Abstract
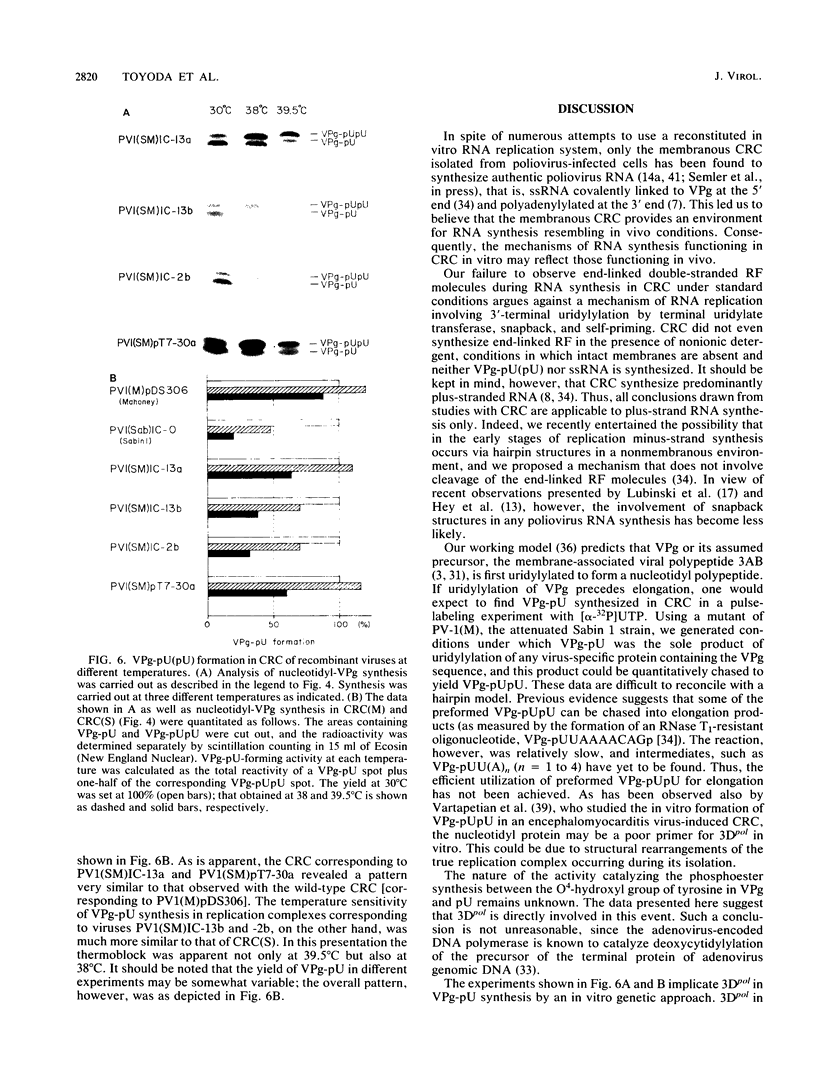
Membranous crude replication complexes (CRC) were isolated from poliovirus-infected HeLa cells as recently described (N. Takeda, R.J. Kuhn, C.-F. Yang, T. Takegami, and E. Wimmer, J. Virol. 60:43-53, 1986). Viruses used to produce the CRC were poliovirus type 1 (Mahoney), [PV-1(M)], poliovirus type 1 (Sabin) [PV-1(S)], and four in vitro recombinants that were constructed from infectious cDNA clones. RNA synthesis in CRC was studied. No end-linked, full-length double-stranded poliovirus RNA was detected in CRC regardless of whether nonionic detergent (Nonidet P-40) was added prior to incubation. Synthesis of VPg-pU and VPg-pUpU, two nucleotidyl proteins presumed to be involved in the initiation of RNA synthesis, was slower at 30 degrees C in CRC induced by PV-1(S) than by PV-1(M). This observation was used to design a pulse-chase experiment whose result suggested that synthesis of VPg-pUpU occurred by uridylylation of VPg-pU. Synthesis of VPg-pU(pU) was thermosensitive in CRC induced by PV-1(S). With CRC of recombinant viruses, the thermosensitive block covaried to nucleotide substitutions in PV-1(S) that mapped to the virus-induced RNA polymerase 3Dpol. We conclude that plus-stranded RNA synthesis in CRC does not proceed via hairpin structures. The results of VPg-pU----VPg-pUpU synthesis are consistent with a model in which VPg-pU is the primer of RNA synthesis mediated by 3Dpol. The data suggest that uridylylation of VPg or a precursor thereof may be catalyzed by 3Dpol itself, a mechanism resembling events occurring in adenovirus DNA replication.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Andrews N. C., Baltimore D. Purification of a terminal uridylyltransferase that acts as host factor in the in vitro poliovirus replicase reaction. Proc Natl Acad Sci U S A. 1986 Jan;83(2):221–225. doi: 10.1073/pnas.83.2.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews N. C., Levin D., Baltimore D. Poliovirus replicase stimulation by terminal uridylyl transferase. J Biol Chem. 1985 Jun 25;260(12):7628–7635. [PubMed] [Google Scholar]
- Baron M. H., Baltimore D. Anti-VPg antibody inhibition of the poliovirus replicase reaction and production of covalent complexes of VPg-related proteins and RNA. Cell. 1982 Oct;30(3):745–752. doi: 10.1016/0092-8674(82)90279-3. [DOI] [PubMed] [Google Scholar]
- Caliguiri L. A., Tamm I. Characterization of poliovirus-specific structures associated with cytoplasmic membranes. Virology. 1970 Sep;42(1):112–122. doi: 10.1016/0042-6822(70)90243-6. [DOI] [PubMed] [Google Scholar]
- Challberg M. D., Kelly T. J. Eukaryotic DNA replication: viral and plasmid model systems. Annu Rev Biochem. 1982;51:901–934. doi: 10.1146/annurev.bi.51.070182.004345. [DOI] [PubMed] [Google Scholar]
