMed] [Google Scholar]
- Guo, M., L.Y. Jan, and Y.N. Jan. 1996. Control of daughter cell fates during asymmetric division: interaction of Numb and Notch. Neuron. 17:27–41. [DOI] [PubMed] [Google Scholar]
- Han, Z., and R. Bodmer. 2003. Myogenic cells fates are antagonized by Notch only in asymmetric lineages of the Drosophila heart, with or without cell division. Development. 130:3039–3051. [DOI] [PubMed] [Google Scholar]
- Herzig, S., S. Hedrick, I. Morantte, S.H. Koo, F. Galimi, and M. Montminy. 2003. CREB controls hepatic lipid metabolism through nuclear hormone receptor PPARgamma. Nature. 426:190–193. [DOI] [PubMed] [Google Scholar]
- Hierlihy, A.M., P. Seale, C.G. Lobe, M.A. Rudnicki, and L.A. Megeney. 2002. The postnatal heart contains a myocardial stem cell population. FEBS Lett. 530:239–243. [DOI] [PubMed] [Google Scholar]
- High, F.A., and J.A. Epstein. 2008. The multifaceted role of Notch in cardiac development and disease. Nat. Rev. Genet. 9:49–61. [DOI] [PubMed] [Google Scholar]
- Hsieh, J.J., T. Henkel, P. Salmon, E. Robey, M.G. Peterson, and S.D. Hayward. 1996. Truncated mammalian Notch1 activates CBF1/RBPJk-repressed genes by a mechanism resembling that of Epstein-Barr virus EBNA2. Mol. Cell. Biol. 16:952–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurlbut, G.D., M.W. Kankel, R.J. Lake, and S. Artavanis-Tsakonas. 2007. Crossing paths with Notch in the hyper-network. Curr. Opin. Cell Biol. 19:166–175. [DOI] [PubMed] [Google Scholar]
- Iredale, J.P., R.C. Benyon, J. Pickering, M. McCullen, M. Northrop, S. Pawley, C. Hovell, and M.J. Arthur. 1998. Mechanisms of spontaneous resolution of rat liver fibrosis. Hepatic stellate cell apoptosis and reduced hepatic expression of metalloproteinase inhibitors. J. Clin. Invest. 102:538–549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iso, T., L. Kedes, and Y. Hamamori. 2003. HES and HERP families: multiple effectors of the Notch signaling pathway. J. Cell. Physiol. 194:237–255. [DOI] [PubMed] [Google Scholar]
- Jarriault, S., C. Brou, F. Logeat, E.H. Schroeter, R. Kopan, and A. Israel. 1995. Signalling downstream of activated mammalian Notch. Nature. 377:355–358. [DOI] [PubMed] [Google Scholar]
- Kajstura, J., B. Pertoldi, A. Leri, C.A. Beltrami, A. Deptala, Z. Darzynkiewicz, and P. Anversa. 2000. Telomere shortening is an in vivo marker of myocyte replication and aging. Am. J. Pathol. 156:813–819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang, M.J., and G.Y. Koh. 1997. Differential and dramatic changes of cyclin-dependent kinase activities in cardiomyocytes during the neonatal period. J. Mol. Cell. Cardiol. 29:1767–1777. [DOI] [PubMed] [Google Scholar]
- Koyanagi, M., P. Bushoven, M. Iwasaki, C. Urbich, A.M. Zeiher, and S. Dimmeler. 2007. Notch signaling contributes to the expression of cardiac markers in human circulating progenitor cells. Circ. Res. 101:1139–1145. [DOI] [PubMed] [Google Scholar]
- Laugwitz, K.L., A. Moretti, J. Lam, P. Gruber, Y. Chen, S. Woodard, L.Z. Lin, C.L. Cai, M.M. Lu, M. Reth, et al. 2005. Postnatal isl1+ cardioblasts enter fully differentiated cardiomyocyte lineages. Nature. 433:647–653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, L., I.D. Krantz, Y. Deng, A. Genin, A.B. Banta, C.C. Collins, M. Qi, B.J. Trask, W.L. Kuo, J. Cochran, et al. 1997. Alagille syndrome is caused by mutations in human Jagged1, which encodes a ligand for Notch1. Nat. Genet. 16:243–251. [DOI] [PubMed] [Google Scholar]
- Loomes, K.M., L.A. Underkoffler, J. Morabito, S. Gottlieb, D.A. Piccoli, N.B. Spinner, H.S. Baldwin, and R.J. Oakey. 1999. The expression of Jagged1 in the developing mammalian heart correlates with cardiovascular disease in Alagille syndrome. Hum. Mol. Genet. 8:2443–2449. [DOI] [PubMed] [Google Scholar]
- Loomes, K.M., D.B. Taichman, C.L. Glover, P.T. Williams, J.E. Markowitz, D.A. Piccoli, H.S. Baldwin, and R.J. Oakey. 2002. Characterization of Notch receptor expression in the developing mammalian heart and liver. Am. J. Med. Genet. 112:181–189. [DOI] [PubMed] [Google Scholar]
