Abstract
The inability of heart muscle to regenerate by replication of existing cardiomyocytes has engendered considerable interest in identifying developmental or other stimuli capable of sustaining the proliferative capacity of immature cardiomyocytes or stimulating division of postmitotic cardiomyocytes. Here, we demonstrate that reactivation of Notch signaling causes embryonic stem cell–derived and neonatal ventricular cardiomyocytes to enter the cell cycle. The proliferative response of neonatal ventricular cardiomyocytes declines as they mature, such that late activation of Notch triggers the DNA damage checkpoint and G2/M interphase arrest. Notch induces recombination signal-binding protein 1 for Jκ (RBP-Jκ)-dependent expression of cyclin D1 but, unlike other inducers, also shifts its subcellular distribution from the cytosol to the nucleus. Nuclear localization of cyclin D1 is independent of RBP-Jκ. Thus, the influence of Notch on nucleocytoplasmic localization of cyclin D1 is an unanticipated property of the Notch intracellular domain that is likely to regulate the cell cycle in multiple contexts, including tumorigenesis as well as cardiogenesis.
Introduction
Cardiomyocytes actively divide during embryonic development but exit the cell cycle and stop proliferating shortly after birth (Winick and Noble, 1965; Li et al., 1996; Burton et al., 1999). The incapacity of adult cardiomyocytes to divide underlies the inability of the heart muscle to regenerate after an injury, such as a myocardial infarction, that frequently leads to overload and heart failure. Notch signaling is normally active in the embryonic heart of multiple species, where it influences cell fate and morphogenesis, including within the early myocardial field, atrioventricular canal myocardium and cushions, and in the ventricular wall (Rones et al., 2000; Loomes et al., 2002; Rutenberg et al., 2006; Watanabe et al., 2006; Grego-Bessa et al., 2007; Niessen and Karsan, 2007). In addition to controlling cell fate, several studies indicate that Notch influences tissue formation and morphogenesis during development by regulating the balance between a progenitor or precursor pool and differentiating progeny (Dallas et al., 2005; Fre et al., 2005; Mammucari et al., 2005; van Es et al., 2005). In principal, a similar process could contribute to tissue regeneration (Kohler et al., 2004; Raya et al., 2004; Su et al., 2006; Nakamura and Chiba, 2007; Poss, 2007). Because the signals that control fetal and early neonatal cardiomyocyte replication and the subsequent postnatal withdrawal from cell cycle are unclear, we explored whether Notch reactivation might trigger and sustain the cell cycle of neonatal dividing and quiescent cardiomyocytes.
Binding to the transmembrane Delta or Jagged ligands on a signaling cell triggers a γ-secretase–dependent proteolytic cleavage of transmembrane Notch receptors on the responding cell (Schweisguth, 2004). Cleavage releases the Notch intracellular domain (ICD) that complexes with the transcription factor recombination signal-binding protein 1 for Jκ (RBP-Jκ; also known as CSL [CBF1, Su(H), LAG-1]) to activate transcription of downstream targets. In addition to this canonical cascade, RBP-Jκ–independent signaling has recently been demonstrated to occur through various intermediaries, including the Dishevelled (Dsh) protein required for Wnt signaling (Bush et al., 2001; Ramain et al., 2001). Despite its involvement in tumorigenesis and tissue morphogenesis, details of the mechanisms that might link Notch signaling to cell cycle control are unclear, although the complex containing the Notch ICD and RBP-Jκ directly activates transcription of the cyclin D1 gene promoter (Ronchini and Capobianco, 2001).
We found that activation of the Notch pathway in mouse embryonic stem cell (mESC)-derived and early neonatal cardiomyocytes induced cyclin D expression and stimulated cell cycle entry, mitosis, and cell division. Cell cycle progression involved the RBP-Jκ–dependent induction of cyclin D1, which led to its cytosolic accumulation. Translocation to the nucleus, retinoblastoma protein (Rb) phosphorylation, and cell cycle entry, however, specifically required the Notch ICD. This constitutes a novel function for the ICD that might account for aspects of RBP-Jκ–independent signaling that have been implicated recently in tumorigenesis (for reviews see Leong and Karsan, 2006; Miele et al., 2006). Interestingly, N2ICD induced older cardiomyocytes to enter cell cycle; however, DNA damage checkpoint activation and G2/M interphase arrest followed, most likely reflecting an intrinsic barrier to replication of mature cardiomyocytes that might serve a beneficial role in the mature heart by preventing mitotic catastrophe and apoptosis in response to mitogenic stimuli. Together, these results point to a role for Notch in the expansion of cardiomyocyte progenitor or precursor pools needed for heart growth during development and potentially during regeneration after injury in the adult.
Results
Notch induces quiescent cardiomyocytes to reenter the cell cycle
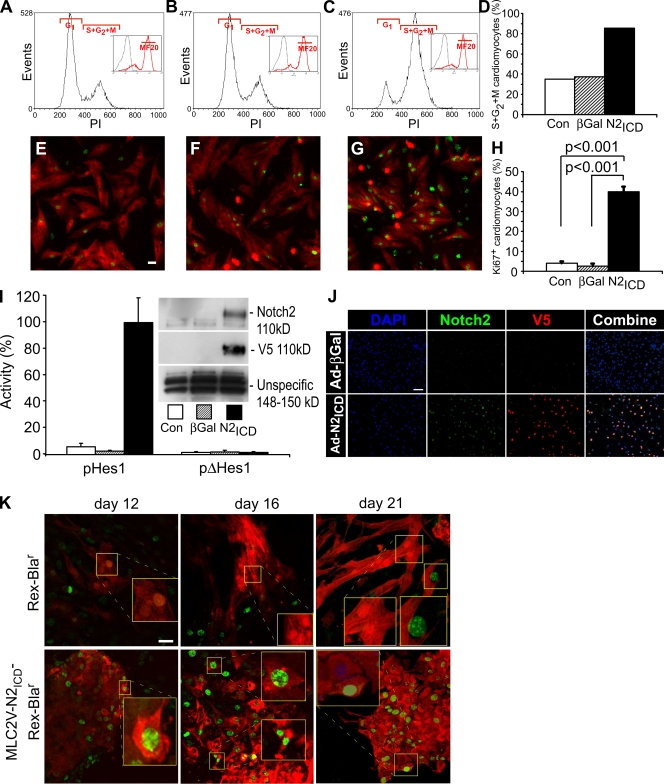
Neonatal rat ventricular cardiomyocytes (NRVCs) normally exit the cell cycle and enter G0 (Burton et al., 1999), but infection with a recombinant adenovirus encoding the N2ICD (Ad-N2ICD) dramatically changed the cell cycle profile of cardiomyocytes, increasing the percentage of NRVCs in S/G2/M phase 2.4-fold 48 h after infection (which corresponds to postnatal day 5 [P5]; Fig. 1, A, C, and D). In contrast, control NRVCs infected with an adenovirus encoding β-galactosidase (Ad-βGal) had no effect over background (Fig. 1, B and D). Ad-N2ICD also increased the incidence of Ki67+ NRVCs nearly 10-fold (Fig. 1, E, G, and H), whereas Ad-βGal had no significant effect (Fig. 1 F). Notch signaling was confirmed by the induction of hairy and enhancer of split 1 (Hes1) promoter–dependent luciferase activity in Ad-N2ICD–infected cells (Fig. 1 I) and the expression of high levels of recombinant N2ICD (V5-tagged) protein in the nuclei of Ad-N2ICD–infected cardiomyocytes (Fig. 1 I, inset; and Fig. 1 J). Endogenous Notch2 in cardiomyocytes declined postnatally (not depicted) such that it was no longer detectable by P5 (Fig. 1 J), which indicates that exogenous reactivation of Notch2 can direct cell cycle entry.
Figure 1.
Activated Notch induces cell cycle reentry in quiescent cardiomyocytes. (A–D) NRVCs were either uninfected (A and E) or infected with Ad-βGal (B and F) or Ad-N2ICD (C and G). 48 h after infection, cells were stained with mouse anti-MF20 (green, Alexa 488) and propidium iodide (PI; A–C). The number of MF20+ cells (green, Alexa 488) was >90% in all cases. DNA content was analyzed for the MF20+ population (insets) and the percentage in S/G2/M phase for each condition was calculated (D). The examples shown are representative of more than five experiments with similar outcomes. (E–H) Examples of NRVCs as in A–D stained with MF20 (red, Alexa 594) and Ki67 (green, Alexa 488; E–G) 48 h after infection, and the percentage of Ki67+ cells within the MF20+ population was measured (H). The examples shown are representative of more than five experiments with similar outcomes. (I) 12 h after infection, NRVCs were transfected with 0.6 μg of the pHes1-Luc or pΔHes1-Luc plasmids and 0.2 μg pGL3-Renilla-Luc. Luciferase activities and protein expression of Notch2 and V5 epitope (inset) were determined 12 h later. Firefly activity was normalized using Renilla luciferase activity. (J) Examples of NRVCs as in A–D stained with V5 (red, Alexa 594), Notch2, (green, Alexa 488), and DAPI (blue) 48 h after infection. (K) Confocal images showing the persistence of Ki67+ (green, Alexa 488) and MF20+ (red, Alexa 594) cardiomyocytes in differentiating mESCs expressing N2ICD from the MLC2V promoter compared with typically quiescent cardiomyocytes in cultures harboring only the REX promoter–Blar gene. Error bars indicate ±SD across experimental replicates. Bars: (E–G and K) 10 μm; and (J) 50 μm.
