Abstract
The zinc finger protein ZPR1 translocates from the cytoplasm to the nucleus after treatment of cells with mitogens. The function of nuclear ZPR1 has not been defined. Here we demonstrate that ZPR1 accumulates in the nucleolus of proliferating cells. The role of ZPR1 was examined using a gene disruption strategy. Cells lacking ZPR1 are not viable. Biochemical analysis demonstrated that the loss of ZPR1 caused disruption of nucleolar function, including preribosomal RNA expression. These data establish ZPR1 as an essential protein that is required for normal nucleolar function in proliferating cells.
INTRODUCTION
ZPR1 is a zinc finger protein that is present in the cytoplasm of quiescent cells (Galcheva-Gargova et al., 1996). Coimmunoprecipitation analysis demonstrates that ZPR1 interacts with the cytoplasmic domain of receptor tyrosine kinases, including the epidermal growth factor (EGF) receptor. Deletion analysis indicates that this binding interaction is mediated by the zinc fingers (C-X2-C-X25-C-X2-C) of ZPR1. The interaction between ZPR1 and the cytoplasmic domain of the EGF receptor is disrupted by treatment of cells with EGF. Thus, ZPR1 binds selectively to the inactive EGF receptor. The mechanism used by ZPR1 to discriminate between inactive and active EGF receptors is unclear. However, the observation that EGF does not regulate ZPR1 binding to kinase-negative EGF receptors suggests that tyrosine phosphorylation is required. Measurement of EGF-stimulated Shc phosphorylation in vivo indicates that the overexpression of ZPR1 suppresses EGF receptor tyrosine kinase activity. The cytoplasmic ZPR1 protein may therefore function to repress the basal state of the EGF receptor tyrosine kinase (Galcheva-Gargova et al., 1996).
Treatment with mitogens, including EGF, causes translocation of cytoplasmic ZPR1 into the nucleus (Galcheva-Gargova et al., 1996). The primary sequence of ZPR1 does not contain an obvious nuclear localization signal, and the mechanism of induced nuclear import has not been defined. The binding interaction between ZPR1 and the cytoplasmic domain of tyrosine kinase receptors could serve as a mechanism of cytoplasmic retention. However, analysis of the stoichiometry of the ZPR1–receptor interaction indicates that the amount of ZPR1 exceeds the amount of receptor in most cell types (Galcheva-Gargova et al., 1996). Thus, retention by receptors is insufficient to account for the cytoplasmic sequestration of ZPR1 in quiescent cells. Additional growth factor-regulated mechanisms must therefore contribute to induced nuclear accumulation of ZPR1.
The regulated nuclear accumulation of ZPR1 suggests that this protein may function as a signaling molecule. ZPR1 could participate in many nuclear processes. For example, ZPR1 could be a regulated transcription factor that is sequestered in the cytoplasm in latent form before activation, similar to NF-κB, NFAT, STAT, or SMAD (Hunter, 1997). The purpose of this study was to identify a nuclear function for ZPR1 in proliferating cells. We demonstrate that nuclear ZPR1 accumulates within the nucleolus. Disruption of the ZPR1 gene causes defects in the biochemical properties of the nucleolus. The ZPR1 protein may therefore contribute to the normal function of the nucleolus in proliferating cells.
MATERIALS AND METHODS
Molecular Cloning of ZPR1
The human ZPR1 cDNA was isolated by screening a HeLa cDNA library cloned in λ phage (Stratagene, La Jolla, CA) using the murine ZPR1 cDNA (Galcheva-Gargova et al., 1996) as a probe. Similar procedures were used to isolate the Schizosaccharomyces pombe zpr1+ and the Saccharomyces cerevisiae ZPR1 genes by screening genomic libraries. These clones were characterized by automated sequencing using an Applied Biosystems (Foster City, CA) model 373A machine.
Immunofluorescence Analysis
HEp-2 and A431 cells were examined by immunofluorescence analysis using methods described previously (Galcheva-Gargova et al., 1996). The cells were fixed with methanol (5 min, −20°C) followed by acetone (2 min, −20°C). The cells were permeabilized with 0.1% Triton X-100 and incubated with primary antibodies diluted in PBS. Double-labeling experiments were performed using rabbit anti-ZPR1 and characterized human polyclonal or mouse monoclonal antibodies. Incubations were performed at 25°C (1 h). After extensive washing, the cells were incubated with appropriate secondary reagents (Caltag Laboratories, San Francisco, CA), washed, and mounted on slides using Vectashield (Vector Laboratories, Burlingame, CA). The rabbit polyclonal antibody to ZPR1 has been described (Galcheva-Gargova et al., 1996). Antisera to human p80 coilin (Andrade et al., 1991) and Pol I (Reimer et al., 1987b) were provided by Dr. E.K.L. Chan (The Scripps Research Institute, La Jolla, CA). The mouse monoclonal antibody to fibrillarin 72B9 (Reimer et al., 1987a) was provided by Dr. K.M. Pollard (Scripps Research Institute); the anti-DNA antibody 1.D12 (Kotzin et al., 1984) was provided by Dr. R.L. Rubin (Scripps Research Institute); the anti-SC-35 antibody (Fu and Maniatis, 1990) was provided by Dr. X.-D. Fu (Harvard University, Cambridge, MA); and the anti-B23 monoclonal antibody was provided by Dr. I. Todorov (Desmos Inc., San Diego, CA) (Zatsepina et al., 1997).
