Abstract
The fission yeast Rad3p checkpoint protein is a member of the phosphatidylinositol 3-kinase-related family of protein kinases, which includes human ATMp. Mutation of the ATM gene is responsible for the disease ataxia-telangiectasia. The kinase domain of Rad3p has previously been shown to be essential for function. Here, we show that although this domain is necessary, it is not sufficient, because the isolated kinase domain does not have kinase activity in vitro and cannot complement a rad3 deletion strain. Using dominant negative alleles of rad3, we have identified two sites N-terminal to the conserved kinase domain that are essential for Rad3p function. One of these sites is the putative leucine zipper, which is conserved in other phosphatidylinositol 3-kinase-related family members. The other is a novel motif, which may also mediate Rad3p protein–protein interactions.
INTRODUCTION
Checkpoint pathways ensure the correct temporal order of the cell cycle (Hartwell and Weinert, 1989) and are an evolutionarily conserved feature of eukaryotic cells. The Schizosaccharomyces pombe checkpoint kinase Rad3p (unrelated to the Saccharomyces cerevisiae DNA helicase Rad3p) acts to delay cell cycle events in response to DNA damage or incomplete DNA replication (Jimenez et al., 1992; Seaton et al., 1992; Bentley et al., 1996). Unlike wild-type cells, rad3Δ cells do not arrest the cell cycle in response to DNA damage or blocked DNA replication and lose viability rapidly under these conditions (Al-Khodairy and Carr, 1992). Two human proteins with similarities to fission yeast Rad3p, ATRp (Bentley et al., 1996; Cimprich et al., 1996) and ATMp (Savitsky et al., 1995a,b), also function in checkpoint control (Painter and Young, 1980; Beamish and Lavin, 1994; Cliby et al., 1998). In humans, mutation of the ATM gene causes the autosomal recessive disease ataxia-telangiectasia (A-T), in which patients suffer from a variety of symptoms, including a predisposition to cancer (Harnden, 1994). In mice, deletion of the ATM gene greatly increases the frequency of thymic lymphomas (Barlow et al., 1996; Xu et al., 1996). Thus, normal checkpoints appear to be required to prevent cancer in mammalian cells. Fission yeast provides the opportunity to study these checkpoint kinases in a simple, genetically tractable model system.
Fission yeast Rad3p and human ATMp and ATRp are members of a large family of structurally and functionally similar proteins from diverse organisms (Savitsky et al., 1995a; Bentley et al., 1996, Cimprich et al., 1996). Other members of the family include human DNA-PKcs, the catalytic subunit of DNA-dependent protein kinase (Hartley et al., 1995), and FKBP12–rapamycin-associated protein (Brown et al., 1994; Chiu et al., 1994; Sabatini et al., 1994); S. cerevisiae Mec1p (Weinert, 1992; Kato and Ogawa, 1994), Tel1p (Greenwell et al., 1995; Morrow et al., 1995), Tor1p, and Tor2p (Heitman et al., 1991; Kunz et al., 1993); and Drosophila Mei-41p (Hari et al., 1995). S. pombe Tel1p has also recently been identified (Naito et al., 1998). At their C termini, these large proteins (>200 kDa) all contain a kinase domain related to phosphatidylinositol 3-kinases (PI3Ks). Despite this similarity, none of the PI3-kinase-related (PI3KR) proteins have been shown to phosphorylate lipids. ATMp, ATRp, DNA-PK, and Rad3p are all capable of autophosphorylation (Bentley et al., 1996; Chan and Lees-Miller, 1996; Cliby et al., 1998; Scott et al., 1998), and are also known to directly phosphorylate other protein substrates as well (Lees-Miller et al., 1992; Banin et al., 1998; Canman et al., 1998, Martinho et al., 1998). In addition to the kinase domain, many PI3KR proteins contain a putative leucine zipper motif, which may mediate homo- or heterodimerization.
The PI3KR family can be subdivided on the basis of sequence similarity (Keith and Schreiber, 1995; Bentley et al., 1996). In such a phylogenetic tree, S. pombe Rad3p, human ATRp, Drosophila Mei-41p, and S. cerevisiae Mec1p form one subgroup (the ATR group), whereas human ATMp, S. cerevisiae Tel1p, and S. pombe Tel1p are in another distinct but closely related cluster (the ATM group). In yeast and in higher eukaryotes, ATR and ATM-like proteins appear to have overlapping molecular functions in cell cycle control and telomere regulation. For example, S. cerevisiae mec1 tel1 and S. pombe rad3 tel1 double mutants have more severe defects than either single mutant (Morrow et al., 1995; Naito et al., 1998). Moreover, overexpression of S. cerevisiae tel1+ partially complements the radiation sensitivity of mec1 mutants (Morrow et al., 1995), and human ATR overexpression can suppress A-T cell defects (Cliby et al., 1998). In contrast, the remaining family members are more distantly related and have not been shown to overlap functionally with the ATM and ATR subgroups. S. cerevisiae Tor1p and Tor2p and human FKBP12–rapamycin-associated protein regulate G1 progression, whereas DNA-PKcs is involved in V(D)J recombination and repair of double-stranded DNA breaks.
Although not directly involved in checkpoint control, DNA-PKcs serves as a paradigm for understanding the regulation of other kinases in the family because the biochemistry of its regulation is relatively well understood (Jeggo et al., 1995). In vitro, DNA-PKcs is activated by binding double-stranded DNA ends in the presence of a heterodimer, composed of Ku70 and Ku86 subunits (Lieber et al., 1997). In this way, the activity of this kinase is dependent both on specific DNA structures and on regulatory cofactors.
Analogously, the ATR and ATM-like PI3KR kinases may also require regulatory cofactors and may be activated by DNA structures. Although the activating structures and regulatory cofactors for these kinases have not been identified, it is known that ionizing radiation enhances the ATM-dependent phosphorylation of p53 in mammalian cells (Banin et al., 1998; Canman et al., 1998). Similarly, a number of fission yeast proteins, including Hus1p, Chk1p, and Cds1p, are phosphorylated in response to DNA damage in a Rad3p-dependent manner (Walworth and Bernards, 1996; Kostrub et al., 1998; Lindsay et al., 1998), suggesting that Rad3p activity could also be activated by DNA damage. The phosphorylation of these proteins is also dependent on other fission yeast checkpoint proteins, which do not contain recognizable kinase domains, such as Rad1p, Rad9p, Rad17p, and Rad26p (Sunnerhagen et al., 1990; Enoch et al., 1992; Rowley et al., 1992; Al-Khodairy et al., 1994). These proteins, as well as others known to be involved in checkpoint control (McFarlane et al., 1997; Saka et al., 1997; Willson et al., 1997), are candidate cofactors or regulators of Rad3p. For example, they may stimulate Rad3p kinase activity by directing the localization of Rad3p to activating DNA structures such as sites of DNA damage, just as the Ku proteins regulate DNA-PKcs.
The Rad3p kinase domain, which comprises <15% of the protein, is essential for Rad3p function in cell cycle checkpoint control (Bentley et al., 1996). Little is known about the function of Rad3p sequences outside the kinase domain. In this study we demonstrate the importance of sequences outside the kinase domain of Rad3p using a combination of genetic and biochemical methods. We find that they are required for full complementation of the checkpoint defects of rad3Δ cells and are also required for the catalytic activity of Rad3p. Using a genetic assay we have shown that the Rad3p N terminus contains at least two important sites, the leucine zipper and another novel site. Our results differ strikingly from previous studies of ATMp, which indicated that the isolated ATMp kinase domain is sufficient for complementation of A-T cell defects and for kinase activity (Baskaran et al., 1997; Morgan et al., 1997). Thus, there may be important differences in the regulation of the different classes of proteins within the PI3KR family.
MATERIALS AND METHODS
Strains
The strains used in this study are as follows: TE235 (leu1–32 h−), TE236 (leu1–32 ura4-D18 h−), and TE890 (rad3::ura4+ leu1–32 ura4-D18 h−). TE890 is a derivative of the rad3::ura4+ strain created by Bentley et al. (1996). These strains were transformed with various rad3 plasmids, which are listed in Table 1. To assay complementing activity of rad3 alleles (see Figures 1 and 7), constructs were introduced into strain TE890. To assay dominant negative activity of rad3 alleles (see Figures 3–6), constructs were introduced into strain TE235. To measure kinase activity (see Figures 2 and 7), constructs were introduced into strain TE890. For the coimmunoprecipitation experiments (see Figure 8), strain TE236 was transformed sequentially with two differentially marked plasmids, and leu+ura+ transformants were selected. In all cases, Rad3p expression from the transformed plasmids was controlled by the thiamine-repressible promoter nmt1+ or a modified version of nmt1+ termed the nmt41 promoter (Basi et al., 1993). Under the nmt1+ promoter, proteins are induced ∼80-fold when thiamine is removed from the media (Maundrell, 1990). The nmt41 promoter has a mutated TATA box such that proteins are only induced ∼12-fold (Basi et al., 1993; Forsburg, 1993). Transformations were performed by electroporation (Prentice, 1992). Media and growth conditions were as previously described (Moreno et al., 1991). Transformed strains were routinely maintained in Edinburgh minimal medium containing thiamine. To assay activity of Rad3 proteins, thiamine was washed out of the media to induce expression. It takes ∼16 h to induce full expression of the nmt promoter by removing thiamine and ∼20 h to repress it with the addition of thiamine (Maundrell, 1990). Fluorescence microscopy was performed using a Zeiss (Thornwood, NY) Axiophot microscope and a Photonic C1966 microscope image processor (Hamamatsu Photonic Systems, Bridgewater, NJ).
