Abstract
The Nucleolar Localization Elements (NoLEs) of Xenopus laevis U3 small nucleolar RNA (snoRNA) have been defined. Fluorescein-labeled wild-type U3 snoRNA injected into Xenopus oocyte nuclei localized specifically to nucleoli as shown by fluorescence microscopy. Injection of mutated U3 snoRNA revealed that the 5′ region containing Boxes A and A′, known to be important for rRNA processing, is not essential for nucleolar localization. Nucleolar localization of U3 snoRNA was independent of the presence and nature of the 5′ cap and the terminal stem. In contrast, Boxes C and D, common to the Box C/D snoRNA family, are critical elements for U3 localization. Mutation of the hinge region, Box B, or Box C′ led to reduced U3 nucleolar localization. Results of competition experiments suggested that Boxes C and D act in a cooperative manner. It is proposed that Box B facilitates U3 snoRNA nucleolar localization by the primary NoLEs (Boxes C and D), with the hinge region of U3 subsequently base pairing to the external transcribed spacer of pre-rRNA, thus positioning U3 snoRNA for its roles in rRNA processing.
INTRODUCTION
Many aspects of how macromolecules are targeted to their correct subcellular destination still need to be defined. Although principles governing RNA export to the cytoplasm and import into the nucleus are beginning to be understood, very little is known about signals that direct RNA within the nucleus to the nucleolus. The nucleolus contains a vast array of different RNA species involved in ribosome biogenesis: ribosomal RNA (rRNA) and its precursors and ∼200 small nucleolar RNAs (snoRNAs). Unlike rRNA whose genes are located within the nucleolus, the other transcripts found in the nucleolus must travel there from their sites of synthesis in the nucleoplasm. What are the “zip codes” for targeting snoRNA to the nucleolus?
To address this question, we have studied U3 snoRNA because it is the most abundant snoRNA in the nucleolus, has been sequenced from a large number of animals and plants (Gu and Reddy, 1997), and is well characterized in terms of secondary structure and function in rRNA processing. U3 snoRNA is synthesized in the nucleoplasm in the proximity of coiled bodies (Gao et al., 1997), and formation of its trimethylguanosine cap can occur in the nucleoplasm (Terns and Dahlberg, 1994; Terns et al., 1995), unlike spliceosomal snoRNAs that are exported to the cytoplasm for cap trimethylation (Mattaj, 1986). Once U3 snoRNA is transported to the nucleolus, it is found highly concentrated in the dense fibrillar component (Matera et al., 1994) where initial rRNA processing events are believed to occur, but up to half of the U3 snoRNA is also detected by electron microscopy to be diffuse in the granular component (Fischer et al., 1991; Puvion-Dutilleul et al., 1991, 1992; Azum-Gélade et al., 1994) where later rRNA processing cleavages take place. On the basis of the localization of U3 snoRNA after various actinomycin D treatments (Puvion-Dutilleul et al., 1992; Rivera-León and Gerbi, 1997), a model has been proposed in which U3 snoRNA travels with pre-rRNA in a vectorial manner in the nucleolus from the dense fibrillar component to the granular component. Once the newly formed rRNA is released for export to the cytoplasm, U3 snoRNA may be recycled and stored in the dense fibrillar component for reutilization in rRNA processing (Gerbi and Borovjagin, 1997). The present study identifies the nucleolar localization elements in U3 snoRNA.
U3 was the first snoRNA demonstrated to be essential for rRNA processing in metazoa and yeast (Kass et al., 1990; Savino and Gerbi, 1990; Hughes and Ares, 1991; Hughes, 1996). Xenopus U3 snoRNA is the subject of the present study; it has been cloned and sequenced, and its secondary structure has been experimentally determined (Jeppesen et al., 1988). It contains evolutionarily conserved sequences named Boxes A′, A, B, C′, C, and D (reviewed by Gerbi et al., 1990; Gerbi, 1995), which are of presumed structural and/or functional importance. The present study analyzes whether any of these boxes are needed for nucleolar localization. Boxes A′ and A at the 5′ end of U3 snoRNA are thought to base pair with 18S rRNA and are essential for 18S rRNA formation (Hughes, 1996; Mereau et al., 1997; Borovjagin and Gerbi, unpublished data); however, they do not function as Nucleolar Localization Elements (NoLEs). Boxes C and D are present in all snoRNAs of the Box C/D family (tabulated by Xia et al., 1997), and we have recently shown by analysis of U14 and U8 snoRNAs that they constitute a consensus nucleolar localization signal for this family (Lange et al., 1998b). The data presented here support this finding, as both Boxes C and D are critical for nucleolar localization of U3. In addition, Box B affects nucleolar localization of U3 snoRNA, perhaps by facilitating the binding of the Box C/D protein(s).
Other structural features in U3 snoRNA were also examined for their role in nucleolar localization. The terminal stem structure is not essential for nucleolar localization of U3 snoRNA, nor is the presence or nature of a 5′ cap. In contrast, the “hinge” region does influence nucleolar localization of U3 snoRNA. Results of competition experiments suggest that the hinge region and Box B act at different steps in U3 snoRNA localization. We propose that Boxes C and D, common to many snoRNAs, are the primary nucleolar localization elements and are sites for cooperative binding of proteins. Furthermore, binding of proteins to Boxes C and D in U3 snoRNA may be facilitated by Box B. The hinge region of U3 may stabilize nucleolar localization by base pairing with the external transcribed spacer of pre-rRNA, thus positioning U3 correctly for its roles in rRNA processing.
MATERIALS AND METHODS
Plasmids, Primers, and PCR Reactions
The plasmid pXlU3A, containing a wild-type Xenopus laevis U3 gene subcloned into pBlueScript SK(+) (Stratagene, La Jolla, CA) (Savino et al., 1992), served as the template for mutagenesis using a variety of strategies. The Box C and Box D substitution mutants of U3 snoRNA were created by PCR of the 5′ and 3′ parts of the molecule that were subsequently ligated together at a restriction site placed in the substituted sequence (method as in Zaret et al., 1990). These PCR constructions used the following primers (substituted sequence is in lowercase letters and wild-type U3 sequence is in uppercase letters; vector sequences upstream and downstream of the U3 gene are listed for the universal primers and are also written in uppercase letters; sequences extending beyond the restriction site that were absent in the final construction are shown in italics):
Forward (5′) primers (sense): universal (wild-type U3), 5′-GGA AAC AGC TAT GAC CAT GAT TAC GCC AAG C-3′; Box C, 5′-cgc gga tcc cAC GTT CTG CTC CCC TTT ATT ATT GGG G-3′ (BamHI site underlined); Box D, 5′-cCA tcg atg GTG GTT TTA TTA CTG TTG GTG-3′ (ClaI site underlined).
Reverse (3′) primers (antisense): universal (wild-type U3), 5′-CCA GTG AAT TGT AAT ACG ACT CAC TAT AG-3′; Box C, 5′-cgc gga tcc gAG CAA ACA GCA GCC ACA ATG AAG C-3′ (BamHI site underlined); Box D, 5′-cca tcg aTG TGT TCT CTC CCT CCA TCT CC-3′ (ClaI site underlined).
Fragments amplified by PCR were gel purified, phenol/chloroform extracted, and digested with the appropriate restriction enzyme (see underlining above). The restricted PCR fragments were ligated in 50 mM Tris-HCl (pH 7.6), 10 mM MgCl2, 1 mM ATP, 1 mM dithiothreitol, 5% polyethylene glycol-8000 with 1 U DNA ligase (Life Technologies, Gaithersburg, MD) for 1 h at 25°C for the two mutants above or using TakaRa DNA ligase (PanVera, Madison, WI) at 16°C overnight for all other mutants. For the two mutants above, an aliquot was used from the ligation for reamplification and was phenol/chloroform purified, restricted, ligated into KpnI–NotI-digested pBSK(+), and transformed into Escherichia coli HB101 cells. The mutated U3 clones were verified by DNA sequence analysis.
