Abstract
Coiled bodies (CBs) in the amphibian oocyte nucleus are spherical structures up to 10 μm or more in diameter, much larger than their somatic counterparts, which rarely exceed 1 μm. Oocyte CBs may have smaller granules attached to their surface or embedded within them, which are identical in structure and composition to the many hundreds of B-snurposomes found free in the nucleoplasm. The matrix of the CBs contains the diagnostic protein p80-coilin, which is colocalized with the U7 small nuclear ribonucleoprotein (snRNP), whereas the attached and embedded B-snurposomes contain splicing snRNPs. A few of the 50–100 CBs in the oocyte nucleus are attached to lampbrush chromosomes at the histone gene loci. By coimmunoprecipitation we show that coilin and the U7 snRNP can form a weak but specific complex in the nucleoplasm, which is dependent on the special U7 Sm-binding site. Under the same conditions coilin does not associate with the U1 and U2 snRNPs. Coilin is a nucleic acid-binding protein, as shown by its interaction with single-stranded DNA and with poly r(U) and poly r(G). We suggest that an important function of coilin is to form a transient complex with the U7 snRNP and accompany it to the CBs. In the case of CBs attached to chromosomes at the histone gene loci, the U7 snRNP is thus brought close to the actual site of histone pre-mRNA transcription.
INTRODUCTION
Coiled bodies (CBs)1 are small nuclear organelles, generally 1 μm or less in diameter, found in a variety of animal and plant cells. They are characterized by the presence of a unique protein, p80-coilin (Andrade et al., 1991), along with components involved in three different RNA-processing pathways: splicing, pre-rRNA processing, and histone pre-mRNA processing (reviewed by Lamond and Carmo-Fonseca, 1993; Bohmann et al., 1995; Gall et al., 1995; Roth, 1995; Lamond and Earnshaw, 1998; Matera, 1998). Long before their homology to somatic CBs was recognized, CBs had been described from the oocyte nucleus or germinal vesicle (GV) of insects (Jörgensen, 1913) and amphibians (Gall, 1954; Callan and Lloyd, 1960). In amphibians they have generally been known under the name spheres or sphere organelles (reviewed by Callan, 1986). GVs from mature or nearly mature Xenopus oocytes contain 50–100 CBs, of which the majority are free in the nucleoplasm, with a few specifically attached to lampbrush chromosomes at the histone gene loci (Callan et al., 1991). CBs in the Xenopus GV consist of three parts, easily distinguished by phase contrast or differential interference microscopy (DIC): a nearly perfectly spherical body (hence the original name), one to many rounded structures on the surface of the sphere, and one to many inclusions (Figure 1). The attached structures and inclusions are identical in morphology and composition to the thousands of so-called B-snurposomes found free in the nucleoplasm (Wu et al., 1991). The unusually large size of oocyte CBs, some of which have diameters greater than 10 μm, permits more precise cytological localization of components than is possible with the much smaller somatic CBs. By in situ hybridization and immunofluorescence, we earlier showed that the U7 small nuclear ribonucleoprotein (snRNP), which is required for histone pre-mRNA 3′-end processing, is restricted to the sphere body (Wu and Gall, 1993). On the other hand, components involved in pre-mRNA splicing, including the five splicing snRNPs, U1, U2, U4/U6, and U5, are concentrated in the B-snurposomes on the surface and the B-like inclusions (Wu et al., 1991; Gall et al., 1995). Thus, snRNPs are compartmentalized within oocyte CBs depending on the type of RNA processing in which they are involved.
Figure 1.
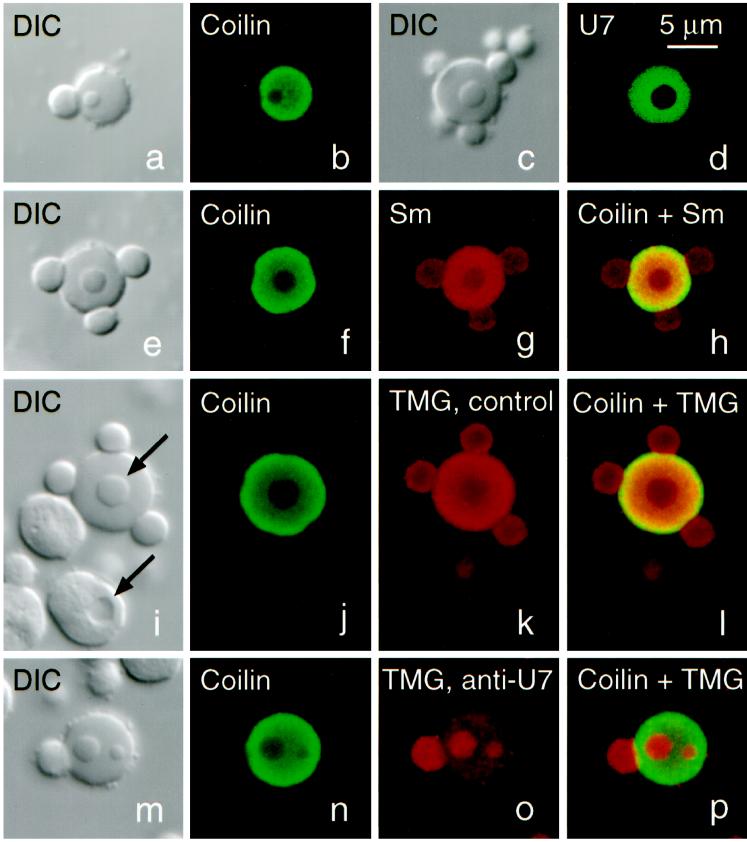
Structure and composition of CBs from Xenopus oocytes (stages IV–VI). (a and b) DIC and immunofluorescence image of a CB stained for coilin (serum C236, fluorescein). Stain is limited to the matrix of the CB and is absent from the attached B-snurposome and the B-like inclusion. (c and d) DIC and fluorescence image of a CB from an oocyte injected 1 d previously with capped, fluorescein-labeled U7 snRNA, showing that U7, like coilin, is strictly limited to the matrix. (e–h) CB stained for coilin (serum C236, fluorescein) and Sm proteins (mAb Y12, Cy3). Overlap shows colocalization of coilin and Sm proteins in the matrix, only Sm proteins in the B-snurposomes and the inclusion. (i–l) CB stained for coilin (serum C236, fluorescein) and the trimethylguanosine (TMG) cap of snRNAs (mAb K121, Cy3). TMG occurs at highest concentration in the matrix but is also strong in the B-snurposomes and the inclusion. Upper arrow in the DIC image points to the inclusion in the CB; lower arrow points to a vacuole in a nucleolus. Note that the shadowing is on opposite sides, demonstrating that the inclusion in the CB is denser than the matrix in which it is embedded, whereas the vacuole is less dense than the body of the nucleolus. (m–p) CB stained exactly as in panels i–l, except that oocyte was injected 1 d previously with an antisense oligodeoxynucleotide against bases 1–16 of U7 snRNA. This treatment results in complete loss of U7 from the GV (Figure 2). Concomitantly, TMG stain in the matrix of the CB is reduced by ∼90% (compare o with k), but stain in the B-snurposome on the surface and the two inclusions is unaffected.
In Xenopus oocytes, the homologue of mammalian p80-coilin, SPH-1, is also limited to the sphere body (Tuma et al., 1993; Wu et al., 1994). Little is known about the structure of coilin, and so far no similar proteins have appeared in the databases. Although the function of coilin is unknown, its colocalization with the U7 snRNP suggested that it might play some role in the processing pathway for histone pre-mRNA. For this reason we looked for molecular interaction between coilin and the U7 snRNP. We took advantage of two special features of the Xenopus oocyte: a pool of free snRNP proteins in the cytoplasm (Forbes et al., 1983) and a pool of “soluble” coilin in the GV, which we describe here. When exogenous splicing small nuclear (sn)RNAs are injected into the oocyte cytoplasm, free snRNP proteins associate with the RNA to form functional snRNPs, which are quickly imported into the GV (reviewed in Mattaj, 1988). The same is true for injected U7 snRNA (Grimm et al., 1993; Stefanovic et al., 1995). After import into the GV, the U7 snRNP is efficiently targeted to the CBs (Wu et al., 1996). However, only a fraction of the imported U7 snRNP actually goes to the CBs, creating an in vivo situation where a large amount of the U7 snRNP occurs along with soluble coilin in the nucleoplasm. Under these conditions, we find that the U7 snRNP can be immunoprecipitated by an antibody against coilin. Similar experiments with U1 and U2 snRNAs fail to demonstrate an association of their snRNPs with coilin. Further biochemical characterization of coilin by chromatography on single-stranded nucleic acids demonstrates that coilin is an RNA-binding protein. We suggest that coilin may normally bind to the U7 snRNP and function as part of the transport system that targets it to the CBs.
