Abstract
The human major histocompatibility complex (MHC), in addition to its role in the regulation of cell-cell interactions in the immune response, also influences reproductive success. Human leukocyte antigen-G (HLA-G) is an MHC class I gene of particular interest in reproductive biology because of its specific expression on fetal cytotrophoblast cells, and its reported involvement both in protection of the developing fetus from destruction by the maternal immune response and in the prevention of maternal pre-ecalampsia. HLA-G has 15 known alleles at the DNA level, and allelic frequency varies among ethnic groups. This study describes the results of an inaugural attempt to correlate an HLA-G genetic polymorphism with pregnancy outcome in a patient population undergoing IVF. The study group was composed of 102 Caucasian women. A maternal HLA-G genetic polymorphism was investigated by polymerase chain reaction (PCR) analysis of DNA collected from granulosa cells surrounding oocytes harvested for the IVF procedure. While no statistically significant correlation was identified in this initial study, larger studies examining DNA from trios of mother, father and offspring are planned.
Keywords: assisted reproduction, HLA, human, major histocompatibility complex, infertility, IVF
Introduction
The major histocompatibility complex (MHC), well known for its role in the regulation of cell–cell interactions in the immune response, also influences reproductive success. The MHC in humans is called the HLA complex and is located on the short arm of chromosome 6. There arc three major classes of proteins encoded by genes in the HLA complex: class I, class II, and class III. The complete DNA sequence of the 3.6 Mb of DNA comprising the HLA complex has recently been reported (Beck et al., 1999). The HLA complex is, to date, the most carefully sequenced and analysed piece of the human genome (Beck and Trowsdale, 2000). Comparative DNA sequence analysis, along with many years of immunological analysis of HLA complex-encoded proteins, has shown that there is tremendous genetic polymorphism in the HLA complex Janer and Geraghty, 1998; Geraghty et al., 1999). The task of deciphering which of these polymorphisms is correlated with reproductive success represents a significant challenge.
It has previously been reported that the MHC affects a variety of reproductive parameters, including spontaneous abortion (Ober 1998; Ober et al. 1998), protection of the fetus from attack by the maternal immune system (King et al., 1997; Rouas-Freiss et al., 1997; Rolstad and Seaman, 1998), and the regulation of preimplantation embryo growth and survival (Warner et al., 1998a, 1998b, 1998c; Warner and Brenner, 2001). Examination of the DNA in the HLA complex has revealed the presence of 224 identified gene loci, 128 of which are predicted to be expressed and 96 of which are thought to be pseudogenes (Beck et al., 1999; Beck and Trowsdale, 2000). About half of these genes were unknown before the DNA sequencing effort was undertaken. This sequencing information now makes it possible to begin a genetic analysis of exactly which genes in the HLA complex are involved in the regulation of reproductive success.
One gene in the HLA complex that has received special attention with respect to reproduction is the class I gene HLA-G. There are 15 known HLA-G alleles (van der Ven et al., 1998a; Hviid et al., 2001), and allelic frequency varies among ethnic groups (van der Ven et al., 1998a, 1998b). Shown in Table 1 are the known allelic frequencies for the Caucasian population. While there is increasing evidence of polymorphisms in the HLA-G gene, there is still very little DNA sequence information for each of the known alleles. Only the full-length HLA-G1 protein of the HLA-G*01011 allele and a soluble version of this isoform (HLA-G1sol) have so far been reported to have a function in pregnancy (Bainbridge et al., 2000). HLA-G expression is co-dominant (Hviid et al., 1998) and HLA-G protein is found in high concentration on extravillous trophoblast cells derived from the trophectoderm of the blastocyst (McMaster et al., 1995). Recent studies looking at a newly described non-conservative amino acid substitution in the α3 domain of HLA-G in women with a history of pre-eclampsia during pregnancy and their partners (Hviid et al., 2001) and HLA-G allele distribution in couples with a history of recurrent spontaneous abortion (Pfeiffer et al., 2001) did not show statistical correlation with pregnancy outcome. Thus, an ideal analysis of the effect of HLA-G polymorphisms on pregnancy outcome should include trio samples of maternal, paternal and offspring DNA.