Table 1.
DNA plasmids used in this paper
| Plasmid number | Plasmid name | Source |
|---|---|---|
| pTE101 | rep3x | |
| pTE119 | rep41-HAtag | Bentley et al., 1996 |
| pTE120 | rep42-myctag | Bentley et al., 1996 |
| pTE157 | rep1-rad3+ | Bentley et al., 1996 |
| pTE165 | pET3His-rad3-Cterm | This study |
| pTE446 | rep1-rad3-C725 | This study |
| pTE521 | rep41-HA-rad3+ | This study |
| pTE541 | rep1-HA-rad3+ | This study |
| pTE672 | rep1-HA-rad3-C725 | This study |
| pTE696 | rep1-rad3-N775 | This study |
| pTE697 | rep1-HA-rad3-N775 | This study |
| pTE698 | rep1-rad3-N690 | This study |
| pTE699 | rep1-HA-rad3-N690 | This study |
| pTE700 | rep1-rad3-N541 | This study |
| pTE701 | rep1-HA-rad3-N541 | This study |
| pTE706 | rep1-rad3-P | This study |
| pTE707 | rep1-HA-rad3-P | This study |
| pTE709 | rep1-HA-rad3-N775-P | This study |
| pTE710 | rep1-rad3-N775-P | This study |
| pTE715 | rep1-HA-rad3-LZ | This study |
| pTE716 | rep1-rad3-LZ | This study |
| pTE717 | rep1-HA-rad3-N775-LZ | This study |
| pTE718 | rep1-rad3-N775-LZ | This study |
| pTE743 | rep1-rad3-LZ-KD | This study |
| pTE745 | rep1-rad3-LZ-P-KD | This study |
| pTE746 | rep41-HA-rad3-LZ | This study |
| pTE747 | rep42-myc-rad3-LZ | This study |
| pTE748 | rep42-myc-rad3+ | This study |
| pTE750 | rep1-rad3-P-KD | This study |
| pTE783 | rep1-HA-rad3-KD | This study |
| pTE784 | rep1-HA-rad3-C328 | This study |
| pTE785 | rep1-HA-rad3-C261 | This study |
| pTE786 | rep1-HA-rad3-C549 | This study |
| pTE787 | rep1-HA-rad3-C488 | This study |
| pTE791 | rep1-rad3-KD | This study |
| pTE792 | rep42-myc-chk1+ | This study |
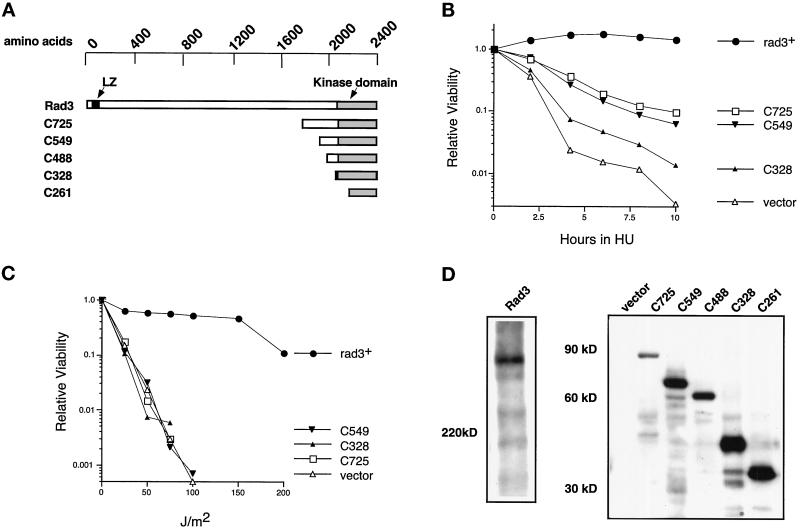
Figure 1.
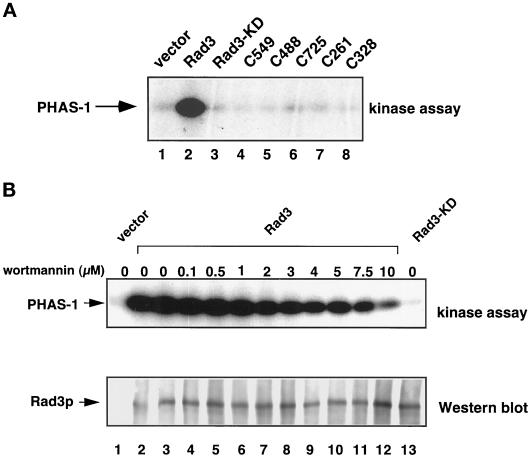
The isolated Rad3p kinase domain does not complement rad3Δ cells. (A) Rad3p kinase domain fragments are diagrammed approximately to scale. The positions of the kinase domain (shaded) and the leucine zipper motif (black) are indicated. (B) Relative viability of rad3Δ cells (TE890) expressing HA-tagged mutant Rad3 proteins, after indicated periods of incubation in 10 mM HU. SE analysis of these curves indicates that their positions are significant. The plasmids used are rep1-HA-rad3+(pTE541), rep1-HA-rad3-C725 (pTE672), rep1-HA-rad3-C549 (pTE786), rep1-HA-rad3-C328 (pTE784), and rep3x (pTE101). These proteins are overexpressed because their expression is controlled by the full-strength nmt1+ promoter. Note that rep1-HA-rad3+ fully complements the rad3Δ strain, exhibiting wild-type levels of viability in both HU and UV assays (our unpublished results). (C) Relative viability of the same strains after irradiation with the indicated doses of UV. The results shown are the average of two independent experiments. No colonies were recovered at doses at which no points are shown. (D) Expression of full-length Rad3p and Rad3p C-terminal fragments. Left panel, Western blot of total extracts prepared from rad3::ura4+ leu1–32 h− cells (TE890) and cells transformed with rep1-HA-rad3+ (pTE541). Right panel, Western blot of total extracts prepared from the transformants used in B and C, as well as rad3Δ cells (TE890) transformed with rep1-HA-rad3-C261 (pTE785) and rep1-HA-rad3-C488 (pTE787). Both blots were probed with monoclonal antibodies specific for the HA epitope tag (12CA5). Note that direct comparison of levels of expression of full-length to truncations is not feasible, because the different proteins are resolved on different percentage gels and are likely to have different transfer efficiencies. However, we estimate that the truncations are three- to fivefold more abundant than the full-length protein.
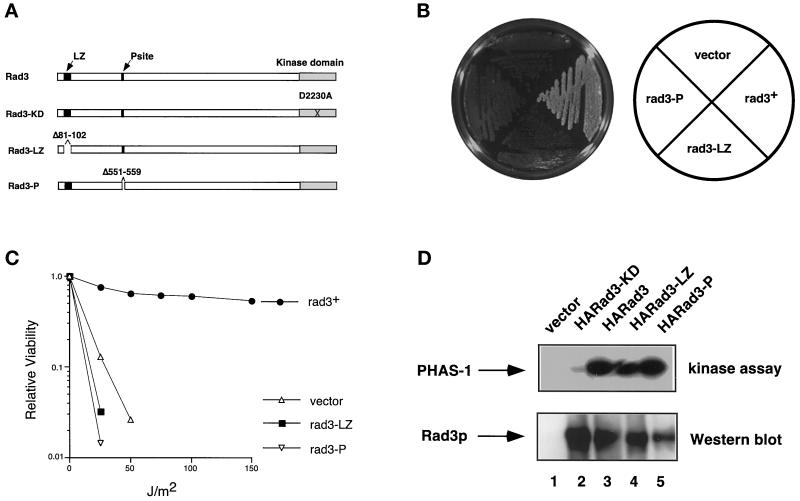
Figure 7.
