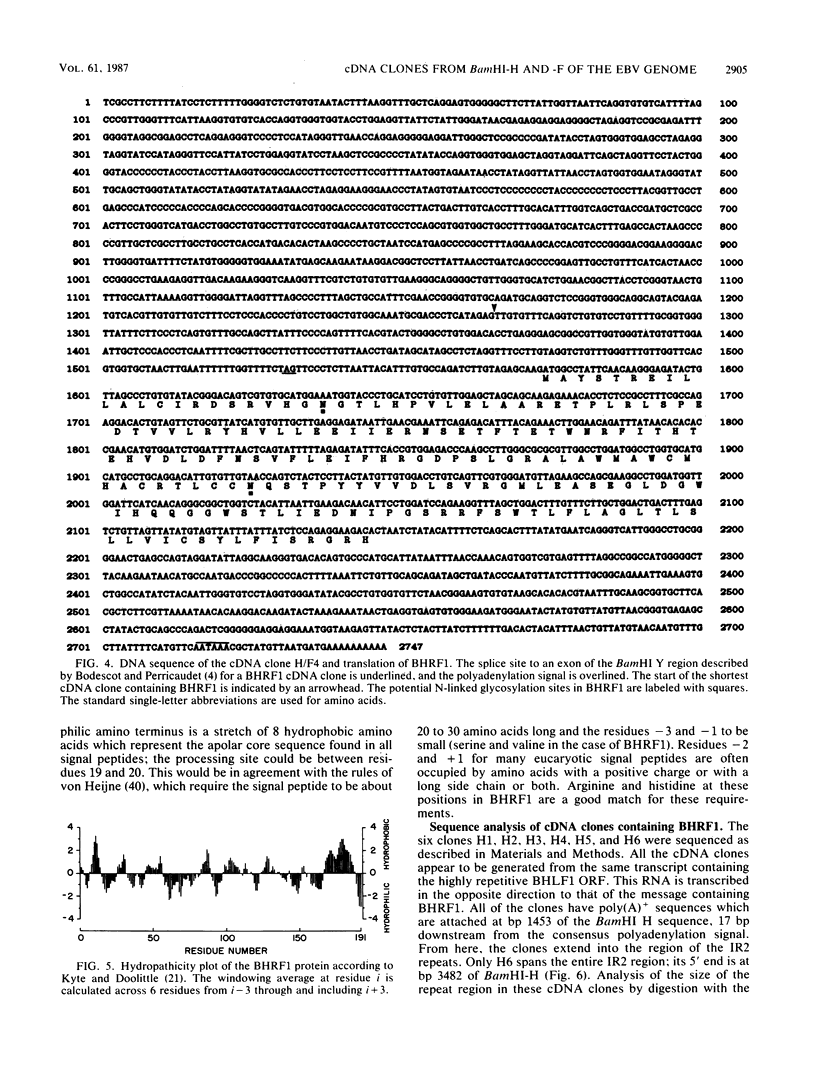
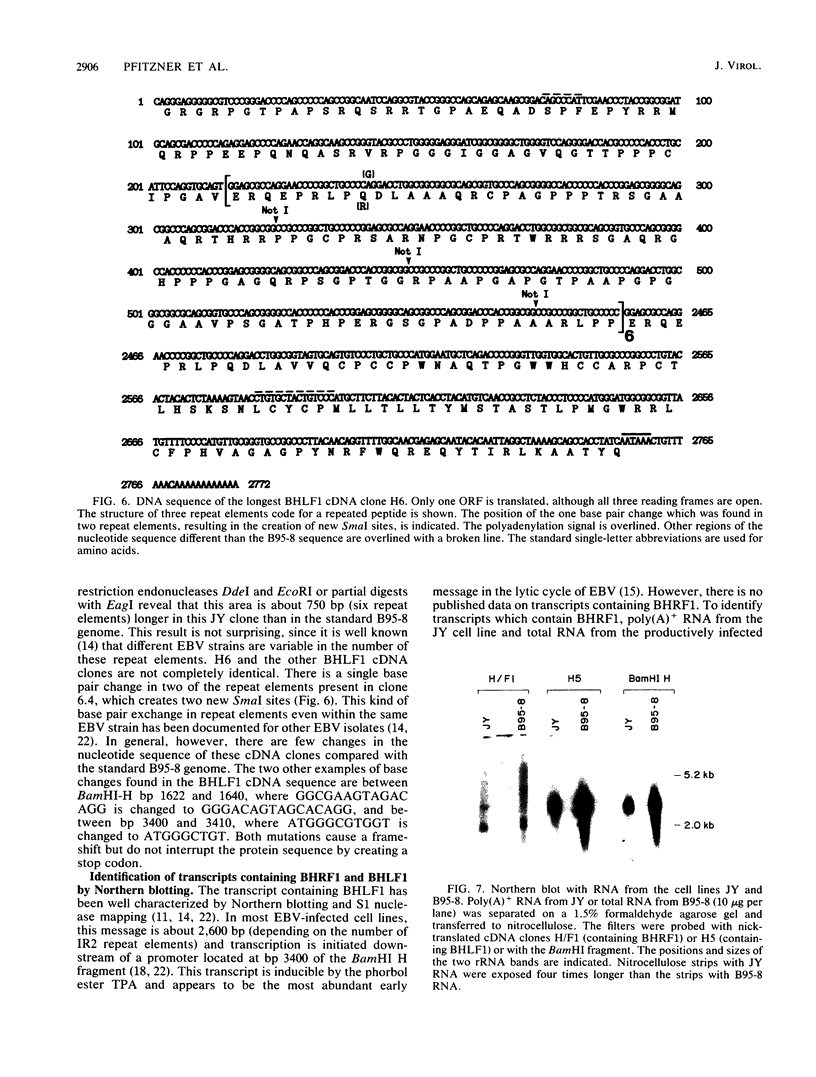
Abstract
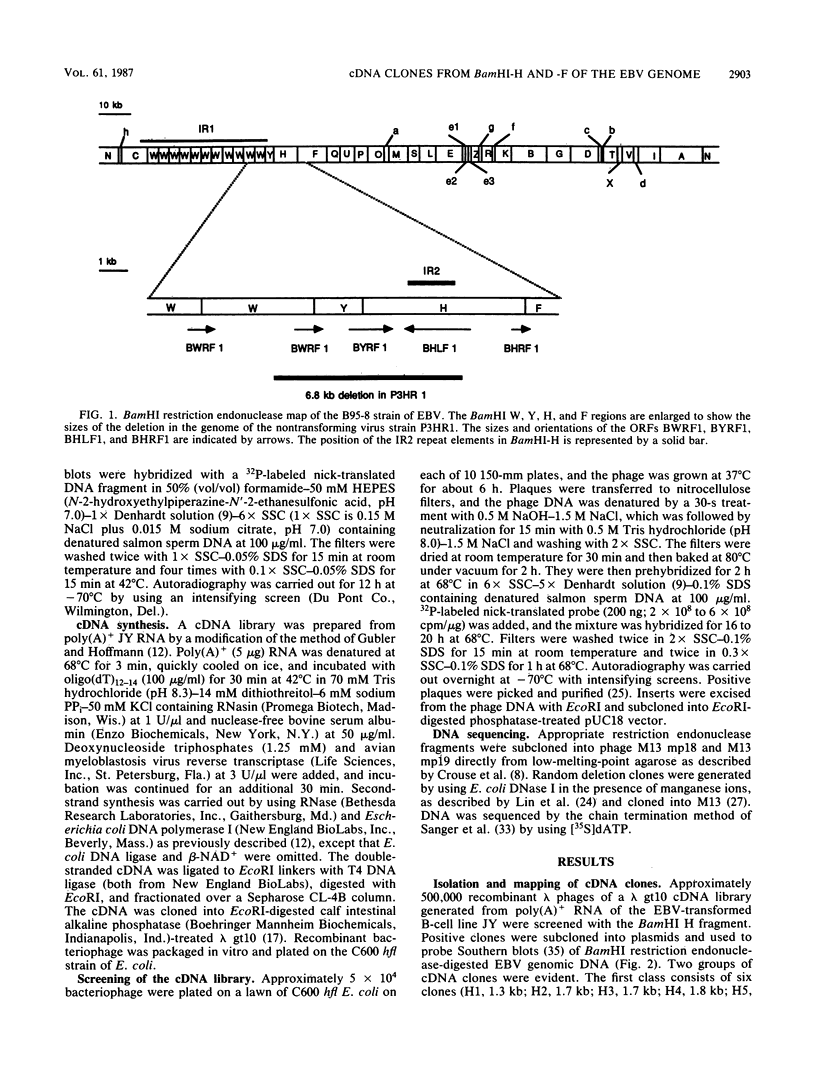
The Epstein-Barr virus (EBV) mutant P3HR1 is incapable of immortalizing B lymphocytes because of a 6.8-kilobase deletion in the BamHI W, Y, and H regions of the viral genome (M. Rabson, L. Gradoville, L. Heston, and G. Miller, J. Virol. 44:834-844, 1982). To characterize transcripts that are encoded in this region, poly(A)+ RNA from the EBV-transformed lymphoblastoid cell line JY was isolated, and this RNA was used to generate a cDNA library in lambda gt10. By screening 500,000 recombinant bacteriophages with the BamHI H fragment, we isolated 10 cDNA clones and characterized them in detail. One group of six cDNA clones was derived from a 2.9-kilobase early transcript encoded by the IR2 repeat element and showed restriction site polymorphism for the enzyme SmaI. The second group consisted of four cDNA clones, all of which contained the BamHI-H right reading frame (BHRF1), and used the polyadenylation signal at base pair 662 in the BamHI F fragment. Computer analysis of the hydrophobicity of the BHRF1 protein revealed that it is likely to be a membrane protein. Northern blotting experiments with RNA from an EBV producer line, B95-8, and a tightly latent lymphoblastoid B-cell line, IB4, revealed that BHRF1 is contained in at least two different mRNA species which can be detected during the latent cycle of EBV. These data and the recent characterization of a spliced transcript (containing five exons in common with other known latent messages [M. Bodescot and M. Perricaudet, Nucleic Acids Res. 14:7103-7113, 1986]) suggest that alternative splicing is used to generate transcripts containing BHRF1, as for the EBV nuclear antigen 1 transcripts. Furthermore, the observation that a potential oncogene activated in human follicular lymphomas is homologous to the BHRF1-encoded polypeptide (M. L. Cleary, S.D. Smith, and J. Sklar, Cell 47:19-28, 1986) suggests a possible role for this putative viral protein in EBV-induced growth transformation of B lymphocytes.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Auffray C., Rougeon F. Purification of mouse immunoglobulin heavy-chain messenger RNAs from total myeloma tumor RNA. Eur J Biochem. 1980 Jun;107(2):303–314. doi: 10.1111/j.1432-1033.1980.tb06030.x. [DOI] [PubMed] [Google Scholar]
