Abstract
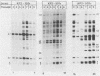
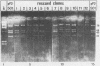
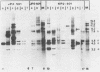
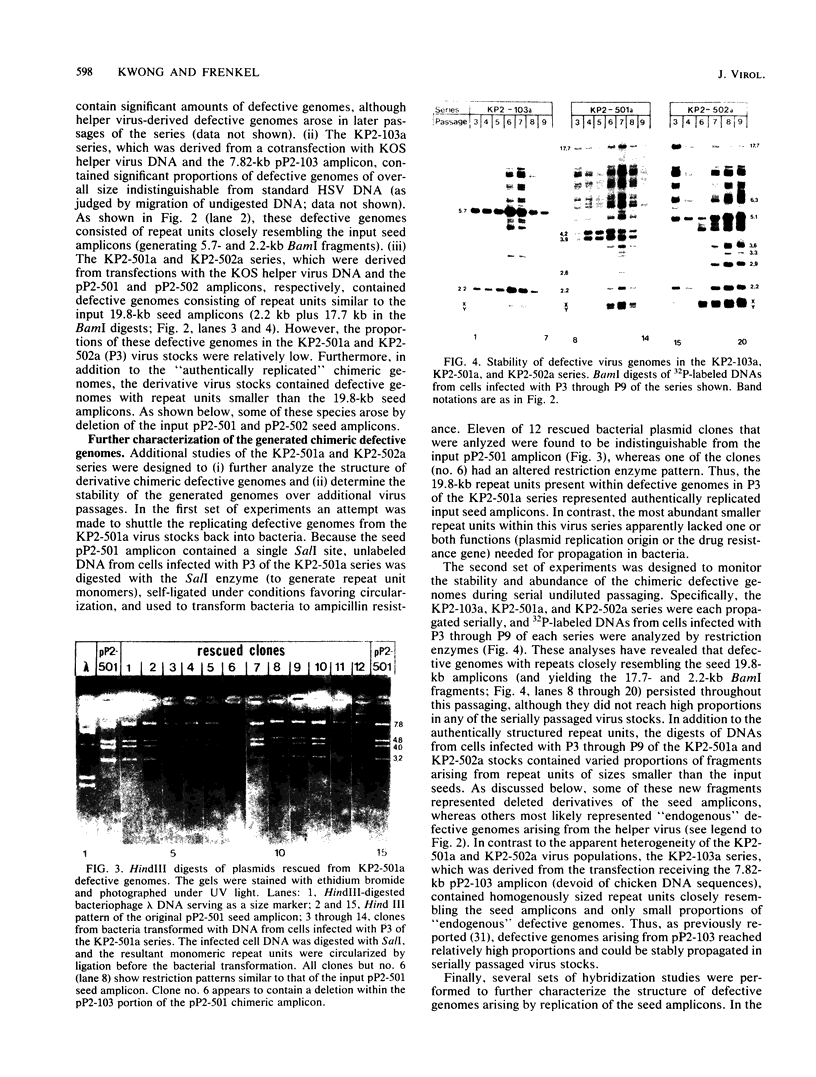
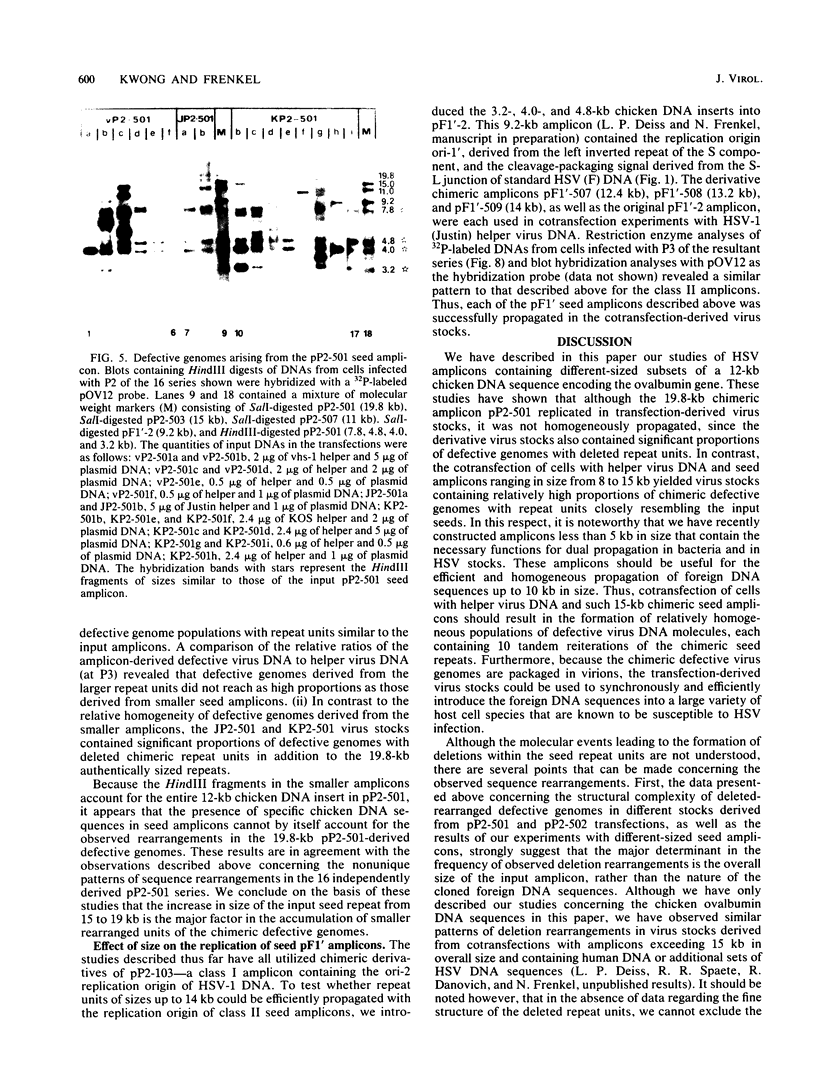
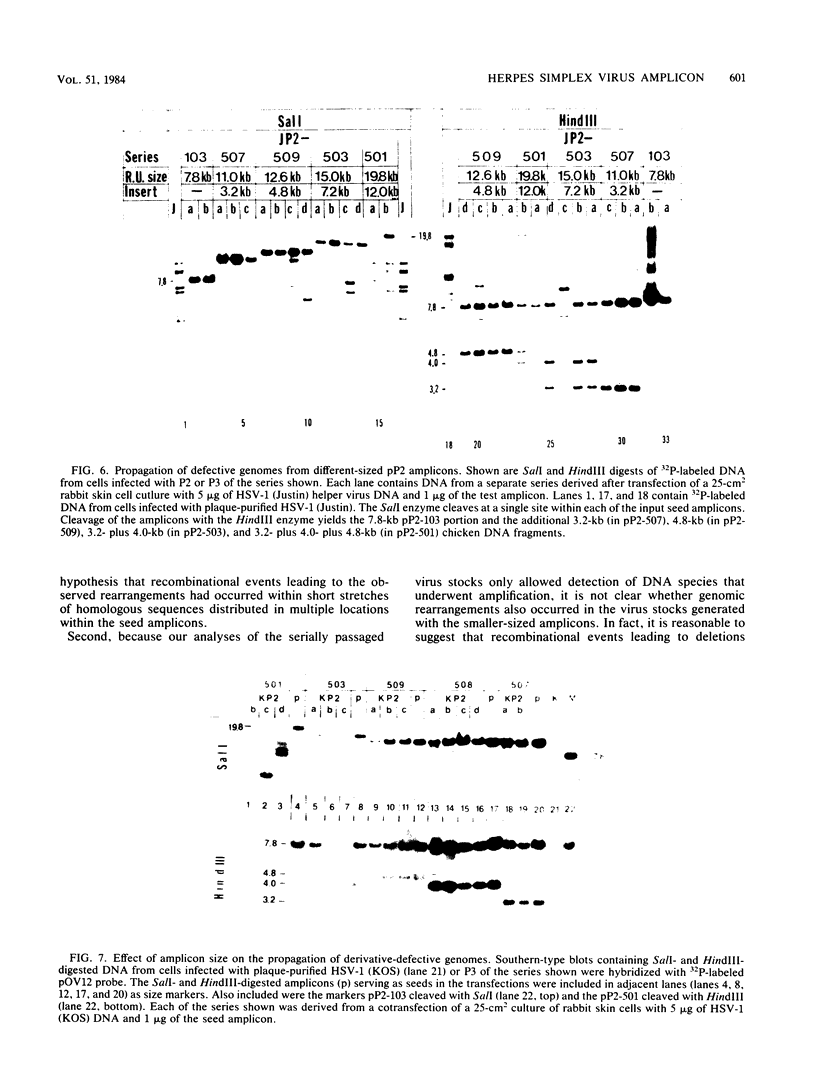
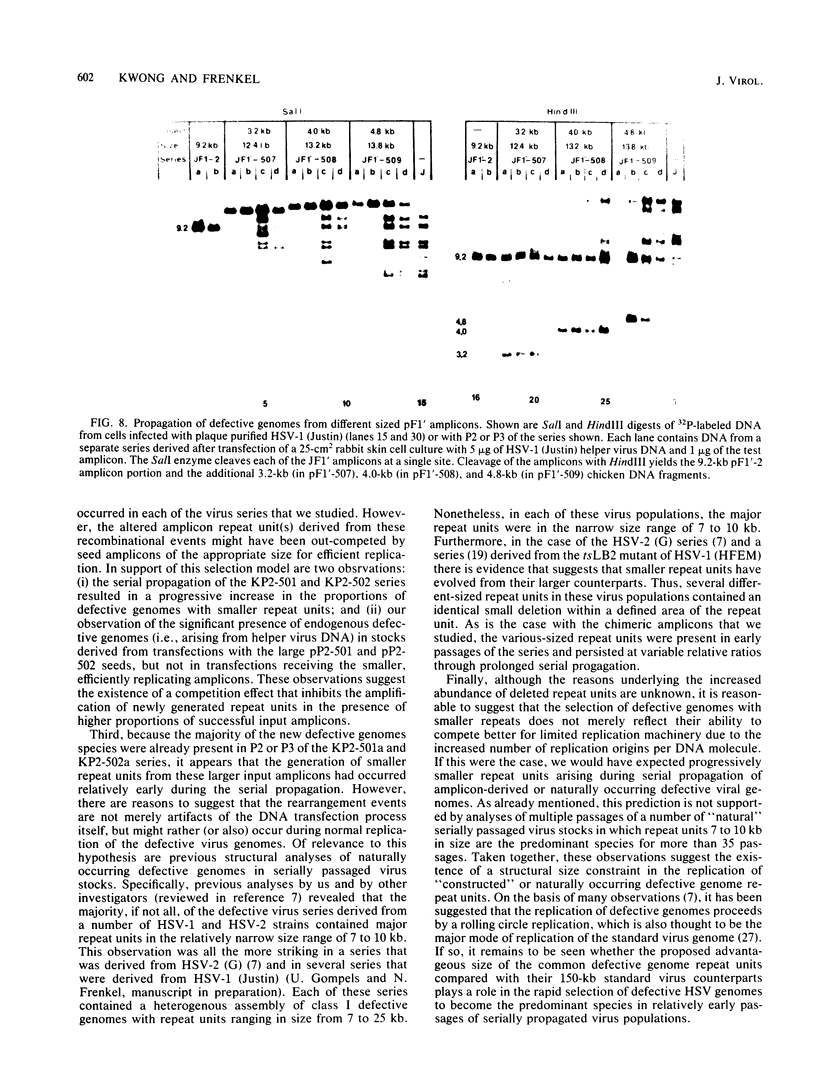
Previous studies (R. R. Spaete and N. Frenkel, Cell 30:295-304, 1982) have documented the potential use of defective virus vectors (amplicons) derived from herpes simplex virus for the efficient introduction of foreign DNA sequences into eucaryotic cells. Specifically, cotransfection of cells with helper virus DNA and cloned amplicons (8 to 10 kilobases [kb]) containing bacterial plasmid DNA sequences linked to a set of herpes simplex virus cis-acting propagation signals (a replication origin and a cleavage-packaging signal) resulted in the generation of virus stocks containing packaged defective genomes that consisted of uniform head-to-tail reiterations of the chimeric seed amplicon sequences. The chimeric defective genomes could be stably propagated in virus stocks and could thus be used to efficiently infect cells. We now report on additional studies designed to propagate relatively large sets of eucaryotic DNA sequences within chimeric packaged defective genomes. These studies have utilized a 12-kb chicken DNA sequence encoding the chicken ovalbumin gene and cloned by Lai et al. (Proc. Natl. Acad. Sci. U.S.A. 77:244-248, 1980) in the plasmid pOV12. Virus stocks derived from cells cotransfected with helper virus DNA and chimeric amplicons (overall size of 19.8 kb, of which 12 kb corresponded to the chicken DNA) contained defective genomes composed of reiterations of the 19.8-kb seed amplicon sequences. However, in addition to the authentically sized repeat units, defective genomes in the derivative virus stocks contained smaller repeat units