Similarly, mESC lines were created with a dual-cassette vector that produced N2ICD under control of the ventricular cardiomyocyte-specific myosin light chain 2V (MLC2V) promoter and the blastocidinr gene for drug selection from the stem cell promoter Rex. N2ICD showed extended replicative capacity of cardiomyocytes to at least day 21 after initiation of differentiation, yielding overgrowths in the cultures (Fig. 1 K), whereas control mESCs harboring only Rex-Blar showed the normal cessation of cell division by day 12–16.
Notch2ICD induces expression and nuclear translocation of cyclin D1
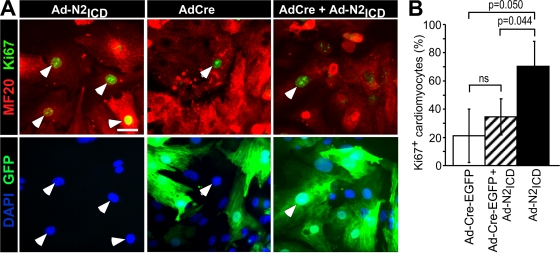
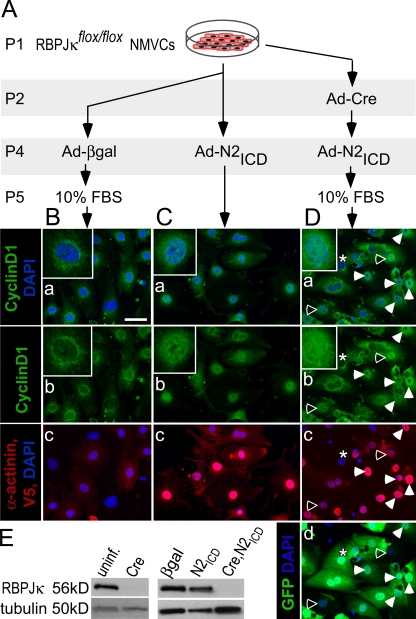
Neonatal mouse ventricular cardiomyocytes (NMVCs) were prepared from RBP-JκFlox/Flox mice to probe whether cell cycle reentry required RBP-Jκ. Excision of exons 6 and 7 of RBP-Jκ by Cre recombinase eliminates function (Tanigaki et al., 2002), and introduction of a Cre-EGFP fusion protein blocked the ability of N2ICD to stimulate cell cycle reentry, demonstrating the dependence on RBP-Jκ (Fig. 2).
Figure 2.
Cell cycle reentry requires RBP-Jκ. (A) Examples of RBP-JκFlox/Flox NMVCs infected with Ad-Cre-EGFP to remove RBP-Jκ, then reinfected 12 h later with Ad-N2ICD, as indicated, cultured 48 h, and stained with MF20 (red, Alexa 594), Ki67 (far red, Cy5, shown as green), and DAPI (blue). Arrows indicate Ki67+ cells. Bar, 10 μm. (B) The percentage of Ki67+ cardiomyocytes (MF20+) in each condition. Only Cre-expressing cells (GFP+, green) were counted in cultures infected with Ad-Cre-EGFP. N2ICD induced KI67 (P = 0.05) but the N2ICD effect following Cre was not significantly different from that of Cre alone (ns). Error bars indicate ±SD across experimental replicates.
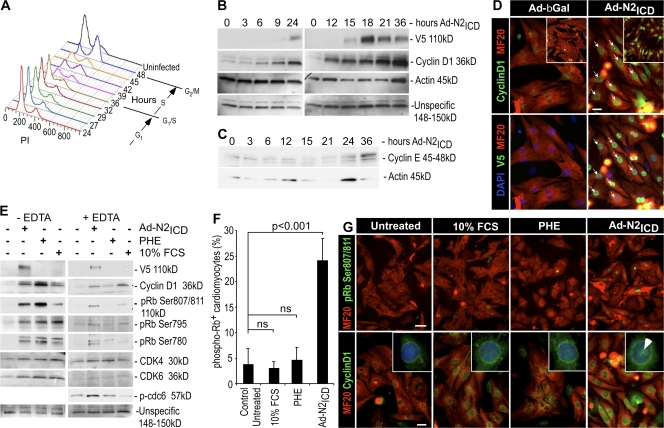
A potential transcriptional target of RBP-Jκ is cyclin D1. Cyclin D1 and other D-type cyclins (D2 and D3) translocate to the nucleus, where they function with Cdk4 and 6 to phosphorylate Rb and cause progression through the cell division cycle (DeGregori, 2004). A time-dependant up-regulation of cyclin D1 was apparent beginning ∼6–9 h after Ad-N2ICD infection (Fig. 3 B). Cyclin D1 induction precedes cell cycle entry (Fig. 3 A) by ∼24 h, which correlates well with the duration of G1 phase observed for rat cells (i.e., 24 h in primary cultures of endothelial cells; Solodushko and Fouty, 2007). Also, the level of the G1/S-phase cyclin, cyclin E, increased at 36 h (Fig. 3 C), which correlates with the observed cell cycle profile (Fig. 3 A) and confirms the S-phase entry of NRVCs.
Figure 3.
Notch induces expression and nuclear localization of cyclin D1 and phosphorylation of Rb before cell cycle reentry. (A) The DNA content of the cardiac cells analyzed by flow cytometry (as in Fig. 1) at the indicated times after infection. One of two experiments with similar outcomes is shown. (B) Cyclin D1 and V5 epitope (N2ICD) expression in NRVCs infected with Ad-N2ICD at the indicated times after infection. (C) Time course of cyclin E expression after Ad-N2ICD infection of NRVCs. (D) Examples of nuclear localization of cyclin D1 (arrows) in NRVCs infected with Ad-N2ICD as compared with Ad-βGal–infected control cultures 48 h after infection. Immunostaining is shown for cyclin D1 (green, Alexa 488), MF20 (red, Alexa 594), V5 epitope tag on N2ICD (far red, Cy5, shown as green), and DAPI (blue). Confocal microscopic images (insets) confirmed nuclear localization. (E) Phosphorylation status of Rb in NRVCs 30 h after infection with Ad-N2ICD or 24 h after treatment with 10% FCS or 20 μM PHE. 5 mM EDTA in the lysis buffer prevents phosphorylation from occurring in the lysates. N2ICD (V5 epitope), cyclin D1, phospho-Rb (Ser807/811, Ser795, and Ser780), Cdk4, Cdk6, and phospho-Cdc6 expression are shown. (F) Incidence of phospho-Rb (Ser807/811) in NRVC cardiomyocytes (MF20+ cells). Note that only N2ICD induced significant levels of phospho-Rb. Error bars indicate ±SD. (G) Immunostaining with MF20 (red, Alexa 594) and for phospho-Rb (Ser807/811; green, Alexa 488) or cyclin D1 (green, Alexa 488). Note that cyclin D1 and phospho-Rb are present in the nuclei of N2ICD-expressing cells (arrow) but not in the nuclei of cells cultured with high serum or PHE. Higher-magnification images are shown in insets with DAPI staining (blue). Bars: (D) 10 μm; (G, top) 50 μm; (G, bottom) 10 μm.
Although low serum was included in the preceding experiments to enhance cell survival, its absence did not prevent cell cycle reentry by N2ICD (18.3% S+G2+M phase cells in uninfected cells vs. 44.4% in infected cells), which indicates that Notch-induced cell cycle reentry is independent of serum factors, consistent with induction of cyclin D1 by RBP-Jκ. Moreover, even high (10%) serum was insufficient to stimulate cell cycle entry, see the following paragraph.
Cyclin D1 levels were elevated within the nuclei of Ad-N2ICD–infected NRVCs, visualized by coimmunofluorescent staining with the V5 epitope on N2ICD, but were absent in nuclei of control Ad-βGal–infected NRVCs (arrows indicate examples in Fig. 3 D). Nuclear localization of cyclin D1 was accompanied by phosphorylation of Rb at Ser807/811, Ser795, and Ser 780 in extracts prepared with 10 mM EDTA to inhibit kinase activity after lysis (Fig. 3 E, right). Although phenylephrine (PHE) and 10% FCS induced cyclin D1 (Fig. 3, E and G), both failed to stimulate its characteristic nuclear localization (Fig. 3 G, inset, arrowhead in the N2ICD panel indicates a characteristic nuclear pattern), Rb phosphorylation (Fig. 3, E–G) or DNA synthesis (Fig. S1, available at http://www.jcb.org/cgi/content/full/jcb.200806104/DC1). Lysates prepared in the absence of EDTA to preserve kinase activity after dissolution of the cell membranes showed that PHE or FCS can stimulate Cdk activity (Fig. 3 E, left). Because Rb phosphorylation occurred under these conditions but not in the presence of EDTA, we concluded that PHE and FCS alone cannot stimulate Rb phosphorylation because the components are not colocalized in the nucleus. This raises the possibility that the N2ICD might have a unique role in localizing the cyclin D1–Cdk complex to the nucleus.
