Abstract
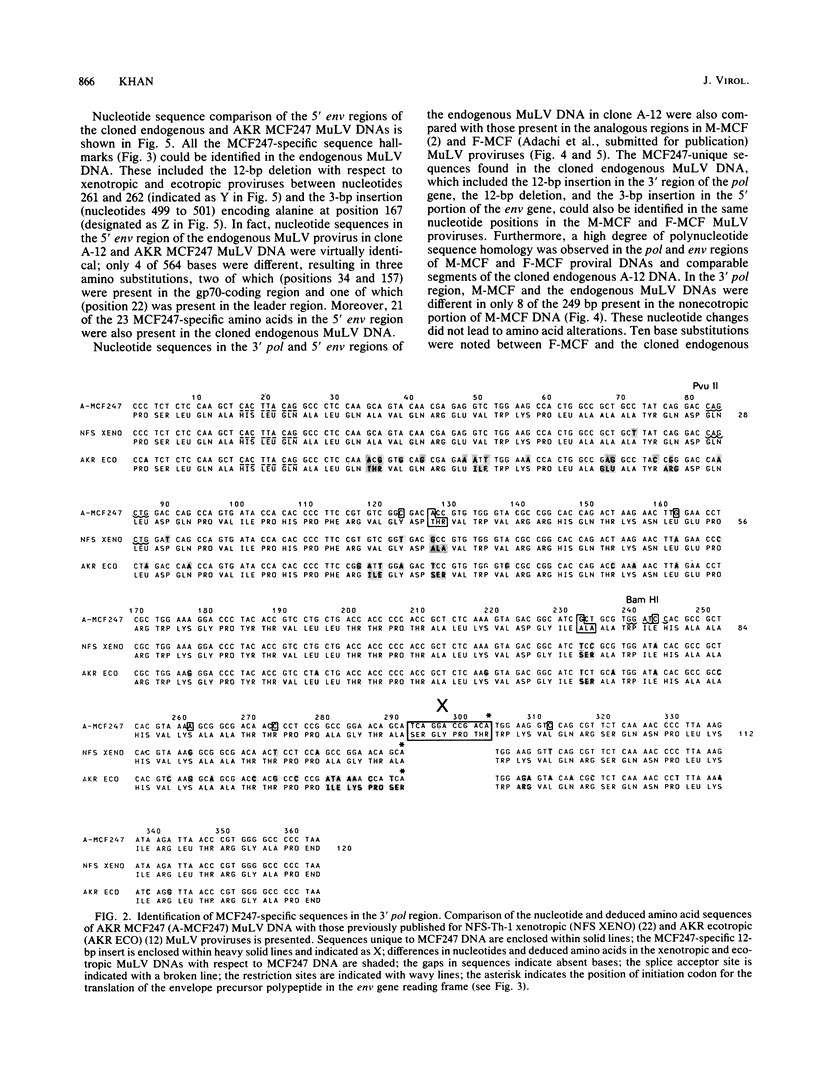
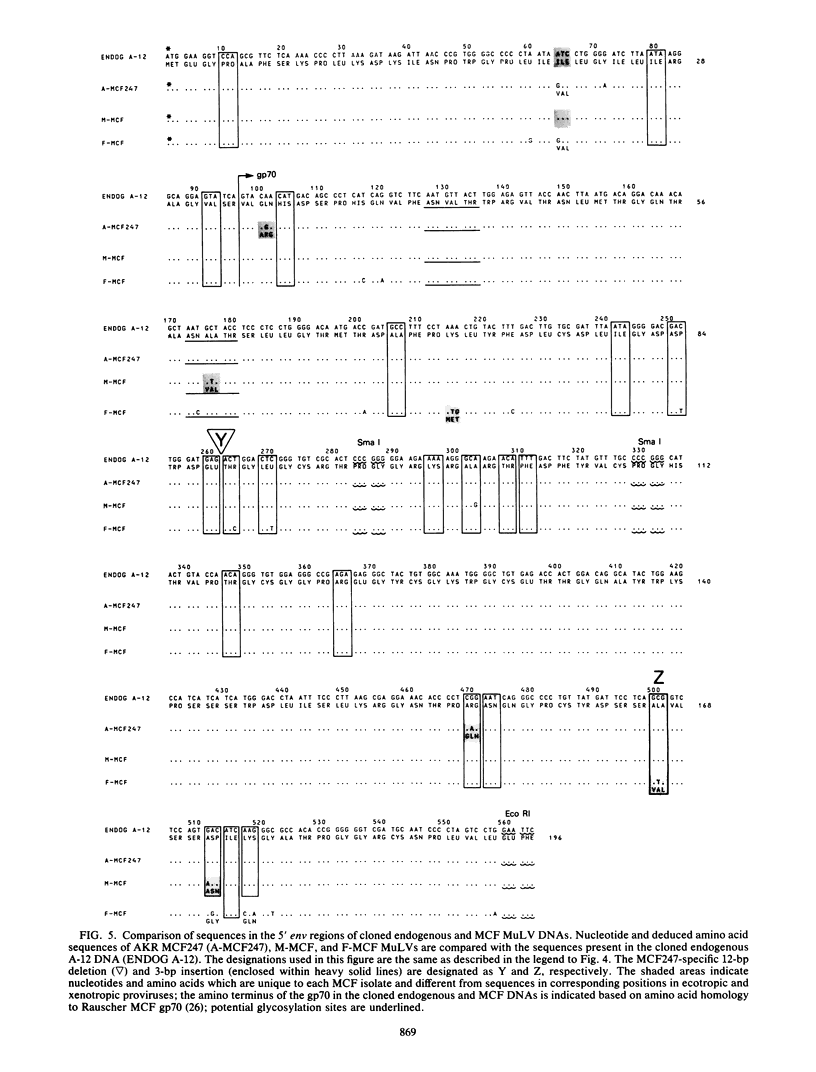
The sequence of 363 nucleotides near the 3' end of the pol gene and 564 nucleotides from the 5' terminus of the env gene in an endogenous murine leukemia viral (MuLV) DNA segment, cloned from AKR/J mouse DNA and designated as A-12, was obtained. For comparison, the nucleotide sequence in an analogous portion of AKR mink cell focus-forming (MCF) 247 MuLV provirus was also determined. Sequence features unique to MCF247 MuLV DNA in the 3' pol and 5' env regions were identified by comparison with nucleotide sequences in analogous regions of NFS -Th-1 xenotropic and AKR ecotropic MuLV proviruses. These included (i) an insertion of 12 base pairs encoding four amino acids located 60 base pairs from the 3' terminus of the pol gene and immediately preceding the env gene, (ii) the deletion of 12 base pairs (encoding four amino acids) and the insertion of 3 base pairs (encoding one amino acid) in the 5' portion of the env gene, and (iii) single base substitutions resulting in 2 MCF247 -specific amino acids in the 3' pol and 23 in the 5' env regions. Nucleotide sequence comparison involving the 3' pol and 5' env regions of AKR MCF247 , NFS xenotropic, and AKR ecotropic MuLV proviruses with the cloned endogenous MuLV DNA indicated that MCF247 proviral DNA sequences were conserved in the cloned endogenous MuLV proviral segment. In fact, total nucleotide sequence identity existed between the endogenous MuLV DNA and the MCF247 MuLV provirus in the 3' portion of the pol gene. In the 5' env region, only 4 of 564 nucleotides were different, resulting in three amino acid changes between AKR MCF247 MuLV DNA and the endogenous MuLV DNA present in clone A-12. In addition, nucleotide sequence comparison indicated that Moloney-and Friend-MCF MuLVs were also highly related in the 3' pol and 5' env regions to the cloned endogenous MuLV DNA. These results establish the role of endogenous MuLV DNA segments in generation of recombinant MCF viruses.