Construction of the Box C′ and Box B mutants followed a similar approach of creating 5′ and 3′ parts of the molecule joined together by a restriction site to create the mutation. The following primers were used:
Forward (5′) primers (sense): Box B, 5′-gac ttt gat cAG TGA GCT CAC AGT GCT GCT TC-3′ (BclI site underlined); Box C′, 5′-ACC ACG tcc ttc tag aag tGT GTT CTC TCC TGA GCG-3′ (XbaI site underlined).
Reverse (3′) primers (antisense): Box B, 5′-gag tgT gat cag aCA GGA GAG AAC ACT GAC GC-3′ (BclI site underlined); Box C′, 5′-GAA CAC act tct aga agg aCG TGG TTT GTG AGT TCA GAC-3′ (XbaI site underlined).
Unlike the Box C and Box D mutants that initially contained DNA sequences flanking the U3 gene, the Box B and Box C′ constructions used 5′ and 3′ primers for PCR that coincided with the beginning and end of the gene. The same 5′ and 3′ primers were also used to create PCR products of just the U3 transcription unit from the Box C and Box D mutants described above. The forward (5′) primer was a 42-nucleotide (nt) oligonucleotide containing the T7 promotor sequence (underlined) preceding positions 1–22 of the wild-type U3 snoRNA: 5′-TAA TAC GAC TCA CTA TAG GGA AGA CTA TAC TTT CAG GGA TCA-3′.
Similarly, for all mutants listed above (except Box D), the reverse (3′) primer was a 31-nt oligonucleotide that contained a 5-nt tail (underlined) at its 3′ end, identical to the genomic sequence beyond the 3′ end of the gene, that was added to give enhanced stability (Terns, personal communication): 5′-TAA AAC CAC TCA GCC TGT GTT CTC TCC CTC C-3′. The 3′ primer used for PCR of the U3 gene of the Box D mutant was 5′-TAA AAC CAC cat cga TGT GTT CTC TCC CTC C-3′.
Box A and Box A′ mutants were created by PCR, without creating two halves of the molecule linked together at a restriction site as done above. A 60-nt forward (5′) primer was used that included the T7 promoter sequence (underlined) and nt 1–40 of U3 snoRNA with sequences substituted at positions 17–28 (Box A) or positions 8–28 (Box A+ that spans Box A and Box A′): Box A, 5′-TAA TAC GAC TCA CTA TAG GGA AGA CTA TAC TTT CAG cct agt aaa gat TAG GTT GTA CCT-3′; Box A+, 5′-TAA TAC GAC TCA CTA TAG GGA AGA CTA atg aaa gtc cct agt aaa gat TAG GTT GTA CCT-3′. For both these mutants, the reverse (3′) primer was the same as that listed above containing the 5-nt tail.
Similarly, the hinge mutation (Tag*) was created by PCR using a forward (5′) primer spanning nt 10–86 with mutation of nt 65–72 (lowercase letters): 5′-CTT TCA GGG ATC ATT TCT ATA GGT TGT ACC TGG TGA AAT GTG CTC GAA AGT GTC Ttt tag ata AAA CCA CGA GGA AG-3′.
The reverse (3′) primer was the same as that listed earlier, containing the 5-nt tail. The resulting PCR fragment that lacked the first 9 nt was then used as the template for a PCR reaction using the same reverse (3′) primer as above and the same 42-nt oligonucleotide containing the T7 promoter and nt 1–22 of wild-type U3 snoRNA as the forward (5′) primer described earlier to create a full-length U3 molecule substituted in nt 65–72 of the hinge region.
The open stem mutant was created by PCR using the forward (5′) primer used for all the other mutants (except Box A and Box A+), which contained the T7 promoter. The reverse (3′) end primer contained a variant of the 5-nt stabilizing tail (underlined) and was: open stem, 5′-TAA ATg ggg TCA GCC TGT GTT CTC TCC-3′.
All PCR products of the wild-type and mutant U3 snoRNA genes containing the T7 promoter and the 5-nt stabilizing tail were cloned into the vector pT7 (Novagen, Madison, WI), except for the Tag* and open stem mutations that were cloned into pCR3.1 (Invitrogen, Carlsbad, CA) and transformed into E. coli (Epicurian coli XL1 Blue competent cells; Stratagene). The inserts were confirmed by sequencing.
U2 small nuclear RNA (snRNA) was used as a control; plasmid pXlU2 that contains the X. laevis U2 snRNA gene (Mattaj and Zeller, 1983) was used as the template for PCR. The T7 promotor sequence (underlined) was added to U2 DNA by PCR: forward (5′) primer (sense), 5′-TAA TAC GAC TCA CTA TAG GGA TCG CTT CTC GGC CTT TTG GC-3′; reverse (3′) primer (antisense), 5′-AAG TGC ACC GGT CCT GGA GG-3′.
In Vitro Transcription
All RNAs were transcribed either from their appropriately digested plasmid DNAs or from PCR-derived templates using a T7 megashortscript in vitro transcription kit (Ambion, Austin, TX) as described previously (Lange et al., 1998b). The transcript was made with no cap or with G-cap by adding 4 mM m7G(5′)ppp(5′)G cap (Ambion) or A-cap by adding 4 mM m7G(5′)ppp(5′)A cap (Boehringer Mannheim, Indianapolis, IN) to the transcription reaction. The G-cap analogue can be inserted in the forward or reverse orientation (Pasquinelli et al., 1995), but the A-cap analogue is inserted only in the reverse orientation with A at the 5′-most end of the synthetic RNA (Jacobson and Pederson, 1998). The integrity of the in vitro transcripts was confirmed by 8% polyacrylamide, 8 M urea gel electrophoresis. The amount of fluorescent transcript or unlabeled transcript for competition was determined by spectrophotometry at 260 nm and confirmed by electrophoresis on a gel subsequently stained with methylene blue. Concentrations were adjusted accordingly so that equivalent amounts of mutated and wild-type U3 snoRNAs were injected.
Oocyte Microinjection
Stage V oocytes from X. laevis (Nasco, Fort Atkinson, WI) were obtained as described previously (Lange et al., 1998b). For snoRNA depletion and rescue experiments, U3 snoRNA was disrupted by two nuclear injections spaced 4 h apart of 9.2 nl each of the antisense oligonucleotide 5′-GAG CAC ATT TCA CCA GG-3′ complementary to nt 39–54 of wild-type U3 snoRNA at a concentration of 3 μg/μl (28 ng/oocyte). To in vivo label newly synthesized rRNA, [α-32P]UTP at 150 μCi/μl was added to the second injection of antisense oligonucleotide. Rescue was achieved by injecting 9.2 nl of in vitro–synthesized snoRNA (with or without fluorescein label) 4 h after the second antisense oligonucleotide injection. To cover the concentration needed for maximum rescue, a range of RNA concentrations was used for the injection (0.125, 0.3, and 1.0 μg/μl); 0.125 μg/μl gave partial rescue, but only the two higher concentrations are shown here). Oocytes were incubated at 20°C for 16 h after the rescue injection. Subsequently, the nuclei were manually isolated, and total RNA was extracted using an RNA extraction kit (5 Prime → 3 Prime, Boulder, CO).
For analysis of U3 snoRNA nucleolar localization by fluorescence microscopy, oocyte nuclei were injected with 9.2 nl of in vitro–transcribed RNA in H2O (0.1 μg/μl; 0.92 ng/oocyte). After subsequent incubation for 2 h at 20°C, the nuclei were isolated as described previously (Lange et al., 1998b).
Analysis by Fluorescence Microscopy
Nucleolar preparations were made following the method of Gall et al. (1991), originally designed to prepare lampbrush chromosomes, and microscopy was performed as described previously (Lange et al., 1998b).
U3 snoRNA Stability Assays
32P-labeled U3 snoRNA and 32P-labeled U2 snRNA were coinjected into oocyte nuclei, and 2 h later the RNA of 6–10 nuclei per sample was purified with an RNA extraction kit (5 Prime → 3 Prime) according to the manufacturer’s instructions. Two oocyte equivalents of nuclear RNA were fractionated on an 8% polyacrylamide, 8 M urea gel and exposed to PPB blue–sensitive x-ray film (Konica Medical, Wayne, NJ).