- Lutolf, S., F. Radtke, M. Aguet, U. Suter, and V. Taylor. 2002. Notch1 is required for neuronal and glial differentiation in the cerebellum. Development. 129:373–385. [DOI] [PubMed] [Google Scholar]
- Mason, J.L., J.J. Jones, M. Taniike, P. Morell, K. Suzuki, and G.K. Matsushima. 2000. Mature oligodendrocyte apoptosis precedes IGF-1 production and oligodendrocyte progenitor accumulation and differentiation during demyelination/remyelination. J. Neurosci. Res. 61:251–262. [DOI] [PubMed] [Google Scholar]
- Matsuura, K., T. Nagai, N. Nishigaki, T. Oyama, J. Nishi, H. Wada, M. Sano, H. Toko, H. Akazawa, T. Sato, et al. 2004. Adult cardiac Sca-1-positive cells differentiate into beating cardiomyocytes. J. Biol. Chem. 279:11384–11391. [DOI] [PubMed] [Google Scholar]
- McCright, B., J. Lozier, and T. Gridley. 2002. A mouse model of Alagille syndrome: Notch2 as a genetic modifier of Jag1 haploinsufficiency. Development. 129:1075–1082. [DOI] [PubMed] [Google Scholar]
- McElhinney, D.B., I.D. Krantz, L. Bason, D.A. Piccoli, K.M. Emerick, N.B. Spinner, and E. Goldmuntz. 2002. Analysis of cardiovascular phenotype and genotype-phenotype correlation in individuals with a JAG1 mutation and/or Alagille syndrome. Circulation. 106:2567–2574. [DOI] [PubMed] [Google Scholar]
- Messina, E., L. De Angelis, G. Frati, S. Morrone, S. Chimenti, F. Fiordaliso, M. Salio, M. Battaglia, M.V. Latronico, M. Coletta, et al. 2004. Isolation and expansion of adult cardiac stem cells from human and murine heart. Circ. Res. 95:911–921. [DOI] [PubMed] [Google Scholar]
- Nemir, M., and T. Pedrazzini. 2008. Functional role of Notch signaling in the developing and postnatal heart. J. Mol. Cell. Cardiol. In press. [DOI] [PubMed]
- Nemir, M., A. Croquelois, T. Pedrazzini, and F. Radtke. 2006. Induction of cardiogenesis in embryonic stem cells via downregulation of Notch1 signaling. Circ. Res. 98:1471–1478. [DOI] [PubMed] [Google Scholar]
- Niessen, K., and A. Karsan. 2008. Notch signaling in cardiac development. Circ. Res. 102:1169–1181. [DOI] [PubMed] [Google Scholar]
- Noseda, M., L. Chang, G. McLean, J.E. Grim, B.E. Clurman, L.L. Smith, and A. Karsan. 2004. Notch activation induces endothelial cell cycle arrest and participates in contact inhibition: role of p21Cip1 repression. Mol. Cell. Biol. 24:8813–8822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oda, T., A.G. Elkahloun, B.L. Pike, K. Okajima, I.D. Krantz, A. Genin, D.A. Piccoli, P.S. Meltzer, N.B. Spinner, F.S. Collins, and S.C. Chandrasekharappa. 1997. Mutations in the human Jagged1 gene are responsible for Alagille syndrome. Nat. Genet. 16:235–242. [DOI] [PubMed] [Google Scholar]
- Oh, H., S.B. Bradfute, T.D. Gallardo, T. Nakamura, V. Gaussin, Y. Mishina, J. Pocius, L.H. Michael, R.R. Behringer, D.J. Garry, et al. 2003. Cardiac progenitor cells from adult myocardium: homing, differentiation, and fusion after infarction. Proc. Natl. Acad. Sci. USA. 100:12313–12318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palomeque, J., E.R. Chemaly, P. Colosi, J.A. Wellman, S. Zhou, F. Del Monte, and R.J. Hajjar. 2007. Efficiency of eight different AAV serotypes in transducing rat myocardium in vivo. Gene Ther. 14:989–997. [DOI] [PubMed] [Google Scholar]
- Park, M., L.E. Yaich, and R. Bodmer. 1998. Mesodermal cell fate decisions in Drosophila are under the control of the lineage genes numb, Notch, and sanpodo. Mech. Dev. 75:117–126. [DOI] [PubMed] [Google Scholar]
- Rhyu, M.S., L.Y. Jan, and Y.N. Jan. 1994. Asymmetric distribution of numb protein during division of the sensory organ precursor cell confers distinct fates to daughter cells. Cell. 76:477–491. [DOI] [PubMed] [Google Scholar]
- Rones, M.S., K.A. McLaughlin, M. Raffin, and M. Mercola. 2000. Serrate and Notch specify cell fates in the heart field by suppressing cardiomyogenesis. Development. 127:3865–3876. [DOI] [PubMed] [Google Scholar]