- Aviv H., Leder P. Purification of biologically active globin messenger RNA by chromatography on oligothymidylic acid-cellulose. Proc Natl Acad Sci U S A. 1972 Jun;69(6):1408–1412. doi: 10.1073/pnas.69.6.1408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baer R., Bankier A. T., Biggin M. D., Deininger P. L., Farrell P. J., Gibson T. J., Hatfull G., Hudson G. S., Satchwell S. C., Séguin C. DNA sequence and expression of the B95-8 Epstein-Barr virus genome. Nature. 1984 Jul 19;310(5974):207–211. doi: 10.1038/310207a0. [DOI] [PubMed] [Google Scholar]
- Bodescot M., Perricaudet M. Epstein-Barr virus mRNAs produced by alternative splicing. Nucleic Acids Res. 1986 Sep 11;14(17):7103–7114. doi: 10.1093/nar/14.17.7103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bornkamm G. W., Hudewentz J., Freese U. K., Zimber U. Deletion of the nontransforming Epstein-Barr virus strain P3HR-1 causes fusion of the large internal repeat to the DSL region. J Virol. 1982 Sep;43(3):952–968. doi: 10.1128/jvi.43.3.952-968.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caput D., Beutler B., Hartog K., Thayer R., Brown-Shimer S., Cerami A. Identification of a common nucleotide sequence in the 3'-untranslated region of mRNA molecules specifying inflammatory mediators. Proc Natl Acad Sci U S A. 1986 Mar;83(6):1670–1674. doi: 10.1073/pnas.83.6.1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleary M. L., Smith S. D., Sklar J. Cloning and structural analysis of cDNAs for bcl-2 and a hybrid bcl-2/immunoglobulin transcript resulting from the t(14;18) translocation. Cell. 1986 Oct 10;47(1):19–28. doi: 10.1016/0092-8674(86)90362-4. [DOI] [PubMed] [Google Scholar]
- Crouse G. F., Frischauf A., Lehrach H. An integrated and simplified approach to cloning into plasmids and single-stranded phages. Methods Enzymol. 1983;101:78–89. doi: 10.1016/0076-6879(83)01006-x. [DOI] [PubMed] [Google Scholar]
- Denhardt D. T. A membrane-filter technique for the detection of complementary DNA. Biochem Biophys Res Commun. 1966 Jun 13;23(5):641–646. doi: 10.1016/0006-291x(66)90447-5. [DOI] [PubMed] [Google Scholar]
- Edson C. M., Cohen L. K., Henle W., Strominger J. L. An unusually high-titer human anti-Epstein Barr virus (EBV) serum and its use in the study of EBV-specific proteins synthesized in vitro and in vivo. J Immunol. 1983 Feb;130(2):919–924. [PubMed] [Google Scholar]