Full text
PDF





Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Besmer P., Baltimore D. Mechanism of restriction of ecotropic and xenotropic murine leukemia viruses and formation of pseudotypes between the two viruses. J Virol. 1977 Mar;21(3):965–973. doi: 10.1128/jvi.21.3.965-973.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosselman R. A., van Straaten F., Van Beveren C., Verma I. M., Vogt M. Analysis of the env gene of a molecularly cloned and biologically active Moloney mink cell focus-forming proviral DNA. J Virol. 1982 Oct;44(1):19–31. doi: 10.1128/jvi.44.1.19-31.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chattopadhyay S. K., Cloyd M. W., Linemeyer D. L., Lander M. R., Rands E., Lowy D. R. Cellular origin and role of mink cell focus-forming viruses in murine thymic lymphomas. Nature. 1982 Jan 7;295(5844):25–31. doi: 10.1038/295025a0. [DOI] [PubMed] [Google Scholar]
- Chattopadhyay S. K., Lander M. R., Gupta S., Rands E., Lowy D. R. Origin of mink cytopathic focus-forming (MCF) viruses:comparison with ecotropic and xenotropic murine leukemia virus genomes. Virology. 1981 Sep;113(2):465–483. doi: 10.1016/0042-6822(81)90175-6. [DOI] [PubMed] [Google Scholar]
- Chien Y. H., Verma I. M., Shih T. Y., Scolnick E. M., Davidson N. Heteroduplex analysis of the sequence relations between the RNAs of mink cell focus-inducing and murine leukemia viruses. J Virol. 1978 Oct;28(1):352–360. doi: 10.1128/jvi.28.1.352-360.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cloyd M. W., Hartley J. W., Rowe W. P. Lymphomagenicity of recombinant mink cell focus-inducing murine leukemia viruses. J Exp Med. 1980 Mar 1;151(3):542–552. doi: 10.1084/jem.151.3.542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donoghue D. J., Rothenberg E., Hopkins N., Baltimore D., Sharp P. A. Heteroduplex analysis of the nonhomology region between Moloney MuLV and the dual host range derivative HIX virus. Cell. 1978 Aug;14(4):959–970. doi: 10.1016/0092-8674(78)90350-1. [DOI] [PubMed] [Google Scholar]
- Engelman D. M., Steitz T. A. The spontaneous insertion of proteins into and across membranes: the helical hairpin hypothesis. Cell. 1981 Feb;23(2):411–422. doi: 10.1016/0092-8674(81)90136-7. [DOI] [PubMed] [Google Scholar]
- Fischinger P. J., Nomura S., Bolognesi D. P. A novel murine oncornavirus with dual eco- and xenotropic properties. Proc Natl Acad Sci U S A. 1975 Dec;72(12):5150–5155. doi: 10.1073/pnas.72.12.5150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartley J. W., Wolford N. K., Old L. J., Rowe W. P. A new class of murine leukemia virus associated with development of spontaneous lymphomas. Proc Natl Acad Sci U S A. 1977 Feb;74(2):789–792. doi: 10.1073/pnas.74.2.789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayward W. S., Neel B. G., Astrin S. M. Activation of a cellular onc gene by promoter insertion in ALV-induced lymphoid leukosis. Nature. 1981 Apr 9;290(5806):475–480. doi: 10.1038/290475a0. [DOI] [PubMed] [Google Scholar]
- Herr W., Corbin V., Gilbert W. Nucleotide sequence of the 3' half of AKV. Nucleic Acids Res. 1982 Nov 11;10(21):6931–6944. doi: 10.1093/nar/10.21.6931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoggan M. D., Buckler C. E., Sears J. F., Rowe W. P., Martin M. A. Organization and stability of endogenous xenotropic murine leukemia virus proviral DNA in mouse genomes. J Virol. 1983 Jan;45(1):473–477. doi: 10.1128/jvi.45.1.473-477.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holland C. A., Wozney J., Hopkins N. Nucleotide sequence of the gp70 gene of murine retrovirus MCF 247. J Virol. 1983 Sep;47(3):413–420. doi: 10.1128/jvi.47.3.413-420.