representing deleted versions of the seed 19.8-kb amplicons. The recombinational events leading to the formation of deleted repeats did not appear to occur at unique sites, as shown by comparative analyses of multiple, independently generated virus series propagated from separate transfections. In contast, seed amplicons ranging in size from 11 to 15 kb and containing subsets of the 12-kb chicken DNA sequences replicated efficiently and could be stably propagated in virus stocks. The results of these studies suggest the existence of size restrictions (up to 15 kb) on the efficient replication of seed herpes simplex virus amplicons.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Barnett J. W., Eppstein D. A., Chan H. W. Class I defective herpes simplex virus DNA as a molecular cloning vehicle in eucaryotic cells. J Virol. 1983 Nov;48(2):384–395. doi: 10.1128/jvi.48.2.384-395.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellard M., Kuo M. T., Dretzen G., Chambon P. Differential nuclease sensitivity of the ovalbumin and beta-globin chromatin regions in erythrocytes and oviduct cells of laying hen. Nucleic Acids Res. 1980 Jun 25;8(12):2737–2750. doi: 10.1093/nar/8.12.2737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benoist C., O'Hare K., Breathnach R., Chambon P. The ovalbumin gene-sequence of putative control regions. Nucleic Acids Res. 1980 Jan 11;8(1):127–142. doi: 10.1093/nar/8.1.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colbert D. A., Knoll B. J., Woo S. L., Mace M. L., Tsai M. J., O'Malley B. W. Differential hormonal responsiveness of the ovalbumin gene and its pseudogenes in the chick oviduct. Biochemistry. 1980 Nov 25;19(24):5586–5592. doi: 10.1021/bi00565a020. [DOI] [PubMed] [Google Scholar]
- Denhardt D. T. A membrane-filter technique for the detection of complementary DNA. Biochem Biophys Res Commun. 1966 Jun 13;23(5):641–646. doi: 10.1016/0006-291x(66)90447-5. [DOI] [PubMed] [Google Scholar]