Analysis of Yeast
Genetic and biochemical manipulation of S. pombe was done using standard techniques (Moreno et al., 1991). Disruption of the zpr1+ gene was done by insertion of the ura4+ gene (1.7-kb BamHI–HindIII fragment) in the BstE2 and EcoRV sites of the zpr1+ coding region (2.9-kb XbaI fragment). The disrupted zpr1+ genomic clone (4.6-kb XbaI fragment) was transformed in the diploid strain 480. Disruption of the zpr1+ gene was confirmed by Southern blot analysis of genomic DNA isolated from transformants. The heterozygous diploid strain was designated TE630 [zpr1::ura4+/zpr1+, ade6-M210/ade6-M216, ura4-d18/ura4-d18, leuI-32/leuI-32, h−/h+]. Control haploid isogenic strains isolated by tetrad analysis were designated TE331 and TE332 [ade6-M210, ura4-d18, leuI-32 h+ (or h−)].
Complementation studies were done using the promoterless vector pIRT2 and the nmt promoter vector pREP41. The S. pombe zpr1+ gene (2.9-kb XbaI fragment) was cloned into the SmaI site of pIRT2. Regulated expression vectors for S. pombe zpr1+, S. cerevisiae ZPR1, and murine ZPR1 were constructed by cloning PCR fragments in the polylinker of pREP41. The yeast strain TE630 was transformed, and haploid yeast were selected on plates supplemented with adenine. The growth of the haploid yeast was examined on agar plates and liquid minimal medium in the absence and presence of thiamine (10 mM). Cells grown to midlog phase in liquid culture were used for RNA isolation, [35S]methionine labeling, and microscopy using standard procedures. The RNA was examined by Northern blot analysis by probing with a random-primed PCR fragment (bp 150-1120) corresponding to the 5′ external transcribed spacer (ETS) region of S. pombe pre-rRNA (GenBank accession number Z19578).
The sequences of the murine ZPR1 cDNA, the human ZPR1 cDNA, the S. pombe zpr1+ gene, and the S. cerevisiae ZPR1 gene have been deposited in GenBank with accession numbers U41287, AF019767, AF019768, and AF019769, respectively.
RESULTS
ZPR1 Is Localized in a Subregion of the Nucleus
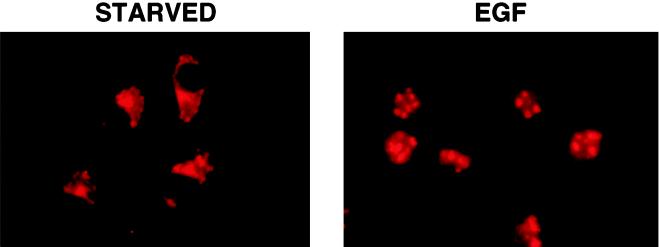
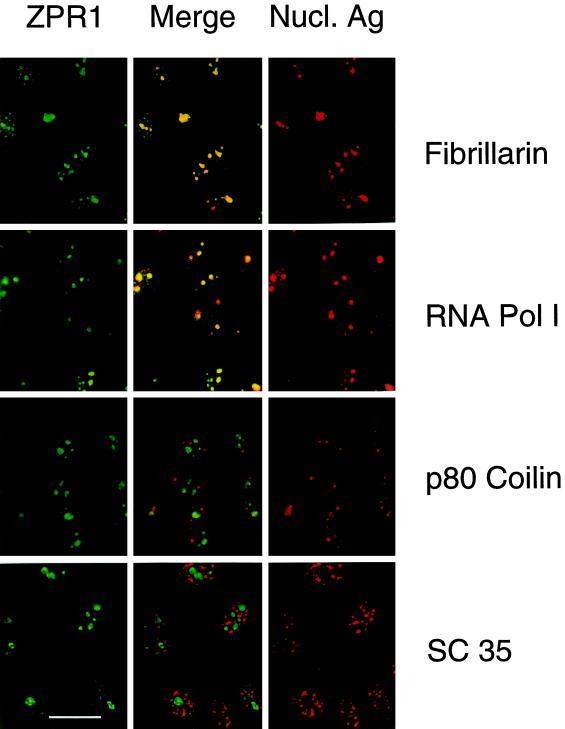
The mammalian ZPR1 protein is located in the cytoplasm of serum-starved cells (Galcheva-Gargova et al., 1996). Treatment with growth factors leads to the accumulation of ZPR1 in the nucleus (GalchevaGargova et al., 1996). This redistribution of ZPR1 from the cytoplasm to the nucleus was detected in cells treated with EGF (Figure 1). The intranuclear distribution of ZPR1 was punctate in appearance. To gain insight into the function of ZPR1, we examined the subnuclear localization of ZPR1. Immunofluorescence analysis of serum-treated human HEp-2 cells demonstrated staining of the nucleoplasm with a distinct punctate appearance. Double-label immunofluorescence analysis demonstrated that the punctate ZPR1 staining colocalized extensively with fibrillarin and RNA Pol I, but not with splicing factor SC35 or p80 coilin (Figure 2). These data indicate that ZPR1 may accumulate within the nucleolus.