- Rutenberg, J.B., A. Fischer, H. Jia, M. Gessler, T.P. Zhong, and M. Mercola. 2006. Developmental patterning of the cardiac atrioventricular canal by Notch and Hairy-related transcription factors. Development. 133:4381–4390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder, T., S.T. Fraser, M. Ogawa, S. Nishikawa, C. Oka, G.W. Bornkamm, S. Nishikawa, T. Honjo, and U. Just. 2003. Recombination signal sequence-binding protein Jkappa alters mesodermal cell fate decisions by suppressing cardiomyogenesis. Proc. Natl. Acad. Sci. USA. 100:4018–4023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder, T., F. Meier-Stiegen, R. Schwanbeck, H. Eilken, S. Nishikawa, R. Hasler, S. Schreiber, G.W. Bornkamm, S. Nishikawa, and U. Just. 2006. Activated Notch1 alters differentiation of embryonic stem cells into mesodermal cell lineages at multiple stages of development. Mech. Dev. 123:570–579. [DOI] [PubMed] [Google Scholar]
- Shawber, C.J., and J. Kitajewski. 2004. Notch function in the vasculature: insights from zebrafish, mouse and man. Bioessays. 26:225–234. [DOI] [PubMed] [Google Scholar]
- Stahelin, B.J., U. Marti, M. Solioz, H. Zimmermann, and J. Reichen. 1998. False positive staining in the TUNEL assay to detect apoptosis in liver and intestine is caused by endogenous nucleases and inhibited by diethyl pyrocarbonate. Mol. Pathol. 51:204–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taipale, J., and J. Keski-Oja. 1997. Growth factors in the extracellular matrix. FASEB J. 11:51–59. [DOI] [PubMed] [Google Scholar]
- Timmerman, L.A., J. Grego-Bessa, A. Raya, E. Bertran, J.M. Perez-Pomares, J. Diez, S. Aranda, S. Palomo, F. McCormick, J.C. Izpisua-Belmonte, and J.L. de la Pompa. 2004. Notch promotes epithelial-mesenchymal transition during cardiac development and oncogenic transformation. Genes Dev. 18:99–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urbanek, K., D. Cesselli, M. Rota, A. Nascimbene, A. De Angelis, T. Hosoda, C. Bearzi, A. Boni, R. Bolli, J. Kajstura, et al. 2006. Stem cell niches in the adult mouse heart. Proc. Natl. Acad. Sci. USA. 103:9226–9231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vas, V., L. Szilagyi, K. Paloczi, and F. Uher. 2004. Soluble Jagged-1 is able to inhibit the function of its multivalent form to induce hematopoietic stem cell self-renewal in a surrogate in vitro assay. J. Leukoc. Biol. 75:714–720. [DOI] [PubMed] [Google Scholar]
- Wakamatsu, Y., T.M. Maynard, S.U. Jones, and J.A. Weston. 1999. NUMB localizes in the basal cortex of mitotic avian neuroepithelial cells and modulates neuronal differentiation by binding to NOTCH-1. Neuron. 23:71–81. [DOI] [PubMed] [Google Scholar]
- Wang, S., A.D. Sdrulla, G. diSibio, G. Bush, D. Nofziger, C. Hicks, G. Weinmaster, and B.A. Barres. 1998. Notch receptor activation inhibits oligodendrocyte differentiation. Neuron. 21:63–75. [DOI] [PubMed] [Google Scholar]
- Watanabe, Y., H. Kokubo, S. Miyagawa-Tomita, M. Endo, K. Igarashi, K. Aisaki, J. Kanno, and Y. Saga. 2006. Activation of Notch1 signaling in cardiogenic mesoderm induces abnormal heart morphogenesis in mouse. Development. 133:1625–1634. [DOI] [PubMed] [Google Scholar]
- Wohlschlegel, J.A., B.T. Dwyer, S.K. Dhar, C. Cvetic, J.C. Walter, and A. Dutta. 2000. Inhibition of eukaryotic DNA replication by geminin binding to Cdt1. Science. 290:2309–2312. [DOI] [PubMed] [Google Scholar]
- Wong, M.K., I. Prudovsky, C. Vary, C. Booth, L. Liaw, S. Mousa, D. Small, and T. Maciag. 2000. A non-transmembrane form of Jagged-1 regulates the formation of matrix-dependent chord-like structures. Biochem. Biophys. Res. Commun. 268:853–859. [DOI] [PubMed] [Google Scholar]
- Zentilin, L., A. Marcello, and M. Giacca. 2001. Involvement of cellular double-strand DNA break-binding proteins in processing of recombinant adeno-associated virus (AAV) genome. J. Virol. 75:12279–12287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zincarelli, C., S. Soltys, G. Rengo, and J.E. Rabinowitz. 2008. Analysis of AAV serotypes 1-9 mediated gene expression and tropism in mice after systemic injection. Mol. Ther. 16:1073–1080. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.