- Dugaiczyk A., Woo S. L., Colbert D. A., Lai E. C., Mace M. L., Jr, O'Malley B. W. The ovalbumin gene: cloning and molecular organization of the entire natural gene. Proc Natl Acad Sci U S A. 1979 May;76(5):2253–2257. doi: 10.1073/pnas.76.5.2253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gannon F., Jeltsch J. M., Perrin F. A detailed comparison of the 5'-end of the ovalbumin gene cloned from chicken oviduct and erythrocyte DNA. Nucleic Acids Res. 1980 Oct 10;8(19):4405–4421. doi: 10.1093/nar/8.19.4405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gannon F., O'Hare K., Perrin F., LePennec J. P., Benoist C., Cochet M., Breathnach R., Royal A., Garapin A., Cami B. Organisation and sequences at the 5' end of a cloned complete ovalbumin gene. Nature. 1979 Mar 29;278(5703):428–434. doi: 10.1038/278428a0. [DOI] [PubMed] [Google Scholar]
- Garel A., Axel R. Selective digestion of transcriptionally active ovalbumin genes from oviduct nuclei. Proc Natl Acad Sci U S A. 1976 Nov;73(11):3966–3970. doi: 10.1073/pnas.73.11.3966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham F. L., van der Eb A. J. A new technique for the assay of infectivity of human adenovirus 5 DNA. Virology. 1973 Apr;52(2):456–467. doi: 10.1016/0042-6822(73)90341-3. [DOI] [PubMed] [Google Scholar]
- Kuebbing D., Liarakos C. D. Nucleotide sequence at the 5' end of ovalbumin messenger RNA from chicken. Nucleic Acids Res. 1978 Jul;5(7):2253–2266. doi: 10.1093/nar/5.7.2253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai E. C., Woo S. L., Bordelon-Riser M. E., Fraser T. H., O'Malley B. W. Ovalbumin is synthesized in mouse cells transformed with the natural chicken ovalbumin gene. Proc Natl Acad Sci U S A. 1980 Jan;77(1):244–248. doi: 10.1073/pnas.77.1.244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawson G. M., Tsai M. J., O'Malley B. W. Deoxyribonuclease I sensitivity of the nontranscribed sequences flanking the 5' and 3' ends of the ovomucoid gene and the ovalbumin and its related X and Y genes in hen oviduct nuclei. Biochemistry. 