RESULTS
Wild-type U3 snoRNA Localizes Specifically to Nucleoli
Nucleolar localization of U3 snoRNA was monitored by direct visualization of nucleolar preparations after injection of fluorescein-labeled in vitro transcripts into Xenopus oocyte nuclei, similar to the approach used by others for tissue culture cells (Wang et al., 1991; Jacobson et al., 1995, 1997, 1998; Jacobson and Pederson, 1997, 1998). We have combined the injection of snoRNAs with a procedure originally developed to study lampbrush chromosomes of amphibian oocytes (Gall et al., 1991). Two hours after injection, nuclei were manually dissected from oocytes and the nuclear envelope was removed; the nuclear contents (including lampbrush chromosomes, nucleoli, and snurposomes) were centrifuged onto a microscope slide. The soluble nucleoplasm remains in the supernatant and is discarded from the nucleolar preparations. Oocyte nucleoli are variable in size (Wu and Gall, 1997) and can fuse into multinucleolar clusters (Shah et al., 1996).
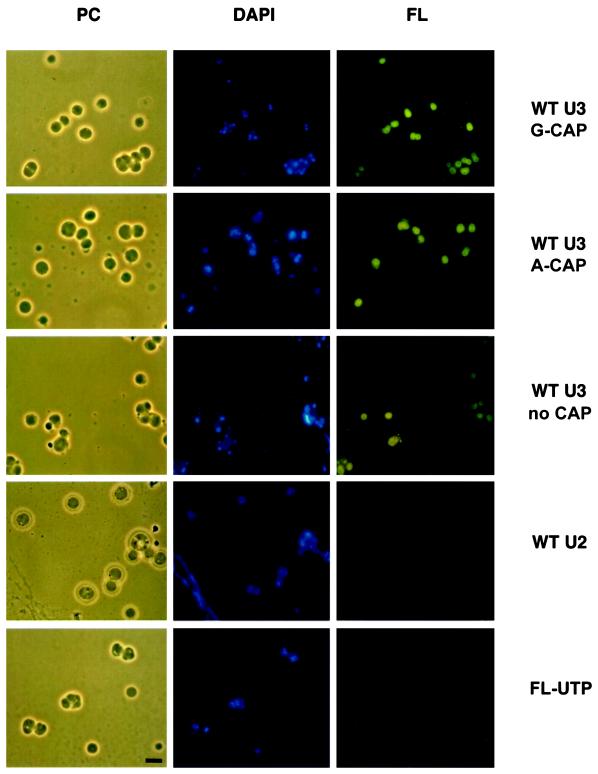
Fluorescein-labeled U3 snoRNA localized in nucleoli, independent of the presence or absence of a cap at the 5′ end of the molecule (Figure 1). Moreover, the nature of the cap has no effect on nucleolar localization in oocytes; synthetic U3 snoRNA made with an A-cap localizes to nucleoli as well as U3 made with a G-cap (Figure 1). When the same amount of fluorescein-labeled U2 snRNA was injected into Xenopus oocyte nuclei, it did not localize to nucleoli after 2 h (Figure 1). U2 is a component of spliceosomes in the nucleoplasm, and endogenous U2 snRNA has not been detected in nucleoli (Gall and Callan, 1989; Carmo-Fonseca et al., 1991; Wu et al., 1991, 1993). As another control, 1.8 pmol of unincorporated fluorescein-UTP was injected into Xenopus oocyte nuclei (∼130-fold molar excess relative to the amount of fluorescein-labeled U3 snoRNA injected above), and 2 h after injection no label was seen in nucleoli (Figure 1). This supports the conclusion that U3 snoRNA is localized to nucleoli after injection because of elements within the molecule and not simply because of the fluorescent label with which it was tagged. Moreover, this control rules out the possibility that the injected U3 snoRNA had been degraded and the label has been reincorporated into newly synthesized RNA in nucleoli. Assays to be summarized below also demonstrated that the injected U3 snoRNA was stable and not degraded.
Figure 1.
U3 wild-type snoRNA localizes to nucleoli regardless of the presence or absence of a cap. Fluorescein-coupled wild-type U3 small nucleolar RNA (WT U3) or the spliceosomal U2 small nuclear RNA (WT U2) or unincorporated fluorescein-UTP (FL-UTP) were injected into the nuclei of Xenopus laevis oocytes. After 2 h, nucleoli were prepared for visualization by phase-contrast (PC) or fluorescence microscopy. The nucleolar rDNA is stained blue by DAPI. In contrast to the control of U2 snRNA, fluorescein-labeled U3 snoRNA (FL, green) with a G-cap or A-cap or without a cap localizes to nucleoli. A lampbrush chromosome is visible in the bottom left of the phase-contrast and DAPI images of injected U2 snRNA. Bar, 10 μm.
The injected U3 snoRNA localized specifically to nucleoli and not to other structures in the oocyte nucleus 2 h after injection. Nucleoli can be identified by DAPI, which stains the amplified ribosomal DNA (rDNA) contained within them (Shah et al., 1996; Wu and Gall, 1997; Lange et al., 1998a,b). In favorable preparations, the injected fluorescent U3 snoRNA forms ring-like structures surrounding the rDNA. The dense fibrillar component surrounds the rDNA (Shah et al., 1996), and the injected U3 snoRNA appears to be localized in this region. Snurposomes do not contain DNA and thus can be visually distinguished from small nucleoli (Wu and Gall, 1997). Injected fluoresceinlabeled U3 snoRNA localizes only in nucleoli and not in snurposomes after 2 h.
Injected Fluorescein-labeled U3 snoRNA Is Functional in rRNA Processing
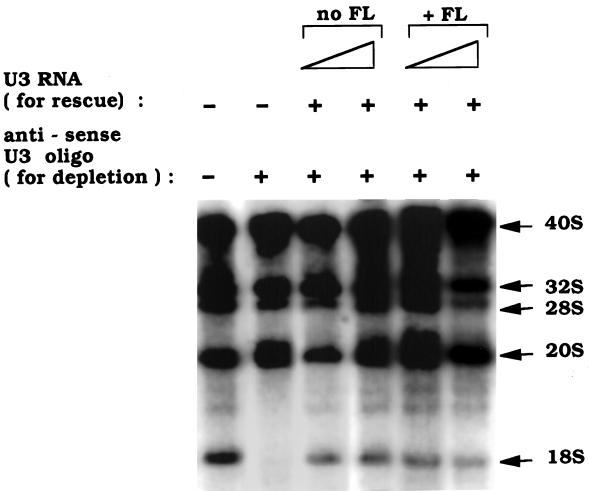
Not only does fluorescein-labeled U3 snoRNA localize specifically to nucleoli, but it is also functional in rRNA processing. This is shown by a U3 depletion and rescue experiment (Figure 2). Depletion of endogenous U3 snoRNA by injection of an antisense oligonucleotide results in the underaccumulation of newly synthesized 18S rRNA (Figure 2) caused by impairment of cleavage of pre-rRNA at sites 1 and 2 (Borovjagin and Gerbi, unpublished data). Production of 18S rRNA can be restored by a subsequent injection of wild-type U3 snoRNA with or without fluorescein label incorporated during its in vitro transcription (Figure 2). Hence, the fluorescein label in U3 snoRNA does not interfere with U3 function in rRNA processing.
Figure 2.
Fluorescein-labeled U3 snoRNA is functional in rRNA processing. rRNA precursors (40, 32, and 20S) and products (28 and 18S) were labeled in vivo with [32P]UTP, and the isolated nuclear RNA was subjected to agarose gel electrophoresis. 18S rRNA is not formed after antisense oligonucleotide depletion of endogenous U3 snoRNA but is formed after subsequent injection of U3 snoRNA (0.3 and 1.0 μg/μl) with or without fluorescein (FL) label to rescue rRNA processing.