Table 1.
Frequency of HLA-G alleles in the Caucasian populationa.
| Allele | Frequency (%) |
|---|---|
| HLA-G*01011 | 32.1 |
| HLA-C*01012 | 36.3 |
| HLA-G*01013 | 6.8 |
| HLA-G*01014 | ND |
| HLA-G*01015 | ND |
| HLA-G*01016 | ND |
| HLA-G*01017 | ND |
| HLA-G*01018 | ND |
| HLA-G*0102 | ND |
| HLA-G*0103 | 2.3 |
| HLA-G*01041 | 6.1 |
| HLA-G*01042 | ND |
| HLA-G*01043 | ND |
| HLA-G*0105N | 2.3 |
| HLA-G*0106 | 4.0 |
Based on van der Ven et al (1998a); Hviid et al. (2001) GenBank; and www.ebi.ac.uk/imgt/hla/align.html. ND= not determined.
Despite the paucity of DNA sequence information for the 15 known alleles of HLA-G and a current, but it is hoped temporary, unavailability of suitable trio DNA for analysis, it was decided to make a beginning by setting up a protocol to examine the effect of a maternal HLA-G polymorphism on pregnancy outcome in a cohort of 102 Caucasian women undergoing IVF. The maternal DNA used in the analysis was extracted from granulosa cells surrounding oocytes used in the IVF procedure.
Materials and methods
DNA isolation from cheek cells
A slightly modified version of the Cold Spring Harbor (New York) procedure to isolate cheek cell DNA was used (http://www.gene.com/ae/AE/AEPC/DNA/detection.html). Briefly, 10 ml of 0.9% NaCl, pH 7.0. was vigorously washed around the inside of the mouth. The saline was expelled into a clean 15 ml centrifuge tube. The tube was spun for 10 min at 756 g at 4°C. The supernatant was removed and the pellet-containing tube placed on ice. The pellet was resuspended in 500 ml of 10% Chelex beads (BioRad, Hercules, CA, USA). A 500 µ1 aliquot of the resuspended pellet was transferred to a clean 1 .5 ml reaction tube and incubated in boiling water for 10 min. The tube was then cooled on ice for one min and spun for 30 s at room temperature in a bench-top microfuge to pellet the Chelex beads. A 200 µl aliquot of supernatant was transferred to a fresh 1.5 ml reaction tube and the sample stored at 4°C. This protocol was undertaken in accordance with regulations of Northeastern University’s Department of Institutional Compliance.
DNA isolation from granulosa cells
Granulosa cells were collected from 102 Caucasian women attending the IVF clinic. Seventy-three of the women were 37 years of age or under and the remaining 29 women were between 38 and 42 years old. DNA from the granulosa cells was isolated using the same procedure as for cheek cells described above, except that the collected granulosa cells were resuspended in 500 µl of 0.9% NaCl as the first step of the procedure. The IVF clinic’s Institutional Review Board (IRB) approved this procedure.
Quantification of isolated DNA
The DNA concentration of each sample was measured using a TKO 100 mini-fluorometer (Hoefer Scientific Instruments, San Francisco, CA, USA) following the manufacturer’s instructions. After quantifying the DNA, the samples were diluted to make 5 ng/µl working stocks and stored at −20°C until needed.