MATERIALS AND METHODS
Oocyte Injection and Protein Extracts
Oocytes from adult Xenopus laevis were defolliculated for 2 h at room temperature in Ca2+-free OR2 (Wallace et al., 1973) containing 0.2% collagenase (type II, C6885; Sigma Chemical, St. Louis, MO). Stage IV-VI oocytes were collected and held in OR2 at 18°C. Wild-type U7, U7(U2), and U7(mut) snRNAs were transcribed in vitro as described by Wu et al. (1996). cDNAs corresponding to full-length Xenopus U1 and U2 snRNAs were amplified by PCR from genomic clones kindly provided by I. Mattaj; the PCR products were cloned directly in the PCR2.1 vector (Invitrogen, San Diego, CA) (primers A and B for U1; C and D for U2). In both cases, the 5′-primer used for amplification contained the sequence of the T3 promoter, allowing direct transcription with T3 polymerase after linearization of the plasmid with EcoRI restriction enzyme. Labeling was performed by incorporating high specific activity 32P-UTP (3000 Ci/mmol; DuPont, Wilmington, DE) during transcription. For wild-type U7, ∼5 × 104 cpm (1 ng) was injected into the oocyte cytoplasm, of which ∼10% was actually imported into the GV. Because the efficiency of import varied from one RNA to another, the number of injected counts was adjusted to obtain roughly equal counts in the GV: 2 × 104 cpm for U7(U2), U1, and U2 snRNA, and 5 × 105 cpm for U7(mut) snRNA. Each RNA was injected into 100 oocytes. After overnight incubation at 18°C in OR2, GVs and cytoplasms were hand isolated and pooled in ice-cold STE (10 mM Tris, pH 7.5, 100 mM NaCl, 1 mM EDTA). After centrifugation at 20,000 × g at 4°C, the supernatant fractions (“soluble nucleoplasm” and “soluble cytoplasm”) were collected and kept at −80°C. In some experiments, GVs were collected in NET-2 (150 mM NaCl, 50 mM Tris, pH 7.4, 0.05% Nonidet P-40).
The primers for U1 and U2 snRNA gene amplifications were
(A): 5′-GCAATTAACCCTCACTAAAGGGATACTTACCTGGCAGGGGAG-3′
(B): 5′-CAGGGGAAAGCGCGAACGCAGTCCCCCAC-3′
(C): 5′-GCAATTAACCCTCACTAAAGGGATCGCTTCTCGGCCTTTTGGC-3′
(D): 5′-AAGTGCACCGGTCCTGGAGGTACTGC-3′
The sequence of the T3 promoter is underlined.
Immunoprecipitation
Cell culture supernates of mAb H1 (Tuma et al., 1993) and mAb Y12 (Lerner et al., 1981) were concentrated 10× using a centrifugal filter device that excluded proteins >30 kDa (Biomax-30 K; Millipore, Bedford, MA) and were incubated with activated agarose beads (Affigel 10; Bio-Rad Laboratories, Hercules, CA) under conditions suggested by the manufacturer. Antibody-coated beads were blocked in 10 mg/ml BSA for 1 h at room temperature and washed with STE. The volume of each sample was adjusted to 80 μl with STE, and 20 μl of washed beads were added. Immunoprecipitation was carried out for 1 h at 4°C. Beads were washed five times for 5 min with 1 ml of STE, and the bound material was eluted with 40 μl of 8 M urea in STE. Aliquots were further processed for Western blot and RNA analysis. BSA-coated beads were used as controls.
Western Blots
Proteins were separated on 10% polyacrylamide/SDS gels and transferred to Immobilon membranes (Millipore, Bedford, MA) for 2 h at 40 V in Tris-glycine buffer containing 20% methanol. Membranes were blocked in 5% dry milk-PBS for 1 h at room temperature, followed by overnight incubation in the primary antibody in PBS–Tween at 4°C. Detection was performed using the ECF kit (Amersham Life Science, Arlington Heights, IL), and films were quantitated with the STORM 860 scanner (Molecular Dynamics, Sunnyvale, CA).
RNA Gels and Northern Blots
Immunoprecipitated RNAs were fractionated on a 10% acrylamide/8 M urea gel. Gels were dried, and the radioactive counts in each band of interest were quantitated with the STORM 860 phosphorimager. RNAs for Northern blotting were transferred after electrophoresis to a GeneScreen nylon membrane (New England Nuclear, Boston, MA) and were linked to the membrane by UV irradiation (Stratalinker; Stratagene, La Jolla, CA). The membrane was incubated in hybridization buffer (0.5 M Na2HPO4, pH 7.5, 7% SDS, 1 mM EDTA) at 65°C for 30 min before the probe was added. Hybridizations were carried out at 65°C overnight in the same buffer with 32P-labeled antisense probes (5 × 106 cpm/ml U7, 5 × 104 cpm/ml U6, or 5 × 103 cpm/ml U2). Nonspecific binding of the probes was removed by two washes of 1× SSC, 0.1% SDS at 65°C for 20 min followed by two washes of 0.1× SSC, 0.2% SDS for 20 min (SSC is 0.15 M NaCl, 0.015 M Na citrate, pH 7.0). Radioactive signals were analyzed with the STORM 860.
Affinity Chromatography on Single-stranded DNA
Soluble nucleoplasm was prepared from 100 GVs as described above and loaded on a 500-μl single-stranded DNA-agarose column equilibrated with STE buffer. The flow through was collected, and the column was washed with 25 ml of STE. Elutions were performed with 5 ml of each of the following solutions: 0.1 M NaCl, 0.2 M NaCl containing 1.0 M heparin, and increasing concentrations of NaCl from 0.3 M to 1.0 M in steps of 0.1 M; all solutions contained 10 mM Tris, pH 7.5. Proteins were precipitated from the column fractions with 25% trichloroacetic acid, rinsed with 5% trichloroacetic acid, and dried from acetone. Coilin was detected on a Western blot with mAb H1.
RNA-binding Assay
Binding assays were carried out with r(A), r(C), r(G), and r(U) homopolymers coupled to agarose beads (P8708, P9827, P1908, and P8563; Sigma, St. Louis, MO). Nucleoplasmic extracts (10 GVs) or purified coilin (100 ng) were incubated with 20 μg of homopolymer on beads for 1 h at 4°C with rocking in 300 μl of RNA binding buffer (25 mM HEPES, pH 7.5, 150 mM NaCl, 5 mM MgCl2, 0.1% NP40, 2% glycerol). Beads were washed five times for 15 min with 1 ml of cold binding buffer and boiled in SDS sample buffer. The eluted proteins were analyzed by Western blots.
Expression of 6 myc-tagged Coilin in Oocytes
Various deletion constructs of coilin are shown diagrammatically in Figure 9. A full-length 6 myc-tagged human coilin cDNA, and constructs A116, A251, and C484 were kindly provided by Z. Wu and C. Murphy. Construct A467 has a short region of the carboxy terminus deleted beyond a PvuII restriction site. Constructs Δ127–407 and Δ408–576 were obtained by PCR amplification (primers E and F for Δ127–407; G and H for Δ408–576) and subsequent cloning at the BamHI and XbaI sites of the MT6 vector (Roth et al., 1991), in which an SV40 nuclear localization signal had been inserted downstream of the 6 myc epitopes (Wu et al., 1994). RNA injections and oocyte preparations were as described (Bellini et al., 1995; Gall, 1998).
Figure 9.
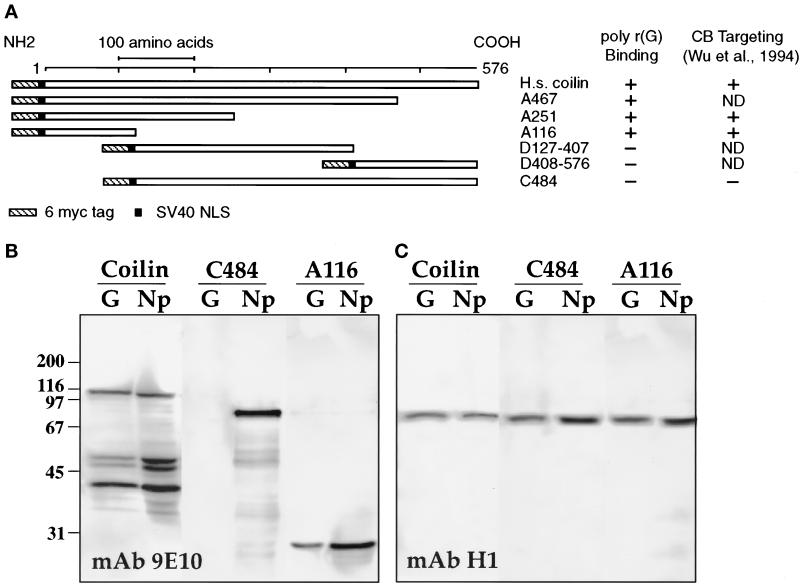
(A) Diagram of full-length myc-tagged human coilin and various deletion constructs. To the right is a summary of poly r(G) binding data for all constructs. Four of these constructs were examined earlier for their ability to target to CBs (Wu et al., 1994). The first 116 amino acids are necessary and sufficient for both poly r(G) binding and for targeting to the CBs. (B) Each of these proteins was expressed in stage VI oocytes by injecting in vitro transcripts from appropriate plasmids into the cytoplasm. After overnight incubation, soluble nuclear proteins were bound to a poly r(G) column, eluted, and electrophoresed. The bound proteins were detected on Western blots with mAb 9E10 against the myc tag. A Western blot for three proteins is shown: full-length coilin, C484 that lacks the first 116 residues at the amino terminus, and A116 that contains only these 116 residues. In each case the first lane (G) contains only material that bound to poly r(G), whereas the second lane (Np) contains total soluble GV proteins. Note that A116 bound to poly r(G) but C484 did not. (C) The same blot reprobed with mAb H1 to demonstrate binding of endogenous coilin.