Figure 1.
The cytoplasmic ZPR1 protein redistributes to the nucleus of mitogen-activated cells. A431 cells were incubated without (STARVED) and with 100 nM EGF for 15 min. The cells were processed for immunofluorescence microscopy using an antibody to ZPR1.
Figure 2.
Nuclear ZPR1 accumulates in the nucleolus. Activated HEp-2 cells were examined by laser scanning confocal microscopy using antibodies to ZPR1 (FITC, green) and nuclear antigens (rhodamine, red). In the merged image, fibrillarin and RNA Pol I show extensive colocalization (yellow), whereas p80 coilin and SC35 splicing factor show extensive segregation (green and red). Bar, 35 μm. Fibrillarin and RNA Pol I are markers for the nucleolus.
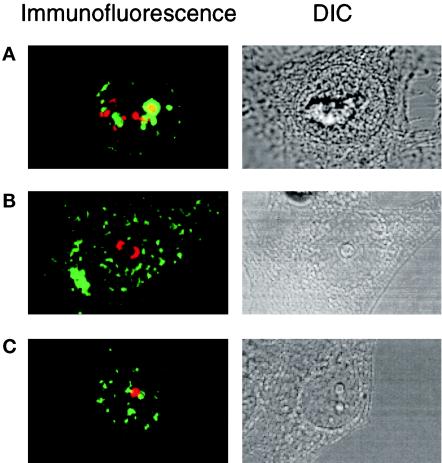
Comparison of the intranuclear staining of fibrillarin and RNA Pol I with ZPR1 indicates that the distribution of these proteins is not identical. For example, fibrillarin is located in both coiled bodies and the nucleolus. In addition, both fibrillarin and RNA Pol I are located within specific subregions of the nucleolus. The nucleolus is composed of separate regions, including fibrillar centers, the dense fibrillar component, and the granular component (Shaw and Jordan, 1995). Each of these components is thought to correspond to a different functional region of the nucleolus. Because these regions are interspersed, it is not possible to determine the subnucleolar localization of ZPR1 by immunofluorescence microscopy. However, the nucleolus can be segregated into fibrillar and granular regions using drugs (Simar and Bernhardt, 1966; Ochs et al., 1985). For example, at low doses, actinomycin D intercalates into GC-rich regions of DNA and markedly inhibits Pol I transcription of rRNA (Abelson and Penman, 1975). Treatment of HEp-2 cells with actinomycin D caused the formation of fibrillar caps (stained with an antibody to fibrillarin) and the rapid dissociation of ZPR1 from the nucleolus (Figure 3A). These data indicated that ZPR1 was not located in the same compartment of the nucleolus as fibrillarin. Because actinomycin D causes some disruption of the granular region of the nucleolus (Yung et al., 1985), it is possible that ZPR1 may be located within the granular component of the nucleolus. To test this hypothesis, we examined the effect of the adenosine analogue 5,6-dichloro-β-d-ribofuranosylbenzimidazole (DRB), which induces segregation of nucleolar components without altering the ultrastructural characteristics of the fibrillar and granular compartments (Granick, 1975). These experiments demonstrated that ZPR1 did not colocalize with either a marker for the fibrillar component (fibrillarin) (Figure 3B) or the granular component (B23) of the segregated nucleolus (Figure 3C). In the absence of DRB, it is possible that ZPR1 may associate with either the fibrillar or the granular component of the nucleolus in proliferating cells. Further studies are required to resolve this question.
Figure 3.
ZPR1 is not an integral component of the fibrillar or granular components of the nucleolus. Proliferating HEp-2 cells were treated (4 h) with 0.1 μg/ml actinomycin D or with 25 μg/ml of the adenosine analogue DRB, fixed, and processed for immunofluorescence microscopy using antibodies to ZPR1 (FITC, green) and antibodies that stain the fibrillar component (fibrillarin; rhodamine, red) and the granular component (B23; rhodamine, red) of the nucleolus. Cells treated with actinomycin D were stained with ZPR1 and fibrillarin (A). Cells treated with DRB were stained with antibodies to ZPR1 and fibrillarin (B) or ZPR1 and B23 (C). A photomicrograph of the differential interference contrast (DIC) image is presented in each panel.