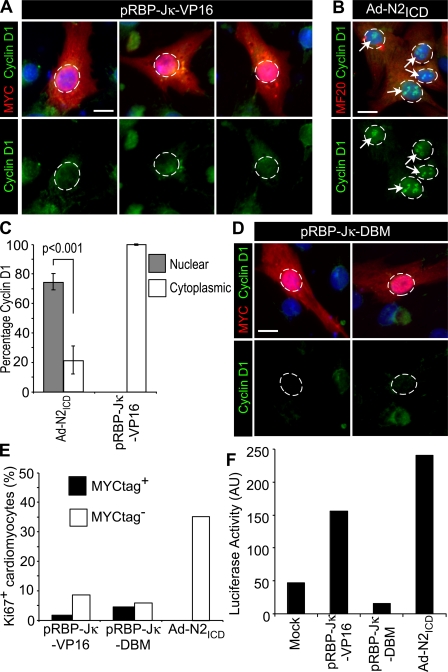
Cyclin D1 nuclear localization requires notch ICD
To distinguish induction of cyclin D1 gene expression from nuclear localization, we transfected NRVCs to express a constitutively active form of RBP-Jκ made by fusion to the VP16 transactivation domain and a MYC epitope tag (pRBP-Jκ-VP16). RBP-Jκ-VP16 activates transcription of Notch-dependent genes, including pHes1-luciferase in cardiomyocytes (Fig. 4 F). RBP-Jκ-VP16 induced the accumulation of cyclin D1 in the cytosol but not in the nucleus, as had been seen with N2ICD (Fig. 4, A–C), and did not stimulate cell cycle entry (Fig. 4 E), which confirms that elevated cyclin D1 expression is insufficient to promote cell cycle entry. As a control, a DNA-binding mutant (DBM) control of RBP-Jκ (pRBP-Jκ-DBM) that does not activate pHes1-luciferase (Fig. 3 F) did not induce cyclin D1 (Fig. 4 D). PHE or 10% FCS also induced only cytosolic accumulation of cyclin D1 (Fig. 3 G). The failure of activated RBP-Jκ, PHE, or 10% FCS to localize cyclin D1 to the nucleus indicated a specific requirement for N2ICD.
Figure 4.
RBP-Jκ is insufficient to promote nuclear localization of cyclin D1. (A and B) Examples of NRVCs were transfected with 0.8 μg of plasmids encoding a myc epitope-tagged, constitutively active version of RBP-Jκ in pCDNA3 (pRBP-Jκ-VP16; A) or infected with Ad-N2ICD (B) and stained 48 h later for cyclin D1 (green, Alexa 488), c-myc epitope (red, Alexa 594), MF20 (red, Alexa 594), and DAPI (blue). Note the absence of nuclear localized cyclin D1 in c-myc+ (RBP-Jκ-VP16+) cells. Circles indicate nuclear regions and arrows indicate nuclear localized cyclin D1. (C) Percentage of cardiomyocytes treated as in A and B showing nuclear versus cytosolic localization of cyclin D1. Error bars indicate ±SD. (D) NRVCs transfected to express a control DNA-binding mutant of RBP-Jκ (pRBP-Jκ-DBM) and stained as in A. Only a basal level of cyclin D1 was detected. Circles indicate nuclear regions. (E) Percentage of c-myc+/Ki67+ (transfected) and c-myc−/Ki67+ (untransfected) cells for pRBP-Jκ-VP16 (n = 69 and 107, respectively) and pRBP-Jκ-DBM (n = 57 and 106, respectively) as compared with Ad-N2ICD infected (n = 100); results are representative of four trials. (F) RBP-Jκ and N2ICD (0.5 μg of plasmid per transfection), as in A–E, showed expected activities on the pHes1-luciferase reporter (0.25 μg of plasmid per transfection); results are representative of two trials. Bars, 5 μm.
N2ICD also stimulated nuclear localization of cyclin D1 in RBP-JκFlox/Flox NMVCs after Cre excision (Fig. 5). Ad-Cre-EGFP on P1 effectively excised RBP-Jκ 1 d later (Fig. 5, A and E). As shown in Fig. 2, N2ICD cannot induce cyclin D1 in the absence of RBP-Jκ, therefore 10% FCS was added 1 d before harvesting the cells to induce cyclin D1 (Fig. 5 A). Ad-βgal–treated cells showed the characteristic cytoplasmic and perinuclear localization of cyclin D1 (Fig. 5 B), whereas Ad-N2ICD (Fig. 5 B) and the sequential treatment with Ad-Cre-EGFP and Ad-N2ICD (Fig. 5, C and D) both induced nuclear localization. Closed arrows (Fig. 5 D) indicate examples of Cre-EGFP+, N2ICD+ cells with nuclear localization of cyclin D1. Prominent nuclear expression of cyclin D1 was seen in the rare cells that expressed N2ICD but had not received Cre-EGFP (indicated by open arrows). In contrast, cyclin D1 was cytoplasmic and perinuclear in cells that had Cre-EGFP but did not express N2ICD (Fig. 5 D, indicated by asterisks). Collectively, RBP-Jκ is required for Notch to induce cyclin D1 transcription but not for nuclear localization.
Figure 5.
Nuclear localization of cyclin D1 is RBP-Jκ independent. (A) NMVCs were isolated from RBP-JκFlox/Flox pups on the day of birth (P1) and treated as indicated until processing for immunostaining (the equivalent of P6). 10% FCS was added to induce cyclin D1 in the absence of RBP-Jk. (B–D) Confocal images of NMVCs showing nuclear localization of cyclin D1 (green, Alexa 568; “a” and “b” panels) overlapping with DAPI (blue; “a” panels). α-actinin cytosolic immunostaining identified cardiomyocytes, and nuclear V5 epitope immunostaining identified cells expressing transduced N2ICD (both visualized with a far red [Alexa 680] secondary antibody and shown as red in “c” panels). EGFP (D, d) shows Cre-EGFP fusion protein expression. Closed arrows indicate examples of Cre-EGFP+, N2ICD+ cells with nuclear localization of cyclin D1. Open arrows indicate the rare cells that expressed N2ICD but did not express Cre-EGFP; note the prominent nuclear presence of cyclin D1. The asterisks indicate a cell that expressed Cre-EGFP but not N2ICD; note that cyclin D1 was cytoplasmic and perinuclear. (E) RBP-Jκ protein expression 1 and 3 d after Ad-Cre-EGFP treatment (corresponding to P3 and P5), showing efficient depletion by Cre.
Notch-regulated nuclear export versus import of cyclin D1
Nuclear localization of cyclin D1 is controlled by a fine balance between import, through an unknown mechanism, and export through the chromosome region maintenance 1 (CRM1)/exportin-1 transporter. Cyclin D1 phosphorylation by glycogen synthase kinase-3β (GSK3β) at Thr286 is thought to trigger export to the cytosol and degradation by the proteasome (Diehl et al., 1998).
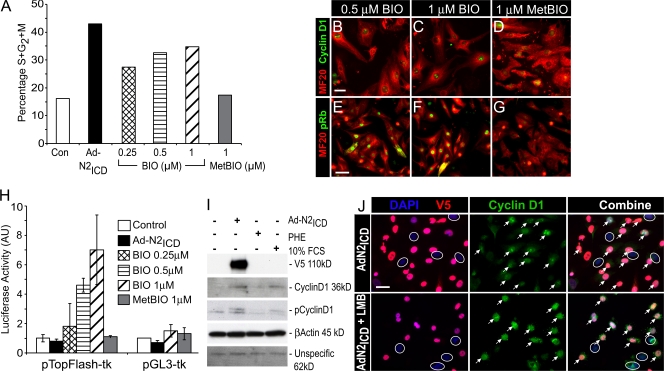
To test if N2ICD regulates export, we first compared its action to 6-bromoindirubin-3′-oxime (BIO), a small molecule inhibitor of GSK3β. BIO induced cell cycle reentry in a dose-dependent manner, albeit less dramatically than what is seen with N2ICD (Fig. 6 A), as previously described (Tseng et al., 2006). The inactive control compound MetBIO failed to promote cell cycle reentry of NRVCs. BIO, but not MetBIO, also caused nuclear accumulation of cyclin D1 (Fig. 6, B–D) and phosphorylation of nuclear Rb (Fig. 6, E–G), which is suggestive that retention of cyclin D1 in the nucleus contributes to cell cycle entry by BIO.
Figure 6.