Polymerase chain reaction
Primers
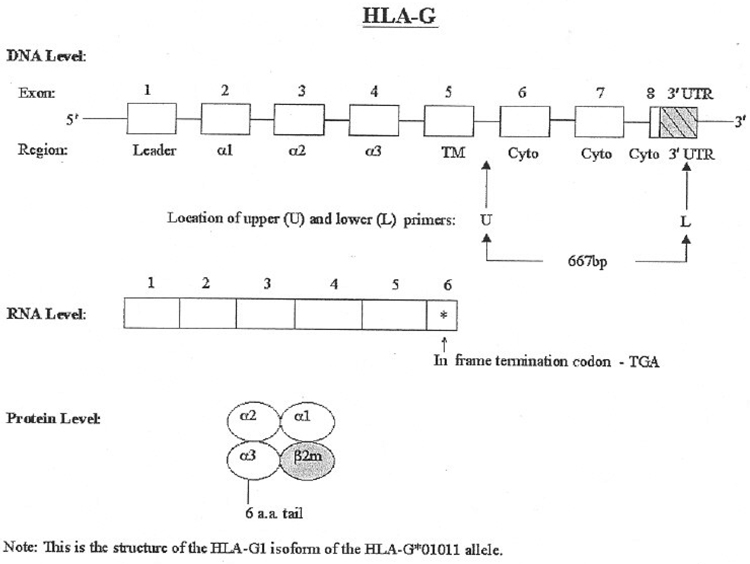
Diagrams showing the locations of the HLA-G primer pair are shown in Figure 1 and Figure 2. The primer sequences are based on the published sequence of allele HLA-G*01011 (Janer and Geraghty, 1998). The HLA-G primers and the internal control β-actin primers, together with the expected bands and a summary of the PCR conditions, are summarized in Table 2. Cheek cell DNA was used as a positive control and in the negative-control samples the DNA was replaced with nuclease free water, The HLA-G primers were purchased froth. Sigma/Genosys (St Louis. MO, USA), and the β-actin primers from Stratagene (La Jolla, CA, USA).
Figure 1.
Structure of HLA-G at the DNA, RNA, and protein levels. At the DNA level, the exons are open boxes and the 3 ′UTR is a shaded, hatched box. The primary RNA transcript is larger than shown; the RNA transcript is shown only up to the stop codon in exon 6. Also shown is the location of the upper (U) and lower (L) primers used in this study.
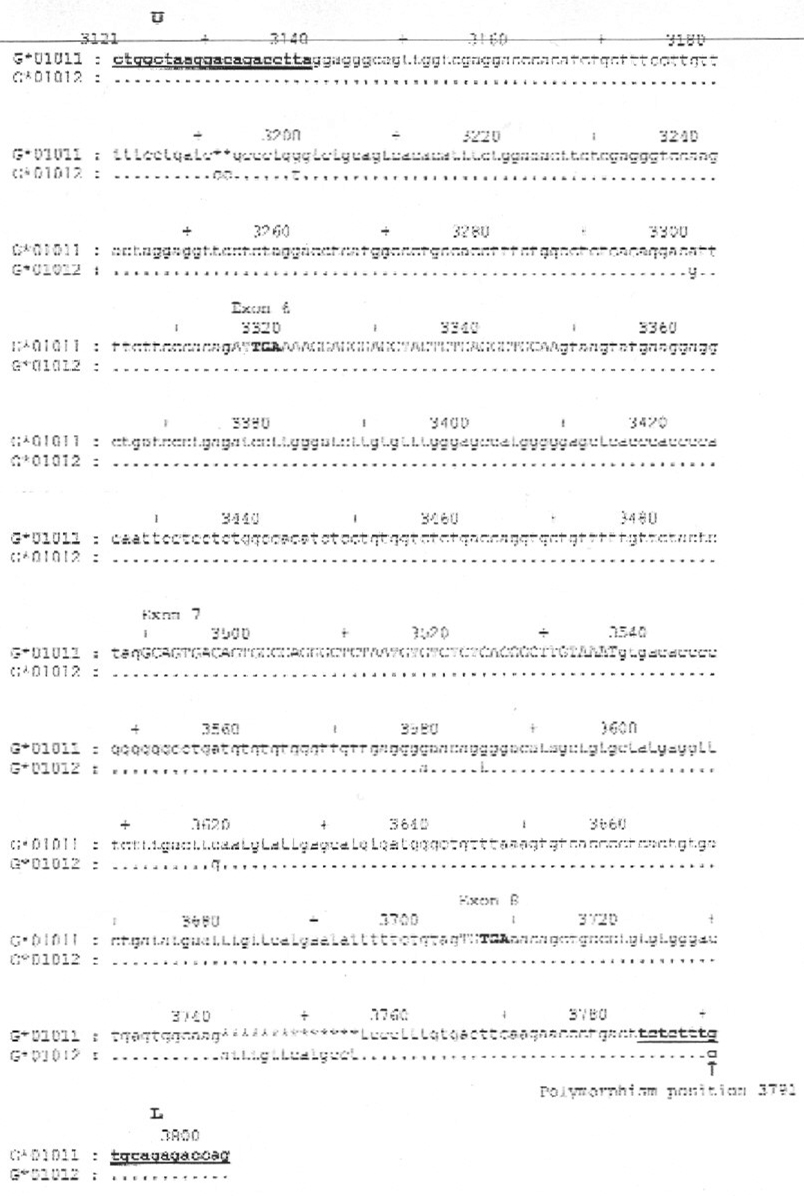
Figure 2.