Evolutionarily Conserved Boxes C and D Are Necessary for U3 snoRNA Localization to Nucleoli
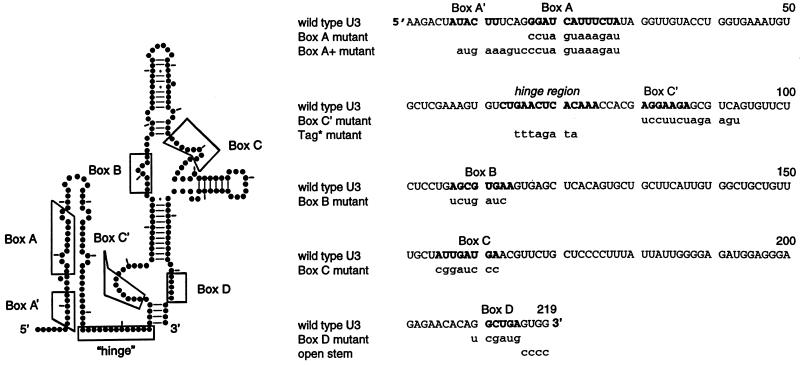
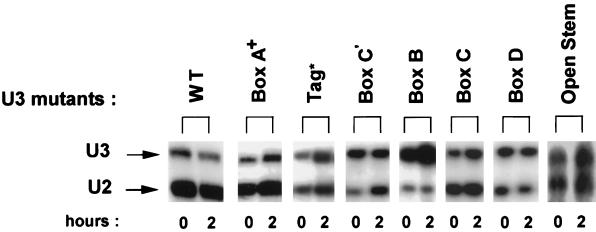
To determine whether any of the evolutionarily conserved boxes and other features in U3 snoRNA are necessary for nucleolar localization, mutations were created (Figure 3) and assayed. It was important to ascertain the stability of each of these mutant U3 molecules. 32P-labeled U3 was coinjected into oocyte nuclei with 32P-labeled U2 snRNA as an internal control and analyzed by gel electrophoresis 2 h after injection. As seen in Figure 4, all of the mutant U3 snoRNAs were stable 2 h after injection, except the Box C′ mutant that had begun to degrade but was still present. Therefore, failure of the fluorescein-labeled U3 snoRNAs to stain nucleoli can be interpreted as an inability to localize there rather than degradation. In the case of the Box C′ mutant, the variability in its strength of nucleolar labeling may reflect varying amounts of degradation.
Figure 3.
Structure and mutations of Xenopus U3 snoRNA. The experimentally derived secondary structure (left) as well as the sequence (right) is from Jeppesen et al. (1988), with sequence corrections at positions 99 and 194 by Savino et al. (1992). Bold letters in the sequences (right) indicate the hinge region or the evolutionarily conserved boxes, using the locations of these sequences as in Gerbi (1995). The consensus definition presented by Xia et al. (1997) does not include the two nucleotides at the 5′ end of Box C as shown here and adds one nucleotide at the 5′ end of Box D as shown here. The sequence substitutions in the boxes, the hinge region, or the 3′ terminal stem are given in lowercase letters; the three A residues at the 3′ end of the hinge region were not mutated because they do not participate in the proposed base pairing with the external transcribed spacer of pre-rRNA.
Figure 4.
Mutant U3 snoRNAs are stable 2 h after injection. 32P-labeled U3 snoRNA (mutants or wild type) were injected into oocyte nuclei, and the isolated RNA was analyzed by gel electrophoresis 2 h after injection. 32P-labeled U2 snRNA was coinjected as an internal control to normalize for any differences in injection or recovery of the samples. The stability results are from different experiments for the various mutants, and the amounts of coinjected U3 vs. U2 varied between experiments. For any given mutation, the ratio of U3 to U2 between 0 and 2 h shows that all the mutants are stable at the 2-h time point used for analysis of nucleolar localization, except for the Box C′ mutant, which has partially degraded relative to the U2 snRNA internal control.
Figure 5 shows nucleolar localization results of the various mutant U3 snoRNAs tested. The Box A plus Box A′ mutant (Box A+) was able to localize as well as wild-type U3 snoRNA to nucleoli (Figure 5). Boxes A and A′ are essential for 18S rRNA formation (Hughes, 1996; Mereau et al., 1997; Borovjagin and Gerbi, unpublished data), yet these regions are not critical for nucleolar localization. Therefore, signals for nucleolar localization are separable from sequences in the 5′ area of U3 needed for rRNA processing. Because Box A is a subset of Box A+, it is not surprising that U3 mutated in just Box A was also able to localize to nucleoli.
Figure 5.
Role of evolutionarily conserved boxes and certain structural features on U3 snoRNA nucleolar localization. U3 snoRNA carrying substitutions in Boxes A plus A′ (Box A+) localized as well as wild-type U3 snoRNA to nucleoli. U3 snoRNA with a substitution for Box B or the hinge region (Tag*) labeled nucleoli weakly, whereas U3 with substitutions in Box C or Box D did not label nucleoli at all. Results with the Box C′ mutant were variable, ranging from strong to weak nucleolar localization, but generally were moderate as shown here. PC, DAPI, and FL as in Figure 1. Bar, 10 μm.
Similarly, U3 snoRNA with substitutions in Box B or Box C′ was able to localize to nucleoli (Figure 5), although the nucleolar labeling by Box B mutants was consistently fainter than that of wild-type U3 snoRNA, and the degree of labeling by Box C′ mutants was variable. This suggests that the sequences normally present in Box B have an effect on nucleolar localization, but they are not a prerequisite. The nucleolar labeling generally seen for U3 Box C′ mutants also suggests that Box C′ is not an essential nucleolar localization element.
In contrast, nucleolar localization of injected U3 snoRNA was abolished by substitutions in Boxes C and D (Figure 5). Boxes C and D are found in many different snoRNA species (summarized by Xia et al., 1997) and are implicated here as critical signals for nucleolar localization of U3 snoRNA.
Secondary Structure Features Not Essential for Nucleolar Localization
Certain features of U3 snoRNA secondary structure were examined. The most highly exposed region of the Xenopus U3 snoRNA molecule is the 3′ end of the “hinge” region (Jeppesen et al., 1988). When the hinge region is replaced by another sequence called Tag*, the resulting U3 snoRNA mutant is still able to localize to nucleoli, although the signal is generally weak (Figure 5). Hence, the hinge region is of some importance for nucleolar localization of U3 snoRNA.
The 3′-terminal stem of U3 snoRNA is needed for nuclear localization (Baserga et al., 1992), and we have inquired whether it is also essential for nucleolar localization. Sequences at the 3′ end of U3 snoRNA were replaced (Figure 3) such that base pairing to form the terminal stem was no longer possible. This open stem U3 mutant is stable 2 h after injection (Figure 4) and was still able to localize to nucleoli (Figure 5). Therefore, the terminal stem of U3 snoRNA is not recognized as a signal for nucleolar localization.
Some Mutant U3 snoRNAs Are Effective Competitors of Wild-Type U3 snoRNA Nucleolar Localization
Fluorescein-labeled wild-type U3 snoRNA was coinjected with a 100-fold molar excess of various other unlabeled RNA in vitro transcripts. To keep the amount of injected RNA as low as feasible, only one-half the amount of fluorescein-labeled U3 was used here, compared with the experiments described above. As can be seen in Figure 6, this is still sufficient to give visible staining of nucleoli, although the signal is somewhat less intense. U2 snRNA (which is normally not found in nucleoli) does not compete for the nucleolar localization of U3 snoRNA. Similarly, U3 mutated in Box C or Box D is unable to compete for the nucleolar localization of wild-type U3 snoRNA. Partial competition is achieved by U3 mutated in Box B, and full competition is achieved by wild-type U3 or U3 mutated in Box A plus Box A′ (U3 Box A+) or in Box C′. The ability of the latter group of molecules to compete for wild-type U3 snoRNA localization to nucleoli indicates that the mechanism used for localization can be saturated. U3 mutated in the hinge region (U3 Tag*) generally provided full competition of wild-type U3 nucleolar location, but occasionally faint signals were seen in the nucleoli.