RNA Is Required for the Nucleolar Localization of ZPR1
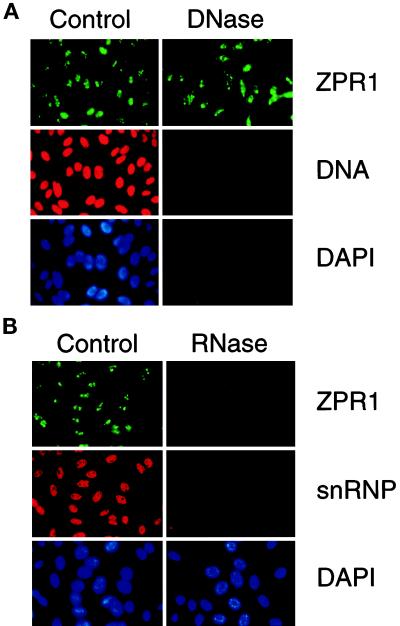
The nucleolus is the major site of transcription of rRNA genes and the processing of rRNA into preribosomal particles (Woolford and Warner, 1991; Shaw and Jordan, 1995). The nucleic acid composition of the nucleolus differs from other regions of the nucleus because of the abundance of rRNA genes and rRNA transcripts. This distinctive nucleic acid composition (both DNA and RNA) may contribute to the accumulation of ZPR1 in the nucleolus. To examine the possible role of nucleic acids, we investigated the effect of nuclease digestion on ZPR1 localization by immunofluorescence microscopy. Digestion of permeabilized cells with DNase I caused a marked decrease in nuclear DNA, which was detected using a monoclonal antibody to DNA (Figure 4A). However, this treatment did not affect the nucleolar location of ZPR1 (Figure 4A). In contrast, digestion with RNase A caused a marked decrease in the nucleolar location of ZPR1 (Figure 4B). Control studies demonstrated that RNase A digestion did not alter nuclear DNA (our unpublished results) but did reduce the nuclear accumulation of small nuclear ribonuclear protein (snRNP) (Figure 4B). Together, these data indicate that the nucleolar localization of ZPR1 requires RNA but not DNA. The requirement for RNA suggests that ZPR1 nucleolar localization may be mediated by the interaction of ZPR1 (directly or indirectly) with RNA.
Figure 4.
RNA is required for the nucleolar localization of ZPR1. HEp-2 cells were grown on microscopic slides, permeabilized with 0.1% Triton X-100 for 3 min on ice, washed, and digested (60 min at 37°C) with 0.1 mg/ml DNase I in PBS containing 5 mM MgCl2 or with 0.1 mg/ml RNase A in PBS. The cells were washed in PBS and processed for indirect immunofluorescence. Buffers without enzymes served as negative controls. A human antibody to snRNP or a monoclonal antibody to DNA was used to monitor the efficiency of DNase I and RNase A digestions, respectively. The effect of DNase I (A) and RNase A (B) is presented.
The Zinc Finger Protein ZPR1 Is Conserved in Mammals and Yeast
The ZPR1 protein was first identified in mice (GalchevaGargova et al., 1996). A human homologue was identified by screening a cDNA library for sequences related to ZPR1. The human ZPR1 protein deduced from the sequence of cDNA clones is similar to mouse ZPR1 (Figure 5). Homologues of ZPR1 were also identified in yeast, including the budding yeast S. cerevisiae and the fission yeast S. pombe (Figure 5). Comparison of the sequence of the mammalian and yeast ZPR1 proteins demonstrates that they share conserved structural motifs, including the presence of two zinc fingers (C-X2-C-X25-C-X2-C). The presence of two molecules of zinc per molecule of ZPR1 was confirmed by atomic absorbtion spectroscopy (Galcheva-Gargova et al., 1996). The high level of conservation of ZPR1 between mammals and yeast indicates that ZPR1 is likely to serve an important basic physiological function.
Figure 5.
The ZPR1 gene is highly conserved in mammals and yeast. The sequences of the human, mouse, and yeast (S. pombe and S. cerevisiae) ZPR1 proteins were compared using the PILE-UP program (version 7.2; Wisconsin Genetics Computer Group, Madison, WI). Residues that are identical with human ZPR1 are indicated with a period. Gaps were introduced to optimize the alignment (–). The two zinc fingers are overlined, and the Cys residues are indicated with asterisks. The sequence of mouse ZPR1 (GenBank accession number U41287) has been reported (Galcheva-Gargova et al., 1996). The human and yeast ZPR1 protein sequences were deduced from the nucleotide sequence of HeLa cDNA clones isolated from a λZAP II phage library (Stratagene) and from the sequence of yeast genomic clones. The sequences of the human ZPR1 cDNA, the S. pombe zpr1+ gene, and the S. cerevisiae ZPR1 gene have been deposited in GenBank with accession numbers AF019767, AF019768, and AF019769, respectively.