Sequence alignment of the two most common Caucasian HLA-G alleles. The locations of the upper (U) and lower (L) primers are bold and underlined. (The actual primer sequences are shown in Table 2.) Similarities between the two alleles, G*01011 and G*01012, are shown by a dot. A two bp deletion and a 14 bp deletion in the G*01011 allele are denoted by asterisks (*). Ten by markers are denoted by a plus sign (+). The exon sequences are shown in upper case letters and the introns and 3′UT region sequences are shown in lower case letters. The termination codons in exons 6 and 8 are shown in bold letters. A single nucleotide polymorphism (SNP) is present at the location of the lower primer (position 3791) and is indicated by an arrow. The GenBank accession numbers for G*01011 and G*01012 are J03027 and S50740 respectively.
Table 2.
Primers used to detect an HLA-G polymorphism.
| Marker | Primers | Expected band (bp) | Observed band (bp) | TAa °C | [MgCl2] mM | PCRb No. of cycles | Primer sequence (5′-3′) | %DNA sequence identityc |
|---|---|---|---|---|---|---|---|---|
| HLA-G | Upper | 667 | 667 | 60 | 2.0 | 35 | CTG GCT AAG GAC AGA CCT TA | 98.0 |
| Lower | CTG GTC TCT GCA CAA AGA GA | |||||||
| β-Actin | Upper | 109 | 109 | 55 | 2.0 | 35 | ATG GGT CAG AAG GAT TCC TA | 100 |
| Lower | TCC ATG TCG TCC CAG TT |
TA = annealing temperature.
PCR programme is described in the text.
Identity = percentage identity of the product DNA sequence compared with the BLAST entry.
PCR conditions
The PCR experiments were set up and the amplified products analysed on a 6% polyacrylamide gel stained with ethidium bromide. Each reaction was set up in a 0.8 µl Hot Start reaction tube to give a total volume of 50 µ1 (Molecular Bio-Products, San Diego, CA, USA). The reaction tubes contained a lower layer that consisted of the following: 1 µl 20 µmol/1 of each primer; 4 µl 10 mmol/1 mixed dNTP (Perkin Elmer, Foster City, CA, USA); 2.5 µl 10x PCR buffer (Perkin Elmer), 3.5 µl 25 mmol/1 MgCl2 (Perkin Elmer) and 13 µl nuclease-free water. The tube was then heated to 80°C for 5 min followed by 25°C for 1 min to melt the wax bead. Then the following were placed on top of the melted wax: 17.25 µl nuclease free water, 2.5 µl 10x buffer, 0.25 µl 5 IU/µl AmpliTaq® DNA polymerase (Perkin Elmer), and 5 µl of DNA (5 ng/µl). The PCR experiments were all run using a DNA Thermal Cycler 480 (Perkin Elmer). The PCR programme used was one cycle at 96°C for 5 min; 35 cycles at 94°C for 20 s, at the annealing temperature (TA) for 30 s, and at 72°C for 45 s; one cycle at 72°C for 5 min (Janer and Geraghty, 1998).
Sequencing of PCR product
The identity of the product from the PCR reactions was confirmed by sequencing, The appropriate band from each reaction was excised from an agarose gel using a clean razor blade, and the DNA extracted using the QIAquick Gel Extraction Kit (Qiagen, Germany) following the manufacturer’s instructions. The final product of the isolation was resuspended in 30 µl of elution buffer (Qiagen) and sent at ambient temperature to The University of Maine Sequencing Facility.