Notch-induced nuclear accumulation of cyclin D1 in NRVCs is not caused by inhibition of export. (A) NRVCs were treated for 36 h with the GSK3β inhibitor BIO or control MetBIO before staining with MF20 and analysis of DNA content of cardiomyocytes (MF20+ population) by flow cytometry, as for Fig. 1. One of two experiments with identical outcomes is shown. (B–G) NRVCs were treated for 24 h with the indicated concentrations of BIO (B, C, E, and F) or MetBIO (D and G) before staining with MF20 (red, Alexa 594) and for cyclin D1 (green, Alexa 488; B–D) or phospho-Rb (Ser807/811; green, Alexa 488; E–G). BIO induced both nuclear accumulation of cyclin D1 and phosphorylation of Rb. (H) Ad-N2ICD–infected or control NRVCs were transfected with 0.6 μg pTopFlash-tk-Luc or pGL3-tk-Luc, as indicated, and 0.2 μg of pGL3-Renilla-Luc before exposure with BIO (0.25 μM, 0.5 μM, or 1 μM) for an additional 36 h before luciferase activities were determined. Firefly luciferase activity was normalized using Renilla luciferase activity. N2ICD did not modulate the β-catenin/T cell factor–dependent transcription. Error bars indicate ±SD. (I) Total and phospho–Thr286–cyclin D1 in NRVCs infected with Ad-N2ICD or treated with 10% FCS or PHE (20 μM). N2ICD did not prevent Thr286 phosphorylation of cyclin D1. (J) LMB enhanced nuclear localization of cyclin D1 (green, Alexa 488) by N2ICD (red, Alexa 594; arrows) but did not act on uninfected cells (circled nuclei), which is consistent with N2ICD regulating nuclear localization upstream of export by the CRM1–exportin-1 complex. Bars: (B–D) 10 μm; (E–G) 25 μm; (J) 10 μm.
Unlike BIO, N2ICD did not inhibit GSK3β, which was determined by monitoring luciferase activity from the T cell factor–dependent luciferase activity from the pTOPflash reporter plasmid that senses stabilized β-catenin, a direct target of GSK3β (Fig. 6 H). Moreover, the levels of phospho–Thr286–cyclin D1 levels were not inhibited in Ad-N2ICD–infected NRVCs but were in fact up-regulated compared with controls and, more importantly, with PHE- or 10% FCS-treated NRVCs, which also expressed high levels of Cyclin D1 (Fig. 6 I). The increase in phospho–Thr286–cyclin D1 correlated with increased levels of nuclear cyclin D1 available for phosphorylation by GSK3β.
As an independent test for regulated export, cells were treated with leptomycin B (LMB) to specifically inhibit CRM1/exportin-1. LMB enhanced nuclear localization of cyclin D1 beyond that achieved by N2ICD alone (Fig. 6 J). Furthermore, LMB stimulated nuclear cyclin D1 only in cells that expressed N2ICD and not in control, uninfected cells (compare N2ICD+ cells [Fig. 6 J, indicated by arrows] to uninfected cells [Fig. 6 J, indicated by circles]). These data are consistent with the idea that N2ICD acts upstream of CRM1/exportin-1. LMB did not increase the number of S/G2/M cells (unpublished data) because inhibition of CRM1/exportin-1 inhibition prevents cell cycle progression (Yoshida et al., 1990). The results with LMB, together with the lack of an inhibitory effect of N2ICD on GSK3β or the phosphorylation of cyclin D1, suggest that N2ICD does not control export or degradation but might stimulate nuclear import.
Activation of the DNA damage checkpoint in older NRVCs
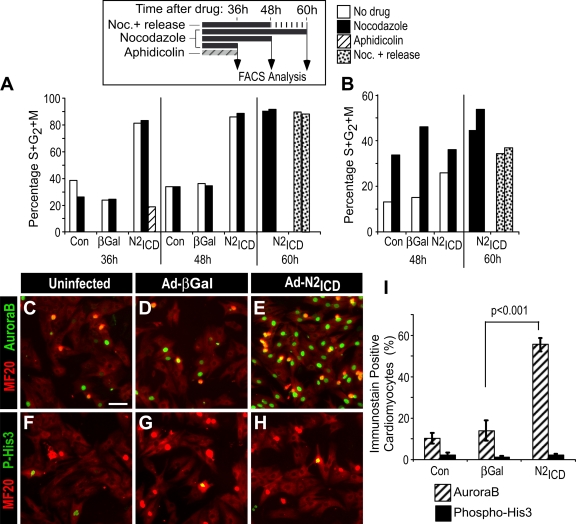
Activated Notch effectively stimulated P2 NRVCs to enter mitosis, as revealed by phosphorylation of histone 3 (His3) on Ser10 (62.1% phospho-Ser10-His3+ in AdN2ICD-infected P2 NRVCs vs. 44.5% for uninfected; P < 0.001, two-tailed Student's t test). The presence of phospho-Ser10 is a hallmark for entry into mitosis and is required for chromatin condensation (e.g., Adams et al., 2001; Crosio et al., 2002). In striking contrast, older P5 NRVCs completely failed to phosphorylate His3 (0.57% phospho-Ser10-His3+; Fig. 7, H and I), which revealed a postnatal decline in the ability to traverse the G2/M interphase. A similar postnatal decline in cell division was noted by Collesi et al. (see p. 117 of this issue) in response to stimulation by exogenous Jagged.
Figure 7.
Ad-N2ICD–infected NRVCs arrest at the G2/M interphase. (A and B) 3 d after birth, NRVCs were infected with Ad-N2ICD or Ad-β-gal, or not infected, and, 12 h later, treated with aphidicolin or nocodazole for an additional 36, 48, or 60 h. For nocodazole+ release, nocodazole was removed during the last 12 h of culture (see schematic in A, top). The percentages of cells in S/G2/M phase in MF20+ (cardiomyocytes; A) and MF20− (noncardiomyocytes; B) populations were determined from flow cytometry. Nocodazole did not increase the incidence of G2/M cells among the N2ICD-treated cardiomyocyte population, which is indicative of a block at the onset of M phase. The example shown is representative of four experiments with similar outcomes. (C–I) NRVCs were either left uninfected (C and F) or infected with Ad-βGal (D and G) or Ad-N2ICD (E and H), cultured for 48 h, then stained with MF20 (red, Alexa 594) and for AuroraB (green, Alexa 488; C–E) or for phospho-His3 (Ser10; green, Alexa 488; F–H). The percentages of positive cells within the MF20+ population was then determined (I). Nuclear-localized AuroraB but not phospho-His3 was detected in response to N2ICD. Error bars indicate ±SD. Bar, 25 μm.
To understand the nature of the block, we probed the cell cycle status of treated cells using the DNA polymerase inhibitor aphidicolin or the mitotic spindle assembly inhibitor nocodazole for various times to block the cell cycle (see Fig. 7 A, inset, for experimental design). As expected, aphidicolin, which blocks cells at the G1/S interphase, reduced the percentage of Ad-N2ICD–expressing NRVCs that had progressed into S, G2, or M phases of the cell cycle (Fig. 7 A), confirming entry into S phase. Nocodazole interferes with microtubules and causes cells to arrest at the G2/M interphase. Importantly, nocodazole did not alter the percentage of cardiomyocytes in S/G2/M phase (Fig. 7 A), which is suggestive of the fact that cells were already arrested at this point. To test this idea, the nocodazole was washed out during the last 12 h of culture. Normally, cycling cells resume progression after removal of nocodazole; however, this had no effect on the P5 NRVCs, which indicates that they were likely arrested at or before G2/M. In contrast, the residual noncardiomyocytes (MF20− cells) in the preparations arrested at G2/M upon nocodazole treatment and were released by its removal (Fig. 7 B), which demonstrates that the Notch-induced block was specific to the cardiomyocytes in the cell preparation.
AuroraB directly phosphorylates Ser10 of His3. As shown in Fig. 7 (C–I), Ad-N2ICD dramatically increased AuroraB expression in the nuclei of P5 NRVCs but completely failed to induce His3 Ser10 phosphorylation in control uninfected or Ad-βGal–infected NRVCs. A blockade at the level of AuroraB confirmed the G2/M arrest and suggested activation of the DNA damage checkpoint.
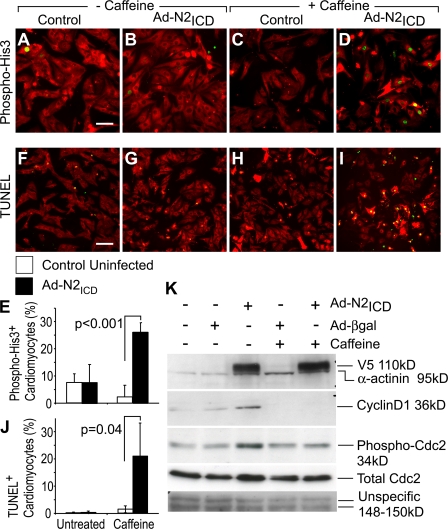
Caffeine effectively overrides DNA damage G2/M checkpoint activation and can permit entry of arrested cells into M phase by inhibiting the ataxia telangiectasia mutated (ATM) and ATM and Rad-3 related (ATR) kinases, and most likely other DNA checkpoint components (e.g., Kastan et al., 1991; Sarkaria et al., 1999). Entry into M phase in the presence of caffeine is therefore diagnostic of checkpoint activation. Caffeine treatment alone did not increase the level of phospho-Ser10-His3 in uninfected P5 NRVCs and in fact caused a minor reduction (Fig. 8, A–E). In contrast, N2ICD increased the number of phospho-His3+ NRVCs by 3.5-fold. Overriding checkpoint activation pushed cells into mitosis and resulted in cell death, visualized by an 87-fold increase in the number of TUNEL-positive cardiomyocytes (Fig. 8, I and J), which is consistent with induction of programmed cell death when the checkpoint is overridden in cells with DNA replication defects (Belka, 2006). The mitosis-inducing kinase Cdc2 is a key component of the DNA damage checkpoint pathway downstream of ATM/ATR and Chk1, and it shows inhibitory Tyr15 phosphorylation with checkpoint activation. Fig. 8 K shows an increase in phospho-Tyr15-Cdc2 in N2ICD-stimulated NRVCs, as well as a possible increase in the total Cdc2 protein levels, providing further evidence of DNA damage checkpoint activation after the N2ICD-induced cell cycle. As expected, phospho-Tyr15-Cdc2 returned to basal levels after caffeine treatment of N2ICD-stimulated P5 cardiomyocytes. We conclude that activation of DNA damage checkpoint causes the G2/M arrest and is triggered as a consequence of improper or incomplete DNA synthesis during S phase.