1980 Sep 16;19(19):4403–4441. doi: 10.1021/bi00560a004. [DOI] [PubMed] [Google Scholar]
- LeMeur M., Glanville N., Mandel J. L., Gerlinger P., Palmiter R., Chambon P. The ovalbumin gene family: hormonal control of X and Y gene transcription and mRNA accumulation. Cell. 1981 Feb;23(2):561–571. doi: 10.1016/0092-8674(81)90152-5. [DOI] [PubMed] [Google Scholar]
- Locker H., Frenkel N. BamI, KpnI, and SalI restriction enzyme maps of the DNAs of herpes simplex virus strains Justin and F: occurrence of heterogeneities in defined regions of the viral DNA. J Virol. 1979 Nov;32(2):429–441. doi: 10.1128/jvi.32.2.429-441.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locker H., Frenkel N., Halliburton I. Structure and expression of class II defective herpes simplex virus genomes encoding infected cell polypeptide number 8. J Virol. 1982 Aug;43(2):574–593. doi: 10.1128/jvi.43.2.574-593.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locker H., Frenkel N. Structure and origin of defective genomes contained in serially passaged herpes simplex virus type 1 (Justin). J Virol. 1979 Mar;29(3):1065–1077. doi: 10.1128/jvi.29.3.1065-1077.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKnight G. S. The induction of ovalbumin and conalbumin mRNA by estrogen and progesterone in chick oviduct explant cultures. Cell. 1978 Jun;14(2):403–413. doi: 10.1016/0092-8674(78)90125-3. [DOI] [PubMed] [Google Scholar]