Leucine zipper and P-site are required for normal function of Rad3 protein. (A) Rad3 proteins are diagrammed approximately to scale. See Figure 5 for explanation of symbols. (B) rad3Δ (TE890) cells transformed with rep3x (pTE101), rep1-HA-rad3+ (pTE541), rep1-HA-rad3-LZ (pTE716), or rep1-HA-rad3-P (pTE706) were grown on plates containing 10 mM HU. (C) Relative viability of the same strains after exposure to the indicated doses of UV irradiation. Results shown are the average of two independent experiments. (D) Rad3-LZp and Rad3-Pp are active kinases. Anti HA-immunopreciptates were prepared from rad3Δ (TE890) cells transformed with the plasmids indicated above. Half of each immunoprecipitate was assayed for PHAS-1 kinase activity, and the other half was subjected to SDS-PAGE, Western blotted, and probed with HA monoclonal antibody.
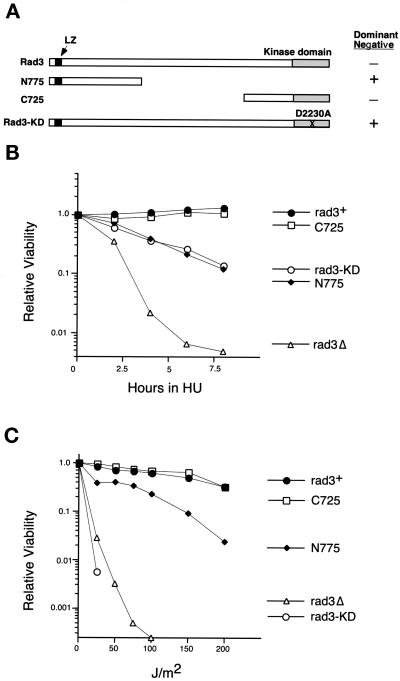
Figure 3.
Overexpression of an N-terminal fragment of Rad3p disrupts the response of wild-type cells to HU and UV. (A) Diagram of mutant Rad3 proteins, drawn approximately to scale. See Figure 1 for explanation of symbols. Dominant negative activity of each mutant protein is indicated. (B) Relative viability after treatment with 10 mM HU of wild-type cells (TE235) expressing rad3+ (pTE157), rad3-C725 (pTE446), rad3-KD (pTE791), rad3-N775 (pTE696), and rep3x (pTE101). (C) Relative viability of the same strains with indicated doses of UV irradiation. No colonies were recovered at doses at which no points are shown. The results shown are the average of two independent experiments.
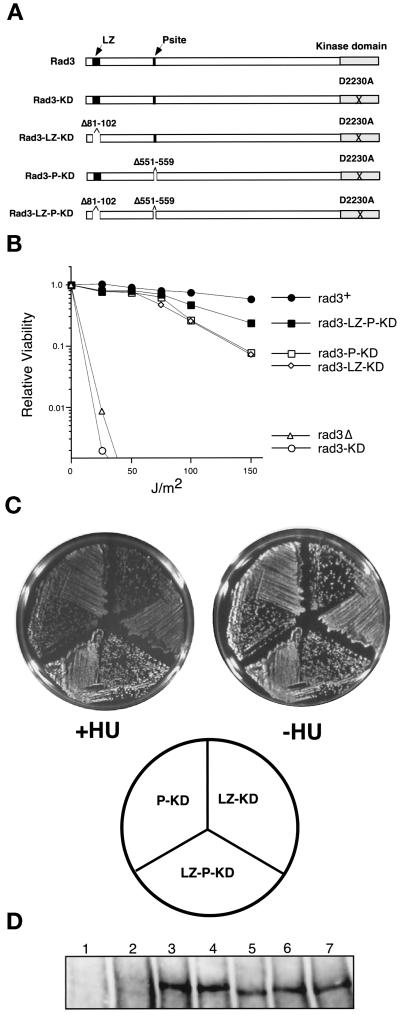
Figure 6.
LZ and P-site mutations are additive. (A) Diagram of mutant Rad3 proteins. See Figures 1 and 4 for explanation of symbols. The position of the mutation (D2230A) is indicated with a black X. (B) The LZ and P mutations substantially reduce the dominant negative activity of the Rad3-KD allele. Relative viability after indicated doses of UV irradiation of rad3Δ cells (TE890) expressing empty vector (pTE101) and wild-type cells (TE235) overexpressing rad3+ (pTE157), rad3-KD (pTE791), rad3-LZ-KD (pTE743), rad3-P-KD (pTE750), and rad3-LZ-P-KD (pTE745). The results shown are the average of two independent experiments. Note that the difference between the double and triple mutants is significant at 150 J/m2. (C) The triple mutant Rad3-LZ-P-KD is a weaker dominant negative than the double mutants Rad3-LZ-KD and Rad3-P-KD on plates containing HU. Wild-type (TE235) cells overexpressing rad3-LZ-KD, rad3-P-KD, and rad3-LZ-P-KD were grown for 3 d on plates with (+HU) or without (−HU) 10 mM HU. (D) Expression of the Rad3p proteins. Rad3p polyclonal antibodies (HM126) were used to Western blot extracts made from rad3Δ cells transformed with rep3x vector control (lane 1); wild-type cells transformed with rep3x vector control (lane 2), rep1-rad3+ (lane 3), rep1-rad3-KD (lane 4), rep1-rad3-LZ-KD (lane 5), rep1-rad3-P-KD (lane 6), and rep1-rad3-LZ-P-KD (lane 7). Note that endogenous Rad3p is not detected under these conditions (lane 2).
Figure 2.
Characterization of Rad3p kinase activity. (A) The isolated kinase domain of Rad3p is not catalytically active. HA-tagged Rad3 proteins were immunoprecipitated from rad3Δ (TE890) transformed with rep3x (pTE101), rep1-HA-rad3+ (pTE541), rep1-HA-rad3-C549 (pTE786), rep1-HA-rad3-C488 (pTE787), rep1-HA-rad3-C725 (pTE672), rep1-HA-rad3-C261 (pTE785), rep1-HA-rad3-C328 (pTE784), or rep1-HA-rad3-KD (pTE783). These proteins are overexpressed because their expression is under the control of the nmt1+ promoter. Immunoprecipitates were divided in half and either assayed for their ability to phosphorylate PHAS-1 (top panel) or Western-blotted and probed with anti-HA antibody (our unpublished data). (B) Rad3p is relatively insensitive to wortmannin. HA-tagged Rad3 proteins were immunoprecipitated from rad3Δ (TE890) transformed with rep3x (pTE101), rep1-HA-rad3+ (pTE541), or rep1-HA-rad3-KD (pTE783). HA immunoprecipitates from the indicated strains were assayed for their ability to phosphorylate PHAS-1 in the presence of increasing concentrations of wortmannin.
Figure 8.
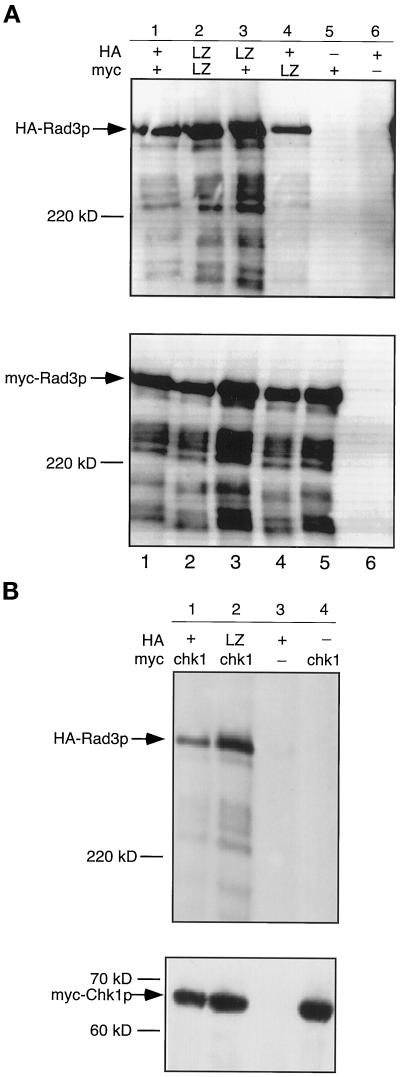
Rad3p leucine zipper motif is not required for self-association or for interaction with Chk1p. Wild-type cells (TE236) were cotransformed with the indicated rad3 and/or chk1 alleles, and myc immunoprecipitations were performed on extracts from the indicated strains. Samples were split, resolved on SDS-PAGE gels, blotted, and probed with either anti-HA antibodies (top) or anti-myc antibodies (bottom). (A) Self-association assay. Lane 1, rep41-HA-rad3+ (pTE521) and rep42-myc-rad3+ (pTE748); lane 2, rep41-HA-rad3-LZ (pTE746) and rep42-myc-rad3-LZ (pTE747); lane 3, rep41-HA-rad3-LZ and rep42-myc-rad3+; lane 4, rep41-HA-rad3+ and rep42-myc-rad3-LZ; lane 5, rep41-HAtag vector (control) (pTE119) and rep42-myc-rad3+; lane 6, rep41-HA-rad3+ and rep42-myctag vector (control) (pTE120). (B) Chk1p/Rad3p coimmunoprecipitation assay. Lane 1, rep41-HA-rad3+ and rep42-myc-chk1+ (pTE792); lane 2, rep41-HA-rad3-LZ and rep42-myc-chk1+; lane 3, rep41-HA-rad3+and rep42-myctag vector (control); lane 4, rep41-HAtag vector and rep42-myc-chk1+.