Primers used for PCR were
E: 5′-ACTGAACCGGATCCCAAATATTCAAAGAAG-3′
F: 5′-GCATGCCCCGTCTAGAAGCTCCCTTCCAAG-3′
G: 5′-GCATGCGGGATCCAGGTCGAGGACGAGGGC-3′
H: 5′-GGAGAGGTCTAGATCAGGCAGGTTCTGTAC-3′
Fusion Protein Purification
A full-length Xenopus coilin cDNA clone was kindly provided by Z. Wu. The complete open reading frame was subcloned between the unique BamHI and XbaI sites of the pRSETB vector (Invitrogen, San Diego, CA). After subcloning, the coilin open reading frame was downstream of and in frame with sequences encoding the T7 tag and six histidines. Expression of the fusion protein was induced in 300 ml of BL21-DE3 bacteria. An overnight culture was grown in LB broth with 100 μg/ml of ampicillin. It was diluted 100× in LB broth with 50 μg/ml of ampicillin, grown to mid-log phase, and induced with 1 mM isopropylthiogalactoside. The induced culture was grown an additional 4 h before the cells were harvested. Cells were pelleted, washed twice with PBS, and resuspended in 20 ml of PBS. Cells were sonicated at 4°C (four times for 30 s), and the lysate was centrifuged at 25,000 × g for 20 min at 4°C. The fusion protein was found in the pellet. The pellet was washed twice with PBS and resuspended in buffer A (10 mM Tris, pH 8, 100 mM NaCl, 8 M urea). The fusion protein was bound to a 2 ml Ni2+ column in buffer A. The column was washed with 20 ml of buffer B (10 mM Tris, pH 5.8, 100 mM NaCl, 8 M urea) and the fusion protein was eluted with 4 ml of buffer C (10 mM Tris, pH 4.5, 100 mM NaCl, 8 M urea). The fusion protein was then dialyzed overnight at 4°C against 5 L of buffer D (25 mM Tris, pH 7.5, 75 mM KCl, 25 mM NaCl, 1 mM DTT, 10% glycerol) and stored in buffer D at −80°C.
Immunofluorescent Staining and Microscopy
The contents of single Xenopus GVs were spread and attached to glass microscope slides as described (Gall, 1998). Fixation was in 2% paraformaldehyde in PBS for 1 h or more. After fixation, preparations were rinsed in PBS, blocked in 10% horse serum, and stained for 1 h with antibody in 10% horse serum. Antibodies used in this study were: mAb Y12 against the Sm epitope (Lerner et al., 1981), mAb H1 against Xenopus coilin (SPH-1; Tuma et al., 1993), mAb K121 against the trimethylguanosine (TMG) cap of snRNAs (Krainer, 1988), affinity-purified rabbit polyclonal serum C236 against Xenopus coilin (Z. Wu, unpublished). Secondary antibodies were goat anti-mouse immunoglobulin G (IgG) or goat anti-rabbit IgG labeled with fluorescein or Cy3 (Jackson ImmunoResearch Laboratories, West Grove, PA). Slides were mounted in 50% glycerol containing 1 mg/ml phenylenediamine, and 1 μg/ml 4′,6-diamidino-2-phenylindole (DAPI). Confocal laser scanning microscopy was carried out with the Leica TCS NT System.
RESULTS
Coilin and the U7 snRNP Are Colocalized in CBs
CBs (or spheres) in the Xenopus GV have a characteristic tripartite structure consisting of a spherical body with B-snurposomes frequently, but not invariably, attached to the surface and B-like inclusions in the interior. Hundreds to thousands of B-snurposomes are also found free in the nucleoplasm. These three components are readily distinguished by phase contrast or DIC microscopy (Figure 1). Earlier studies demonstrated that coilin (Tuma et al., 1993; Wu et al., 1994) and U7 snRNA (Wu and Gall, 1993) are limited to the main body or matrix of the sphere but are absent from the B-snurposomes and inclusions. Figure 1, a and b, shows a CB stained with polyclonal serum C236 against Xenopus coilin, originally called SPH-1 (Tuma et al., 1993). Figure 1, c and d, shows a CB from an oocyte injected with fluorescein-labeled U7 snRNA. In both cases only the main body of the CB is stained.
All three parts of the CB stain with mAb Y12 (Lerner et al., 1981) against the Sm proteins (Figure 1g) and with mAb K121 (Krainer, 1988) against the TMG cap found on mature U7 snRNA and on four mature splicing snRNAs (U1, U2, U4, and U5) (Figure 1k). These staining results imply that the snRNAs in the CB exist in the form of mature snRNP complexes, although in principle some immature snRNAs could also be present. Quantitative scans of the confocal images show that the staining intensity is about twice as high in the matrix as in the inclusions or snurposomes. A similar staining ratio is found in small CBs that consist of a main body associated with a B-snurposome of the same size. Whether this staining difference reflects a true difference in snRNP concentration or unequal access of the antibody is not certain. Arguing for unequal access is the fact that the absolute density of the inclusions and the B-snurposomes is higher than that of the matrix. This conclusion is based on the DIC images, in which the shadows of the inclusions lie on the same side as the shadow of the matrix in which they are embedded. If the inclusions were of lower density than the matrix, their shadows would lie on the opposite side, as is true for vacuoles in the nucleoli (Figure 1i, arrows). The same conclusion can be drawn from phase contrast images, which show that B-snurposomes are darker than CBs of the same diameter.
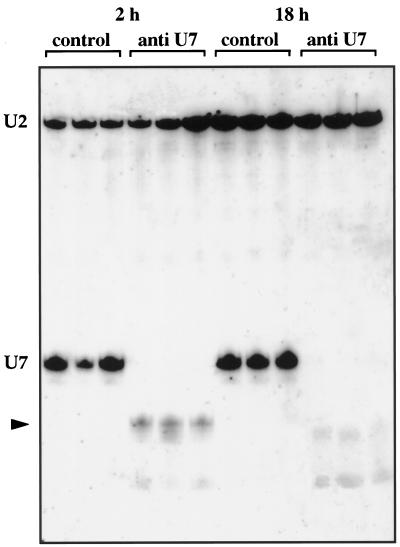
We addressed the question whether U7 is the only snRNA in the matrix of the CB by injecting an antisense oligodeoxynucleotide against the first 16 bases of U7. When injected into the GV, this oligo effectively degrades U7 snRNA (Grimm et al., 1993), presumably by binding to it and activating endogenous RNase H activity. We found that injection into the oocyte cytoplasm was equally effective in targeting U7. GVs were isolated from oocytes that had been injected 2 h or 18 h previously with the antisense oligo or with an unrelated control oligo. Figure 2 shows a Northern blot of RNAs from single GVs; the antisense oligo caused shortening of the U7 snRNA, leaving a truncated 3′-fragment that was detectable after 2 h (lanes 4–6) but had disappeared by 18 h (lanes 10–12). The antisense oligo had no effect on U2 snRNA in the same GVs (lanes 4–6 and 10–12), and the control oligo had no effect on either U7 or U2 snRNA (lanes 1–3 and 7–9). Figure 1, i–p, shows CBs from oocytes that had been injected with either the antisense or control oligos, and then double stained with mAb K121 for the TMG cap and serum C236 for coilin. In oocytes treated with the antisense oligo, staining was markedly reduced in the matrix of the CB but was essentially unchanged in the B-snurposomes and the inclusions (Figure 1o). Quantitative scans showed that the matrix now stained ∼0.2 as strongly as the inclusions and B-snurposomes; i.e., a 10-fold decrease in relative staining intensity. The simplest interpretation is that U7 snRNA is the major 5′-TMG–capped snRNA in the matrix, so that removal of its cap causes most of the staining with mAb K121 to disappear. On the other hand, TMG caps on the splicing snRNAs remain in the B-snurposomes and in the inclusions, which under these conditions are quite similar in appearance (Figure 1o). The control oligo had no effect on staining with mAb K121 (Figure 1k).
Figure 2.
Truncation of U7 snRNA by an antisense oligodeoxynucleotide. Xenopus oocytes were injected with an antisense U7 or a control oligonucleotide. After 2 h or 18 h incubation at 19°C, RNA was purified from single GVs, separated on a polyacrylamide gel, and hybridized with anti-U7 and anti-U2 snRNA probes (ratio 1000:1). Truncated U7 is detected at 2 h, corresponding to loss of 16 nucleotides from the 5′-end. The truncated product was unstable and was not detected at 18 h. U2 snRNA was unaffected by either the anti-U7 or the control oligonucleotide.
Along with our earlier in situ hybridization data, the immunostaining results lead to the following conclusions:
1) The U7 snRNP and coilin are colocalized in the matrix of the CB.
2) U7 snRNA is the major TMG-capped snRNA in the matrix of the CB.
3) Other capped snRNPs occur in the B-snurposomes and in the B-like inclusions of the CB.
These localizations suggested that coilin might play a role in the pathway for histone pre-mRNA processing rather than in splicing and led us to examine possible molecular interaction between coilin and the U7 snRNP.
Most Coilin in the GV Is “Soluble” and Not Associated with CBs
In a first attempt to demonstrate an association between coilin and the U7 snRNP, we injected 32P-labeled U7 snRNA into oocytes, isolated GVs, solubilized them in NET-2 (a standard snRNP buffer that contains the detergent NP40), and immunoprecipitated with anti-coilin mAb H1. Under these circumstances we found no U7 snRNA in the immunoprecipitate. We show later that NP40 disrupts the association between coilin and the U7 snRNP. We used NET-2 buffer for two reasons. First, many snRNP proteins remain associated with their respective snRNAs in this buffer (Steitz, 1989). Second, and more importantly, we knew from cytological observations that CBs were soluble in NET-2 but remained intact in several buffered salines, even in the absence of divalent ions and the presence of EDTA.