- Crawford N. M., Baltimore D. Genome-linked protein VPg of poliovirus is present as free VPg and VPg-pUpU in poliovirus-infected cells. Proc Natl Acad Sci U S A. 1983 Dec;80(24):7452–7455. doi: 10.1073/pnas.80.24.7452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorsch-Häsler K., Yogo Y., Wimmer E. Replication of picornaviruses. I. Evidence from in vitro RNA synthesis that poly(A) of the poliovirus genome is genetically coded. J Virol. 1975 Dec;16(6):1512–1517. doi: 10.1128/jvi.16.6.1512-1517.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etchison D., Ehrenfeld E. Comparison of replication complexes synthesizing poliovirus RNA. Virology. 1981 May;111(1):33–46. doi: 10.1016/0042-6822(81)90651-6. [DOI] [PubMed] [Google Scholar]
- Fiszman M., Reynier M., Bucchini D., Girard M. Thermosensitive block of the Sabin strain of poliovirus type I. J Virol. 1972 Dec;10(6):1143–1151. doi: 10.1128/jvi.10.6.1143-1151.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flanegan J. B., Baltimore D. Poliovirus polyuridylic acid polymerase and RNA replicase have the same viral polypeptide. J Virol. 1979 Jan;29(1):352–360. doi: 10.1128/jvi.29.1.352-360.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flanegan J. B., Baltimore D. Poliovirus-specific primer-dependent RNA polymerase able to copy poly(A). Proc Natl Acad Sci U S A. 1977 Sep;74(9):3677–3680. doi: 10.1073/pnas.74.9.3677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girard M. In vitro synthesis of poliovirus ribonucleic acid: role of the replicative intermediate. J Virol. 1969 Apr;3(4):376–384. doi: 10.1128/jvi.3.4.376-384.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hey T. D., Richards O. C., Ehrenfeld E. Host factor-induced template modification during synthesis of poliovirus RNA in vitro. J Virol. 1987 Mar;61(3):802–811. doi: 10.1128/jvi.61.3.802-811.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitamura N., Semler B. L., Rothberg P. G., Larsen G. R., Adler C. J., Dorner A. J., Emini E. A., Hanecak R., Lee J. J., van der Werf S. Primary structure, gene organization and polypeptide expression of poliovirus RNA. Nature. 1981 Jun 18;291(5816):547–553. doi: 10.1038/291547a0. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lee Y. F., Nomoto A., Detjen B. M., Wimmer E. A protein covalently linked to poliovirus genome RNA. Proc Natl Acad Sci U S A. 1977 Jan;74(1):59–63. doi: 10.1073/pnas.74.1.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lubinski J. M., Kaplan G., Racaniello V. R., Dasgupta A. Mechanism of in vitro synthesis of covalently linked dimeric RNA molecules by the poliovirus replicase. J Virol. 1986 May;58(2):459–467. doi: 10.1128/jvi.58.2.459-467.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonnell J. P., Levintow L. Kinetics of appearance of the products of poliovirus-induced RNA polymerase. Virology. 1970 Dec;42(4):999–1006. doi: 10.1016/0042-6822(70)90348-x. [DOI] [PubMed] [Google Scholar]