Description of Plasmids
The plasmids used in this study are listed in Table 1, and oligonucleotides used in their construction are listed in Table 2. The following is a brief description of the plasmids used in this study; detailed construction methods are available on request. Standard subcloning methods were used for all constructions. PCR reactions were carried out using Pfu polymerase according manufacturer’s instructions (Stratagene, La Jolla, CA), and all PCR-amplified regions were sequenced. Protein expression from these plasmids was driven by either the nmt1+ promoter (rep1) or the nmt41 (rep41 or rep42) promoter (Basi et al., 1993). The rep1 and rep41 plasmids are marked with the S. cerevisiae LEU2 gene, which complements S. pombe leu1–32 mutants, and the rep42 plasmids are marked with the S. pombe ura4+ gene. Site positions are provided for restriction enzymes, using the A of the starting rad3 ATG codon as position 1. rep1-rad3+ was described previously (Bentley et al., 1996) and contains an NdeI site at position −2, and PstI, SalI, and BamHI in the 3′ multicloning site. The following rad3+ sites were used in cloning: BamHI (4428), BsaBI (1106), BsmI (6391), DraIII (5534), EarI (6188), EcoRV (5711), MluI (2062), NsiI (2321), and XhoI (4987). A BstEII site in the rep vector was also used. To generate rep42-myc-chk1+, the NdeI–SacI piece of rep1-chk1+ (Carr et al., 1995) was cloned into the rep42-myctag vector backbone (pTE120).
Table 2.
Oligonucleotides used in this study
| Oligo number | Oligo sequence |
|---|---|
| 54 | GGAATCCTGGCATATCATCAATTG |
| 55 | GCAGCTTGAATGGGCTTCCATAGT |
| 154 | TAATCATATGTATC |
| 155 | TCGAGATACATATGAT |
| 196 | TATGTTAAATGAAGGATCCG |
| 197 | GATCCTTCATTTAACA |
| 198 | TATGATGAATTCTCTTCA |
| 199 | TTTTGAAGAGAATTCATCA |
| 200 | CGGTGGAGCCAGCATTGCTCAAA |
| 201 | TTTAACAGAGCTCGAAATTATGTCGGG |
| 210 | GCATTAATCCCATGTACCCGTACGATGTTCCTGAC |
| 217 | TAAATAGTAGCTCGAG |
| 225 | CGCGTCATTAACTCGAG |
| 226 | TCGACACGAGTTAATGA |
| 227 | TCGACTCGAGCTACTATTTATGCA |
| 234 | TATGGCCTTAAGCCAAATGAT |
| 235 | ATCATTTGGCTTAAGGCCA |
| 236 | TATGAGTATGCCAAAATTGCTCACACT |
| 237 | GTGAGCAATTTTGGCATACTCA |
| 263 | GAAGAGGAAATTATGTCGACTTATTTTAGCAATGC |
| 264 | GAATTTATCTCTCCTAAGATGCGGGAGATTAAATCATCTGT |
| 265 | ACAGATGATTTAATCTCCCGCATCTTAGGAGAGATAAATTC |
Tagged Wild-Type Rad3.
To construct tagged versions of wild-type rad3+ under control of the moderate strength nmt promoter, the NdeI–SalI piece from rep1-rad3+ (pTE157) was ligated into different vectors; for pTE521, rep41-HA-rad3+, the rep41-HAtag vector was used (pTE119); for pTE748, the rep42-myctag vector (pTE120) was used. To construct pTE541, rep1-HA-rad3+, in which hemagglutinin (HA)-tagged Rad3 expression is controlled by the wild-type nmt1+ promoter, the HA tag from pTE119 was PCR amplified using oligos 210 and 55, such that the HA tag was flanked on the 5′ end with an AseI site and at the 3′ end with an NdeI site. The resulting fragment was digested with AseI and NdeI and cloned into the NdeI site of rep1-rad3+ (pTE157).
C-terminal Fragments.
pTE446, rep1-C725, is a deletion of rad3+ sequences that encode amino acids 1–1660. Sequences between NdeI and XhoI were replaced with an NdeI–XhoI linker (oligos 154 and 155), which adds a starting methionine such that the sequence starts MYLE (Y = aa 1661). pTE672, rep1-HA-C725, contains the NdeI–SalI fragment of pTE446 in the rep1-HA tag backbone of pTE541. pTE786, rep1-HA-C549, is a deletion of rad3+ sequences that encode amino acids 1–1837. Sequences between NdeI and DraIII were replaced with an NdeI–DraIII linker (oligos 236 and 237), which adds a starting methionine such that the sequence after the HA tag starts MSMPK (S = aa 1838). pTE787, rep1-HA-C488, is a deletion of rad3+ sequences that encode the first 1898 amino acids of Rad3p such that the sequence after the N-terminal HA tag starts MALSQ (A = aa 1899). It was constructed by using an NdeI–EcoRV linker (oligos 234 and 235) to replace rad3+ sequences from NdeI to EcoRV by performing a three-way ligation with the linker, the EcoRV–SalI fragment of pTE791, and the SalI–NdeI fragment of the rep1HA tagging vector from pTE541. pTE784, rep1-HA-C328, is a deletion of rad3+ sequences that encode amino acids 1–2058. Sequences between NdeI and EarI were replaced with an NdeI–EarI linker (oligos 198 and 199), which adds a starting methionine such that the sequence after the HA tag begins MMNSL (second M = aa 2059). pTE785, rep1-HA-C261, contains a deletion of rad3+ sequences that encode amino acids 1–2125. Sequences between NdeI and BsmI were replaced with an NdeI–BsmI linker (oligos 196 and 197) such that the sequence after the HA tag starts MLNEGS (L = aa 2126, also GS is mutated from EC in wild-type Rad3p).
N-terminal Fragments.
pTE696, rep1-rad3-N775, is a deletion of rad3+ sequences that encode amino acids 776-2386. Sequences between NsiI and SalI of rep1-rad3+ (pTE157) were replaced with an NsiI-SalI linker (oligos 217 and 227), which created an in-frame stop site after amino acid 775. pTE697, rep1-HA-rad3-N775, contains the same 3′ truncation as pTE696, in the rep1-HA backbone from pTE541. pTE698, rep1-rad3-N690, is a deletion of rad3+ sequences 691-2386. Sequences between MluI and SalI were replaced with a MluI–SalI linker (oligos 225 and 226), which created a stop codon after amino acid 690. pTE699, rep1-HA-rad3-N690, is the same 3′ truncation as in pTE698, in the rep1-HA backbone from pTE541. pTE700, rep1-rad3-N541, is a deletion of rad3+ sequences 542-2386. A stop codon and a SalI site were inserted after amino acid 541 using PCR amplification with oligos 263 and 54, followed by subcloning. pTE701, rep1-HA-N541, contains the same 3′ truncation as pTE700 in the rep1-HA backbone from pTE541.
Leucine Zipper Deletion and Derivatives.
pTE718, rep1-rad3-N775-LZ, contains the N775 C-terminal truncation with a 22 (Δ81–102)-amino-acid deletion of the leucine zipper. It was made by two-step PCR, using primers 264 and 55 and 265 and 54. Oligos 264 and 265 are primers that created the 22-amino-acid deletion of the leucine zipper. These products were then amplified again in the same tube with primers 54 and 55. A piece containing the deletion was then subcloned into pTE696, rep1-rad3-N775. pTE717, rep1-HA-rad3-N775-LZ, contains the same 22-amino-acid deletion (Δ81–102) as pTE718 but in the rep1-HA backbone of pTE541. pTE716, rep1-rad3-LZ, contains the leucine zipper deletion (pTE718) in full-length rad3 in the rep1 vector. pTE715, rep1-HA-rad3-LZ, contains the leucine zipper deletion (from pTE718) in full-length rad3 in the rep1 HA tagging vector. pTE746, rep41-HA-rad3-LZ, contains the leucine zipper deletion (from pTE718) in full-length rad3 in the rep41 HA tagging vector. pTE747, rep42-myc-rad3-LZ, contains the full-length rad3 sequence with the leucine zipper deletion in the rep42 myc tagging vector (pTE120).