Gene Disruption Studies Demonstrate That ZPR1 Is Essential for Cell Viability
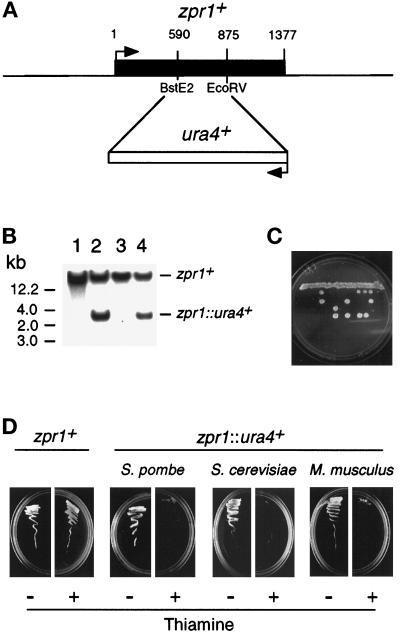
We examined the effect of disruption of the ZPR1 gene. These studies were facilitated by the identification of ZPR1 in fission and budding yeast (Figure 5). We focused our analysis on the zpr1+ gene of the fission yeast S. pombe. We used homologous recombination to disrupt the zpr1+ gene by replacement with the ura4+ gene (Figure 6, A and B). These heterozygous diploid yeast were sporulated and examined by tetrad analysis (Figure 6C). Viability segregated 2:2, and none of the viable haploid colonies were ura4+. Thus, zpr1+ is an essential gene in S. pombe. Similar studies demonstrated that the ZPR1 gene was also essential for viability in S. cerevisiae (our unpublished results).
Figure 6.
The zpr1+, gene is essential for cell viability. (A) The S. pombe zpr1+ gene was disrupted by homologous recombination. The structure of the zpr1+ genomic locus and the disrupted gene (zpr1::ura4+) is presented schematically. (B) Southern blot analysis of the diploid yeast transformants. The genomic DNA was restricted with BglII and probed with a random-primed fragment of the zpr1+ genomic locus (2.9-kb XbaI fragment). The wild-type zpr1+ allele was identified in wild-type yeast (strain 480; lane 1). The disrupted zpr1::ura4+ allele (3 kb) was identified in some (lanes 2 and 4) but not all (lane 3) transformants. (C) The heterozygous diploid yeast strain TE630 (zpr1+/zpr1::ura4+) was sporulated, and the tetrads were dissected. The viability of the spores was examined by growth on agar plates supplemented with uracil. (D) The heterozygous (zpr1+/zpr1::ura4+) diploid yeast strain TE630 was transformed with the plasmid pREP41 or the plasmid pREP41-zpr1, selected on minimal agar plates without leucine and uracil, and sporulated, and haploid yeast were selected on minimal media supplemented with adenine. No viable haploid yeast (zpr1::ura4+) were obtained from diploid yeast transformed with pREP41. However, the zpr1 expression vector pREP41-zpr1 complemented the lethal phenotype of the disrupted zpr1+ gene. Complementation was observed in experiments using S. pombe zpr1+, S. cerevisiae ZPR1, and murine ZPR1. Repression of the nmt promoter in the pREP plasmid with thiamine decreased the growth of the complemented ZPR::ura4+ haploid strains, but not the wild-type zpr1+ haploid strain transformed with pREP41-zpr1.
To demonstrate that the loss of viability was caused by the disruption of the zpr1+ gene, we performed complementation analysis using plasmid vectors that express zpr1+. We found that a 2.9-kb XbaI genomic fragment that contained the zpr1+ gene complemented the loss of viability caused by the disrupted zpr1+ allele (our unpublished results). Furthermore, complementation was observed in experiments using a plasmid vector (pREP41-zpr1) in which the zpr1+ coding sequence was expressed under the control of the nmt promoter (Figure 6D). Repression of the nmt promoter with thiamine did not affect the growth of the control (zpr1+) strain transformed with pREP41-zpr1. In contrast, thiamine markedly decreased the growth of the zpr1::ura4+ strain (Figure 6D). Similar results were obtained in complementation studies using S. cerevisiae and murine ZPR1 (Figure 6D). The observation of complementation by both mammalian and yeast ZPR1 indicates that the biological function of ZPR1 has been conserved during evolution.