Figure 6.
Some mutant U3 snoRNAs can compete for wild-type U3 nucleolar localization. One-half (0.46 ng/oocyte) of the usual amount of fluorescein-labeled wild-type U3 snoRNA was coinjected with a 100-fold molar excess of various unlabeled RNAs as competitor. No competition is seen by U2 snRNA or U3 Box C or U3 Box D mutants, partial competition is seen by the U3 Box B mutant, and full competition is seen by the other samples. PC, DAPI, and FL as in Figure 1. Lampbrush chromosomes are seen in the PC and DAPI panels of the U3 Box A+ and U3 Box C competitor samples. Bar, 10 μm.
DISCUSSION
The method used here to follow the localization of fluorescent RNA injected into Xenopus oocytes is widely applicable. It provides a tool to determine signals that guide RNA molecules to their correct destination in the cell, which has been studied in only a few other examples so far in Box C/D family snoRNAs (U8: Lange et al., 1998a, U14: Lange et al., 1998b) or other snoRNAs (7-2/MRP RNA: Jacobson et al., 1995; RNase P RNA: Jacobson et al., 1997). Principles governing the nucleolar localization of U3 snoRNA have emerged from the present study, as will be discussed below.
Sequences at the 5′ End of U3 snoRNA That Are Critical for rRNA Processing Are Not Needed for Nucleolar Localization of U3
Nucleolar localization of snoRNAs could occur passively by diffusion through the nucleoplasm to the nucleolus where snoRNAs may become trapped by base pairing with pre-rRNA. One possible base-pairing interaction is that between Boxes A and A′ of U3 snoRNA and 18S rRNA. This interaction has been proposed as a way to prevent premature formation of a pseudoknot in 18S rRNA and is important for 18S rRNA formation (Hughes, 1996; Mereau et al., 1997; Borovjagin and Gerbi, unpublished data). However, in the present study we show that the Box A+ mutant (containing Box A and A′ mutations) is not important for nucleolar localization of U3 snoRNA. Mutations in this region do not affect nucleolar localization of U3 (Figure 5), and these mutated molecules effectively compete for nucleolar localization of wild-type U3 snoRNA (Figure 6). Therefore, interactions of Boxes A and A′ in U3 snoRNA with rRNA are not essential for nucleolar localization of U3 snoRNA. Similarly, the 5′ region of U8 snoRNA needed for rRNA processing and hypothesized to bind to the 5′ end of 28S rRNA is not essential for nucleolar localization (Lange et al., 1998a). Moreover, the middle part of U14 snoRNA that contains regions of complementarity to 18S rRNA, crucial for rRNA processing and 18S rRNA methylation, is unimportant for U14 snoRNA nucleolar localization (Lange et al., 1998b). These data support the conclusion that the signals for nucleolar localization are often distinct from the sequences necessary for snoRNA function in rRNA processing.
Boxes C and D Are Nucleolar Localization Elements Common to Many snoRNAs
Another model for nucleolar localization of snoRNA molecules is that specific regions of the molecule are recognized as nucleolar targeting sequences, perhaps acting via protein carriers that bind to these sequences. Our observations are in agreement with this model, because we found that Boxes C and D are important for nucleolar localization of U3 snoRNA (Figure 5) as well as for U8 and U14 snoRNAs in Xenopus (Lange et al., 1998b); this also applies to U14 snoRNA in mammalian cells (Samarsky et al., 1998). Boxes C and D are common to a large number of snoRNA species (tabulated by Xia et al., 1997). Even a mini-construct of U14 snoRNA carrying only Boxes C and D but no other U14 sequences can localize to nucleoli (Lange et al., 1998b). Our data support the conclusion that Boxes C and D are NoLEs for the Box C/D family of snoRNAs. Box D is also known to be necessary for the nuclear retention of U3 snoRNA (Terns et al., 1995), perhaps because U3 is anchored in the nucleolus by the NoLE.
What is the mechanism by which Boxes C and D target snoRNAs to the nucleolus? Proteins may mediate the transport of snoRNAs to the nucleolus, similar to the role played by Sm proteins for nuclear import of snRNAs (Mattaj and DeRobertis, 1985; Hamm et al., 1990; Fischer et al., 1991, 1993; Marshallsay and Lührmann, 1994; Grimm et al., 1997). Boxes C and D are implicated in binding the nucleolar protein fibrillarin (Baserga et al., 1991; Peculis and Steitz, 1994; Watkins et al., 1996), suggesting that fibrillarin could be a carrier protein to transport several different snoRNA species to the nucleolus. However, this might not be the case, because yeast strains genetically depleted of fibrillarin have nucleoli that stain with antibody against the trimethylguanosine cap found on snoRNAs (Tollervey et al., 1991), although in situ hybridization was not performed to directly assay snoRNA location in these strains. Moreover, U14 snoRNA carrying a large internal deletion (ΔA/V/B) can still localize to nucleoli (Lange et al., 1998b), although it apparently cannot bind fibrillarin (Watkins et al., 1996). Other proteins that may bind to Boxes C and D could also serve to localize Box C/D snoRNAs to the nucleolus. One such protein has been identified and is 65 kDa in mouse extracts (Watkins et al., 1998) and 68 kDa in Xenopus oocytes (Caffarelli et al., 1998). However, in addition to Boxes C and D, the terminal stem is also needed for binding of this protein. Because a terminal stem is not essential for nucleolar localization of U3 (Figure 5) or U14 snoRNAs (Lange et al., 1998b), this 65/68-kDa protein is unlikely to mediate nucleolar localization. Future studies have to determine which, if any, protein(s) may mediate the transport of Box C/D snoRNAs to the nucleolus.
Box C and Box D are each important, and neither box by itself in the absence of the other box is sufficient for nucleolar localization of U3 (Figure 5). In addition, U3 mutated in either Box C or Box D cannot compete for nucleolar localization of wild-type U3 snoRNA (Figure 6), suggesting that the binding of protein(s) to Boxes C and D is cooperative. It can be hypothesized that each box may bind a separate protein, both of which are needed for nucleolar localization. Thus, for example, if a competitor U3 containing a mutated Box C cannot bind its protein, the cooperative binding of the putative protein that should bind to the wild-type Box D region will also be impaired, and therefore this protein will be available to bind to Box D of the wild-type fluorescein-labeled U3 probe. The net result is failure of Box C or Box D mutants to compete for nucleolar localization of wild-type U3 snoRNA. Another explanation of the competition data is that a single protein may bind to both Boxes C and D, with distinct binding sites for each, and both sites are needed to anchor the single protein. In addition, Box B, which lies opposite Box C, may assist but not be crucial for binding of the Box C/D protein(s), because the Box B mutation reduces U3 snoRNA localization to nucleoli (Figure 5) and partially competes for nucleolar localization of wild-type U3 snoRNA (Figure 6).