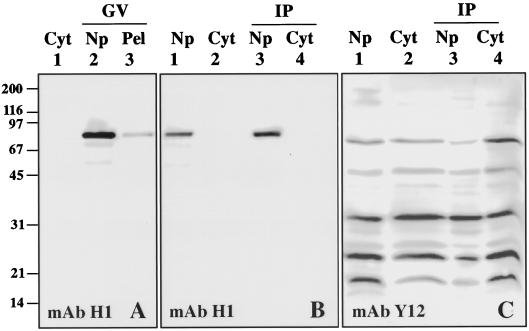
During the course of our experiments, however, it became clear that most coilin in the GV is not actually in CBs, but rather in the soluble nucleoplasm that remains after low-speed centrifugation. Figure 3 illustrates this fact. GVs were isolated in “5:1” saline solution (83 mM KCl, 17 mM NaCl, 6.5 mM Na2HPO4, 3.5 mM KH2PO4, 1 mM MgCl2, 1 mM DTT, pH 7.0), disrupted briefly by pipetting, and centrifuged at 15,000 × g for 15 min. Under these circumstances all cytologically visible structures in the GV are pelleted, including the lampbrush chromosomes, nucleoli, CBs, and B-snurposomes. A Western blot of the pellet and supernate (soluble nucleoplasm) shows that most of the coilin is in the supernate (Figure 3A). This observation indicated that we could study the soluble coilin fraction without detergent. However, endogenous U7 snRNA in the GV behaves differently, ∼90% of it remaining in the pellet fraction after a similar centrifugation. Nevertheless, we knew that it was possible to increase the amount of soluble U7 snRNP in the GV by injecting an excess of exogenous U7 snRNA into the cytoplasm (Wu et al., 1996). We therefore asked whether an interaction could be demonstrated between soluble coilin normally resident in the nucleoplasm and soluble U7 snRNP derived by injection.
Figure 3.
(A) Western blot of Xenopus oocyte proteins probed with mAb H1 to determine the intracellular distribution of coilin. Lane 1 contains total proteins from three cytoplasms (Cyt). Lanes 2 and 3 contain soluble nucleoplasmic (Np) and insoluble pellet (Pel) proteins from 10 GVs. Coilin is detected exclusively in the GV, where most is in the soluble nucleoplasmic fraction. (B) Western blot probed with mAb H1 against coilin. Lanes 1 and 2 contain total soluble proteins from 10 GVs and 3 cytoplasms, respectively. Lanes 3 and 4 contain proteins immunoprecipitated by mAb H1 from similar numbers of GVs and cytoplasms. Coilin is efficiently immunoprecipitated from the soluble nucleoplasmic fraction. (C) Western blot similar to that in panel B, except immunoprecipitated with mAb Y12 against the Sm proteins and probed with mAb Y12. Sm proteins are efficiently immunoprecipitated from both cytoplasmic and nucleoplasmic fractions.
Constructs and Injections
We examined the behavior of five snRNA constructs after injection into the cytoplasm of Xenopus oocytes: wild-type U1, U2, and U7 snRNAs and two mutants of U7 designated U7(U2) and U7(mut). The wild-type snRNAs each contain a U-rich Sm-binding site, required for association of the Sm proteins in the cytoplasm and for import of the mature snRNP into the nucleus (Mattaj, 1988; Grimm et al., 1993; Stefanovic et al., 1995). The U7(U2) and U7(mut) constructs consist of U7 snRNA in which the U7 Sm-binding site has been replaced by that of U2 snRNA or by an unrelated sequence, respectively (Wu et al., 1996). The specific Sm-binding site of U7 is required for the efficient targeting of U7 snRNA to CBs, as demonstrated by the fact that the U7(U2) construct is poorly targeted to CBs, even though it is imported into the nucleus more efficiently than wild-type U7 (Wu et al., 1996). The U7(mut) construct does not associate with Sm proteins when injected into the cytoplasm of oocytes and is not imported as a snRNP particle into the GV. However, if a large amount of U7(mut) is injected, some does get to the GV, possibly by diffusion.
Using T7 polymerase, we transcribed capped 32P-labeled RNAs from wild-type and mutant U7 clones and injected them into the cytoplasm of stage VI oocytes. The amount of injected RNA was adjusted to obtain roughly similar counts in the GV. After the oocytes had been incubated in OR2 saline for 18 h at 18°C, GVs and cytoplasm were manually isolated in STE buffer and further separated into pellet and soluble fractions by centrifugation at 20,000 × g for 15 min at 4°C. Under these conditions, the chromosomes and all other cytologically identifiable structures, including the CBs, were pelleted out of the “soluble nucleoplasm.” Similarly, yolk granules were separated from the “soluble cytoplasm.” The soluble fractions of the nucleus and cytoplasm were then immunoprecipitated with two different mAbs linked to agarose beads.
mAb Y12 (Lerner et al., 1981) recognizes the Sm epitope on proteins that are common to the U7 snRNP and to four of the major splicing snRNPs (U1, U2, U4, and U5). It was used here to test whether snRNP complexes were formed when snRNAs were injected into the oocyte. mAb H1 (Tuma et al., 1993) specifically recognizes Xenopus coilin (SPH-1). After the soluble nucleoplasmic and cytoplasmic fractions had been incubated with antibody-coated beads, bound and unbound 32P-labeled RNAs were extracted and electrophoresed on 10% acrylamide gels. Although the clones were designed to produce full-length U7 snRNAs, the products of the in vitro transcriptions resolved into three to four closely spaced bands (Figure 4A); the lowest is the expected 58 nucleotides, whereas the others probably correspond to transcripts with extra nucleotides at the 3′-end.
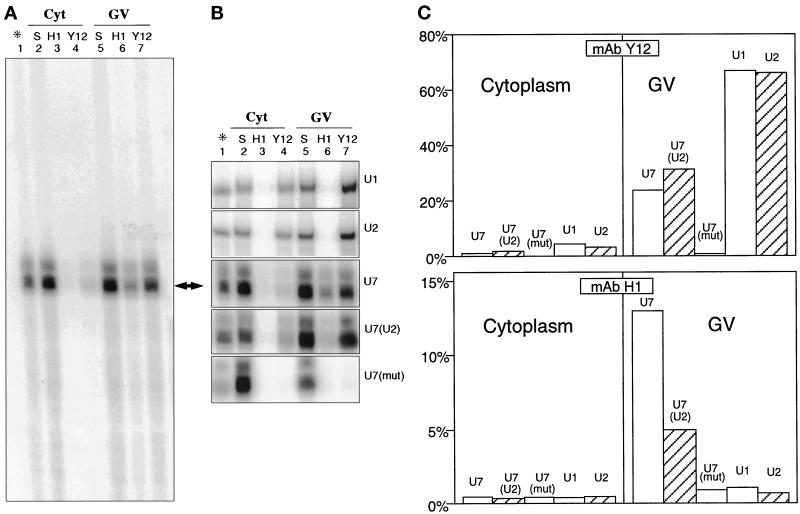
Figure 4.
Capped 32P-labeled snRNA transcripts were injected into the cytoplasm of stage VI Xenopus oocytes. After 18 h incubation, GVs and cytoplasm were isolated and immunoprecipitated with either mAb H1 (anti-coilin) or mAb Y12 (anti-Sm). RNA was isolated from the immunoprecipitates and from untreated control fractions and separated on a polyacrylamide gel. (A) Autoradiograph of entire gel from experiment in which wild-type U7 snRNA was injected. Lane 1 contains a sample of the injected snRNA. Lane 2 contains a sample of total cytoplasmic RNA before immunoprecipitation. Lanes 3 and 4 contain RNA immunoprecipitated from cytoplasm by mAbs H1 and Y12, respectively. Lanes 5, 6, and 7 contain a sample of total GV RNA and the RNA immunoprecipitated by mAbs H1 and Y12. (B) Autoradiograph of relevant portion of gels from experiments in which various constructs were injected; lanes as in panel A. U1 and U2 are wild-type U1 and U2 snRNA. U7, U7(U2), and U7(mut) are wild-type U7, U7 with its Sm site replaced by that of U2, and U7 with an unrelated sequence at the Sm site (wild-type U7 lane is the same as in panel A). Note that all constructs except U7(mut) are immunoprecipitated from the GV by mAb Y12, demonstrating that they exist as Sm complexes. However, only wild-type U7 and, to a lesser extent, U7(U2) are immunoprecipitated from the GV by mAb H1, indicating an association with coilin. (C) The lanes in panel B were quantitated with a phosphorimager, and lanes 3, 4, 6, and 7 were plotted as % immunoprecipitated.
The U7 snRNP Coimmunoprecipitates with Coilin
As expected, mAb Y12 (anti-Sm) efficiently immunoprecipitated U1 and U2 snRNAs from the soluble fraction of the GV, confirming that these snRNAs exist as Sm complexes in the nucleus (Figure 4, panels B [lane 7] and C). Wild-type U7 and U7(U2) were also precipitated from the nuclear fraction, although less efficiently than U1 and U2, but U7(mut) was not precipitated at all (Figure 4, panels B [lane 7] and C). In fact, U7(U2) was precipitated slightly better than wild-type U7 (Figure 4C), confirming earlier observations of Schümperli’s group (Grimm et al., 1993; Stefanovic et al., 1995) on a similar construct (their U7 Sm OPT). The failure of U7(mut) to be immunoprecipitated was presumably due to its inability to bind Sm proteins.