P-Site Deletion and Derivatives.
pTE706, rep1-rad3-P, which harbors a nine-amino-acid deletion (Δ551–559) marked by a silent SacI site (1642), was created in a series of steps, which included whole-plasmid PCR mutagenesis with primers 200 and 201 using the protocol described in Stratagene’s QuikChange site-directed mutagenesis kit instruction manual. pTE707, rep1-HA-rad3-P, consists of the NdeI–SalI piece of pTE706 in the rep1-HA backbone of pTE541. pTE710, rep1-rad3-N775-P, which contains the P-site deletion in the N775 truncation construct, was created by ligating the NdeI–NsiI insert piece from pTE707 into the similarly cut backbone of pTE696.
Kinase-dead Alleles of rad3.
pTE791, rep1-rad3-KD, which encodes a kinase-dead form of Rad3p under the control of the nmt1+ promoter, was constructed by replacing the BamHI–PstI fragment of pTE157 (rep1-rad3) with the equivalent piece from a vector containing the rad3.a allele (Bentley et al., 1996), which contains an AT→CG mutation at 6689–6690 and thus encodes a kinase-dead (D2230A) form of Rad3p. pTE783, rep1-HA-rad3-KD, which encodes an N-terminally HA-tagged kinase-dead form of Rad3p, was constructed by cloning the BamHI–BamHI piece of pTE791 into the similarly cut backbone of pTE541, rep1-HA-rad3. pTE743, rep1-rad3-LZ-KD, which contains the leucine zipper deletion in the context of the full-length rad3 kinase-dead D2230A allele, was constructed by subcloning the NdeI–NsiI insert piece of pTE715 into the similarly cut backbone of pTE791. pTE750 contains the P-site deletion in full-length rad3 dominant negative D2230A allele and was made by subcloning the NdeI–NsiI insert from pTE710 into the similarly cut backbone of pTE791. pTE745, rep1-rad3-LZ-P-KD, contains the leucine zipper deletion, the P-site deletion, and the kinase-dead D2230A mutation in the context of full-length rad3. This construct was made by using a three-way ligation to combine the following appropriate pieces: BstEII–NsiI of pTE791, BstEII–BsmI of pTE718, and BsmI–NsiI of pTE707.
Hydroxyurea Sensitivity Assays
Liquid Assays.
In these assays, rad3 alleles were overexpressed from the full-strength nmt1+ promoter. Overnight cultures of cells were grown in the presence of thiamine to repress transcription of rad3 alleles. To induce transcription, these cultures were diluted into media lacking thiamine and grown for 20 h to an OD595 between 0.1 and 0.2. At this time, 10 mM hydroxyurea (HU) was added, and the cultures were incubated at 29°C with shaking. Samples were taken every 2 h, plated in duplicate on solid media containing thiamine, and incubated at 29°C for 3–5 d. Two time courses were done for each experiment, and the data points on the graphs represent the average of the results of these time courses.
Plate Assays.
As in the liquid assays, rad3 alleles were expressed from the full-strength nmt1+ promoter. Strains were initially streaked onto plates of media containing thiamine. These plates were incubated at 29°C for 2–3 d and replica plated onto media lacking thiamine to induce expression of the rad3 alleles. After 1–2 d of growth at 29°C, these plates were replica plated again, to plates lacking thiamine and containing 10 mM HU. Pictures of the plates were taken after 3 d of growth on HU at 29°C.
UV Sensitivity Assays
For UV sensitivity assays, Rad3p expression was induced as described above for the HU liquid assays. After 20 h of induction, samples were plated on solid media containing thiamine. The plates were dried at room temperature for 1–2 h, irradiated in a UV Stratalinker 2400 with doses of UV irradiation from 0 to 200 J/m2, and then incubated at 29°C for 3–5 d. It takes at least 20 h to repress the nmt promoter under these conditions so at the time of irradiation Rad3 proteins are still expressed at high levels; however, for most of growth afterward at 29°C, the thiamine in the plates represses expression of Rad3p from the nmt promoter. The numbers of colonies on duplicate plates for each UV dose were averaged, and viability for each dose was calculated using the number of colonies on the mock-irradiated (0 J/m2) plate as a viability of 1.0. The graphs presented in this paper are each the average of results from two dosage series.
Fluorescence-activated Cell-sorting (FACS) Analysis
Samples for FACS analysis were prepared as described (Sazer and Sherwood, 1990), except that ethanol-fixed cells were stored at −20°C before processing for propidium iodide staining. FACS analysis was performed with a FACSCalibur cytometer and Cell Quest version 3.1f software (Becton Dickinson, San Jose, CA). Ten thousand events were counted for each sample. FACS data were gated to plot cells with an FL2 area of 68–520 and smoothed by a factor of 5.
Polyclonal Antibodies to Rad3p
To produce Rad3p in bacteria, the C-terminal region of Rad3p (the BamHI–BamHI piece of rep1-rad3+ [pTE157]) was cloned in-frame into the BamHI site of the pET3-His Escherichia coli histidine-tagging vector (Chen and Hai, 1994) and transformed into BL21 cells. Expression was induced by adding 0.5 mM isopropyl-1-thio-α-d-galactopyranoside to an exponentially growing culture for 3 h at 25°C. The bacterial pellet was resuspended in IMAC5 (20 mM Tris-Cl, pH 8.0, and 0.5 M NaCl, with 5 mM imidazole) containing 1 mg/ml lysozyme and 0.1% NP-40, sonicated, and centrifuged. The insoluble pellet was solubilized with IMAC5 containing 6 M GuHCl and bound by batch method to Talon Ni2+ agarose (Clontech, Palo Alto, CA) and washed with IMAC5, IMAC10, and IMAC20 all containing 6 M GuHCl, followed by H2O. Rad3p protein was eluted from the Ni2+ beads by boiling into 2% SDS and 10 mM EDTA, dialyzed against 1% SDS, 0.025 M Tris, 0.192 M glycine, and 10 mM EDTA, and injected into rabbits (Cocalico Biologicals. Reamstown, PA).
Protein Electrophoresis and Western Blotting
All full-length Rad3p samples were resolved on 5% gels for ∼16 h at 100 V (as described by Scully et al., 1997). Smaller proteins were resolved on 10 or 12% gels. A BenchMark prestained protein ladder (Life Technologies, Gaithersburg, MD) was used for molecular weight markers. Transfer to Immobilon-P (Millipore, Bedford, MA) was performed in a semidry apparatus (Owl Scientific, Woburn, MA) using the transfer buffer described by Scully et al. (1997). Blots were then dried after brief immersion in methanol. Once dry, the blots were briefly resoaked in methanol, washed with H2O, and blocked with 1× Tris-buffered saline (TBS) containing 1% dry milk and 1% BSA. Primary antibody was added for one h in 1× TBS and 0.05% Tween 20 (TBST). For HA blots, monoclonal anti-HA antibody, clone 12CA5, was diluted 1:5000 (a generous gift from Ed Harlow, Harvard Medical School, Boston, MA); for myc blots, the 9E10 monoclonal antibody was diluted 1:200 (M5546; Sigma, St. Louis, MO). The Rad3p HM126 polyclonal antibody serum was diluted 1:5000. After washing, secondary antibody (HRP-conjugated anti-mouse or anti-rabbit; Amersham, Arlington Heights, IL) was added at a 1:5000 dilution in 1× TBST. After washing with 1× TBST, blots were developed using ECL (Amersham).
Immunoprecipation Assays
Yeast protein extracts were prepared from early log phase cultures, grown in the absence of thiamine for 20–24 h, to induce protein expression from the attenuated nmt promoter (rep41/rep42) for the coimmunoprecipitation assays, or from nmt1+ for the kinase assay experiments. Immunoprecipitations were performed as described by Bentley et al. (1996) with the following modifications. Cells (4 × 108) were lysed with glass beads into immunoprecipitation buffer (50 mM Tris, pH 8.0, 120 mM NaCl, 0.5% NP-40, 50 mM NaF, 60 mM β-glycerophosphate, 1 mM NaVO4, 2 mM PMSF, 20 μg/ml aprotinin, 10 μg/ml leupeptin) in a BIO 101 (La Jolla, CA) Fast-Prep. Primary antibody (anti-myc clone 9E10, Sigma M5546, or anti-HA 12CA5, a generous gift from Ed Harlow) was preincubated with protein G (anti-myc)- or protein A (anti-HA)-Sepharose beads in lysis buffer at 4°C for 1 h on a rotating wheel and then washed in lysis buffer. Beads were then divided, combined with extract, and incubated on a rotating wheel for 1 h at 4°C. After washing the beads five times with lysis buffer, the beads were split, and 2× SDS sample buffer was added. Samples were then boiled and loaded onto SDS-PAGE gels, as described above in Protein Electrophoresis and Western Blotting. To check expression of various proteins in total cell extracts, cells were lysed with acid-washed glass beads directly into 2× SDS sample buffer in a BIO 101 Fast-Prep.