The Trimethylguanosine Cap of U3 snoRNA Is Not Needed for Nucleolar Localization in Xenopus Oocytes
The monomethylguanosine cap of U3 snoRNA can be converted to a trimethyguanosine cap upon injection into Xenopus oocytes, and Box D of U3 snoRNA is needed for cap trimethylation of the molecule (Baserga et al., 1992; Terns et al., 1995). Hence, it is possible that Box D acts as a NoLE indirectly because of its role in cap trimethylation. We have ruled out this possibility, demonstrating that U3 snoRNA is localized in Xenopus oocyte nucleoli regardless of the presence of a 5′ cap (Figure 1). Moreover, the nature of the cap does not seem to matter, because U3 with an A-cap, which cannot be trimethylated (Baserga et al., 1992; Peculis and Steitz, 1994), localized to nucleoli as well as U3 with a G-cap (Figure 1). Similarly, U8 snoRNA is localized to Xenopus oocyte nucleoli regardless of the presence or nature of its cap (Lange et al., 1998a), and the cap is not important for U8 function in rRNA processing (Peculis and Steitz, 1994). It has recently been reported that an m7G-cap commits U3 and U8 snoRNAs to the nucleolar localization pathway in mammalian cells in culture (Jacobson and Pederson, 1998), perhaps reflecting a difference between that model system and Xenopus oocytes. Moreover, U3 snoRNA of higher plants is transcribed by RNA polymerase III rather than RNA polymerase II, and it lacks a monomethyl (and trimethyl) guanosine cap (Shimba et al., 1992) yet can localize to nucleoli. In addition, the intronic members of the Box C/D family of snoRNAs naturally lack a cap but localize to nucleoli nonetheless (e.g., U14 snoRNA) (Lange et al., 1998b). Finally, most snRNAs of the spliceosome have a trimethylguanosine cap and spliced mRNAs contain a monomethylguanosine cap, yet they are not localized in nucleoli, so this cap structure cannot be a general determinant for nucleolar localization.
Role of Other Regions Besides the Primary NoLEs in Nucleolar Localization of U3 snoRNA
The sequences of Boxes C and D are essential for the nucleolar localization of U3 (Figure 5) and other Box C/D snoRNAs, such as U8 and U14 (Lange et al., 1998b). However, in certain snoRNAs other regions also affect nucleolar localization. In U8 snoRNA, the region just upstream of Box C (called Cup) is needed for nucleolar localization (Lange et al., 1998a). Similarly, in U3 snoRNA, Box B and the hinge region influence nucleolar localization. Fluorescein-labeled U3 snoRNA with mutations in Box B or the hinge region (Tag*) localizes weakly to nucleoli (Figure 5). However, these two mutations differ in their ability to compete wild-type U3 nucleolar localization (Figure 6), suggesting that they act differently from one another as accessory NoLEs. The Box B mutant partially competed wild-type U3 for its nucleolar localization (Figure 6). As suggested above, Box B may facilitate but not be essential for binding Box C/D proteins. In contrast, a mutant of the hinge region (Tag*) fully competed wild-type U3 for its nucleolar localization (Figure 6), as might be expected, because this mutant retains wild-type sequences for Boxes C and D that are the primary NoLEs. The reduced nucleolar labeling by the U3 hinge mutant (Tag*) when it is used as the probe (Figure 5) may reflect a destabilization of nucleolar localization normally provided by base pairing between the U3 hinge region (nt 65–72 in Xenopus U3 snoRNA; Jeppesen et al., 1988) and sequences in the external transcribed spacer (ETS of 40S pre-rRNA; Xenopus nt 311–318) (Furlong et al., 1983). The possibility for base pairing between the U3 hinge region and the ETS was first noted by Maser and Calvet (1989), is phylogenetically conserved by compensatory mutations (our unpublished results), and has been substantiated by mutagenesis and cross-linking studies in yeast (Beltrame and Tollervey, 1992, 1995; Beltrame et al., 1994). The site in the ETS that is complementary to the hinge region of U3 may act as a loading site for U3 snoRNA on the rRNA precursor to help position U3 snoRNA correctly for rRNA processing.
Most snoRNA species of the Box C/D family can be drawn as a Y-shaped structure (“terminal core structure”), with Boxes C and D flanking the 3′-terminal stem (Tycowski et al., 1993). This terminal core structure is necessary and sufficient for excision of intron-encoded snoRNAs such as U14 (Watkins et al., 1996), perhaps acting as the binding site for proteins (Caffarelli et al., 1998; Watkins et al., 1998), which may serve as roadblocks against exonucleolytic digestion. Other snoRNAs, such as Xenopus U3, are transcribed from their own genes and not from introns (Savino et al., 1992). U3 snoRNA does not have its 5′ end base paired with its 3′ end in a classical terminal core structure characteristic of many other Box C/D snoRNAs, but it does have a base paired stem at its 3′ end (Figure 3) that is important for nuclear import (Baserga et al., 1992) and stability of the molecule (Terns et al., 1995). In U3 snoRNA, Box C is not found adjacent to the terminal base paired stem, but a related sequence called Box C′ is present there (Tycowski et al., 1993). Box C′ is critical for stability of U3 in yeast (Mereau et al., 1997; Samarsky and Fournier, 1998) and in Xenopus oocytes 12 h after injection (Borovjagin and Gerbi, unpublished data). However, some of the Box C′ mutant of U3 is still present 2 h after injection (Figure 4), which allows its nucleolar localization to be assayed at that time point. U3 mutated in Box C′ generally localized to nucleoli 2 h after injection, although the extent was variable (Figure 5), and it provided almost full competition for nucleolar localization of wild-type U3 snoRNA (Figure 6). Two of the six nucleotides of Box C′ (GGAAGA) differ from the Box C consensus UGAUGA (tabulated by Xia et al., 1997), which may explain why Box C′ in Xenopus U3 snoRNA does not act as a primary NoLE. Thus, Box C′ is not equivalent to Box C either in sequence or as a NoLE.
Similar to Box C′, the base paired terminal stem does not seem to play a primary role in nucleolar localization of U3 snoRNA (Figure 5). Hence, although Box D is necessary for nucleolar localization of U3 snoRNA, the rest of the terminal structure (Box C′ and the terminal stem) is not essential for this purpose. If nucleolar localization is mediated by proteins, then only the protein that recognizes Box D for association with U3 seems likely to function in nucleolar localization, and not other proteins that may be associated with the terminal structure (Parker and Steitz, 1987; Hartshorne and Agabian, 1994; Mereau et al., 1997).
In summary, Boxes C and D are the primary NoLEs of U3 snoRNA. Box B also plays a role in U3 nucleolar localization, perhaps by facilitating the binding of Box C/D protein(s) to U3 snoRNA. The hinge region of U3 snoRNA may stabilize the nucleolar localization of U3 by binding to the ETS and thereby position U3 snoRNA on the pre-rRNA so that it can carry out its role in rRNA processing.
ACKNOWLEDGMENTS
We thank M. Jacobson and T. Pederson for sharing details of fluorescein labeling and injection of snoRNAs, J. Gall for use of his centrifuge rotor, J. Dahlberg for a gift of the Xenopus U2 clone, A.W. Coleman for generous use of her fluorescence microscope, A.-K. Bielinsky for comments on this article, and C. González for assistance in typing. This research was supported by CTR-3983 and by National Institutes of Health GM 20261 research grants to S.A.G.
Footnotes
Corresponding author. E-mail address: Susan_Gerbi@Brown.EDU.