Neither U1 nor U2 snRNA was immunoprecipitated from the soluble GV fraction by mAb H1 against coilin (Figure 4, panels B [lane 6] and C). By contrast, 13% of wild-type U7 was precipitated under the same conditions (Figure 4, panels A, B [lane 6], and C). Because mAb H1 is specific for Xenopus coilin, some soluble coilin in the GV must be associated as a complex with U7 snRNA. Mutations of the Sm-binding site dramatically affected the interaction of U7 snRNA with coilin: only 5% of U7(U2) was precipitated from the nucleoplasm and <1% of U7(mut) (Figure 4, panels B [lane 6] and C). The failure of U7(mut) to coimmunoprecipitate with coilin suggests that U7 must exist as an Sm snRNP to interact with coilin. However, an Sm site is not sufficient for this interaction, since neither U1 nor U2 shows any association with coilin, although both exist as Sm snRNPs in the GV. Of particular interest is the fact that U2 cannot be precipitated with mAb H1, whereas U7(U2), which has an identical Sm site, is weakly precipitated. Taken together, the data suggest that coilin does not interact with U7 snRNA as such, but with the U7 snRNP. Furthermore, specific features of the U7 snRNP, not shared by other Sm snRNPs, are required for the strongest interaction. Likely candidates for such specificity could be U7-specific proteins (Smith et al., 1991; Stefanovic et al., 1995). Each fraction was also tested on Western blots with mAb H1 and mAb Y12 to ensure that coilin and the Sm proteins were being precipitated in these experiments. The patterns of Figure 3, B and C, were reproducibly obtained.
In one set of experiments, 0.5% NP40 was added to the binding buffer during the immunoprecipitations. In this case, coilin bound to the mAb H1-coated beads as usual, but the U7 snRNP was not coimmunoprecipitated. NP40 is routinely used in studies on snRNPs (Steitz, 1989), because it does not disrupt the association of the snRNAs with their Sm proteins. This experiment demonstrates that the association of coilin with the U7 snRNP is relatively weak, certainly less strong than that of the Sm proteins.
Endogenous U7 snRNA Associates with Coilin
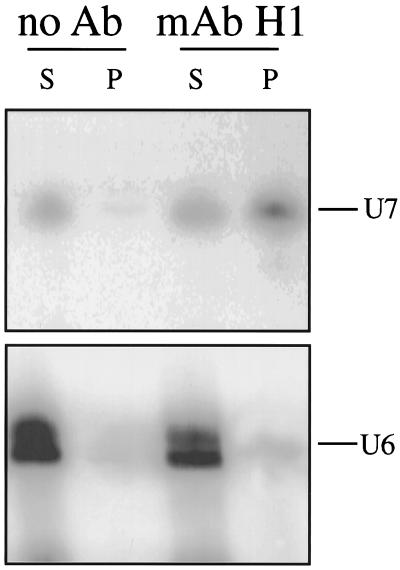
The experiments described in the preceding section demonstrate that soluble coilin in the GV can form a weak association with U7 snRNPs derived from injected U7 RNA. The experiments were designed to take advantage of the fact that a large fraction of injected U7 snRNA remains in the soluble fraction of the GV after centrifugation at 15,000 × g. On the other hand, 85–90% of endogenous U7 pellets under these conditions, presumably because it is in the CBs (Wu et al., 1996). Essentially all U7 in the GV can be solubilized with 0.5% NP40, but under these conditions the association of coilin with the U7 snRNP is disrupted. We carried out an experiment to determine whether the small fraction of non-sedimentable U7 normally present in the GV is associated with coilin. A soluble supernate containing ∼10% of the endogenous U7 snRNA was prepared from 200 GVs by centrifuging and discarding the pellet. This supernate should contain ∼200 pg of U7 snRNA compared with ∼1800 pg in the discarded pellet (Wu et al., 1996). An immunoprecipitation (without detergent) was carried out with mAb H1 against coilin. Approximately 20% of the soluble U7 snRNA was immunoprecipitated in this way, an amount easily detected and quantitated by Northern blotting (Figure 5). Thus, at least part of the endogenous soluble U7 snRNA in the GV is associated with coilin.
Figure 5.
Association of endogenous U7 snRNA in the GV with endogenous coilin. Soluble nucleoplasm from 200 GVs was prepared without detergent by centrifuging and discarding the pellet. The discarded pellet contained ∼5–10% of the nuclear coilin and 90% of the U7 snRNA. Soluble coilin in the nucleoplasm was immunoprecipitated with mAb H1 (anti-coilin). A Northern blot of one fifth of the supernate (S) and all of the immunoprecipitate (P) was probed with antisense U7 snRNA (upper panel). Quantitation showed that ∼20% of the soluble U7 snRNA was immunoprecipitated by the anti-coilin antibody. When the blot was reprobed with antisense U6 snRNA, no significant signal was seen in the precipitate (lower panel). BSA-coated beads served as control.
Coilin Is an RNA-binding Protein in Vitro
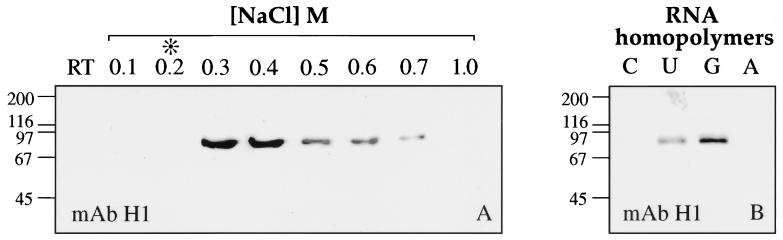
Having found that coilin and the U7 snRNP can associate in the GV, we wanted to determine whether coilin exhibited any general features of an RNP protein. We first examined coilin’s ability to bind to nucleic acids in vitro. Soluble nucleoplasm from Xenopus GVs was subjected to affinity chromatography on a single-stranded DNA-agarose column. As shown in Figure 6A, coilin bound to the column even in the presence of the polyanion heparin (1 M) and could be eluted only in the presence of high-salt concentrations (>0.3 M NaCl). Because of its negative charge, heparin competes with nucleic acids to reduce the nonspecific binding of proteins. Only proteins that interact directly with the bases of the nucleic acid are expected to remain associated after heparin treatment. We next investigated the binding of coilin to RNA homopolymers and found strong binding to poly r(G) and poly r(U) but little or no affinity for poly r(C) or poly r(A) (Figure 6B). Like the binding to single-stranded DNA, the association with poly r(G) and poly r(U) was resistant to heparin and high-salt concentrations and was not competed with a large excess of tRNA (Figure 7).
Figure 6.
(A) Western blot of proteins purified from the nucleoplasm of Xenopus GVs by affinity chromatography on single-stranded DNA agarose. Coilin was detected by mAb H1. Upon addition of 1 M heparin (*), coilin remained on the column and could be eluted only at high-salt concentrations (0.3–0.7 M NaCl), suggesting that it is a single-stranded nucleic acid-binding protein. RT, Run-through. (B) Western blot of nucleoplasmic proteins purified by affinity chromatography on RNA homopolymers. Coilin bound to poly r(G) and poly r(U) but had no affinity for poly r(C) or poly r(A). Coilin was detected by mAb H1. Mobility of molecular weight markers in kilodaltons.
Figure 7.
Soluble nucleoplasm from Xenopus GVs was incubated with poly r(G)-coated agarose beads. After washing with increasing salt concentration (A) or tRNA (B), bound coilin was monitored on a Western blot with mAb H1. The binding of coilin to poly r(G) is resistant to high salt and to an excess of tRNA competitor, as is its binding to single-stranded DNA. Mobility of molecular weight markers in kilodaltons.
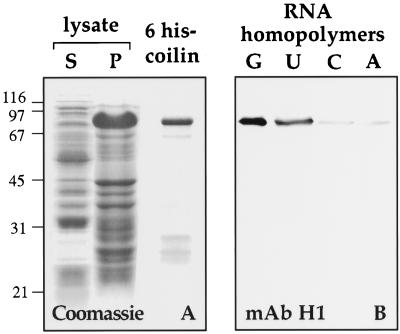
Because a crude nuclear extract was used in the RNA-binding assays, other factors in the nucleoplasm might have acted as intermediates between coilin and the RNA. To test this possibility, we examined the nucleic acid-binding properties of bacterially expressed coilin. We expressed a 6-histidine–tagged version of Xenopus coilin in Escherichia coli and purified it on a Ni2+ column (Figure 8A). As shown in Figure 8B, 6-histidine coilin bound to RNA homopolymers with the same specificity as endogenous coilin. Thus, coilin by itself is an RNA-binding protein in vitro.
Figure 8.
Bacterially expressed Xenopus coilin binds to RNA homopolymers. (A) E. coli expressing 6 histidine-tagged Xenopus coilin were lysed and centrifuged to yield supernatant and pellet fractions. Proteins from these fractions were electrophoresed and stained with Coomassie blue (lanes S and P). The pellet fraction was solubilized with 8 M urea and bound to a Ni2+ column. Nearly pure 6 his-coilin was recovered from the column (6 his-coilin). (B) Purified 6 his-coilin was incubated with beads coated with RNA homopolymers. A Western blot of proteins that bound to the beads was probed with mAb H1. Purified 6 his-coilin bound to poly r(G) and poly r(U) but not to poly r(C) or poly r(A).