- Freese U. K., Laux G., Hudewentz J., Schwarz E., Bornkamm G. W. Two distant clusters of partially homologous small repeats of Epstein-Barr virus are transcribed upon induction of an abortive or lytic cycle of the virus. J Virol. 1983 Dec;48(3):731–743. doi: 10.1128/jvi.48.3.731-743.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gubler U., Hoffman B. J. A simple and very efficient method for generating cDNA libraries. Gene. 1983 Nov;25(2-3):263–269. doi: 10.1016/0378-1119(83)90230-5. [DOI] [PubMed] [Google Scholar]
- Henderson E., Miller G., Robinson J., Heston L. Efficiency of transformation of lymphocytes by Epstein-Barr virus. Virology. 1977 Jan;76(1):152–163. doi: 10.1016/0042-6822(77)90292-6. [DOI] [PubMed] [Google Scholar]
- Hudewentz J., Delius H., Freese U. K., Zimber U., Bornkamm G. W. Two distant regions of the Epstein-Barr virus genome with sequence homologies have the same orientation and involve small tandem repeats. EMBO J. 1982;1(1):21–26. doi: 10.1002/j.1460-2075.1982.tb01118.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hummel M., Kieff E. Epstein-Barr virus RNA. VIII. Viral RNA in permissively infected B95-8 cells. J Virol. 1982 Jul;43(1):262–272. doi: 10.1128/jvi.43.1.262-272.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hummel M., Kieff E. Mapping of polypeptides encoded by the Epstein-Barr virus genome in productive infection. Proc Natl Acad Sci U S A. 1982 Sep;79(18):5698–5702. doi: 10.1073/pnas.79.18.5698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeang K. T., Hayward S. D. Organization of the Epstein-Barr virus DNA molecule. III. Location of the P3HR-1 deletion junction and characterization of the NotI repeat units that form part of the template for an abundant 12-O-tetradecanoylphorbol-13-acetate-induced mRNA transcript. J Virol. 1983 Oct;48(1):135–148. doi: 10.1128/jvi.48.1.135-148.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King W., Dambaugh T., Heller M., Dowling J., Kieff E. Epstein-Barr virus DNA XII. A variable region of the Epstein-Barr virus genome is included in the P3HR-1 deletion. J Virol. 1982 Sep;43(3):979–986. doi: 10.1128/jvi.43.3.979-986.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King W., Thomas-Powell A. L., Raab-Traub N., Hawke M., Kieff E. Epstein-Barr virus RNA. V. Viral RNA in a restringently infected, growth-transformed cell line. J Virol. 1980 Nov;36(2):506–518. doi: 10.1128/jvi.36.2.506-518.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyte J., Doolittle R. F. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982 May 5;157(1):105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- Laux G., Freese U. K., Bornkamm G. W. Structure and evolution of two related transcription units of Epstein-Barr virus carrying small tandem repeats. J Virol. 1985 Dec;56(3):987–995. doi: 10.1128/jvi.56.3.987-995.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehrach H., Diamond D., Wozney J. M., Boedtker H. RNA molecular weight determinations by gel electrophoresis under denaturing conditions, a critical reexamination. Biochemistry. 1977 Oct 18;16(21):4743–4751. doi: 10.1021/bi00640a033. [DOI] [PubMed] [Google Scholar]
- Lin H. C., Lei S. P., Wilcox G. An improved DNA sequencing strategy. Anal Biochem. 1985 May 15;147(1):114–119. doi: 10.1016/0003-2697(85)90016-8. [DOI] [PubMed] [Google Scholar]
- Miller G., Lipman M. Release of infectious Epstein-Barr virus by transformed marmoset leukocytes. Proc Natl Acad Sci U S A. 1973 Jan;70(1):190–194. doi: 10.1073/pnas.70.1.190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norrander J., Kempe T., Messing J. Construction of improved M13 vectors using oligodeoxynucleotide-directed mutagenesis. Gene. 1983 Dec;26(1):101–106. doi: 10.1016/0378-1119(83)90040-9. [DOI] [PubMed] [Google Scholar]
- Ploegh H. L., Orr H. T., Strominger J. L. Major histocompatibility antigens: the human (HLA-A, -B, -C) and murine (H-2K, H-2D) class I molecules. Cell. 1981 May;24(2):287–299. doi: 10.1016/0092-8674(81)90318-4. [DOI] [PubMed] [Google Scholar]
- Rabson M., Gradoville L., Heston L., Miller G. Non-immortalizing P3J-HR-1 Epstein-Barr virus: a deletion mutant of its transforming parent, Jijoye. J Virol. 1982 Dec;44(3):834–844. doi: 10.1128/jvi.44.3.834-844.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowe D., Heston L., Metlay J., Miller G. Identification and expression of a nuclear antigen from the genomic region of the Jijoye strain of Epstein-Barr virus that is missing in its nonimmortalizing deletion mutant, P3HR-1. Proc Natl Acad Sci U S A. 1985 Nov;82(21):7429–7433. doi: 10.1073/pnas.82.21.7429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sample J., Hummel M., Braun D., Birkenbach M., Kieff E. Nucleotide sequences of mRNAs encoding Epstein-Barr virus nuclear proteins: a probable transcriptional initiation site. Proc Natl Acad Sci U S A. 1986 Jul;83(14):5096–5100. doi: 10.1073/pnas.83.14.5096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sample J., Tanaka A., Lancz G., Nonoyama M. Identification of Epstein-Barr virus genes expressed during the early phase of virus replication and during lymphocyte immortalization. Virology. 