REFERENCES
- Azum-Gélade M-C, Noaillac-Depeyre J, Caizergues-Ferrer M, Gas N. Cell cycle redistribution of U3 snRNA and fibrillarin. Presence in the cytoplasmic nucleolus remnant and in the prenucleolar bodies at telophase. J Cell Sci. 1994;107:463–475. doi: 10.1242/jcs.107.2.463. [DOI] [PubMed] [Google Scholar]
- Baserga SJ, Gilmore-Hebert M, Yang HW. Distinct molecular signals for nuclear import of the nucleolar snRNA, U3. Genes Dev. 1992;6:1120–1130. doi: 10.1101/gad.6.6.1120. [DOI] [PubMed] [Google Scholar]
- Baserga SJ, Yang XW, Steitz JA. An intact Box C sequence in the U3 snRNA is required for binding of fibrillarin, the protein common to the major family of nucleolar snRNPs. EMBO J. 1991;10:2645–2651. doi: 10.1002/j.1460-2075.1991.tb07807.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beltrame M, Henry Y, Tollervey D. Mutational analysis of an essential binding site for the U3 snoRNA in the 5′ external transcribed spacer of yeast pre-rRNA. Nucleic Acids Res. 1994;22:5139–5147. doi: 10.1093/nar/22.23.5139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beltrame M, Tollervey D. Identification and functional analysis of two U3 binding sites on yeast pre-ribosomal RNA. EMBO J. 1992;11:1531–1542. doi: 10.1002/j.1460-2075.1992.tb05198.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beltrame M, Tollervey D. Base pairing between U3 and the pre-ribosomal RNA is required for 18S rRNA synthesis. EMBO J. 1995;14:4350–4356. doi: 10.1002/j.1460-2075.1995.tb00109.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caffarelli E, Losito M, Giorgi C, Fatica A, Bozzoni I. In vivo identification of nuclear factors interacting with the conserved elements of box C/D small nucleolar RNAs. Mol Cell Biol. 1998;18:1023–1028. doi: 10.1128/mcb.18.2.1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmo-Fonseca M, Pepperkok R, Sproat BS, Ansorge W, Swanson MS, Lamond A. In vivo detection of snRNP-rich organelles in the nuclei of mammalian cells. EMBO J. 1991;10:1863–1873. doi: 10.1002/j.1460-2075.1991.tb07712.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer D, Weisenberger D, Scheer U. Assigning functions to nucleolar structures. Chromosoma. 1991;101:133–140. doi: 10.1007/BF00355363. [DOI] [PubMed] [Google Scholar]
- Fischer U, Darzynkiewicz E, Tahara SM, Dathan NA, Lührmann R, Mattaj IW. Diversity in the signals required for nuclear accumulation of U snRNPs and variety in the pathways of nuclear transport. J Cell Biol. 1991;113:705–714. doi: 10.1083/jcb.113.4.705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer U, Sumpter V, Sekine M, Satoh T, Lührmann R. Nucleoplasmic transport of U snRNPs; definition of a nuclear localization signal in the Sm core domain that binds a transport receptor independently of the m3G cap. EMBO J. 1993;12:573–583. doi: 10.1002/j.1460-2075.1993.tb05689.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furlong JC, Forbes J, Robertson M, Maden BEH. The external transcribed spacer and preceding region of Xenopus borealis rDNA: comparison with the corresponding region of Xenopus laevis rDNA. Nucleic Acids Res. 1983;11:8183–8196. doi: 10.1093/nar/11.23.8183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gall JG, Callan HG. The sphere organelle contains small nuclear ribonucleoproteins. Proc Natl Acad Sci USA. 1989;86:6635–6639. doi: 10.1073/pnas.86.17.6635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gall JG, Callan HG, Wu Z, Murphy C. Methods in Cell Biology. Vol. 36. B.K. Kay and H.B. Peng, New York: Academic Press; 1991. Lampbrush chromosomes; pp. 149–166. [PubMed] [Google Scholar]
- Gao L, Frey MR, Matera AG. Human genes encoding U3 snRNA associate with coiled bodies in interphase cells and are clustered on chromosome 17p11.2 in a complex inverted repeat structure. Nucleic Acids Res. 1997;25:4740–4747. doi: 10.1093/nar/25.23.4740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerbi SA. Small nucleolar RNA. Biochem Cell Biol. 1995;73:845–858. doi: 10.1139/o95-092. [DOI] [PubMed] [Google Scholar]
- Gerbi SA, Borovjagin A. U3 snoRNA may recycle through different compartments of the nucleolus. Chromosoma. 1997;105:401–406. doi: 10.1007/BF02510476. [DOI] [PubMed] [Google Scholar]
- Gerbi SA, Savino R, Stebbins-Boaz B, Jeppesen C, Rivera-León R. A role for U3 small nuclear ribonucleoprotein in the nucleolus? In: Hill WE, Dahlberg A, Garrett RA, Moore PB, Schlessinger D, Warner JR, editors. The Ribosome: Structure, Function and Evolution. Washington, DC: American Society for Microbiology; 1990. pp. 452–469. [Google Scholar]
- Grimm C, Lund E, Dahlberg JE. In vivo selection of RNAs that localize in the nucleus. EMBO J. 1997;16:793–806. doi: 10.1093/emboj/16.4.793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu J, Reddy R. Small RNA database. Nucleic Acids Res. 1997;25:98–101. doi: 10.1093/nar/25.1.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamm J, Darzynkiewicz E, Tahara SM, Mattaj IW. The trimethylguanosine cap structure of U1 snRNA is a component of a bipartite nuclear targeting signal. Cell. 1990;62:569–577. doi: 10.1016/0092-8674(90)90021-6. [DOI] [PubMed] [Google Scholar]
- Hartshorne T, Agabian N. A common core structure for U3 small nucleolar RNAs. Nucleic Acids Res. 1994;22:3354–3364. doi: 10.1093/nar/22.16.3354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes JMX. Functional base-pairing interaction between highly conserved elements of U3 small nucleolar RNA and the small ribosomal subunit RNA. J Mol Biol. 1996;259:645–654. doi: 10.1006/jmbi.1996.0346. [DOI] [PubMed] [Google Scholar]
- Hughes JMX, Ares M., Jr Depletion of U3 small nucleolar RNA inhibits cleavage in the 5′ external transcribed spacer of yeast pre-ribosomal RNA and impairs formation of 18S ribosomal RNA. EMBO J. 1991;10:4231–4239. doi: 10.1002/j.1460-2075.1991.tb05001.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobson MR, Pederson T. Analysis of mRNA Formation and Function. Methods in Molecular Genetics. J.D. Richter, New York: Academic Press; 1997. RNA traffic and localization reported by fluorescent molecular cytochemistry; pp. 341–359. [Google Scholar]
- Jacobson MR, Pederson T. A 7-methylguanosine cap commits U3 and U8 small nuclear RNAs to the nucleolar localization pathway. Nucleic Acids Res. 1998;26:756–760. doi: 10.1093/nar/26.3.756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobson MR, Cao L-G, Taneja K, Singer RH, Wang Y-L, Pederson T. Nuclear domains of the RNA subunit of RNase P. J Cell Sci. 1997;110:829–837. doi: 10.1242/jcs.110.7.829. [DOI] [PubMed] [Google Scholar]
- Jacobson MR, Cao L-G, Wang Y-L, Pederson T. Dynamic localization of RNase MRP RNA in the nucleolus observed by fluorescent RNA cytochemistry in living cells. J Cell Biol. 1995;131:1649–1658. doi: 10.1083/jcb.131.6.1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobson MR, Pederson T, Wang Y-L. Cell Biology, a Laboratory Handbook. 2nd ed. Vol. 4. J.E. Celis, New York: Academic Press; 1998. Conjugation of fluorescent probes to proteins and nucleic acids; pp. 5–10. [Google Scholar]
- Jeppesen C, Stebbins-Boaz B, Gerbi SA. Nucleotide sequence determination and secondary structure of Xenopus U3 snRNA. Nucleic Acids Res. 1988;16:2127–2148. doi: 10.1093/nar/16.5.2127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kass S, Tyc K, Steitz JA, Sollner-Webb B. The U3 small nucleolar ribonucleoprotein functions in the first step of preribosomal RNA processing. Cell. 1990;60:897–908. doi: 10.1016/0092-8674(90)90338-f. [DOI] [PubMed] [Google Scholar]