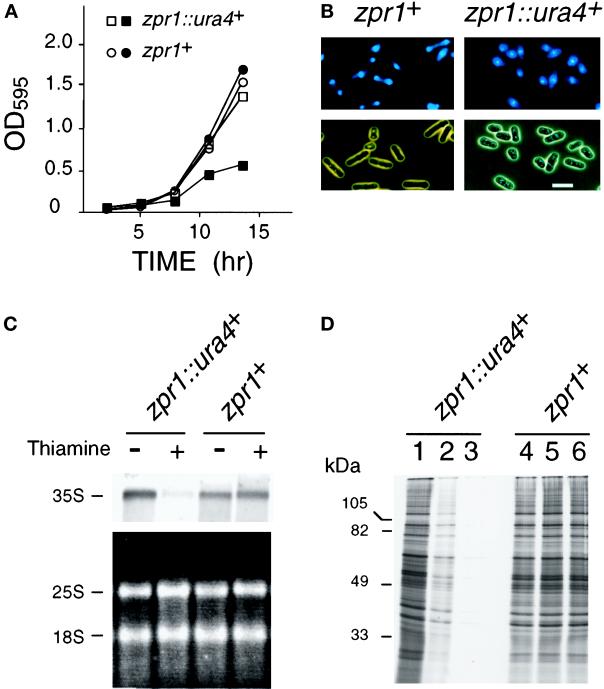
We performed quantitative analysis of the effect of thiamine-induced repression of zpr1+ on the growth of yeast transformed with pREP41-zpr1 in liquid culture. These experiments were performed using haploid yeast (zpr1+ and zpr1::ura4+ strains) that express the zpr1+ gene from the thiamine-repressible nmt promoter. Thiamine caused no change in the growth of the zpr1+ strain in liquid cultures but caused decreased growth of the zpr1::ura4+ strain (Figure 7A). Microscopic analysis demonstrated that the zpr1+ strain consisted of a population of yeast that were distributed throughout the cell cycle, including both large and small yeast (Figure 7B). In contrast, cultures of the zpr1::ura4+ strain contained a more uniform population of small yeast (Figure 7B). The morphology of the zpr1::ura4+ strain is consistent with a growth-arrested phenotype.
Figure 7.
Loss of zpr1+ function causes depletion of the rRNA precursor and decreased protein translation. (A) Wild-type haploid S. pombe (zpr1+) and the zpr1::ura4+ disrupted strain were transformed with the plasmid pREP41-zpr1 and grown in minimal liquid medium. The cultures were divided into two flasks in the absence (open symbols) and presence (closed symbols) of thiamine, respectively. Thiamine is a repressor of the nmt promoter located in the pREP plasmid. The growth of the cultures was monitored by measurement of the optical density at 595 nm. (B) The morphology of the yeast grown in the presence of thiamine (12 h) was examined by phase-contrast microscopy. DNA stained with 4,6-diamidino-2-phenylindole was visualized by epifluorescence. Bar, 10 μm. (C) Northern blot analysis of RNA isolated from the zpr1+ and the zpr1::ura4+ disrupted S. pombe strains transformed with the plasmid pREP41-zpr1. The yeast were grown in the absence and presence of the repressor thiamine (12 h). Ten micrograms of RNA isolated from these yeast were examined by denaturing agarose gel electrophoresis. The 25 and 18S mature rRNA were detected by staining with ethidium bromide (bottom panel). The 35S rRNA precursor was detected by Northern analysis using a 5′-ETS probe. An autoradiogram of the dried blot is shown (top panel). (D) Wild-type and zpr1::ura4+ disrupted strains were grown in the absence (lanes 1 and 4) and presence (lanes 2, 3, 5, and 6) of the repressor thiamine (12 h). The cells were diluted to the same density (0.2 OD595), labeled with [35S]methionine (150 μCi/ml) for 3 h, and harvested. The labeling with [35S]methionine was performed in the absence (lanes 1, 2, 4, and 5) and presence (lanes 3 and 6) of thiamine. Extracts prepared from the yeast were examined by SDS-PAGE and autoradiography.
Repression of zpr1+ Causes Defects in Nucleolar Function
Because zpr1+ is an essential gene and the ZPR1 protein is located in the nucleolus of proliferating cells, we examined whether the repression of zpr1+ expression caused defects in nucleolar function. The nucleolus is the major site of transcription of rRNA genes. We therefore investigated rRNA expression in haploid yeast (zpr1+ and zpr1::ura4+ strains) that express the zpr1+ gene from the thiamine-repressible nmt promoter (Figure 7C). RNA was prepared from these yeast strains cultured in the absence and presence of thiamine. The yield of total RNA from the thiamine-treated zpr1::ura4+ strain was reduced compared with the wild-type zpr1+ strain. The decreased amount of total RNA is accounted for, at least in part, by decreased amounts of rRNA. However, there was no change in the ratio of the mature 18 and 25S rRNA. The decreased amount of mature rRNA suggests that rRNA expression may be altered by repression of zpr1+ gene expression. To test this hypothesis, we examined the 35S rRNA precursor by Northern blot analysis of 10 μg of total RNA (Figure 7C). This analysis demonstrated that the thiamine-induced repression of zpr1+ gene expression in the zpr1::ura4+ strain caused a marked reduction in the accumulation of 35S pre-rRNA. Control experiments using the wild-type strain (zpr1+) demonstrated that thiamine caused no change in the accumulation of 35S pre-rRNA. These data demonstrate that zpr1+ is required for the accumulation of the rRNA precursor.