- Miyamura K., Takeda N., Yamazaki S. Characterization of a temperature-sensitive defect of enterovirus 70: effect of elevated temperature on in vitro transcription. J Virol. 1984 Jul;51(1):192–198. doi: 10.1128/jvi.51.1.192-198.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomoto A., Detjen B., Pozzatti R., Wimmer E. The location of the polio genome protein in viral RNAs and its implication for RNA synthesis. Nature. 1977 Jul 21;268(5617):208–213. doi: 10.1038/268208a0. [DOI] [PubMed] [Google Scholar]
- Nomoto A., Omata T., Toyoda H., Kuge S., Horie H., Kataoka Y., Genba Y., Nakano Y., Imura N. Complete nucleotide sequence of the attenuated poliovirus Sabin 1 strain genome. Proc Natl Acad Sci U S A. 1982 Oct;79(19):5793–5797. doi: 10.1073/pnas.79.19.5793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omata T., Kohara M., Kuge S., Komatsu T., Abe S., Semler B. L., Kameda A., Itoh H., Arita M., Wimmer E. Genetic analysis of the attenuation phenotype of poliovirus type 1. J Virol. 1986 May;58(2):348–358. doi: 10.1128/jvi.58.2.348-358.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omata T., Kohara M., Sakai Y., Kameda A., Imura N., Nomoto A. Cloned infectious complementary DNA of the poliovirus Sabin 1 genome: biochemical and biological properties of the recovered virus. Gene. 1984 Dec;32(1-2):1–10. doi: 10.1016/0378-1119(84)90026-x. [DOI] [PubMed] [Google Scholar]
- Pincus S. E., Diamond D. C., Emini E. A., Wimmer E. Guanidine-selected mutants of poliovirus: mapping of point mutations to polypeptide 2C. J Virol. 1986 Feb;57(2):638–646. doi: 10.1128/jvi.57.2.638-646.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pincus S. E., Wimmer E. Production of guanidine-resistant and -dependent poliovirus mutants from cloned cDNA: mutations in polypeptide 2C are directly responsible for altered guanidine sensitivity. J Virol. 1986 Nov;60(2):793–796. doi: 10.1128/jvi.60.2.793-796.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rueckert R. R., Wimmer E. Systematic nomenclature of picornavirus proteins. J Virol. 1984 Jun;50(3):957–959. doi: 10.1128/jvi.50.3.957-959.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarnow P., Bernstein H. D., Baltimore D. A poliovirus temperature-sensitive RNA synthesis mutant located in a noncoding region of the genome. Proc Natl Acad Sci U S A. 1986 Feb;83(3):571–575. doi: 10.1073/pnas.83.3.571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semler B. L., Anderson C. W., Hanecak R., Dorner L. F., Wimmer E. A membrane-associated precursor to poliovirus VPg identified by immunoprecipitation with antibodies directed against a synthetic heptapeptide. Cell. 1982 Feb;28(2):405–412. doi: 10.1016/0092-8674(82)90358-0. [DOI] [PubMed] [Google Scholar]