- Mocarski E. S., Roizman B. Herpesvirus-dependent amplification and inversion of cell-associated viral thymidine kinase gene flanked by viral a sequences and linked to an origin of viral DNA replication. Proc Natl Acad Sci U S A. 1982 Sep;79(18):5626–5630. doi: 10.1073/pnas.79.18.5626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mocarski E. S., Roizman B. Site-specific inversion sequence of the herpes simplex virus genome: domain and structural features. Proc Natl Acad Sci U S A. 1981 Nov;78(11):7047–7051. doi: 10.1073/pnas.78.11.7047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulvihill E. R., LePennec J. P., Chambon P. Chicken oviduct progesterone receptor: location of specific regions of high-affinity binding in cloned DNA fragments of hormone-responsive genes. Cell. 1982 Mar;28(3):621–632. doi: 10.1016/0092-8674(82)90217-3. [DOI] [PubMed] [Google Scholar]
- Read G. S., Frenkel N. Herpes simplex virus mutants defective in the virion-associated shutoff of host polypeptide synthesis and exhibiting abnormal synthesis of alpha (immediate early) viral polypeptides. J Virol. 1983 May;46(2):498–512. doi: 10.1128/jvi.46.2.498-512.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes G. R., LaFemina R., Hayward S. D., Hayward G. S. Morphological transformation by DNA fragments of human herpesviruses: evidence for two distinct transforming regions in herpes simplex virus types 1 and 2 and lack of correlation with biochemical transfer of the thymidine kinase gene. Cold Spring Harb Symp Quant Biol. 1980;44(Pt 1):629–641. doi: 10.1101/sqb.1980.044.01.066. [DOI] [PubMed] [Google Scholar]
- Robinson S. I., Nelkin B. D., Vogelstein B. The ovalbumin gene is associated with the nuclear matrix of chicken oviduct cells. Cell. 1982 Jan;28(1):99–106. doi: 10.1016/0092-8674(82)90379-8. [DOI] [PubMed] [Google Scholar]
- Roizman B. The structure and isomerization of herpes simplex virus genomes. Cell. 1979 Mar;16(3):481–494. doi: 10.1016/0092-8674(79)90023-0. [DOI] [PubMed] [Google Scholar]