We also tested the binding properties of human coilin, which shares conserved regions at the amino and carboxy termini with Xenopus coilin but differs considerably in the middle of the molecule (Tuma et al., 1993; Chan et al., 1994; Wu et al., 1994). Using T3 RNA polymerase, we synthesized capped, sense-strand RNA from a cDNA clone of wild-type human coilin and from various deletion clones (Figure 9A). All clones contained an SV40 nuclear localization signal (NLS) to direct the resulting proteins into the GV and a 6-myc tag to permit their detection with mAb 9E10 against the myc sequence. Transcripts were injected into the cytoplasm of stage VI oocytes; 18 h later, GVs were isolated and nucleoplasmic extracts prepared. In all cases a nuclear protein of the expected size was detectable on a Western blot using mAb 9E10. The ability of the various expressed proteins to bind to RNA in vitro was then assayed on a poly r(G)-agarose column (Figure 9B). We found that full-length human coilin bound to poly r(G), and that the first 100 residues at the amino terminus were both necessary and sufficient for binding (Figure 9, table). This region shares more than 45% identity with the corresponding amino terminus of Xenopus coilin (Wu et al., 1994).
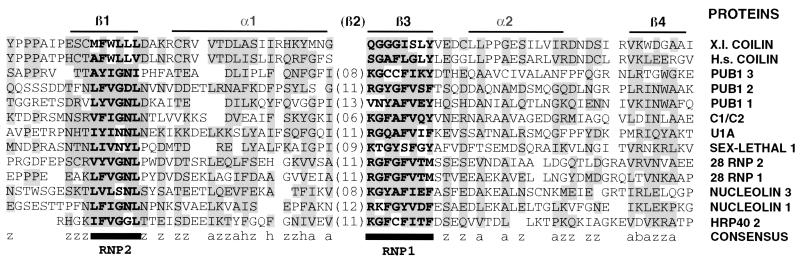
Having determined that the amino terminus of coilin is the region responsible for RNA binding, we examined the first 100 amino acids of Xenopus and human coilin for similarity to the conserved RNA binding-domain (RBD) or RNA recognition motif (RRM) found in a variety of proteins that bind pre-mRNA, mRNA, pre-rRNA, and snRNAs (Bandziulis et al., 1989; Query et al., 1989; Burd and Dreyfuss, 1994). Figure 10 shows Xenopus and human coilin aligned with a selection of RBDs from other proteins. Amino acids of similar properties are boxed, and the consensus property for the most highly conserved residues is listed below the sequences (a, aromatic; b, basic; h, hydrophobic; and z, polar or charged). There is a reasonably good match of the more hydrophobic amino acids over the whole region, and prolines occur upstream of RNP2, as they do in many RBDs. However, several features detract from the alignment suggested here. The two coilins lack a sequence that corresponds to the β2 region, although this region is of variable length in previously defined RBDs. In most RNP2 sequences, at least one of the last three amino acids is hydrophilic (mostly N), whereas this is not the case for the coilins. Similarly, the second from last amino acid in the RNP1 region is generally more hydrophilic than it is in the coilins. Thus, although this region of coilin binds RNA homopolymers, it is only loosely related to a canonical RBD.
Figure 10.
The poly r(G)-binding region of human coilin compared with the corresponding region of Xenopus coilin (45% identity) and with the RNP consensus sequence from a selection of other proteins. Boxed amino acids at a given residue have similar properties: a, aromatic, h, hydrophobic, z, polar or charged, and b, basic. The RNP1 octamer and RNP2 hexamer are shown in bold. Putative α helices (α1, α2) and β sheets (β1, β2, β3) are based on structure determinations by Nagai et al. (1990) and Wittekind et al. (1992). The RNA-binding region of coilin is at best loosely related to the previously defined RNP consensus sequence.
DISCUSSION
The nuclear protein coilin is most highly concentrated in CBs, which also contain a variety of snRNPs involved in splicing, pre-rRNA processing, and histone pre-mRNA 3′-end processing (reviewed by Lamond and Carmo-Fonseca, 1993; Bohmann et al., 1995; Gall et al., 1995; Roth, 1995; Lamond and Earnshaw, 1998; Matera, 1998). The precise spatial relationship between coilin and other components is not resolvable in the small CBs of tissue culture and somatic nuclei but is evident in the much larger CBs (spheres) of the Xenopus GV. Earlier studies showed that coilin occurs exclusively in the matrix of oocyte CBs (Tuma et al., 1993; Wu et al., 1994), where it is colocalized with U7 snRNA (Wu and Gall, 1993). Splicing snRNAs, on the other hand, are limited to the B-snurposomes on the surface of CBs and to the internal B-like inclusions (Wu et al., 1991; Gall et al., 1995). Furthermore, by disrupting U7 snRNA with an antisense oligodeoxynucleotide, we showed that U7 is the major capped snRNA associated with coilin in oocyte CBs. The confocal images of Figure 1 illustrate these features.
The localization data led us to look for molecular interaction between coilin and U7 snRNA. We took advantage of two special features of the GV. First, most coilin in the GV is not in the CBs, but rather in the soluble nucleoplasm. Second, soluble U7 snRNP can be made to accumulate in the GV by injecting excess U7 snRNA into the cytoplasm (Wu et al., 1996). We showed that soluble coilin and soluble U7 snRNP can be coimmunoprecipitated with mAb H1 against Xenopus coilin (SPH-1; Tuma et al., 1993). The association is a relatively weak one, since it is disrupted by 0.5% NP40, a detergent that does not dissociate U7 snRNA or splicing snRNAs from their associated Sm proteins (Steitz, 1989). We also demonstrated that coilin can bind polynucleotides in vitro. Coilin’s ability to bind single-stranded, but not double-stranded, DNA, as well as its specificity for poly r(G) and poly r(U) homopolymers, are features shared with a variety of RNPs (Swanson and Dreyfuss, 1988; Kiledjian et al., 1994).
The in vivo and in vitro binding properties suggest that coilin is an RNP that interacts with U7. Several observations demonstrate that the interaction is not with free U7 snRNA but with the U7 snRNP. Most importantly, we see coimmunoprecipitation of coilin and U7 snRNA only when U7 exists as an Sm complex in the nucleus. Thus, when wild-type U7 snRNA is injected into the cytoplasm, some enters the nucleus as an Sm complex and can be immunoprecipitated with an anti-coilin antibody. By contrast, U7(mut), which does not form an Sm complex, but can be forced into the nucleus, does not coimmunoprecipitate with coilin. Furthermore, in experiments not described here, we were unable to demonstrate a direct interaction in vitro between bacterially expressed coilin and synthetic U7 transcripts. Together these observations suggest that U7 must exist as an Sm snRNP for interaction with coilin.
The Sm proteins are not the entire story, however, as shown clearly by experiments with U7(U2), which has the Sm site of U7 replaced by that of U2. When it is injected into the cytoplasm, it forms an Sm snRNP that is imported into the nucleus even more efficiently than wild-type U7. Nevertheless, it is only poorly immunoprecipitated by the anti-coilin antibody. Wild-type U2 and U1 RNAs likewise form Sm snRNPs, but they fail completely to coimmunoprecipitate. It thus appears that the wild-type U7 snRNP has some specific feature or features that permit its association with coilin. One possibility is that coilin can bind only in the presence of one or more U7-specific proteins. Stefanovic et al. (1995) described a 40-kDa protein that is part of the Xenopus wild-type U7 snRNP, but is not present in a construct designated U7 Sm OPT, whose Sm site was modified for optimal binding of Sm proteins. U7 Sm OPT is very similar to our U7(U2), differing by only two nucleotides in the Sm-binding site. Thus, the reduced binding of coilin to the U7(U2) snRNP might be due to absence of this 40-kDa protein. An additional 60–80 kDa protein was identified by cross-linking of cytoplasmic U7, but its relationship to the nuclear snRNP is unclear. Unfortunately, little additional information is available concerning the composition of the Xenopus U7 snRNP. The mouse U7 particle contains U7 snRNA, the common Sm proteins, and two specific proteins of 14 and 50 kDa (Smith et al., 1991). No protein corresponding in molecular weight to coilin (Mr = 80 kDa) was described.
In an earlier study Wu et al. (1996) examined the nuclear targeting of wild-type U7, U7(U2), and U7(mut) snRNA after injection into Xenopus oocytes. They found that wild-type U7 was rapidly and specifically targeted to the CBs, whereas U7(U2) was poorly targeted, and U7(mut) failed to accumulate at all in CBs. The ability of similar constructs to enter the GV and support cleavage of histone pre-mRNA was studied by Stefanovic et al. (1995). Specifically, they showed that U7 Sm OPT entered the nucleus more efficiently than wild-type U7, but could not support 3′-end processing of histone pre-mRNA, whereas wild-type U7 (U7 Sm WT) entered the GV less efficiently but supported processing. In summary, the wild-type U7 snRNP binds coilin, is efficiently targeted to CBs, and processes histone pre-mRNA, whereas a U7 snRNP with a “splicing” type Sm site binds coilin less efficiently, is targeted poorly to CBs, and does not support processing. Stefanovic et al. (1995) found that a construct with a mutated Sm site (U7 Sm MUT) did not bind Sm proteins and enter the nucleus, and so did not support processing. Our U7(mut) likewise failed to form an Sm snRNP and, even when forced into the GV, failed to bind coilin and was not targeted to CBs.