Kinase Assays
For these assays, rad3 alleles were expressed from the full-strength nmt1+ promoter. Yeast protein extracts for immunoprecipitations were prepared as described above in Immunoprecipitation Assays. After washing the immunoprecipitates, they were split in half. Half was used for kinase assays, whereas the other half was Western blotted to visualize Rad3p as described above. The half of the precipitate to be used for the kinase assay was washed once in kinase buffer without substrate or ATP (25 mM HEPES, pH 7.7, 50 mM KCl, 10 mM MgCl2, 10 mM MnCl2, 0.1% NP-40, 2% glycerol, 1 mM DTT, 1 mM NaVO4). Ten microliters of kinase buffer containing substrate and ATP (kinase buffer containing 10 mM ATP, 5–10 μCi of [γ32P]ATP, 1 μg of PHAS-1) was added to each immunoprecipitate, and the reactions were incubated for 15 min at 30°C at which point 10 μl 2× SDS sample buffer containing 10 mM EDTA was added to each to stop the reactions. PHAS-1 protein was a generous gift from Merl Hoekstra (Signal Pharmaceuticals, San Diego, CA). Each sample was heated at 95°C for 2–5 min and then resolved by SDS-PAGE on a 12 or 15% gel. Gels were Coomassie blue stained, fixed, and dried under vacuum for 2 h with low levels of heat (50–60°C). Bands were visualized by autoradiography of the dried gels for 3–12 h at −70°C. Assays for wortmannin sensitivity were done as described with the following exceptions. After the immunoprecipitates were split, they were washed twice in wash buffer (25 mM HEPES, pH 7.7, 50 mM KCl, 10 mM MgCl2). Wortmannin diluted in the above wash buffer was then added in a volume of 100 μl in various concentrations, and the precipitates were incubated at room temperature for 20 min. The wortmannin was inactivated with the addition of kinase buffer without substrate or ATP, and the precipitates were then washed once with kinase buffer without substrate or ATP.
RESULTS
The Rad3p Kinase Domain Is Not Sufficient for Checkpoint Function
Rad3p is a large, 2386-amino-acid protein. Thus far, the only region of Rad3p shown to be important for function is the C-terminal PI3-like kinase domain that makes up <15% of the Rad3 protein (Bentley et al., 1996). To determine whether sites outside the kinase domain are required for Rad3p activity, we examined the biological activity of a series of C-terminal fragments of Rad3p shown in Figure 1A. All but one of these HA epitope-tagged truncations contains the full kinase domain (Bentley et al. 1996). Of these constructs, rad3-C328 encodes a protein that is most similar to the kinase domain fragment used in studies of ATMp (Morgan et al., 1997). The ability of these constructs to rescue a rad3Δ strain was determined. Because rad3Δ cells are checkpoint defective, they are exquisitely sensitive to the DNA replication inhibitor HU and to DNA damage induced by UV radiation.
As shown in Figure 1B, rad3-C328 exhibits very little complementing activity in the HU assay, because cells expressing this construct were almost as sensitive to HU as cells transformed with empty vector. Although the larger fragments rad3-C549 and rad3-C725 more significantly improved the viability of the rad3Δ strain after HU treatment (Figure 1B), the transformants were still 10 times more HU sensitive than cells transformed with full-length rad3+. Two other fragments, rad3-C488 and rad3-C261, also exhibited complementing activity significantly below that of full-length rad3+ (our unpublished results). Radiation sensitivity of transformants was also analyzed as shown in Figure 1C. None of the truncated alleles showed any complementing activity in this assay (Figure 1C and our unpublished results), although they all directed high levels of protein expression (Figure 1D). We conclude that the isolated kinase domain cannot perform wild-type Rad3p function. This may be because N-terminal sequences are crucial for biological activity. Alternatively, we may have eliminated sequences necessary for correct cellular localization of the protein. In contrast, a 390-amino-acid kinase domain fragment of ATMp, equivalent to the last 355 amino acids of Rad3p, is sufficient for at least some activities (Baskaran et al., 1997; Morgan et al., 1997).
The C-terminal fragments cannot fully rescue the sensitivities of the rad3Δ mutants in either the HU or UV assays. However, several of the mutant proteins are able to partially restore resistance to HU but not to UV. A similar phenomenon is described below for the Rad3-KD protein (see Figure 3). It is possible that the observed differences reflect quantitative or qualitative differences in the assays. For example, the cells are arrested in S phase in the HU assay, whereas most of the cells are in G2 when they are irradiated with UV. An interesting alternative is that the mutant proteins exhibit varying abilities to complement (or disrupt) the DNA replication and DNA damage responses because these two responses have different requirements for certain Rad3p functions.
Sequences Outside the Kinase Domain of Rad3p Are Required for Kinase Activity
The isolated kinase domain may not complement rad3Δ cells, because it is unable to interact with cofactors or substrates. Alternatively, sequences outside the kinase domain may be directly required for Rad3p catalytic activity. To investigate this possibility, we examined the catalytic activity of the Rad3p C-terminal fragments examined above. Rad3p kinase activity was measured by in vitro phosphorylation of an exogenous substrate, the eukaryotic initiation factor-4E (eIF-4E) binding protein, PHAS-1, a human translational regulatory protein also phosphorylated by the Rad3p homologues ATMp and ATRp (Banin et al., 1998; Canman et al., 1998; Sarkaria et al., 1998). High levels of PHAS-1 phosphorylation were readily observed when full-length Rad3p was incubated with the substrate (Figure 2, A, lane 2, and B, lane 2) so that a qualitative assessment of Rad3p activity could be easily made visually. As previously reported (Bentley et al., 1996), autophosphorylation of Rad3p was also observed under these conditions (our unpublished results); however, because the levels of autophosphorylation were much lower than the levels of PHAS-1 phosphorylation in this assay, PHAS-1 phosphorylation was used to measure Rad3p activity. Importantly, the kinase activity observed is likely to be due specifically to the Rad3 protein and not another coimmunoprecipitating protein, because phosphorylation of PHAS-1 was not observed in immunoprecipitates of extracts from cells expressing rad3-KD, which encodes a protein carrying the mutation, D2230A, predicted to abolish kinase activity (Bentley et al., 1996) (Figure 2, A and B). No phosphorylation of PHAS-1 was observed in immunoprecipitates from any of the strains transformed with C-terminal truncations of rad3+ (Figure 2A, lanes 4–8), although Western blot analysis showed that the fragments were expressed and precipitated at levels equal to or greater than wild-type Rad3p (Figure 1D and our unpublished results). Because several of the C-terminal fragments of Rad3p are able to slightly improve the viability of the rad3Δ strain in the presence of HU, we have also examined the kinase activity of the Rad3-C725p, Rad3-C488p, and Rad3-C328p C-terminal fragments in the presence of HU. Cells overexpressing these fragments were treated with 10 mM HU for 3 and 6 h. Rad3p C-terminal fragments immunoprecipitated from these extracts did not exhibit any activity toward PHAS-1 (our unpublished results). We conclude that sequences N-terminal to the kinase domain are required for Rad3p catalytic activity. In contrast, the isolated kinase domain of ATMp (a 390-amino-acid C-terminal fragment) has been shown to phosphorylate c-Abl (Baskaran et al., 1997).
Rad3p Is Relatively Resistant to the PI3-Kinase Inhibitor Wortmannin
Although rad3Δ cells share some phenotypes with A-T cells, suggesting that there may be some functional overlap between Rad3p and ATMp, the primary amino acid sequence of Rad3p is more similar to human ATRp than to ATMp. In contrast to ATMp, regions outside of the kinase domain are absolutely required for Rad3p kinase activity, suggesting that the conformation of the catalytic sites of these proteins may be different. The drug wortmannin, which irreversibly inhibits PI3-kinases by modifying an invariant lysine residue in the catalytic site, has been used to further characterize the relationships among the PI3KR kinases. ATMp is 10 times more sensitive to wortmannin (IC50, 150 nM) than ATRp (IC50, 1.8 μM) (Banin et al., 1998; Sarkaria et al., 1998). Using the kinase assay described above, the sensitivity of Rad3p to wortmannin was examined. As shown in Figure 2B, Rad3p retained close to wild-type levels of activity in the presence of 1 μM wortmannin (lane 6). Approximately 50% inhibition of Rad3p activity occurs when concentrations of 2–4 μM of wortmannin are added. In the presence of a 10 μM concentration of wortmannin, Rad3p activity is decreased ∼10-fold, indicating that the wortmannin is active. Although the kinase assay is a qualitative assay in which activity is assessed visually, it allows us to distinguish between inhibition in the micromolar and nanomolar range. In contrast to the relatively high resistance of Rad3p to wortmannin, ATM and DNA-PK protein kinase activity was completely abolished by a 1 μM concentration of this lot of wortmannin (Rathbun, unpublished observations). Thus, the conformation of the catalytic site of Rad3p likely resembles that of ATRp more than that of ATMp.