The ribosome is the cellular machine that is used for protein synthesis. If zpr1+ is required for rRNA expression, then the repression of zpr1+ gene expression should have consequences on protein synthesis. We therefore examined the effect of repression of zpr1+ gene expression on protein synthesis. Liquid cultures of these yeast were incubated with [35S]methionine for 3 h, and the incorporation of radioactivity into protein was examined after SDS-PAGE by autoradiography (Figure 7D). Addition of thiamine to the zpr1+ strain caused no change in the incorporation of [35S]methionine (Figure 7D, lanes 4–6). The extent of [35S]methionine incorporation was similar to that observed in experiments using the zpr1::ura4+ strain grown in the absence of thiamine (Figure 7D, lane 1). In contrast, addition of thiamine to the zpr1::ura4+ strain caused a marked reduction in protein synthesis (Figure 7D, lane 3). A partial recovery of protein synthesis was observed if thiamine was omitted from the culture during the incubation with [35S]methionine (Figure 7D, lane 2). Together, these data demonstrate that the loss of zpr1+ expression interferes with protein biosynthesis. The marked reduction in protein synthesis may account for the small size and reduced growth of the zpr1::ura4+ strain (Figure 7, A and B).
DISCUSSION
ZPR1 was initially identified in the mouse (Galcheva-Gargova et al., 1996). Human ZPR1 was found to be very similar to the mouse protein (Figure 5). Examination of ZPR1 expression in other organisms identified homologues of ZPR1 in the budding yeast S. cerevisiae and the fission yeast S. pombe (Figure 5). Gene disruption studies demonstrated that ZPR1 is an essential gene in fission yeast (Figure 6) and budding yeast (our unpublished results). The lethal phenotype in fission yeast was complemented by expression of yeast or mammalian ZPR1 proteins (Figure 6). The identification of ZPR1 as an essential gene that has been conserved during evolution indicates that ZPR1 serves a similar function in many cell types.
The ZPR1 protein is located in the cytoplasm of quiescent mammalian cells. However, in proliferating cells, ZPR1 translocates to the nucleus (GalchevaGargova et al., 1996). The mechanism that controls the nuclear redistribution of ZPR1 in proliferating cells is not understood. However, it is possible that the interaction of ZPR1 with cell surface receptor tyrosine kinases may contribute to the cytoplasmic sequestration of ZPR1 in quiescent cells (Galcheva-Gargova et al., 1996). Treatment of mammalian cells with mitogens causes the dissociation of ZPR1 from receptor tyrosine kinases and the translocation of ZPR1 to the nucleus. Whether the binding of ZPR1 to tyrosine kinase receptors in quiescent cells is mechanistically related to the cytoplasmic retention of ZPR1 is unclear. Interestingly, the nuclear redistribution of ZPR1 observed in mammalian cells is also observed in yeast. Thus, ZPR1 is a nuclear protein in proliferating yeast and is a cytoplasmic protein in growth-arrested yeast (our unpublished results). This observation indicates that the growth-associated redistribution of ZPR1 from the cytoplasm to the nucleus is conserved in mammals and yeast. This conservation of function suggests that the mechanism of nuclear redistribution of ZPR1 may also be conserved in yeast and mammals. It is likely that a conserved mechanism would not involve cytoplasmic retention by cell surface receptor tyrosine kinases because such molecules are not expressed in yeast. Clearly, further studies are required to define the mechanism of proliferation-associated nuclear redistribution of ZPR1 and to determine the significance of the interaction of ZPR1 with receptor tyrosine kinases in mammalian cells.
Immunofluorescence analysis demonstrated that ZPR1 is accumulated within a subnuclear compartment, the nucleolus (Figure 2). Fractionation experiments demonstrated that ZPR1 was not an integral component of the fibrillar or granular compartments of the nucleolous (Figure 3), suggesting that either ZPR1 is peripherally associated with one of these nucleolar compartments or is located in a distinct subregion of the nucleolus. However, the nucleolar location of ZPR1 required RNA, suggesting that nucleolar integrity was required for the accumulation of ZPR1 (Figure 4). The movement of ZPR1 from the cytoplasm to the nucleolus in response to proliferative signals indicates that ZPR1 may have a signaling role. However, the nuclear function of ZPR1 has not been established by previous studies (Galcheva-Gargova et al., 1996).
The major functions of the nucleolus are thought to be the transcription of rRNA genes, the processing of the pre-rRNA precursor to form mature rRNA, and the assembly of rRNA into preribosomal particles (Shaw and Jordan, 1995). Because ZPR1 is a nucleolar protein that is essential for cell viability, a plausible role for ZPR1 is to contribute to one step in the process of rRNA expression. Consistent with this hypothesis, we found that the repression of zpr1+ gene expression in fission yeast caused a marked decrease in the level of pre-rRNA (Figure 7C). This decreased rRNA is likely to account for the growth arrest (Figure 7A) and decreased protein translation (Figure 7D) observed after repression of zpr1+ gene expression.