Figure 8.
DNA damage checkpoint activation implicated in G2/M arrest. (A–E) NRVCs were either infected with Ad-N2ICD or left uninfected. 24 h later, 10 mM caffeine (C, D, H, and I) or control (A, B, F, and G) media were added. Cells were then cultured for an additional 24 h, by which time they were at P5, and stained with MF20 (red, Alexa 594) and for phospho-His3 (Ser10; green, Alexa488; A–D). The incidence of phospho-His3+, MF20+cardiomyocytes is shown (E). (F–J) Alternatively, a TUNEL assay (green, fluorescein) was performed before immunostaining with MF20 (red, Alexa594). Note that caffeine permitted the induction of phospho-His3 but triggered TUNEL reactivity. (K) N2ICD (anti-V5 epitope) and cyclin D1 expression, and the corresponding levels of phosphorylated Cdc2 are shown for NRVCs prepared as in A–E. Elevated phospho-Cdc2 was observed with activated N2ICD and reduced by caffeine to basal levels, which together is indicative that N2ICD treatment activates the DNA damage checkpoint. α-actinin and nonspecific proteins are shown as loading controls. Error bars indicate ±SD. Bars, 25 μm.
Discussion
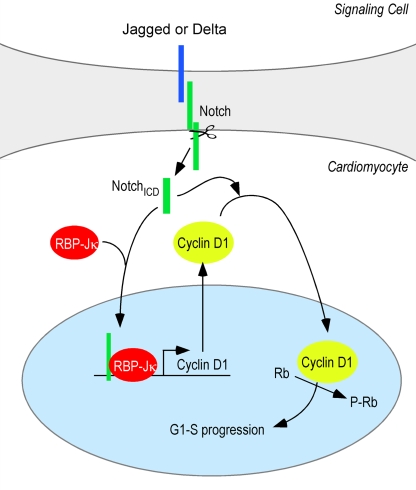
We found that cell cycle entry of mESC-derived and quiescent cardiomyocytes could be profoundly enhanced by Notch activation, which controlled expression and nuclear localization of cyclin D1. Transcriptional induction of the cyclin D1 gene required canonical Notch signaling through RBP-Jκ. Nuclear localization however was a distinct property of the Notch ICD that is essential for cell cycle entry (Fig. 9). The implications of these findings for regulating expansion of cardiomyogenic progenitor or precursor cells and in the control of other normal and pathological processes, such as tumorigenesis, are discussed in the following paragraphs.
Figure 9.
Summary of Notch2-induced cell cycle entry. RBP-Jκ–dependent transcription leads to accumulation of cyclin D1 in the cytosol. Notch ICD regulates entry into the cell cycle by controlling nuclear localization of cyclin D1 independently of RBP-Jκ.
Notch and cardiomyocyte cell cycle reentry
Our finding that RBP-Jκ–dependent Notch signaling induced expression of cyclin D1 in quiescent cardiomyocytes is consistent with the presence of a functional RBP-Jκ–binding site in the cyclin D1 promoter (Ronchini and Capobianco, 2001). FCS and PHE also induced cyclin D1 and, like activated RBP-Jκ, were insufficient to promote nuclear translocation and cell cycle reentry. Nuclear localization of cyclin D1 by Notch ICD coupled with the developmental decline in endogenous Notch during early postnatal development could explain the results of Tamamori-Adachi et al. (2003), who showed that although ectopically expressed cyclin D1 accumulates in the nucleus of still-proliferating fetal cardiomyocytes, it is sequestered in the cytoplasm of normally quiescent NRVCs. Moreover, fusion to a nuclear localization sequence that targeted cyclin D1 to the nucleus was sufficient to induce Rb phosphorylation and cell cycle reentry of quiescent NRVCs.
The localization of cyclin D1 to the nucleus probably occurs as a balance between import, for which the mechanism is poorly understood, and regulated export of phospho–Thr286–cyclin D1 through the transporter CRM1 into the cytosol, where it is degraded by the proteosome (Diehl et al., 1998). Two lines of evidence indicate that N2ICD acts upstream of export: first, we noted a superaccumulation of cyclin D1 in the presence of N2ICD and LMB, which inhibits CRM1-mediated export, indicating that N2ICD acts upstream of CRM1. Second, N2ICD did not modulate GSK3β activity nor reduce the levels of phospho–Thr286–cyclin D1, as did BIO. Thus, inhibition of export seems unlikely to be responsible for N2ICD-directed nuclear accumulation of cyclin D1, and we favor the model in which N2ICD regulates import. In contrast, BIO itself is sufficient to promote cardiomyocyte cell cycle entry (Tseng et al., 2006), and we propose that inhibition of cyclin D1 Thr286 phosphorylation, leading to protein stabilization and nuclear localization, is the likely mechanism.
How might the Notch ICD promote nuclear translocation? We did not detect a physical interaction between N2ICD and cyclin D1, Cdk4, or Cdk6 by coimmunoprecipitation from NRVCs (unpublished data). Moreover, the spatially localized immunostaining pattern of cyclin D1 within the nucleus differed from the diffuse pattern seen for N2ICD or its V5 epitope tag; thus, Notch might not participate directly in a cyclin D complex. In terminally differentiated neurons, cyclin D1 becomes cytoplasmic, and forced expression of p21WAF1 (Cip1) induces nuclear accumulation (Sumrejkanchanakij et al., 2003), so one possibility is that Notch modulates the interaction of cyclin D1 with proteins such as p21. Little is known about the mechanism that keeps cyclin D1 from entering the nuclei of terminally differentiated cells, but activated Notch is seen in several tumors, such as breast, colon, and pancreas (Miyamoto et al., 2003; van Es et al., 2005; Stylianou et al., 2006), and it will be interesting to evaluate whether it controls cyclin D1 trafficking in cancers.
Progression through the cell cycle: implications of the neonatal decline in mitotic response to Notch
Between days 4 and 7 after birth, NRVCs lose most of their ability to respond to exogenous Notch, first because of the decline in Notch 1 and Notch 2 receptors (unpublished data; Collesi et al., 2008) but also because of a pronounced activation of DNA damage checkpoint in response to activated Notch (Fig. 8). As expected, overriding the checkpoint pushed the cardiomyocytes into mitosis, resulting in mitotic catastrophe and apoptosis.
Several studies have shown that manipulation of cell cycle proteins can induce NRVCs to reenter the cell cycle (e.g., Agah et al., 1997; Huh et al., 2001; Datwyler et al., 2003; Tamamori-Adachi et al., 2003; Engel et al., 2005; Jung et al., 2005; Tseng et al., 2006). These studies used reagents (e.g., BrdU incorporation or Ki67 immunostaining) that reflect entry into the cell cycle or S phase but not entry into mitosis or cell division. Based on our results that DNA damage checkpoint activation limits cell division of late-stage quiescent cardiomyocytes, it is important to quantitatively evaluate whether treatment with these or other reagents can indeed direct cells to enter mitosis and divide.
Checkpoint activation is not solely a consequence of Notch activation. The majority of BIO-treated P5 NRVCs also arrested at G2/M and showed DNA damage checkpoint activation, as demonstrated by caffeine sensitivity and Cdc2 phosphorylation (unpublished data). In the context of the adult heart, a beneficial function of DNA damage checkpoint activation might be to protect mature cardiomyocytes from apoptosis and cell death after exposure to stimuli that support replication of nonmyocytes during normal heart function or pathologically as a response to injury or remodeling.
Although most cardiomyocytes after P3–5 arrested at the G2/M interphase, a minor fraction of Ad-N2ICD–infected NRVCs could be found at all mitotic phases, including cytokinesis (Fig. S2, available at http://www.jcb.org/cgi/content/full/jcb.200806104/DC1); however, the incidence was the same as in uninfected NRVCs. The complex structure of sarcomeric apparatus has been proposed to block chromosomal segregation and cytokinesis should mature cardiomyocytes be stimulated to reenter in the cell cycle (Li et al., 1997; Ahuja et al., 2004). The normally striated staining of MF20 in nondividing cardiomyocytes (see G0 and G1 in Fig. S2 A) was diffuse in the dividing cells, and the distinction of whether dividing cells underwent some degree of disassembly or reflected a less mature subpopulation should be investigated. The dividing cardiomyocytes showed normal hallmarks of M phase. For instance, AuroraB was associated with chromosomes during anaphase and localized at the midbody during cytokinesis (Fig. S2, B and C), as expected for dividing cells. Regulation of p21WAF1/Cip1 and p57Kip2 also appeared normal. p21WAF1/Cip1 was expressed normally in the nuclei of both Ki67+ and Ki67− cells and disappeared during mitosis, and the normal expression of p57Kip2 also disappeared during mitosis (Fig. S2, D–F). Interestingly, once the nuclear membrane was disintegrated, staining for the V5 epitope on N2ICD became cytoplasmic, which indicates cytosolic relocalization of the protein (Fig. S2 G).