Overexpression of the N-terminal 775 Amino Acids of Rad3p Disrupts the Checkpoint Response of Wild-Type Cells
The above studies indicate that sequences N-terminal to the kinase domain of Rad3p are required for its catalytic activity. However, they do not address the function of the N terminus in interactions with regulators or substrates. To explore this possible role for the N-terminal sequences, we examined the ability of fragments of Rad3p to function as dominant negative alleles, that is, to disrupt checkpoint control of wild-type cells when overexpressed. It has been shown previously that rad3-KD acts as a dominant negative when overexpressed in wild-type cells, presumably because the catalytically incompetent protein sequesters key regulators or substrates into nonfunctional complexes (Bentley et al., 1996). To define the regions of Rad3p that compete with the wild-type protein, rad3+ truncations were overexpressed in wild-type cells, and their ability to disrupt HU and UV resistance was determined (Figure 3). Overexpression of rad3-N775, which encodes the N-terminal 775 amino acids of Rad3p, greatly increased the sensitivity of wild-type cells to both HU and UV (Figure 3, B and C). These data parallel results with the ATM protein in which an N-terminal fragment can disrupt checkpoint function of human cells (Morgan et al., 1997). Expression of a C-terminal truncation that includes the full kinase domain, rad3-C725, did not affect the HU or UV resistance of wild-type cells.
Although overexpression of rad3-N775 significantly disrupted the UV and HU responses, it behaves differently from the dominant negative rad3-KD allele in a few notable ways. For example, cells overexpressing rad3-N775 are much less UV sensitive than cells overexpressing rad3-KD. In addition, cells overexpressing rad3-N775 are more varied in cell length (Figure 4B), even in the absence of HU (our unpublished results). As previously reported, rad3-KD overexpression is lethal to cells over many generations (Bentley et al., 1996); in contrast, overexpression of rad3-N775 does not grossly affect cell viability.
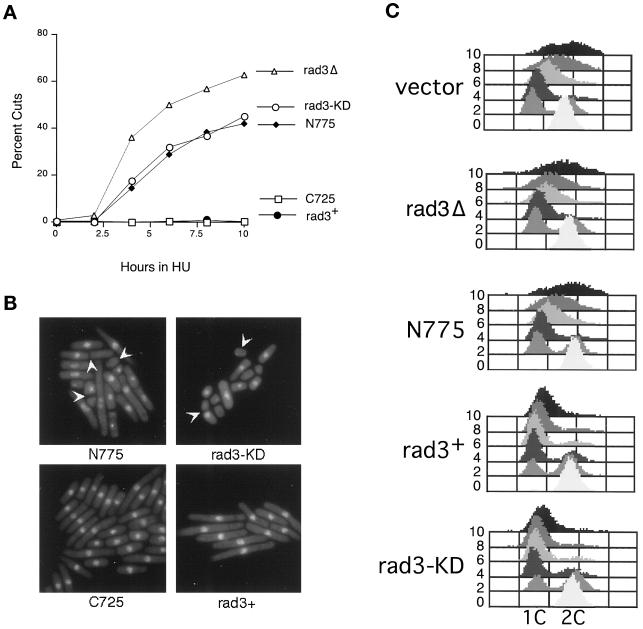
Figure 4.
Rad3p dominant negative alleles disrupt the checkpoint coupling mitosis to completion of DNA replication. (A) Percent aberrant mitoses after treatment with 10 mM HU of wild-type cells (TE235) expressing rad3+ (pTE157), rad3-C725 (pTE446), rad3-KD (pTE791), rad3-N775 (pTE696), and rep3x (pTE101). The results shown are the average of two independent experiments. (B) Photomicrographs of cells cultured in 10 mM HU for 10 h and then fixed and stained with the DNA-specific dye DAPI. Cells were wild-type (TE235) expressing rad3-N775, rad3-KD, rad3-C725, and rad3+. Aberrant mitoses are indicated with arrows. (C) FACS analysis of cells incubated with HU for indicated times (at left). Wild-type cells expressing rep3x control (vector), rad3-N775 (N775), rad3+, and rad3-KD are shown, as well as rad3Δ cells expressing the rep3x vector (rad3Δ). Note that overexpression of rad3+ or rad3-KD inhibits DNA replication after exposure to HU. See text for more details.
To determine whether the HU sensitivity caused by overexpression of these constructs is due to disruption of cell cycle checkpoints, we examined mitosis and DNA replication in the presence of HU in these strains and in rad3Δ and wild-type cells. As shown in Figure 4A, rad3Δ cells and cells overexpressing rad3-KD and rad3-N775 undergo abnormal mitoses (“cuts”) in which the nucleus is cleaved by a septum, or anucleate cells are generated; this is typically observed when cells enter mitosis with less than fully replicated DNA. The rad3-KD and rad3-N775 cultures continue to accumulate cuts for the next 6 h with identical kinetics, although ∼20% fewer cuts are observed in cells overexpressing the mutants compared with rad3Δ cells. In contrast, no cuts are observed in cells transformed with rad3+ or rad3-C725. Photographs of cells treated with HU for 10 h are shown in Figure 4B.
To determine whether the abnormal mitoses are due to cells entering mitosis with unreplicated DNA, we also examined DNA content by FACS analysis during a 10-h HU exposure. As shown in Figure 4C, before HU treatment, most of the cells have a 2C DNA content, as is characteristic of fission yeast cultures, because G2 constitutes 80% of the fission yeast cell cycle. After 2 h in HU, a 1C peak is evident, and by 4 and 6 h the majority of the cells in all the cultures show a 1C DNA content indicating that they are blocked from completing S-phase by HU. Notably, by 4 and 6 h significant numbers of cuts are observed in rad3Δ, rad3-KD, and rad3-N775 cultures, although FACS analysis indicates that they have not completed DNA replication by this time. From these results we conclude that mutant overexpression, like the loss of rad3, causes defects in the checkpoint that makes mitosis dependent on completion of DNA replication. The similarity in the kinetics of the appearance of cuts in all the affected cultures suggests that defects in checkpoint control caused by mutant overexpression are qualitatively similar to the well-characterized defect of rad3Δ cells, although they are quantitatively less severe. As previously noted (Figure 3B), rad3Δ cells are significantly more sensitive to HU. Thus, although the overexpressed mutant proteins significantly disrupt the mitotic checkpoint, other Rad3p functions that are missing in the deletion, such as regulation of S-phase (Lindsay et al., 1998) and recovery (Enoch et al., 1992), may still be intact.
The FACS analysis also reveals that wild-type cells and rad3Δ cells begin to synthesize DNA after 6 h of HU exposure, and synthesis continues inefficiently for the next 4 h. This slow escape from HU arrest has previously been described by others (Sazer and Nurse, 1994). In remarkable contrast, DNA synthesis in the presence of HU is significantly inhibited by overexpression of rad3+ or rad3-KD, because even by 10 h the majority of the cells in the culture still have a 1C DNA content. In contrast, overexpression of rad3-N775 has no effect on this aspect of the HU response. These results suggest that overexpression of rad3+ or rad3-KD inhibits DNA replication under some circumstances. Additional studies of the effects of rad3+ overexpression on DNA replication are under way and will be presented elsewhere (Chapman and Enoch, unpublished data).
The Leucine Zipper Is Required but Not Sufficient for Dominant Negative Activity of rad3-N775
Expression of rad3-N775 may disrupt the checkpoint response by sequestering Rad3p-interacting proteins into nonproductive complexes. Such a model predicts that the dominant negative activity of rad3-N775 will be abolished by mutations that eliminate binding sites for Rad3p-interacting proteins. Thus we hypothesized that by examining the dominant negative activity of rad3-N775 mutants, it should be possible to identify regions of Rad3p that mediate critical interactions with other cellular proteins.