- Semler B. L., Dorner A. J., Wimmer E. Production of infectious poliovirus from cloned cDNA is dramatically increased by SV40 transcription and replication signals. Nucleic Acids Res. 1984 Jun 25;12(12):5123–5141. doi: 10.1093/nar/12.12.5123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stillman B. W. The replication of adenovirus DNA with purified proteins. Cell. 1983 Nov;35(1):7–9. doi: 10.1016/0092-8674(83)90201-5. [DOI] [PubMed] [Google Scholar]
- Takeda N., Kuhn R. J., Yang C. F., Takegami T., Wimmer E. Initiation of poliovirus plus-strand RNA synthesis in a membrane complex of infected HeLa cells. J Virol. 1986 Oct;60(1):43–53. doi: 10.1128/jvi.60.1.43-53.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takegami T., Kuhn R. J., Anderson C. W., Wimmer E. Membrane-dependent uridylylation of the genome-linked protein VPg of poliovirus. Proc Natl Acad Sci U S A. 1983 Dec;80(24):7447–7451. doi: 10.1073/pnas.80.24.7447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takegami T., Semler B. L., Anderson C. W., Wimmer E. Membrane fractions active in poliovirus RNA replication contain VPg precursor polypeptides. Virology. 1983 Jul 15;128(1):33–47. doi: 10.1016/0042-6822(83)90316-1. [DOI] [PubMed] [Google Scholar]
- Toyoda H., Kohara M., Kataoka Y., Suganuma T., Omata T., Imura N., Nomoto A. Complete nucleotide sequences of all three poliovirus serotype genomes. Implication for genetic relationship, gene function and antigenic determinants. J Mol Biol. 1984 Apr 25;174(4):561–585. doi: 10.1016/0022-2836(84)90084-6. [DOI] [PubMed] [Google Scholar]
- Wimmer E. Genome-linked proteins of viruses. Cell. 1982 Feb;28(2):199–201. doi: 10.1016/0092-8674(82)90335-x. [DOI] [PubMed] [Google Scholar]
- Young D. C., Dunn B. M., Tobin G. J., Flanegan J. B. Anti-VPg antibody precipitation of product RNA synthesized in vitro by the poliovirus polymerase and host factor is mediated by VPg on the poliovirion RNA template. J Virol. 1986 Jun;58(3):715–723. doi: 10.1128/jvi.58.3.715-723.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young D. C., Tobin G. J., Flanegan J. B. Characterization of product RNAs synthesized in vitro by poliovirus RNA polymerase purified by chromatography on hydroxylapatite or poly(U) Sepharose. J Virol. 1987 Feb;61(2):611–614. doi: 10.1128/jvi.61.2.611-614.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young D. C., Tuschall D. M., Flanegan J. B. Poliovirus RNA-dependent RNA polymerase and host cell protein synthesize product RNA twice the size of poliovirion RNA in vitro. J Virol. 1985 May;54(2):256–264. doi: 10.1128/jvi.54.2.256-264.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Werf S., Bradley J., Wimmer E., Studier F. W., Dunn J. J. Synthesis of infectious poliovirus RNA by purified T7 RNA polymerase. Proc Natl Acad Sci U S A. 1986 Apr;83(8):2330–2334. doi: 10.1073/pnas.83.8.2330. [DOI] [PMC free article] [PubMed] [Google Scholar]