Notch in ventricular development and disease
We evaluated Notch2 because it is the predominant isoform present on cardiomyocytes while the chamber myocardium is growing during mid-to-late gestation. An implication of our data is that Notch2 in the fetal heart might prevent differentiation and sustain replication of newly formed cardiomyocytes to permit the dramatic growth of ventricular myocardium, which expresses Jagged1, Jagged2, and Notch2 in overlapping patterns (Loomes et al., 2002; Rutenberg et al., 2006; Grego-Bessa et al., 2007; Niessen and Karsan, 2007). Although the phenotypes caused by systemic deletions of Notch signaling components are consistent with such a model, the cardiac defects are likely to also be caused by secondary consequences of the loss of Notch, such as in the endocardium (Grego-Bessa et al., 2007). All 3D-type cyclins are present in the developing heart, and, interestingly, the triple knockout mouse exhibited severely hypoplastic ventricular myocardium and died by embryonic day 16.5 (Kozar et al., 2004). Thus, we speculate that myocyte-autonomous Notch–cyclin D interactions might underlie ventricular growth. Myocardial conditional knockouts of RBP-Jk and Notch2 using appropriate Cre lines are being analyzed to address this issue.
An important implication of this study is that Notch might play a similar role after myocardial injury by sustaining proliferation of a committed precursor or progenitor pool, although the origin and prevalence of such cells remains unclear (e.g., Hsieh et al., 2007) and a direct role for Notch remains to be tested. Regeneration of the adult zebrafish heart is preceded by Notch reactivation (Raya et al., 2003). The zebrafish Notch isoform characterized as being involved, Notch1b, is expressed by endothelial cells of the endocardium rather than by the myocardium, and, therefore, would be expected to influence cardiomyocyte regeneration only indirectly. Thus, whether Notch signaling exerts a cell-autonomous effect in regenerating cardiomyocytes needs to be resolved using appropriate transgenic models and development of specific agonists to stimulate such cells in situ.
Materials and methods
Cell culture
Ventricular cardiomyocytes were isolated with a neonatal rat/mouse cardiomyocyte isolation kit (Cellutron) and cultured at 37°C in humidified air with 5% CO2. In brief, ventricles were dissected from 1–2-d-old Hsd:SD rats (Sprage Dawley) or mice, then digested five times for 15 min each with the enzyme cocktail at 37°C. Cells were pooled, preplated for 90 min on an uncoated dish to remove fibroblasts, and plated on 1% gelatin-coated cell culture plastic dishes in high-serum media (DME/F12 [1:1], 0.2% BSA, 3 mM sodium-pyruvate, 0.1 mM ascorbic acid, 4 mg/liter transferrin, 2 mM l-glutamine, and 5 mg/liter ciprofloxacin supplemented with 10% horse serum and 5% FCS) at 1.25 × 105 cells/cm2. After 24 h, media was changed to low-serum medium (same but with 0.25% FCS) and cultured until use. Nearly all cardiomyocytes were quiescent by P3. The purity of cultures was routinely determined by immunofluorescence staining with an anti-muscle myosins antibody (MF20; Developmental Biology Hybridoma Bank) and Alexa 594–conjugated anti–mouse immunoglobulin secondary antibodies (Invitrogen), and only preps with >90% of cardiomyocytes were used for analyses. CGR8 mESCs were created by stable HIV lentiviral transduction to express Blar from the Rex promoter and N2ICD from the MLC2V promoter (Barcova et al., 2007), and embryoid bodies were prepared by removal of leukemia inhibitory factor and aggregation in serum-containing medium.
Adenoviral infection
Ad-N2ICD (provided by T. Maciag, Maine Medical Institute, Scarborough, ME; Small et al., 2003), Ad-βGal (Clontech Laboratories, Inc.), and Ad-Cre-EGFP (a gift from S.V. Kozlov, National Cancer Institute, National Institutes of Health, Bethesda, MD) are replication-deficient, serotype 5 adenoviruses. Viruses were propagated and concentrated using an Adenopure kit (Puresyn, Inc.), and viral titers were determined by using the Adeno-X Rapid Titer kit (Clontech Laboratories, Inc.). Cardiomyocytes were infected overnight in low-serum media at a multiplicity of infection of four to five viruses per cell. The media was replaced 12–14 h after infection with fresh low-serum media. Optimal titers for infection were determined empirically for each batch of virus based on cyclin D1 expression and cell cycle entry.
Transfection and luciferase assay
Cardiomyocytes were transfected for 6–8 h in serum-free media with 0.8 μg of DNA using Lipofectamine 2000 (Invitrogen) with pHes1-Luc and pΔHes1-Luc (provided by J. Griffin, Dana-Farber Cancer Institute, Boston, MA); TOPFlash-tk-Luc (Luckow and Schutz, 1987) and pGL3-tk-Luc (Promega); pRS2-RBP-Jκ-VP16, pRS2-RBP-Jκ-DBM, pCDN3-RBP-Jκ-VP16, and pCDN3-RBP-Jκ-DBM (Rutenberg et al., 2006); and pRL-tk (Promega) to normalize for transfection efficiency. The media was replaced after transfection with fresh low-serum media and cultured for 36 h before staining or determination of luciferase activities using the Dual-Luciferase system (Promega). For the engineered RBP-Jκ-VP16 and DBM variants, the results were similar whether the promoter used was cytomegalovirus, as used in adenovirus vectors (Fig. 4), or Rous sarcoma virus, which is more potent in cardiomyocytes.
Immunofluorescence staining
After cardiomyocytes were fixed in 4% PFA for 15 min at room temperature, nonspecific binding was blocked for at least 1 h with blocking buffer (PBS × 1, 50 mM glycine, 2% BSA, 2% goat serum, and 0.01% sodium-azide). Next, cells were incubated overnight at 4°C with primary antibodies (Table I) in washing buffer (blocking buffer diluted 1:10 in PBS × 1), and immune complexes were detected with Alexa 488, Alexa 594 (Invitrogen), and Cy5-conjugated (Jackson ImmunoResearch Laboratories) secondary antibodies diluted 1:250 in washing buffer and counterstained with 0.5 μg/ml DAPI. TUNEL staining was performed with the in situ Cell Death Detection kit (Roche). Statistical significance was indicated by p-value as calculated from a two-tailed Student's t test.
Table I. Antibodies and dilutions used for immunofluorescent detection of proteins.
| Antibody | Dilution | Source |
|---|---|---|
| Anti-muscle myosins, MF20 mouse Ab | 1:100 | Developmental Studies Hybridoma Bank, University of Iowa |
| Anti–α-actinin, mouse Ab | 1:200 | Sigma-Aldrich |
| Anti–phospho-His3 (Ser10) rabbit Ab | 1:100 | Millipore |
| Anti-AuroraB rabbit Ab | 1:250 | BD Biosciences |
| Anti-Ki67 rabbit Ab | 1:100 | Novus Biologicals |
| Anti-p21 mouse Ab | 1:100 | Millipore |
| Anti-p57 rabbit Ab | 1:200 | Santa Cruz Biotechnology, Inc. |
| Anti-V5 mouse Ab | 1:200 | Invitrogen |
| Anti-Notch2 rabbit Ab | 1:200 | Abcam |
| Anti-cyclin D1 rabbit Ab | 1:200 | W. Jiang (Burnham Institute for Medical Research, La Jolla, CA) |
| Anti–phospho-Rb (Ser807/811) rabbit Ab | 1:200 | Cell Signaling Technology |
| Anti–c-myc mouse Ab | 1:100 | Santa Cruz Biotechnology, Inc. |
After immunostaining, the cultures were mounted in Dako medium (Dako), and wide-field images were acquired at room temperature on an inverted fluorescence microscope (SX71; Olympus) with a 20× 0.40 NA or 40× 0.60 NA objective (Olympus) using a camera (C47425; Hamamatsu) and MetaMorph version 6 acquisition software (MDS Analytical Technologies). For high-magnification and confocal images, cells were plated on 1% gelatine-coated chamber slides (Permanox; Nunc). Wide-field images were acquired at room temperature on a microscope (Axioplan; Carl Zeiss, Inc.) with a 40× 0.74 NA or 63× 1.4 NA objective (Carl Zeiss, Inc.) using a Spot RT camera (Diagnostic Instruments, Inc.) and MetaMorph version 6 acquisition software. Confocal images were acquired at room temperature on a multiphoton microscope (Radiance 2100/AGR-3Q; Bio-Rad Laboratories) with a 40× 0.6 NA objective using LS2000 software (Bio-Rad Laboratories). Images are presented after digital adjustment of curve levels (gamma) to maximize signal. In all cases, exposure times and digital manipulation were the same for control and experimental samples. Fluorochromes and color are as indicated in the figure legends.