Statistical analysis
Pregnancy outcome was classified into two groups according to whether or not a live birth resulted after IVF. This outcome was correlated with the presence or absence of a PCR band amplified by the HLA-G primers. Maternal age did not affect the results so data from all the women were pooled. The probability of the relationship of marker frequency with pregnancy success was found by using the definitions and procedures listed in Table 3.
Table 3.
Definitions and procedures used in statistical analysis.
| Number of patients in the study: 102 Caucasian women |
|---|
| Pregnancy success: 57 patients who had a live birth |
| Pregnancy failure: 45 patients who did not have a live birth |
| Step 1. Probability of pregnancy success (S) was calculated by dividing the number of patients with the presence of the PCR band who had a live birth by 57. |
| Step 2. The variance of S was calculated as Var(S) [S ×(1−S)]/57. |
| Step 3. Probability of pregnancy failure (F) was calculated by dividing the number of patients with the presence of the PCR band who did not have a live birth by 45. |
| Step 4. The variance of F was calculated as Var(F) = [F×(1−F)]/45. |
| Step 5. The difference (D) of success (S) and failure (F) was found by D = S − F. |
| Step 6. The standard deviation (SD) of the difference was calculated by: SD = square root of [Var(S) + Var(F)]. |
| Step 7. The probability of the difference, P(z), was found by dividing D by SD and the P-value was found by looking up P(z) in a normal distribution probability table (Weiss, 1995). |
Results
In a preliminary set of experiments the optimal DNA concentration was determined for the PCR reactions using DNA isolated from the cheek cells of a donor. The titration for the optimal template DNA concentration showed that a concentration of 5 ng/µl was optimal (data not shown). The products from the optimized PCR conditions were all found to be of the expected size and all had the correct DNA sequence (Table 2). The slightly less than 100% DNA sequence identity for the HLA-G primer pair may be from experimental error in the sequencing reactions or from the presence of a Polymorphic HLA-G allele in the cheek cell donor, as discussed below.
In the next set of experiments, DNA from 102 granulosa cell samples, representing 102 different Caucasian women, was collected. Table 4 is a summary of the outcome of the PCR Performed on these samples using (β-actin and HLA-G primers. The PCR results were classified into two groups: (i) A band of the appropriate size on the polyacrylamide gel was designated as present (+); (ii) An absent band was termed polymorphic (−). In all of the PCR that resulted in a product, the product was of the correct size. A band for the positive control, β-actin, was always present, and a band for the negative control, water, was always absent in each sample tested. A sample gel is shown in Figure 3. A mixture of present and absent bands is shown.
Table 4.
Summary of pregnancy outcome dataa (PCR results).
| Pregnancy Outcome | β-Actin | HLA-G | HLA-G |
|---|---|---|---|
| (PCR band+) | (PCR band +) | (PCR band−) | |
| No. (%) | No. (%) | No. (%) | |
| Success (n = 57) | 57(100) | 24(42) | 33(58) |
| Failure (n = 45) | 45(100) | 24(53) | 21(47) |
P = 0.13 for comparison of presence of PCR band for HLA-G in success group versus failure group.
Figure 3.
Sample gel showing the PCR products using primers for HLA-G. M = Molecular weight markers with an arrow pointing to the 700 bp marker; Lanes 11–18 represent patient samples that either had the presence or absence of a band for HLA-G.
The results show that in this limited sample of maternal DNA, HLA-G does not reveal a statistically significant effect on pregnancy outcome (Table 4). Absence of a PCR band for HLA-G is an indication of a polymorphism at the tested locus and is discussed in detail in the next section.