These relationships suggest a model in which the U7 snRNP binds coilin and is transported to the CB before it is involved in histone pre-mRNA processing. Coilin itself is rapidly targeted to CBs in the oocyte (Wu et al., 1994) and in transfected tissue culture cells (Bohmann et al., 1995), suggesting that it might be the specific carrier protein. The relatively weak interaction between coilin and the U7 snRNP is consistent with a transitory role in which coilin would bind to the U7 snRNP, travel with it to the CB, and release it there. This model is also supported by the well-known association between CBs and the histone genes themselves. Many years ago it was found that CBs (then known as spheres) occur not only free in the nucleoplasm but also attached to the lampbrush chromosomes at a few specific sites (Gall, 1954; Callan, 1986), which were later identified as the histone gene loci (Gall et al., 1981; Callan et al., 1991). In effect, therefore, targeting of the U7 snRNP to the CB targets it to the chromosomal sites of histone pre-mRNA transcription. In the oocyte, transcription does not take place in the CBs themselves but in chromosomal loops immediately adjacent to them (Diaz and Gall, 1985). Just exactly when and where the U7 snRNP engages the pre-mRNA is not known. It is possible that the U7 snRNP associates with the nascent transcript before termination, and that the U7 cleavage reaction releases the nascent transcript from the DNA template. Such a model would help explain why the U7 snRNP is targeted to the site of transcription in the first place.
CBs in HeLa nuclei likewise contain U7 snRNA and associate preferentially with histone genes (Frey and Matera, 1995), suggesting that the model derived from the oocyte applies to other cell types as well. Association of CBs with the genes coding for U1, U2, and U3 snRNA has also been demonstrated in cultured cells (Frey and Matera, 1995; Smith et al., 1995; Gao et al., 1997). These latter associations have led to interesting speculation concerning the involvement of CBs in snRNP biogenesis (Matera, 1998), although the function of coilin itself in these processes remains uncertain.
A role for coilin in targeting snRNPs to the CB was demonstrated in an earlier study on nuclei assembled in Xenopus egg extract from demembranated sperm heads. These nuclei lack a nucleolus but display multiple small granules up to ∼1 μm diameter. The granules have been referred to as prenucleolar bodies, because they contain several nucleolar proteins (Bell et al., 1992). However, they also contain coilin and a variety of snRNPs, suggesting that they are, in fact, closely related to CBs (Bauer et al., 1994). To examine the role of coilin in the biogenesis of these bodies, coilin was removed from the egg extract by immunoprecipitation before addition of sperm heads. Under these conditions, sperm heads formed nuclei of normal appearance, including the small granules. The granules lacked coilin, although they still stained well for nucleolar components (fibrillarin and nucleolin). Surprisingly, they lacked Sm proteins. The converse experiment, in which the majority of Sm proteins were removed from the extract before addition of sperm heads, resulted in bodies with much reduced staining for both Sm proteins and coilin. These experiments suggested that coilin is involved either in the import of one or more snRNPs into the nucleus or targeting of snRNPs to the CB after import. Such functions are consistent with our finding of a direct association between coilin and the U7 snRNP.
Although our experimental work suggests a role for coilin in transport of the U7 snRNP, it is probable that coilin has additional functions in the nucleus. It is noteworthy that most of the coilin in the GV is in the soluble nucleoplasm, whereas most of the U7 is in the CBs. Thus, from a simple quantitative standpoint, only a small fraction of coilin in the GV can be associated with U7 at any time. Immunofluorescent staining suggests that most coilin in nuclei of cultured cells is also widely dispersed (Matera, 1998), although its high concentration in discrete CBs is the feature that has attracted most attention. Moreover, several lines of evidence implicate a close relationship between CBs and nucleoli. Beginning with Cajal’s original description (Cajal, 1903), physical proximity has often been noted, and in some cases CBs actually occur within the nucleolus (Malatesta et al., 1994a, 1994b; Ochs et al., 1994; Lyon et al., 1997). CBs contain several typical nucleolar components, especially fibrillarin (Raska et al., 1991), Nopp140 and NAP57 (Meier and Blobel, 1994), U3 snoRNA (Bauer et al., 1994; Jiménez-García et al., 1994), and ribosomal protein S6 (Jiménez-García et al., 1994). In a recent study, Isaac et al. (1998) found that Nopp140 and coilin interact in a yeast two-hybrid system and can be coimmunoprecipitated with an antibody against Nopp140. They suggest that Nopp140 may accompany coilin and other molecules as they move between the nucleolus and CBs. The interaction of coilin with both Nopp140 and the U7 snRNP suggests that coilin may play a more general role in trafficking of molecules between compartments of the nucleus.
ACKNOWLEDGMENTS
Antibodies were kindly supplied by the following: X.-D. Fu and T. Maniatis (anti-SC35); J. Steitz (Y12); R. Tuma and M. Roth (H1); and Z. Wu (C236). We thank Matthias Görlach for helpful discussion of RNA-binding domains. This work was supported by research grant GM-33397 from the National Institute of General Medical Sciences. J.G.G. is American Cancer Society Professor of Developmental Genetics.
Abbreviations used:
- CB
coiled body
- DIC
differential interference contrast
- GV
germinal vesicle
- RNP
ribonucleoprotein
- sn
small nuclear
- TMG
trimethylguanosine
REFERENCES
- Andrade LEC, Chan EKL, Raška I, Peebles CL, Roos G, Tan EM. Human autoantibody to a novel protein of the nuclear coiled body: immunological characterization and cDNA cloning of p80-coilin. J Exp Med. 1991;173:1407–1419. doi: 10.1084/jem.173.6.1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandziulis RJ, Swanson MS, Dreyfuss G. RNA-binding proteins as developmental regulators. Genes Dev. 1989;3:431–437. doi: 10.1101/gad.3.4.431. [DOI] [PubMed] [Google Scholar]
- Bauer DW, Murphy C, Wu Z, Wu C-HH, Gall JG. In vitro assembly of coiled bodies in Xenopus egg extract. Mol Biol Cell. 1994;5:633–644. doi: 10.1091/mbc.5.6.633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell P, Dabauvalle M-C, Scheer U. In vitro assembly of prenucleolar bodies in Xenopus egg extract. J Cell Biol. 1992;118:1297–1304. doi: 10.1083/jcb.118.6.1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellini M, Lacroix J-C, Gall JG. A zinc-binding domain is required for targeting the maternal nuclear protein PwA33 to lampbrush chromosome loops. J Cell Biol. 1995;131:563–570. doi: 10.1083/jcb.131.3.563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohmann, K., Ferreira, J., Santama, N., Weis, K., and Lamond, A.I. (1995). Molecular analysis of the coiled body. J. Cell Sci.19(suppl), 107–113. [DOI] [PubMed]
- Burd CG, Dreyfuss G. Conserved structures and diversity of functions of RNA-binding proteins. Science. 1994;265:615–621. doi: 10.1126/science.8036511. [DOI] [PubMed] [Google Scholar]
- Cajal SR. Un sencillo metodo de coloracion seletiva del reticulo protoplasmatico y sus efectos en los diversos organos nerviosos de vertebrados e invertebrados. Trab Lab Invest Biol (Madrid) 1903;2:129–221. [Google Scholar]
- Callan HG. Lampbrush chromosomes. In: Solioz M, editor. Molecular Biology, Biochemistry, and Biophysics. Vol. 36. Berlin: Springer-Verlag; 1986. pp. 85–92. [PubMed] [Google Scholar]
- Callan HG, Gall JG, Murphy C. Histone genes are located at the sphere loci of Xenopus lampbrush chromosomes. Chromosoma. 1991;101:245–251. doi: 10.1007/BF00365156. [DOI] [PubMed] [Google Scholar]
- Callan HG, Lloyd L. Lampbrush chromosomes of crested newts Triturus cristatus (Laurenti) Philos Trans R Soc Lond B Biol Sci. 1960;243:135–219. [Google Scholar]
- Chan EKL, Takano S, Andrade LEC, Hamel JC, Matera GA. Structure, expression and chromosomal localization of human p80-coilin gene. Nucleic Acids Res. 1994;22:4462–4469. doi: 10.1093/nar/22.21.4462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz MO, Gall JG. Giant readthrough transcription units at the histone loci on lampbrush chromosomes of the newt Notophthalmus. Chromosoma. 1985;92:243–253. doi: 10.1007/BF00329807. [DOI] [PubMed] [Google Scholar]