Western blot analysis
Lysates were prepared by incubating cells in 20 mM Tris HCl, pH 8, 137 mM NaCl, 10% glycerol, 1% NP-40, and 10 mM EDTA on ice for 15 min, cleared by centrifugation, frozen in dry ice, and thawed at 4°C. 20 μg per lane of lysates were resolved by SDS-PAGE (4–12% or 8–16% gradient gels; Invitrogen), transferred onto polyvinylidene fluoride membranes (Immobilon-P; Millipore), and probed overnight at 4°C with primary antibodies; specific bands were then detected using HRP-conjugated secondary antibodies (1:20,000; GE Healthcare) and the ECL detection system (GE Healthcare). Primary antibodies are listed in Table I.
Cell cycle analysis
The cell cycle profile of cardiac cells was analyzed as described previously (Schmid et al., 1991). In brief, cells were fixed for 1 h at 4°C with 0.25% PFA and then permeabilized for 15 min at 37°C with 0.2% Tween 20 before incubation with MF20 and secondary antibodies (Alexa 488–conjugated anti–mouse Ig) diluted 1:100 in FACS buffer (PBS with 1% goat serum and 0.1% NaN3). Cells were then stained with 10 μg/ml propidium iodide in FACS buffer supplemented with 500 μg/ml of RNAase A.
Online supplemental material
Fig. S1 shows the incidences of cell cycle entry in response to N2ICD, PHE, and 10% FCS. Fig. S2 shows the presence of mitotic figures in cardiomyocytes treated with Ad-N2ICD. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200806104/DC1.
Supplementary Material
Acknowledgments
We thank Yoav Altman and Ed Monosov (Burnham Institute for Medical Research Shared Resources), and Maria Luisa de la Puerta and Maria del Carmen Rodríguez (Universidad de Valladolid) for expert advice and technical support. The Ad-N2ICD virus was provided by Tom Maciag, whose passing we regret.
M. Mercola gratefully acknowledges support from National Heart Lung and Blood Institute (grants R37 HL059502-10 and R01 HL083463-01) and a sponsored research agreement from Exelixis, Inc.
Víctor M. Campa and Raquel Gutiérrez-Lanza's present address is Instituto de Biología y Genética Molecular, Universidad de Valladolid, 47003 Valladolid, Spain.
Abbreviations used in this paper: βGal, β-galactosidase; BIO, 6-bromoindirubin-3′-oxime; CRM1, chromosome region maintenance 1; DBM, DNA-binding mutant; GSK3β, glycogen synthase kinase-3β; Hes1, hairy and enhancer of split 1; His3, histone 3; ICD, intracellular domain; LMB, leptomycin B; mESC, mouse embryonic stem cell; MLC2V, myosin light chain 2V; NMVC, neonatal mouse ventricular cardiomyocyte; NRVC, neonatal rat ventricular cardiomyocyte; P, postnatal day; PHE, phenylephrine; Rb, retinoblastoma protein; RBP-Jκ, recombination signal-binding protein 1 for Jκ.
References
- Adams, R.R., H. Maiato, W.C. Earnshaw, and M. Carmena. 2001. Essential roles of Drosophila inner centromere protein (INCENP) and aurora B in histone H3 phosphorylation, metaphase chromosome alignment, kinetochore disjunction, and chromosome segregation. J. Cell Biol. 153:865–880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agah, R., L.A. Kirshenbaum, M. Abdellatif, L.D. Truong, S. Chakraborty, L.H. Michael, and M.D. Schneider. 1997. Adenoviral delivery of E2F-1 directs cell cycle reentry and p53-independent apoptosis in postmitotic adult myocardium in vivo. J. Clin. Invest. 100:2722–2728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahuja, P., E. Perriard, J.C. Perriard, and E. Ehler. 2004. Sequential myofibrillar breakdown accompanies mitotic division of mammalian cardiomyocytes. J. Cell Sci. 117:3295–3306. [DOI] [PubMed] [Google Scholar]
- Barcova, M., V.M. Campa, and M. Mercola. 2007. Human embryonic stem cell cardiogenesis. In Human Stem Cell Manual: a Laboratory Guide. J.F. Loring, R.L. Wesselschmidt, and P.H. Schwartz, editors. Elsevier/Academic Press, Amsterdam. 227–237.
- Belka, C. 2006. The fate of irradiated tumor cells. Oncogene. 25:969–971. [DOI] [PubMed] [Google Scholar]
- Burton, P.B., M.C. Raff, P. Kerr, M.H. Yacoub, and P.J. Barton. 1999. An intrinsic timer that controls cell-cycle withdrawal in cultured cardiac myocytes. Dev. Biol. 216:659–670. [DOI] [PubMed] [Google Scholar]
- Bush, G., G. diSibio, A. Miyamoto, J.B. Denault, R. Leduc, and G. Weinmaster. 2001. Ligand-induced signaling in the absence of furin processing of Notch1. Dev. Biol. 229:494–502. [DOI] [PubMed] [Google Scholar]
- Crosio, C., G.M. Fimia, R. Loury, M. Kimura, Y. Okano, H. Zhou, S. Sen, C.D. Allis, and P. Sassone-Corsi. 2002. Mitotic phosphorylation of histone H3: spatio-temporal regulation by mammalian Aurora kinases. Mol. Cell. Biol. 22:874–885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dallas, M.H., B. Varnum-Finney, C. Delaney, K. Kato, and I.D. Bernstein. 2005. Density of the Notch ligand Delta1 determines generation of B and T cell precursors from hematopoietic stem cells. J. Exp. Med. 201:1361–1366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datwyler, D.A., J.P. Magyar, C. Weikert, L. Wightman, E. Wagner, and H.M. Eppenberger. 2003. Reactivation of the mitosis-promoting factor in postmitotic cardiomyocytes. Cells Tissues Organs. 175:61–71. [DOI] [PubMed] [Google Scholar]
- DeGregori, J. 2004. The Rb network. J. Cell Sci. 117:3411–3413. [DOI] [PubMed] [Google Scholar]
- Diehl, J.A., M. Cheng, M.F. Roussel, and C.J. Sherr. 1998. Glycogen synthase kinase-3beta regulates cyclin D1 proteolysis and subcellular localization. Genes Dev. 12:3499–3511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engel, F.B., M. Schebesta, M.T. Duong, G. Lu, S. Ren, J.B. Madwed, H. Jiang, Y. Wang, and M.T. Keating. 2005. p38 MAP kinase inhibition enables proliferation of adult mammalian cardiomyocytes. Genes Dev. 19:1175–1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fre, S., M. Huyghe, P. Mourikis, S. Robine, D. Louvard, and S. Artavanis-Tsakonas. 2005. Notch signals control the fate of immature progenitor cells in the intestine. Nature. 435:964–968. [DOI] [PubMed] [Google Scholar]
- Grego-Bessa, J., L. Luna-Zurita, G. del Monte, V. Bolos, P. Melgar, A. Arandilla, A.N. Garratt, H. Zang, Y.S. Mukouyama, H. Chen, et al. 2007. Notch signaling is essential for ventricular chamber development. Dev. Cell. 12:415–429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh, P.C., V.F. Segers, M.E. Davis, C. MacGillivray, J. Gannon, J.D. Molkentin, J. Robbins, and R.T. Lee. 2007. Evidence from a genetic fate-mapping study that stem cells refresh adult mammalian cardiomyocytes after injury. Nat. Med. 13:970–974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huh, N.E., K.B. Pasumarthi, M.H. Soonpaa, S. Jing, B. Patton, and L.J. Field. 2001. Functional abrogation of p53 is required for T-Ag induced proliferation in cardiomyocytes. J. Mol. Cell. Cardiol. 33:1405–1419. [DOI] [PubMed] [Google Scholar]
- Jung, J., T.G. Kim, G.E. Lyons, H.R. Kim, and Y. Lee. 2005. Jumonji regulates cardiomyocyte proliferation via interaction with retinoblastoma protein. J. Biol. Chem. 280:30916–30923. [DOI] [PubMed] [Google Scholar]
- Kastan, M.B., O. Onyekwere, D. Sidransky, B. Vogelstein, and R.W. Craig. 1991. Participation of p53 protein in the cellular response to DNA damage. Cancer Res. 51:6304–6311. [PubMed] [Google Scholar]
- Kohler, C., A.W. Bell, W.C. Bowen, S.P. Monga, W. Fleig, and G.K. Michalopoulos. 2004. Expression of Notch-1 and its ligand Jagged-1 in rat liver during liver regeneration. Hepatology. 39:1056–1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozar, K., M.A. Ciemerych, V.I. Rebel, H. Shigematsu, A. Zagozdzon, E. Sicinska, Y. Geng, Q. Yu, S. Bhattacharya, R.T. Bronson, et al. 2004. Mouse development and cell proliferation in the absence of D-cyclins. Cell. 118:477–491. [DOI] [PubMed] [Google Scholar]