Discussion
The present analysis of the relationship of a genetic polymorphism in the maternal HLA-G gene to pregnancy outcome in Caucasian women undergoing IVF indicates that the presence of the HLA-G*01011 allele does not have a statistically significant association with an enhanced chance of reproductive success However, it is possible that the study cohort comprised too small a sample to reach significance, or that the existence of many alleles of the HLA-G gene complicated the analysis of the data. In our experimental design, a positive band upon PCR means that at least one HLA-G allele in each tested woman had the same sequence in the regions of the upper and lower primers as the G*01011 allele (Figure 1–Figure 3).However, since the reported gene frequency of G*01011 in the Caucasian population is 32% (Table 1), and 41–53% (depending on pregnancy success or failure) HLA-G positive PCR bands was found in our population (Table 4), it is logical to conclude that some of the other alleles shown in Table 1 have the identical sequence to G*01011 in the primer regions. A known single nucleotide polymorphism (SNP) is present at the location of the lower primer (position 3791) where guanine in allele G*01011 is replaced by cytosine in allele G*01012. Since the frequency of G*01012 in the Caucasian population is 36% (Table 1), and 47–58% (depending on pregnancy success or failure) HLA-G negative PCR bands was found in our population (Table 4), it is also Logical to conclude that some of the other alleles shown in Table 1 have nucleotide differences in the region of the primers that lead to Lack of amplification of the HLA-G gene with our primers. The full understanding of the positive and negative PCR results reported in this study will only be possible when DNA sequence information is available for each of the HLA-G alleles.
In spite of the fact that the correlation of pregnancy outcome was not statistically significant in this study, further analysis of the role of HLA-G in pregnancy outcome after IVF seems warranted. First, although the P-value in the study is not <0.05, it is low (P = 0.13). This may mean that a large enough sample size was not analysed. Second, as discussed above, the DNA sequence of each of the HLA-G alleles is not yet known. When this information becomes available it should be possible to do a more comprehensive analysis of the data. Third. it should be possible to separate the two alleles in each sample so that a full analysis of the genotype of each of the women is carried out. Thus, our study is an attempt to analyse the effect of maternal HLA-G genotype on pregnancy outcome.
HLA-G expression has been proposed to affect pregnancy outcome in several ways, with extensive literature suggesting potential mechanisms. First, there are a number of studies suggesting that HLA-G protects the developing fetus from destruction by maternal immune cells, including NK cells (King et al., 1997; Rouas-Freiss et al., 1997; Rolstad and Seaman, 1998) T cells (Le Gal et al., 1999), and myelomonocytic cells (Allan et al., 1999). Second, there is extensive evidence that HLA-G may be the functional homologue of mouse Qa-2, a protein that is responsible for the Ped gene phenotype (reviewed in Loke et al., 1999; Warner and Brenner, 2001). The mouse Ped gene regulates preimplantation growth rate and subsequent embryo survival and the first paper reporting the existence of HLA-G suggested that Q7, a gene encoding Qa-2 protein, was the mouse gene most homologous to human HLA-G (Geraghty et al., 1987), and it has even been suggested that human HLA-G and mouse Qa-2 developed similar functions through convergent evolution (Allcock et al., 2000). Verifying all of the suggested roles of HLA-G will require extensive investigation and establishing allelic correlation with pregnancy outcome is but one line of research, complicated by the existence of many alleles.
Clearly more studies need to he performed to determine exactly which HLA-G alleles are expressed by women attending the IVF clinic, and to ascertain whether particular HLA-G alleles are advantageous or disadvantageous to pregnancy outcome after IVF. As more allelic DNA sequence information becomes available, it is planned to extend the current preliminary study to a larger population of women undergoing IVF. At present, there is only about a 25% success rate after IVF. Many parameters have been examined, including maternal age, aneuploidy, and in-vitro culture conditions, in an attempt to increase the success rate after IVF. One area that has received little attention so far is the genetic make-up of the women attending the IVF clinic. With the advent of new methodologies using DNA chips and micro-arrays, it should be possible in the near future to obtain a genetic profile of the women attending IVF clinics, as well as genetic profiles of the sperm donors and resulting babies (trio analysis). Pending availability of complete trio samples, however, preliminary work on maternal and duo (maternal and sperm donor) profiles contributes to the global understanding of the role of HLA-G in reproductive success. It is expected that DNA chips will soon be available that will allow the analysis of all the genes in the HLA-G complex (Geraghty et al., 1999). It may then be possible to use genetic polymorphisms in the HLA complex, including polymorphisms in HLA-G, as a diagnostic tool to predict the chance of pregnancy success after IVF.
Acknowledgements
We thank Caryn Mack for help in collecting the granulosa cell samples and Dr A Ding for help with the statistical analyses. This study was supported by the Institute for Reproductive Medicine and Science of Saint Barnabas, West Orange, NJ, USA and NIH grant HD39215.