- Forbes DJ, Kornberg TB, Kirschner MW. Small nuclear RNA transcription and ribonucleoprotein assembly in early Xenopus development. J Cell Biol. 1983;97:62–72. doi: 10.1083/jcb.97.1.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frey MR, Matera AG. Coiled bodies contain U7 small nuclear RNA and associate with specific DNA sequences in interphase human cells. Proc Natl Acad Sci USA. 1995;92:5915–5919. doi: 10.1073/pnas.92.13.5915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gall JG. Lampbrush chromosomes from oocyte nuclei of the newt. J Morphol. 1954;94:283–352. [Google Scholar]
- Gall JG. Spread preparations of Xenopus germinal vesicle contents. In: Spector D, Goldman R, Leinwand L, editors. Cell Biology: A Laboratory Manual. Vol. 3. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1998. pp. 3.1–3.3. [Google Scholar]
- Gall JG, Stephenson EC, Erba HP, Diaz MO, Barsacchi-Pilone G. Histone genes are located at the sphere loci of newt lampbrush chromosomes. Chromosoma. 1981;84:159–171. doi: 10.1007/BF00399128. [DOI] [PubMed] [Google Scholar]
- Gall JG, Tsvetkov A, Wu Z, Murphy C. Is the sphere organelle/coiled body a universal nuclear component? Dev Genet. 1995;16:25–35. doi: 10.1002/dvg.1020160107. [DOI] [PubMed] [Google Scholar]
- Gao L, Frey MR, Matera AG. Human genes encoding U3 snRNA associate with coiled bodies in interphase cells and are clustered on chromosome 17p11.2 in a complex inverted repeat structure. Nucleic Acids Res. 1997;25:4740–4747. doi: 10.1093/nar/25.23.4740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimm C, Stefanovic B, Schümperli D. The low abundance of U7 snRNA is partly determined by its Sm binding site. EMBO J. 1993;12:1229–1238. doi: 10.1002/j.1460-2075.1993.tb05764.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isaac C, Yang Y, Meier UT. Nopp140 functions as a molecular link between the nucleolus and the coiled bodies. J Cell Biol. 1998;142:319–329. doi: 10.1083/jcb.142.2.319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiménez-García LF, Segura-Valdez M dL, Ochs RL, Rothblum LI, Hannan R, Spector DL. Nucleologenesis: U3 snRNA-containing prenucleolar bodies move to sites of active pre-mRNA transcription after mitosis. Mol Biol Cell. 1994;5:955–966. doi: 10.1091/mbc.5.9.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jörgensen M. Zellenstudien I. Morphologische Beiträge zum Problem des Eiwachstums. Arch Zellforsch. 1913;10:1–126. [Google Scholar]
- Kiledjian M, Burd CG, Görlach M, Portman DS, Dreyfuss G. Structure and function of hnRNP proteins. In: Nagai K, Mattaj I W, editors. RNA-Protein Interactions. Oxford: IRL Press; 1994. pp. 127–149. [Google Scholar]
- Krainer A. Pre-mRNA splicing by complementation with purified human U1, U2, U4/U6 and U5 snRNPs. Nucleic Acids Res. 1988;16:9415–9429. doi: 10.1093/nar/16.20.9415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamond AI, Carmo-Fonseca M. The coiled body. Trends Cell Biol. 1993;3:198–204. doi: 10.1016/0962-8924(93)90214-l. [DOI] [PubMed] [Google Scholar]
- Lamond AI, Earnshaw WC. Structure and function in the nucleus. Science. 1998;280:547–553. doi: 10.1126/science.280.5363.547. [DOI] [PubMed] [Google Scholar]
- Lerner EA, Lerner MR, Janeway CA, Steitz JA. Monoclonal antibodies to nucleic acid-containing cellular constituents: Probes for molecular biology and autoimmune disease. Proc Natl Acad Sci USA. 1981;78:2737–2741. doi: 10.1073/pnas.78.5.2737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyon CE, Bohmann K, Lamond AI. Inhibition of protein dephosphorylation results in the accumulation of splicing snRNPs and coiled bodies within the nucleolus. Exp Cell Res. 1997;230:84–93. doi: 10.1006/excr.1996.3380. [DOI] [PubMed] [Google Scholar]
- Malatesta M, Zancanaro C, Martin TE, Chan EKL, Amalric F, Lührmann R, Vogel P, Fakan S. Cytochemical and immunocytochemical characterization of nuclear bodies during hibernation. Eur J Cell Biol. 1994a;65:82–93. [PubMed] [Google Scholar]
- Malatesta M, Zancanaro C, Martin TE, Chan EKL, Amalric F, Lührmann R, Vogel P, Fakan S. Is the coiled body involved in nucleolar functions? Exp Cell Res. 1994b;211:415–419. doi: 10.1006/excr.1994.1106. [DOI] [PubMed] [Google Scholar]
- Matera AG. Of coiled bodies, gems, and salmon. J Cell Biochem. 1998;70:181–192. [PubMed] [Google Scholar]
- Mattaj IW. UsnRNP assembly and transport. In: Birnstiel M L, editor. Structure and Function of Major and Minor Small Nuclear Ribonucleoprotein Particles. Berlin: Springer-Verlag; 1988. pp. 100–114. [Google Scholar]
- Meier UT, Blobel G. NAP57, a mammalian nucleolar protein with a putative homolog in yeast and bacteria. J Cell Biol. 1994;127:1505–1514. doi: 10.1083/jcb.127.6.1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagai K, Oubridge C, Jessen TH, Li J, Evans PR. Crystal structure of the RNA-binding domain of the U1 small nuclear ribonucleoprotein A. Nature. 1990;348:515–520. doi: 10.1038/348515a0. [DOI] [PubMed] [Google Scholar]
- Ochs RL, Stein TW, Tan EM. Coiled bodies in the nucleolus of breast cancer cells. J Cell Sci. 1994;107:385–399. doi: 10.1242/jcs.107.2.385. [DOI] [PubMed] [Google Scholar]
- Query CC, Bentley RC, Keene JD. A common RNA recognition motif identified within a defined U1 RNA binding domain of the 70K U1 protein. Cell. 1989;57:89–101. doi: 10.1016/0092-8674(89)90175-x. [DOI] [PubMed] [Google Scholar]
- Raška I, Andrade LEC, Ochs RL, Chan EKL, Chang C-M, Roos G, Tan EM. Immunological and ultrastructural studies of the nuclear coiled body with autoimmune antibodies. Exp Cell Res. 1991;195:27–37. doi: 10.1016/0014-4827(91)90496-h. [DOI] [PubMed] [Google Scholar]
- Roth MB. Spheres, coiled bodies and nuclear bodies. Curr Opin Cell Biol. 1995;7:325–328. doi: 10.1016/0955-0674(95)80086-7. [DOI] [PubMed] [Google Scholar]
- Roth MB, Zahler AM, Stolk JA. A conserved family of nuclear phosphoproteins localized to sites of polymerase II transcription. J Cell Biol. 1991;115:587–596. doi: 10.1083/jcb.115.3.587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith HO, Tabiti K, Schaffner G, Soldati D, Albrecht U, Birnstiel ML. Two-step affinity purification of U7 small nuclear ribonucleoprotein particles using complementary biotinylated 2′-O-methyl oligoribonucleotides. Proc Natl Acad Sci USA. 1991;88:9784–9788. doi: 10.1073/pnas.88.21.9784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith KP, Carter KC, Johnson CV, Lawrence JB. U2 and U1 snRNA gene loci associate with coiled bodies. J Cell Biochem. 1995;59:473–485. doi: 10.1002/jcb.240590408. [DOI] [PubMed] [Google Scholar]
- Stefanovic B, Hackl W, Lührmann R, Schümperli D. Assembly, nuclear import and function of U7 snRNPs studied by microinjection of synthetic U7 RNA into Xenopus oocytes. Nucleic Acids Res. 1995;23:3141–3151. doi: 10.1093/nar/23.16.3141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steitz JA. Immunoprecipitation of ribonucleoproteins using autoantibodies. Methods Enzymol. 1989;180:468–481. doi: 10.1016/0076-6879(89)80118-1. [DOI] [PubMed] [Google Scholar]
- Swanson MS, Dreyfuss G. Classification and purification of proteins of heterogeneous nuclear ribonucleoprotein particles by RNA-binding specificities. Mol Cell Biol. 1988;8:2237–2241. doi: 10.1128/mcb.8.5.2237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuma R, Stolk JA, Roth MB. Identification and characterization of a sphere organelle protein. J Cell Biol. 1993;122:767–773. doi: 10.1083/jcb.122.4.767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace RA, Jared DW, Dumont JN, Sega MW. Protein incorporation by isolated amphibian oocytes: III. Optimum incubation conditions. J Exp Zool. 1973;184:321–333. doi: 10.1002/jez.1401840305. [DOI] [PubMed] [Google Scholar]
- Wittekind M, Görlach M, Friedrichs M, Dreyfuss G, Mueller L. 1H,13C,15N NMR assignments and global folding pattern of the RNA-binding domain of the human hnRNP C proteins. Biochemistry. 1992;31:6254–6265. doi: 10.1021/bi00142a013. [DOI] [PubMed] [Google Scholar]
- Wu C-HH, Gall JG. U7 small nuclear RNA in C snurposomes of the Xenopus germinal vesicle. Proc Natl Acad Sci USA. 1993;90:6257–6259. doi: 10.1073/pnas.90.13.6257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C-HH, Murphy C, Gall JG. The Sm binding site targets U7 snRNA to coiled bodies (spheres) of amphibian oocytes. RNA. 1996;2:811–823. [PMC free article] [PubMed] [Google Scholar]
- Wu Z, Murphy C, Callan HG, Gall JG. Small nuclear ribonucleoproteins and heterogeneous nuclear ribonucleoproteins in the amphibian germinal vesicle: loops, spheres, and snurposomes. J Cell Biol. 1991;113:465–483. doi: 10.1083/jcb.113.3.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Z, Murphy C, Gall JG. Human p80-coilin is targeted to sphere organelles in the amphibian germinal vesicle. Mol Biol Cell. 1994;5:1119–1127. doi: 10.1091/mbc.5.10.1119. [DOI] [PMC free article] [PubMed] [Google Scholar]