The results of the present study provide direct experimental evidence that the cytoplasmic protein ZPR1 redistributes to the nucleolus in proliferating cells and that ZPR1 is required for normal nucleolar function. Further studies are required to determine the molecular basis for the role of ZPR1 in these processes.
ACKNOWLEDGMENTS
We thank M. Caizergues-Ferrer and D. Muris for providing plasmids; E.K.L. Chan, K.M. Pollard, R.L. Rubin, E.M. Tan, I. Todorov, and X.-D. Fu for providing antibodies; S. Gupta, R. Ochs, and R. Singh for discussions; F.G. Klier for assistance with microscopy; T. Barrett, J. Cavanagh, and I.-H. Wu for technical assistance; and K. Gemme for administrative assistance. These studies were supported by grant CA58396 from the National Cancer Institute. R.J.D. is an investigator of the Howard Hughes Medical Institute.
REFERENCES
- Abelson HT, Penman S. Selective interruption of RNA metabolism by chemotherapeutic agents. Handbook Exp Pharmacol. 1975;38:571–581. [Google Scholar]
- Andrade LEC, Chan EKL, Raska I, Peebles CL, Roos G, Tan EM. Human autoantibody to a novel protein of the nucleolar coiled body: immunological characterization and cDNA cloning of p80 coilin. J Exp Med. 1991;173:1407–1419. doi: 10.1084/jem.173.6.1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu X-D, Maniatis T. Factor required for mammalian spliceosome assembly is localized to discrete regions in the nucleus. Nature. 1990;343:437–441. doi: 10.1038/343437a0. [DOI] [PubMed] [Google Scholar]
- Galcheva-Gargova Z, Konstantinov KN, Wu I-H, Klier FG, Barrett T, Davis RJ. Binding of zinc finger protein ZPR1 to the epidermal growth factor receptor. Science. 1996;272:1797–1802. doi: 10.1126/science.272.5269.1797. [DOI] [PubMed] [Google Scholar]
- Granick D. Nucleolar necklaces in chick embryo fibroblast cells. II. Microscopic observations of the effect of adenosine analogues on nucleolar necklace formation. J Cell Biol. 1975;65:418–427. doi: 10.1083/jcb.65.2.418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter T. Oncoprotein networks. Cell. 1997;88:333–346. doi: 10.1016/s0092-8674(00)81872-3. [DOI] [PubMed] [Google Scholar]
- Kotzin BL, Lafferty JA, Portanova JP, Rubin RL, Tan EM. Monoclonal anti-histone autoantibodies derived from urine models of lupus. J Immunol. 1984;133:2554–2559. [PubMed] [Google Scholar]
- Moreno S, Klar A, Nurse P. Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol. 1991;194:795–826. doi: 10.1016/0076-6879(91)94059-l. [DOI] [PubMed] [Google Scholar]
- Ochs RL, Lischwe MA, Shen E, Carrol RE, Busch H. Fibrillarin: a new protein of the nucleolus identified by autoimmune sera. Biol Cell. 1985;54:123–134. doi: 10.1111/j.1768-322x.1985.tb00387.x. [DOI] [PubMed] [Google Scholar]
- Reimer G, Pollard KM, Penning CA, Ochs RL, Lischwe MA, Busch H, Tan EM. Monoclonal autoantibody from a (New Zealand black × New Zealand white) F1 mouse and human scleroderma sera target an Mr 34,000 nucleolar protein of the U3 RNP particle. Arthritis Rheum. 1987a;30:793–800. doi: 10.1002/art.1780300709. [DOI] [PubMed] [Google Scholar]
- Reimer G, Rose KM, Scheer U, Tan EM. Autoantibody to RNA polymerase I in scleroderma sera. J Clin Invest. 1987b;79:65–72. doi: 10.1172/JCI112809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw PJ, Jordan EG. The nucleolus. Annu Rev Cell Dev Biol. 1995;11:93–121. doi: 10.1146/annurev.cb.11.110195.000521. [DOI] [PubMed] [Google Scholar]
- Simar R, Bernhardt W. The phenomenon of nucleolar segregation: specific action of certain anti-metabolites. Int J Cancer. 1966;1:463–479. doi: 10.1002/ijc.2910010506. [DOI] [PubMed] [Google Scholar]
- Woolford JLJ, Warner JR. The Ribosome and Its Synthesis. Vol. 1. New York: Cold Spring Harbor Laboratory Press; 1991. [Google Scholar]
- Yung BY, Busch H, Chan PK. Translocation of nucleolar phosphoprotein B23 (37 kDa/pI 5.1) induced by selective inhibitors of ribosome synthesis. Biochim Biophys Acta. 1985;826:167–173. doi: 10.1016/0167-4781(85)90002-8. [DOI] [PubMed] [Google Scholar]
- Zatsepina OV, Todorov IT, Philipova RN, Krachmarov CP. Cell cycle-dependent translocations of a major nucleolar phosphoprotein, B23, and some characteristics of its variants. Eur J Cell Biol. 1997;73:58–70. [PubMed] [Google Scholar]