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan A. S., Martin M. A. Endogenous murine leukemia proviral long terminal repeats contain a unique 190-base-pair insert. Proc Natl Acad Sci U S A. 1983 May;80(9):2699–2703. doi: 10.1073/pnas.80.9.2699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan A. S., Repaske R., Garon C. F., Chan H. W., Rowe W. P., Martin M. A. Characterization of proviruses cloned from mink cell focus-forming virus-infected cellular DNA. J Virol. 1982 Feb;41(2):435–448. doi: 10.1128/jvi.41.2.435-448.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan A. S., Rowe W. P., Martin M. A. Cloning of endogenous murine leukemia virus-related sequences from chromosomal DNA of BALB/c and AKR/J mice: identification of an env progenitor of AKR-247 mink cell focus-forming proviral DNA. J Virol. 1982 Nov;44(2):625–636. doi: 10.1128/jvi.44.2.625-636.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch W., Hunsmann G., Friedrich R. Nucleotide sequence of the envelope gene of Friend murine leukemia virus. J Virol. 1983 Jan;45(1):1–9. doi: 10.1128/jvi.45.1.1-9.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. A new method for sequencing DNA. Proc Natl Acad Sci U S A. 1977 Feb;74(2):560–564. doi: 10.1073/pnas.74.2.560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montreuil J. Primary structure of glycoprotein glycans: basis for the molecular biology of glycoproteins. Adv Carbohydr Chem Biochem. 1980;37:157–223. doi: 10.1016/s0065-2318(08)60021-9. [DOI] [PubMed] [Google Scholar]
- Rein A. Interference grouping of murine leukemia viruses: a distinct receptor for the MCF-recombinant viruses in mouse cells. Virology. 1982 Jul 15;120(1):251–257. doi: 10.1016/0042-6822(82)90024-1. [DOI] [PubMed] [Google Scholar]
- Repaske R., O'Neill R. R., Khan A. S., Martin M. A. Nucleotide sequence of the env-specific segment of NFS-Th-1 xenotropic murine leukemia virus. J Virol. 1983 Apr;46(1):204–211. doi: 10.1128/jvi.46.1.204-211.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rommelaere J., Faller D. V., Hopkins N. Characterization and mapping of RNase T1-resistant oligonucleotides derived from the genomes of Akv and MCF murine leukemia viruses. Proc Natl Acad Sci U S A. 1978 Jan;75(1):495–499. doi: 10.1073/pnas.75.1.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosner M. R., Grinna L. S., Robbins P. W. Differences in glycosylation patterns of closely related murine leukemia viruses. Proc Natl Acad Sci U S A. 1980 Jan;77(1):67–71. doi: 10.1073/pnas.77.1.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowe W. P., Cloyd M. W., Hartley J. W. Status of the association of mink cell focus-forming viruses with leukemogenesis. Cold Spring Harb Symp Quant Biol. 1980;44(Pt 2):1265–1268. doi: 10.1101/sqb.1980.044.01.137. [DOI] [PubMed] [Google Scholar]
- Schultz A., Rein A., Henderson L., Oroszlan S. Biological, chemical, and immunological studies of Rauscher ecotropic and mink cell focus-forming viruses from JLS-V9 cells. J Virol. 1983 Mar;45(3):995–1003. doi: 10.1128/jvi.45.3.995-1003.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp P. A. Speculations on RNA splicing. Cell. 1981 Mar;23(3):643–646. doi: 10.1016/0092-8674(81)90425-6. [DOI] [PubMed] [Google Scholar]
- Shih T. Y., Weeks M. O., Troxler D. H., Coffin J. M., Scolnick E. M. Mapping host range-specific oligonucleotides within genomes of the ecotropic and mink cell focus-inducing strains of Moloney murine leukemia virus. J Virol. 1978 Apr;26(1):71–83. doi: 10.1128/jvi.26.1.71-83.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinnick T. M., Lerner R. A., Sutcliffe J. G. Nucleotide sequence of Moloney murine leukaemia virus. Nature. 1981 Oct 15;293(5833):543–548. doi: 10.1038/293543a0. [DOI] [PubMed] [Google Scholar]