1984 Nov;139(1):1–10. doi: 10.1016/0042-6822(84)90324-6. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skare J., Farley J., Strominger J. L., Fresen K. O., Cho M. S., zur Hausen H. Transformation by Epstein-Barr virus requires DNA sequences in the region of BamHI fragments Y and H. J Virol. 1985 Aug;55(2):286–297. doi: 10.1128/jvi.55.2.286-297.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Speck S. H., Pfitzner A., Strominger J. L. An Epstein-Barr virus transcript from a latently infected, growth-transformed B-cell line encodes a highly repetitive polypeptide. Proc Natl Acad Sci U S A. 1986 Dec;83(24):9298–9302. doi: 10.1073/pnas.83.24.9298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speck S. H., Strominger J. L. Analysis of the transcript encoding the latent Epstein-Barr virus nuclear antigen I: a potentially polycistronic message generated by long-range splicing of several exons. Proc Natl Acad Sci U S A. 1985 Dec;82(24):8305–8309. doi: 10.1073/pnas.82.24.8305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terhorst C., Parham P., Mann D. L., Strominger J. L. Structure of HLA antigens: amino-acid and carbohydrate compositions and NH2-terminal sequences of four antigen preparations. Proc Natl Acad Sci U S A. 1976 Mar;73(3):910–914. doi: 10.1073/pnas.73.3.910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D., Liebowitz D., Kieff E. An EBV membrane protein expressed in immortalized lymphocytes transforms established rodent cells. Cell. 1985 Dec;43(3 Pt 2):831–840. doi: 10.1016/0092-8674(85)90256-9. [DOI] [PubMed] [Google Scholar]
- Weigel R., Miller G. Major EB virus-specific cytoplasmic transcripts in a cellular clone of the HR-1 Burkitt lymphoma line during latency and after induction of viral replicative cycle by phorbol esters. Virology. 1983 Mar;125(2):287–298. doi: 10.1016/0042-6822(83)90202-7. [DOI] [PubMed] [Google Scholar]
- van Santen V., Cheung A., Kieff E. Epstein-Barr virus RNA VII: size and direction of transcription of virus-specified cytoplasmic RNAs in a transformed cell line. Proc Natl Acad Sci U S A. 1981 Mar;78(3):1930–1934. doi: 10.1073/pnas.78.3.1930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Heijne G. A new method for predicting signal sequence cleavage sites. Nucleic Acids Res. 1986 Jun 11;14(11):4683–4690. doi: 10.1093/nar/14.11.4683. [DOI] [PMC free article] [PubMed] [Google Scholar]