Biography
Carol Warner received her PhD in Biochemistry in 1970 from UCLA after which she spent a postdoctoral year at Yale University. From 1971–1988 she was a faculty member in the Department of Biochemistry and Biophysics at Iowa State University. She moved to Northeastern University in 1988 where she is now Matthews Distinguished Professor of Biology. During the past 30 years she has worked in a variety of research areas with the main emphasis on the role of the major histocompatibility complex (MHC) in disease resistance and reproduction in mice, chickens, pigs, and humans. Her current research interests focus on genes that mediate preimplantation embryo survival in mice and humans. One gene of particular interest is the mouse Ped gene, located in the MHC. The Ped gene encodes a protein product, Qa-2 protein, which mediates the rate of preimplantation cleavage division and subsequent embryo survival. Her working hypothesis is that the human homologue of the Ped gene product is the human MHC protein HLA-G. She is also conducting research on the expression of genes that mediate apoptosis in preimplantation embryos. Finally, she is working on new methods of imaging of preimplantation embryos to be able to use morphological as well as genetic criteria to assess preimplantation embryo health.

Footnotes
Paper based on contribution presented at the Alpha meeting in New York, USA, September 2001.
References
- Allan OS, Colonna M, Lanier LL, et al. Tetrameric complexes of human histocompatibility leukocyte antigen (HLA)-G bind to peripheral blood myelomonocytic cells. Journal of Experimental Medicine. 1999;189:1149–1156. doi: 10.1084/jem.189.7.1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allcock RJ, Martin AM, Price P. The mouse as a model for the effects of MHC genes on human disease. Immunology Today. 2000;21:328–332. doi: 10.1016/s0167-5699(00)01654-6. [DOI] [PubMed] [Google Scholar]
- Bainbridge DR, Ellis SA, Sargent IL. The short forms of HLA-G are unlikely to play a role in pregnancy because they are not expressed at the cell surface. Journal of Reproductive Immunology. 2000;47:1–16. doi: 10.1016/s0165-0378(00)00056-5. [DOI] [PubMed] [Google Scholar]
- Beck S, Trowsdale J. The human major histocompatibility complex: lessons from the DNA sequence. Annual Review of Genomics and Human Gentiles. 2000;1:117–137. doi: 10.1146/annurev.genom.1.1.117. [DOI] [PubMed] [Google Scholar]
- Beck S, Geraghty D, Inoko H, Rowen L. Complete sequence and gene map of a human major histocompatibility complex. The MHC sequencing consortium. Nature. 1999;401:921–923. doi: 10.1038/44853. [DOI] [PubMed] [Google Scholar]
- Geraghty DE, Koller BH, Orr HT. A human major histocompatibility complex class I gene that encodes a protein with a shortened cytoplasmic segment. Proceedings of the National Academy of Sciences of the USA. 1987;84:9l45–9149. doi: 10.1073/pnas.84.24.9145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geraghty DE, Vu Q, Williams L, et al. Mapping HLA for single nucleotide polymorphisms. Reviews in Immunogenetics. 1999;1:231–238. [PubMed] [Google Scholar]
- Hviid TV, Moller C, Sorensen S, Morling N. Co-dominant expression of the HLA-G gene and various forms of alternatively spliced HLA-G mRNA in human first trimester trophoblast. Human Immunology. 1998;59:87–98. doi: 10.1016/s0198-8859(97)00259-0. [DOI] [PubMed] [Google Scholar]
- Hviid TV, Christiansen OB, Johansen JK, et al. Characterization of a new HL A-G allele encoding a nonconservative amino acid substitution in the alpha3 domain (exon 4) and its relevance to certain complications in pregnancy. Immunogenetics. 2001;53:48–53. doi: 10.1007/s002510100296. [DOI] [PubMed] [Google Scholar]