- Leong, K.G., and A. Karsan. 2006. Recent insights into the role of Notch signaling in tumorigenesis. Blood. 107:2223–2233. [DOI] [PubMed] [Google Scholar]
- Li, F., X. Wang, J.M. Capasso, and A.M. Gerdes. 1996. Rapid transition of cardiac myocytes from hyperplasia to hypertrophy during postnatal development. J. Mol. Cell. Cardiol. 28:1737–1746. [DOI] [PubMed] [Google Scholar]
- Li, F., X. Wang, P.C. Bunger, and A.M. Gerdes. 1997. Formation of binucleated cardiac myocytes in rat heart: I. Role of actin-myosin contractile ring. J. Mol. Cell. Cardiol. 29:1541–1551. [DOI] [PubMed] [Google Scholar]
- Loomes, K.M., D.B. Taichman, C.L. Glover, P.T. Williams, J.E. Markowitz, D.A. Piccoli, H.S. Baldwin, and R.J. Oakey. 2002. Characterization of Notch receptor expression in the developing mammalian heart and liver. Am. J. Med. Genet. 112:181–189. [DOI] [PubMed] [Google Scholar]
- Luckow, B., and G. Schutz. 1987. CAT constructions with multiple unique restriction sites for the functional analysis of eukaryotic promoters and regulatory elements. Nucleic Acids Res. 15:5490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mammucari, C., A. Tommasi di Vignano, A.A. Sharov, J. Neilson, M.C. Havrda, D.R. Roop, V.A. Botchkarev, G.R. Crabtree, and G.P. Dotto. 2005. Integration of Notch 1 and calcineurin/NFAT signaling pathways in keratinocyte growth and differentiation control. Dev. Cell. 8:665–676. [DOI] [PubMed] [Google Scholar]
- Miele, L., T. Golde, and B. Osborne. 2006. Notch signaling in cancer. Curr. Mol. Med. 6:905–918. [DOI] [PubMed] [Google Scholar]
- Miyamoto, Y., A. Maitra, B. Ghosh, U. Zechner, P. Argani, C.A. Iacobuzio-Donahue, V. Sriuranpong, T. Iso, I.M. Meszoely, M.S. Wolfe, et al. 2003. Notch mediates TGF alpha-induced changes in epithelial differentiation during pancreatic tumorigenesis. Cancer Cell. 3:565–576. [DOI] [PubMed] [Google Scholar]
- Nakamura, K., and C. Chiba. 2007. Evidence for Notch signaling involvement in retinal regeneration of adult newt. Brain Res. 1136:28–42. [DOI] [PubMed] [Google Scholar]
- Niessen, K., and A. Karsan. 2007. Notch signaling in the developing cardiovascular system. Am. J. Physiol. Cell Physiol. 293:C1–C11. [DOI] [PubMed] [Google Scholar]
- Poss, K.D. 2007. Getting to the heart of regeneration in zebrafish. Semin. Cell Dev. Biol. 18:36–45. [DOI] [PubMed] [Google Scholar]
- Ramain, P., K. Khechumian, L. Seugnet, N. Arbogast, C. Ackermann, and P. Heitzler. 2001. Novel Notch alleles reveal a Deltex-dependent pathway repressing neural fate. Curr. Biol. 11:1729–1738. [DOI] [PubMed] [Google Scholar]
- Raya, A., C.M. Koth, D. Buscher, Y. Kawakami, T. Itoh, R.M. Raya, G. Sternik, H.J. Tsai, C. Rodriguez-Esteban, and J.C. Izpisua-Belmonte. 2003. Activation of Notch signaling pathway precedes heart regeneration in zebrafish. Proc. Natl. Acad. Sci. USA. 100:11889–11895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raya, A., A. Consiglio, Y. Kawakami, C. Rodriguez-Esteban, and J.C. Izpisua-Belmonte. 2004. The zebrafish as a model of heart regeneration. Cloning Stem Cells. 6:345–351. [DOI] [PubMed] [Google Scholar]
- Ronchini, C., and A.J. Capobianco. 2001. Induction of cyclin D1 transcription and CDK2 activity by Notch(ic): implication for cell cycle disruption in transformation by Notch(ic). Mol. Cell. Biol. 21:5925–5934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rones, M.S., K.A. McLaughlin, M. Raffin, and M. Mercola. 2000. Serrate and Notch specify cell fates in the heart field by suppressing cardiomyogenesis. Development. 127:3865–3876. [DOI] [PubMed] [Google Scholar]
- Rutenberg, J.B., A. Fischer, H. Jia, M. Gessler, T.P. Zhong, and M. Mercola. 2006. Developmental patterning of the cardiac atrioventricular canal by Notch and Hairy-related transcription factors. Development. 133:4381–4390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkaria, J.N., E.C. Busby, R.S. Tibbetts, P. Roos, Y. Taya, L.M. Karnitz, and R.T. Abraham. 1999. Inhibition of ATM and ATR kinase activities by the radiosensitizing agent, caffeine. Cancer Res. 59:4375–4382. [PubMed] [Google Scholar]
- Schmid, I., C.H. Uittenbogaart, and J.V. Giorgi. 1991. A gentle fixation and permeabilization method for combined cell surface and intracellular staining with improved precision in DNA quantification. Cytometry. 12:279–285. [DOI] [PubMed] [Google Scholar]
- Schweisguth, F. 2004. Regulation of notch signaling activity. Curr. Biol. 14:R129–R138. [PubMed] [Google Scholar]
- Small, D., D. Kovalenko, R. Soldi, A. Mandinova, V. Kolev, R. Trifonova, C. Bagala, D. Kacer, C. Battelli, L. Liaw, et al. 2003. Notch activation suppresses fibroblast growth factor-dependent cellular transformation. J. Biol. Chem. 278:16405–16413. [DOI] [PubMed] [Google Scholar]
- Solodushko, V., and B. Fouty. 2007. Proproliferative phenotype of pulmonary microvascular endothelial cells. Am. J. Physiol. Lung Cell. Mol. Physiol. 292:L671–L677. [DOI] [PubMed] [Google Scholar]
- Stylianou, S., R.B. Clarke, and K. Brennan. 2006. Aberrant activation of notch signaling in human breast cancer. Cancer Res. 66:1517–1525. [DOI] [PubMed] [Google Scholar]
- Su, Y., P. Buchler, A. Gazdhar, N. Giese, H.A. Reber, O.J. Hines, T. Giese, M.W. Buchler, and H. Friess. 2006. Pancreatic regeneration in chronic pancreatitis requires activation of the notch signaling pathway. J. Gastrointest. Surg. 10:1230–1242. [DOI] [PubMed] [Google Scholar]
- Sumrejkanchanakij, P., M. Tamamori-Adachi, Y. Matsunaga, K. Eto, and M.A. Ikeda. 2003. Role of cyclin D1 cytoplasmic sequestration in the survival of postmitotic neurons. Oncogene. 22:8723–8730. [DOI] [PubMed] [Google Scholar]
- Tamamori-Adachi, M., H. Ito, P. Sumrejkanchanakij, S. Adachi, M. Hiroe, M. Shimizu, J. Kawauchi, M. Sunamori, F. Marumo, S. Kitajima, and M.A. Ikeda. 2003. Critical role of cyclin D1 nuclear import in cardiomyocyte proliferation. Circ. Res. 92:e12–e19. [DOI] [PubMed] [Google Scholar]
- Tanigaki, K., H. Han, N. Yamamoto, K. Tashiro, M. Ikegawa, K. Kuroda, A. Suzuki, T. Nakano, and T. Honjo. 2002. Notch-RBP-J signaling is involved in cell fate determination of marginal zone B cells. Nat. Immunol. 3:443–450. [DOI] [PubMed] [Google Scholar]
- Tseng, A.S., F.B. Engel, and M.T. Keating. 2006. The GSK-3 inhibitor BIO promotes proliferation in mammalian cardiomyocytes. Chem. Biol. 13:957–963. [DOI] [PubMed] [Google Scholar]
- van Es, J.H., M.E. van Gijn, O. Riccio, M. van den Born, M. Vooijs, H. Begthel, M. Cozijnsen, S. Robine, D.J. Winton, F. Radtke, and H. Clevers. 2005. Notch/gamma-secretase inhibition turns proliferative cells in intestinal crypts and adenomas into goblet cells. Nature. 435:959–963. [DOI] [PubMed] [Google Scholar]
- Watanabe, Y., H. Kokubo, S. Miyagawa-Tomita, M. Endo, K. Igarashi, K. Aisaki, J. Kanno, and Y. Saga. 2006. Activation of Notch1 signaling in cardiogenic mesoderm induces abnormal heart morphogenesis in mouse. Development. 133:1625–1634. [DOI] [PubMed] [Google Scholar]
- Winick, M., and A. Noble. 1965. Quantitative changes in DNA, RNA, and protein during prenatal and postnatal growth in the rat. Dev. Biol. 12:451–466. [DOI] [PubMed] [Google Scholar]
- Yoshida, M., M. Nishikawa, K. Nishi, K. Abe, S. Horinouchi, and T. Beppu. 1990. Effects of leptomycin B on the cell cycle of fibroblasts and fission yeast cells. Exp. Cell Res. 187:150–156. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.