- Ruyechan W. T., Morse L. S., Knipe D. M., Roizman B. Molecular genetics of herpes simplex virus. II. Mapping of the major viral glycoproteins and of the genetic loci specifying the social behavior of infected cells. J Virol. 1979 Feb;29(2):677–697. doi: 10.1128/jvi.29.2.677-697.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senear A. W., Palmiter R. D. Multiple structural features are responsible for the nuclease sensitivity of the active ovalbumin gene. J Biol Chem. 1981 Feb 10;256(3):1191–1198. [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Spaete R. R., Frenkel N. The herpes simplex virus amplicon: a new eucaryotic defective-virus cloning-amplifying vector. Cell. 1982 Aug;30(1):295–304. doi: 10.1016/0092-8674(82)90035-6. [DOI] [PubMed] [Google Scholar]
- Stow N. D. Localization of an origin of DNA replication within the TRS/IRS repeated region of the herpes simplex virus type 1 genome. EMBO J. 1982;1(7):863–867. doi: 10.1002/j.1460-2075.1982.tb01261.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlazny D. A., Frenkel N. Replication of herpes simplex virus DNA: localization of replication recognition signals within defective virus genomes. Proc Natl Acad Sci U S A. 1981 Feb;78(2):742–746. doi: 10.1073/pnas.78.2.742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlazny D. A., Kwong A., Frenkel N. Site-specific cleavage/packaging of herpes simplex virus DNA and the selective maturation of nucleocapsids containing full-length viral DNA. Proc Natl Acad Sci U S A. 1982 Mar;79(5):1423–1427. doi: 10.1073/pnas.79.5.1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo S. L., Beattie W. G., Catterall J. F., Dugaiczyk A., Staden R., Brownlee G. G., O'Malley B. W. Complete nucleotide sequence of the chicken chromosomal ovalbumin gene and its biological significance. Biochemistry. 1981 Oct 27;20(22):6437–6446. doi: 10.1021/bi00525a024. [DOI] [PubMed] [Google Scholar]