- Janer M, Geraghty DE. The human major histocompatibility complex: 42221 by of genomic sequence, high-density sequence-tagged site map, evolution, and polymorphism for HLA class L. Genomics. 1998;51:35–44. doi: 10.1006/geno.1998.5377. [DOI] [PubMed] [Google Scholar]
- King A, Loke YW, Chaouat G. NK cells and reproduction. Immunology Today. 1997;18:64–66. doi: 10.1016/s0167-5699(97)01001-3. [DOI] [PubMed] [Google Scholar]
- Le Gal FA, Riteau B, Sedlik C, et al. HLA-G-mediated inhibition of antigen-specific cytotoxic T lymphocytes. International Immunology. 1999;11:1351–1356. doi: 10.1093/intimm/11.8.1351. [DOI] [PubMed] [Google Scholar]
- Loke YW, Hiby S, King A. Human leucocyte antigen-G and reproduction. Journal of Reproductive Immunology. 1999;43:235–242. doi: 10.1016/s0165-0378(99)00023-6. [DOI] [PubMed] [Google Scholar]
- McMaster MT, Librach CL, Zhou Y, et al. Human placental HLA-G expression is restricted to differentiated cytotrophoblasts. Journal of Immunology. 1995;154:3771–3778. [PubMed] [Google Scholar]
- Ober C. HLA and pregnancy: the paradox of the fetal allograft. American Journal of Human Generics. 1998;62:1–5. doi: 10.1086/301692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ober C, Hyslop T, Elias S, et al. Human leukocyte antigen matching and fetal loss: results of a 10 year prospective study. Human Reproduction. 1998;13:33–38. doi: 10.1093/humrep/13.1.33. [DOI] [PubMed] [Google Scholar]
- Pfeiffer KA, Fimmers R, Engels G, et al. The HLA-G genotype is potentially associated with idiopathic recurrent spontaneous abortion. Molecular Human Reproduction. 2001;7:373–378. doi: 10.1093/molehr/7.4.373. [DOI] [PubMed] [Google Scholar]
- Rolstad B, Seaman WE. Natural killer cells and recognition of MHC class I molecules: new perspectives and challenges in immunology. Scandinavian Journal of Immunology. 1998;47:412–425. doi: 10.1046/j.1365-3083.1998.00358.x. [DOI] [PubMed] [Google Scholar]
- Rouas-Freiss N, Goncalves RM, Menier C, et al. Direct evidence to support the role of HLA-G in protecting the fetus from maternal uterine natural killer cytolysis. Proceedings of the National Academy of Sciences of the USA. 1997;94:11520–11525. doi: 10.1073/pnas.94.21.11520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Ven K, Skrablin S, Engels G, Krebs D. HLA-G polymorphisms and allele frequencies in Caucasians. Human Immunology. 1998a;59:302–312. doi: 10.1016/s0198-8859(98)00021-4. [DOI] [PubMed] [Google Scholar]
- van der Ven K, Skrablin S, Ober C, Krebs D. HLA-G polymorphisms: ethnic differences and implications for potential molecule function. American Journal of Reproductive Immunology. 1998b;40:145–157. doi: 10.1111/j.1600-0897.1998.tb00406.x. [DOI] [PubMed] [Google Scholar]
- Warner CM, Brenner CA. Genetic regulation of preimplantation embryo survival. Current Topics in Developmental Biology. 2001;52:151–192. doi: 10.1016/s0070-2153(01)52011-6. [DOI] [PubMed] [Google Scholar]
- Warner CM, Cao W, Exley GE, et al. Genetic regulation of egg and embryo survival. Human Reproduction. 1998a;13:178–190. doi: 10.1093/humrep/13.suppl_3.178. discussion 191–196. [DOI] [PubMed] [Google Scholar]
- Warner CM, Exley GE, McElhinny AS, Tang C. Genetic regulation of preimplantation mouse embryo survival. Journal of Experimental Zoology. 1998b;282:272–279. [PubMed] [Google Scholar]
- Warner CM, McElhinny AS, Wu L, et al. Role of the Ped gene and apoptosis genes in control of preimplantation development. Journal of Assisted Reproduction and Generics. 1998c;15:331–337. doi: 10.1023/A:1022560914833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss NA. Introductory Statistics. 4th Edition. Addison-Wesley,: Reading, MA; 1995. p. 939. [Google Scholar]