- Lange TS, Borovjagin A, Gerbi SA. Nucleolar localization elements (NoLEs) in U8 snoRNA differ from sequences required for rRNA processing. RNA. 1998a;4:789–800. doi: 10.1017/s1355838298980438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange TS, Borovjagin A, Maxwell ES, Gerbi SA. Conserved Boxes C and D are essential nucleolar localization elements of U8 and U14 snoRNAs. EMBO J. 1998b;17:3176–3187. doi: 10.1093/emboj/17.11.3176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshallsay C, Lührmann R. In vitro nuclear import of snRNPs, cytosolic factors mediate m3G-cap dependence of U1 and U2 snRNP transport. EMBO J. 1994;13:222–231. doi: 10.1002/j.1460-2075.1994.tb06252.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maser RL, Calvet JP. U3 small nuclear RNA can be psoralen-cross-linked in vivo to the 5′ external transcribed spacer of pre-ribosomal RNA. Proc Natl Acad Sci USA. 1989;86:6523–6527. doi: 10.1073/pnas.86.17.6523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matera AG, Tycowski KT, Steitz JA, Ward DC. Organization of small nucleolar ribonucleoproteins (snoRNPs) by fluorescence in situ hybridization and immunocytochemistry. Mol Biol Cell. 1994;5:1289–1299. doi: 10.1091/mbc.5.12.1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattaj I. Cap trimethylation of U snRNA is cytoplasmic and dependent on U snRNP protein binding. Cell. 1986;46:905–911. doi: 10.1016/0092-8674(86)90072-3. [DOI] [PubMed] [Google Scholar]
- Mattaj I, DeRobertis EM. Nuclear segregation of U2 snRNA requires the binding of specific snRNP proteins. Cell. 1985;40:111–118. doi: 10.1016/0092-8674(85)90314-9. [DOI] [PubMed] [Google Scholar]
- Mattaj IW, Zeller R. Xenopus laevis U2 snRNA genes, tandemly repeated transcription units sharing 5′ and 3′ flanking homology with other RNA polymerase II transcribed genes. EMBO J. 1983;2:1883–1891. doi: 10.1002/j.1460-2075.1983.tb01675.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mereau A, Fournier R, Gregoire A, Mougin A, Fabrizio P, Lührmann R, Branlant C. An in vivo and in vitro structure-function analysis of the Saccharomyces cerevisiae U3A snoRNP protein-RNA contacts and base-pair interaction with the pre-ribosomal RNA. J Mol Biol. 1997;273:552–571. doi: 10.1006/jmbi.1997.1320. [DOI] [PubMed] [Google Scholar]
- Parker KA, Steitz JA. Structural analysis of the human U3 ribonucleoprotein particle reveal a conserved sequence available for base pairing with pre-rRNA. Mol Cell Biol. 1987;7:2899–2913. doi: 10.1128/mcb.7.8.2899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasquinelli AE, Dahlberg JE, Lund E. Reverse 5′ caps in RNAs made in vitro by phage polymerases. RNA. 1995;1:957–967. [PMC free article] [PubMed] [Google Scholar]
- Peculis BA, Steitz JA. Sequence and structural elements critical for U8 snRNP function in Xenopus oocytes are evolutionarily conserved. Genes Dev. 1994;8:2241–2255. doi: 10.1101/gad.8.18.2241. [DOI] [PubMed] [Google Scholar]
- Puvion-Dutilleul F, Mazan S, Nicoloso M, Christensen ME, Bachellerie J-P. Localization of U3 RNA molecules in nucleoli of HeLa and mouse 3T3 cells by high resolution in situ hybridization. Eur J Cell Biol. 1991;56:178–186. [PubMed] [Google Scholar]
- Puvion-Dutilleul F, Mazan S, Nicoloso M, Pichard E, Bachellerie J-P, Puvion E. Alterations of nucleolar ultrastructure and ribosome biogenesis by actinomycin D. Implications for U3 snRNP function. Eur J Cell Biol. 1992;58:149–162. [PubMed] [Google Scholar]
- Rivera-León R, Gerbi SA. Delocalization of some small nucleolar RNPs after actinomycin D treatment to deplete early pre-rRNAs. Chromosoma. 1997;105:506–514. doi: 10.1007/BF02510487. [DOI] [PubMed] [Google Scholar]
- Samarsky DA, Fournier MJ. Functional mapping of the U3 small nucleolar RNA from the yeast Saccharomyces cerevisiae. Mol Cell Biol. 1998;18:3431–3444. doi: 10.1128/mcb.18.6.3431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samarsky DA, Fournier MJ, Singer RH, Bertrand E. The snoRNA box C/D motif directs nucleolar targeting and also couples snoRNA synthesis and localization. EMBO J. 1998;17:3747–3757. doi: 10.1093/emboj/17.13.3747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savino R, Gerbi SA. In vivo disruption of Xenopus U3 snRNA affects ribosomal RNA processing. EMBO J. 1990;9:2299–2308. doi: 10.1002/j.1460-2075.1990.tb07401.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savino R, Hitti Y, Gerbi SA. Genes for Xenopus laevis U3 small nuclear RNA. Nucleic Acids Res. 1992;20:5435–5442. doi: 10.1093/nar/20.20.5435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah SB, Terry CD, Wells DA, DiMario PJ. Structural changes in oocyte nucleoli of Xenopus laevis during oogenesis and meiotic maturation. Chromosoma. 1996;105:111–121. doi: 10.1007/BF02509521. [DOI] [PubMed] [Google Scholar]
- Shimba S, Buckley B, Reddy R, Kiss T, Filipowicz W. Cap structure of U3 small nucleolar RNA in animal and plant cells is different. J Biol Chem. 1992;267:13772–13777. [PubMed] [Google Scholar]
- Terns MP, Dahlberg JE. Retention and 5′ cap trimethylation of U3 RNA in the nucleus. Science. 1994;264:959–961. doi: 10.1126/science.8178154. [DOI] [PubMed] [Google Scholar]
- Terns MP, Grimm C, Lund E, Dahlberg JE. A common maturation pathway for small nucleolar RNAs. EMBO J. 1995;14:4860–4871. doi: 10.1002/j.1460-2075.1995.tb00167.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tollervey D, Lehtonen H, Carmo-Fonseca M, Hurt EC. The small nucleolar RNP protein NOP1 (fibrillarin) is required for pre-rRNA processing in yeast. EMBO J. 1991;10:573–583. doi: 10.1002/j.1460-2075.1991.tb07984.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tycowski K, Shu M-D, Steitz JA. A small nucleolar RNA is processed from an intron of the human gene encoding ribosomal protein S3. Genes Dev. 1993;7:1176–1190. doi: 10.1101/gad.7.7a.1176. [DOI] [PubMed] [Google Scholar]
- Wang J, Cao L-G, Wang Y-L, Pederson T. Localization of pre-messenger RNA at discrete nuclear sites. Proc Natl Acad Sci USA. 1991;88:7391–7395. doi: 10.1073/pnas.88.16.7391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watkins NJ, Leverette RD, Ling X, Andrews MT, Maxwell ES. Elements essential for processing intronic U14 snoRNA are located at the termini of the mature snoRNA sequence and include conserved nucleotide Boxes C and D. RNA. 1996;2:118–133. [PMC free article] [PubMed] [Google Scholar]
- Watkins NJ, Newman DR, Kuhn JF, Maxwell ES. In vitro assembly of the mouse U14 snoRNP core complex and identification of a 65 kDa box C/D-binding protein. RNA. 1998;4:582–593. doi: 10.1017/s1355838298980128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Z, Gall JG. “Micronucleoli” in the Xenopus germinal vesicle. Chromosoma. 1997;105:438–443. doi: 10.1007/BF02510480. [DOI] [PubMed] [Google Scholar]
- Wu Z, Murphy C, Callan HG, Gall JG. Small nuclear ribonucleoproteins and heterogeneous nuclear ribonucleoproteins in the amphibian germinal vesicle, loops, spheres and snurposomes. J Cell Biol. 1991;113:465–483. doi: 10.1083/jcb.113.3.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Z, Murphy C, Wu C-HH, Tsvetkov A, Gall JG. Snurposomes and coiled bodies. Cold Spring Harbor Symp Quant Biol. 1993;58:747–754. doi: 10.1101/sqb.1993.058.01.082. [DOI] [PubMed] [Google Scholar]
- Xia L, Watkins NJ, Maxwell ES. Identification of specific nucleotide sequences and structural elements required for intronic U14 snoRNA processing. RNA. 1997;3:17–26. [PMC free article] [PubMed] [Google Scholar]
- Zaret KS, Liu J-K, DiPersio CM. Site-directed mutagenesis reveals a liver transcription factor essential for the albumin transcriptional enhancer. Proc Natl Acad Sci USA. 1990;87:5469–5473. doi: 10.1073/pnas.87.14.5469. [DOI] [PMC free article] [PubMed] [Google Scholar]