Like other PI3KR kinases, the N terminus of Rad3p contains a putative leucine zipper. Because leucine zipper motifs frequently mediate protein–protein interactions (Phizicky and Fields, 1995), this site in Rad3p was a logical candidate for a domain required for dominant negative activity. To examine the role of this putative leucine zipper, 22 amino acids, from the first leucine to the last leucine, were deleted from the rad3-N775 construct. The dominant negative activity of the resulting allele (rad3-N775-LZ) was tested. As shown in Figure 5A, wild-type cells transformed with rad3-N775-LZ grew almost normally on plates containing HU, in contrast to cells transformed with rad3-N775. For comparison, rad3Δ cells and wild-type cells are also shown. We conclude that the leucine zipper is likely to interact with factors that are limiting for the checkpoint response.
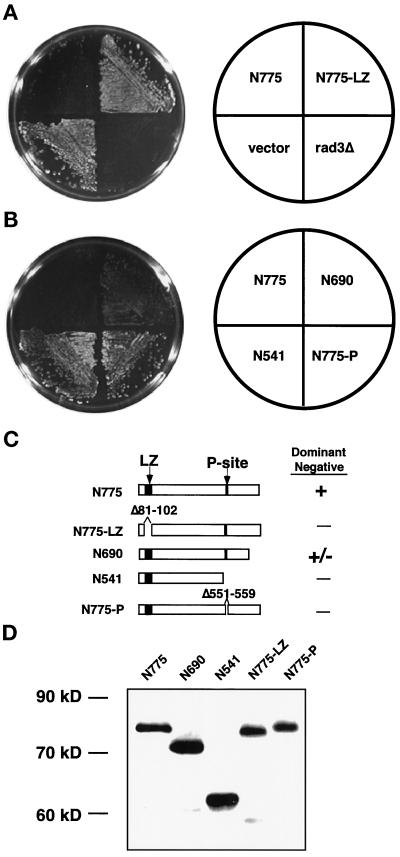
Figure 5.
Identification of regions necessary for dominant negative activity of Rad3-N775p. (A) Leucine zipper motif is required for dominant negative activity of Rad3-N775p. Wild-type (TE235) cells overexpressing rad3-N775 (pTE696), rad3-N775-LZ (pTE718), or empty vector (pTE101) and rad3Δ (TE890) cells with empty vector were grown on plates containing 10 mM HU for 3 d. Left, HU plate. Right, key. (B) Requirement of the P-site for Rad3-N775p dominant negative activity. Wild-type (TE235) cells expressing rad3-N775 (pTE696), rad3-N690 (pTE698), rad3-N541 (pTE700), or rad3-N775-P (pTE710) were grown on plates containing 10 mM HU. Left, HU plate. Right, key. (C) Diagram of mutant Rad3p proteins. The leucine zipper and P-site are indicated by black boxes. Deletions of leucine zipper motif and P-site are indicated by a gap. A plus sign next to the protein indicates it can function as a dominant negative, and a negative sign indicates that it does not have this capability. (D) Expression of Rad3p N-terminal alleles. Anti-HA antibodies were used to Western blot extracts from cells expressing HA-tagged versions of the N-terminal proteins. Lane 1, HA-rad3-N775 (pTE697); lane 2, HA-rad3-N690 (pTE699); lane 3, HA-rad3-N541 (pTE701); lane 4, HA-rad3-N775-LZ (pTE717); lane 5, HA-rad3-N775-P (pTE709).
When a series of smaller N-terminal truncations was examined, evidence for another site involved in the dominant negative activity was found. The deletion series used for this analysis is shown in Figure 5C. We found that a fragment encoding the N-terminal 690 amino acids of Rad3p (rad3-N690) retained dominant negative activity; however, truncation of an additional 150 amino acids (rad3-N541) eliminated this activity. Because rad3-N541 contains the leucine zipper motif, we conclude that overexpression of this motif is not sufficient for dominant negative activity. This result also suggested that another important protein-protein interaction domain might be located in the region between amino acids 541 and 690. Interestingly, this region contains the motif QSLLLDGFF, at amino acids 551–559, which closely resembles the consensus (QXXI/LXXFF) for a domain that mediates an interaction with proliferating cell nuclear antigen (PCNA), the processivity factor for DNA polymerase (Warbrick et al., 1995; Montecucco et al., 1998). To determine whether this site is required for the dominant activity of rad3-N775, a mutant deleted for these nine amino acids of this putative PCNA binding site (rad3-N775-P) was constructed and tested for its ability to render wild-type cells HU sensitive. As shown in Figure 5B, deletion of this site which we call the “P-site” abolishes the dominant negative activity of rad3-N775. We conclude that both the P-site and the leucine zipper may interact with proteins required for the checkpoint response. However, it is possible that mutation of these sites disrupts dominant negative activity for other reasons; for example, the deletions may affect local structure of the protein. To definitively show that these sites are indeed protein-protein interaction sites, a protein that shows a leucine zipper (LZ) or P-site-dependent interaction with Rad3p must be identified.
We also examined the dominant-negative activity of HA-tagged versions of these N-terminal alleles. Again, overexpression of HA-rad3-N775 disrupted the checkpoint response of wild-type cells, however, not as dramatically as the untagged allele, suggesting that the presence of the HA tag may interfere with some Rad3p interactions. The dominant negative activity of the HA-tagged allele could also be abolished by deleting the leucine zipper or the P-site, confirming the significance of these sites. Western blots established that the HA-tagged versions of all these alleles were expressed at relatively equal levels, establishing that the deletions do not affect protein stability (Figure 5D). None of these alleles (tagged or untagged) has gross effects on cell viability when cells are grown in the absence of HU.
Additive Effects of LZ and P-Site Mutations
To ascertain whether the leucine zipper and the P-site affect the same or different functions, the dominant negative activity of rad3-KD constructs lacking either the leucine zipper or the P-site or both sites was examined. If these sites interact with the same protein, we would expect that deletion of both sites would have the same effect as deletion of one of the sites. If these sites interact with different proteins, the effects of deleting both might be additive.
To do these experiments we started with the rad3-KD allele and then deleted either the leucine zipper (rad3-LZ-KD), the P-site (rad3-P-KD), or both (rad3-LZ-P-KD) (Figure 6A). Mutation of either the leucine zipper or the P-site greatly reduced but did not completely abolish the dominant negative activity of the rad3-KD allele in both the UV (Figure 6B) and liquid HU assays (our unpublished results). However, deletion of both sites, rad3-LZ-P-KD, resulted in a further reduction of dominant negative activity. In the UV assay, the difference between the double and triple mutants was only modest, but is significant at the 150-J/m2 time point. In the HU liquid assay, the triple mutant also reduced the dominant negative activity of the rad3-KD allele more than mutation of each site on its own, although the differences were small (our unpublished results). Therefore, to examine the HU response of the double and triple mutants, we used the plate assay, which requires the cells to grow for many generations in HU and thus is a more stringent test of dominant negative activity. Although cells overexpressing the rad3-KD allele grew poorly on plates (Bentley et al., 1996; our unpublished results), cells overexpressing the double and triple mutants grow normally (Figure 6C), indicating that these mutations eliminate the toxicity of the rad3-KD allele as well as its dominant negative activity. In the presence of HU, the triple mutant rad3-LZ-P-KD clearly grows better than either of the double mutants (Figure 6C). All of these proteins are expressed at equal levels (Figure 6D). Thus, the leucine zipper and the P-site mutations work additively to reduce the dominant negative activity of rad3-KD, suggesting that the two sites could affect different Rad3p functions.
The Leucine Zipper and P-Site Are Required for Normal Function of Rad3p
Because deletion of the LZ and P-sites abrogates dominant negative activity, we wished to determine whether these sites are also required for the normal function of rad3+. To do so, we deleted these sites from full-length rad3+ (Figure 7A) and examined the ability of these alleles to complement a rad3Δ strain. Neither rad3-LZ nor rad3-P was able to complement the HU or UV sensitivity of a rad3Δ strain, indicating that the LZ and P-sites are indeed required for the normal function of rad3+ (Figure 7, B and C). To determine whether either the leucine zipper or P-site is required for catalytic activity, the kinase activities of Rad3-LZp and Rad3-Pp were measured. Neither mutant had significantly diminished activity compared with wild-type protein (Figure 7D, lanes 3–5), and all three proteins phosphorylated significantly more PHAS-1 than a kinase-dead form of Rad3p (Figure 7D, lane 2). Because the mutated proteins are expressed well (Figure 7D, bottom panel) and retain catalytic activity, we believe that the structures of the proteins are grossly intact. These data indicate that these sites are not required for kinase activity of Rad3p and, moreover, that kinase activity is not sufficient for full Rad3p function.
The Leucine Zipper Is Not Required for Rad3p Self-Association or for Interaction with Chk1p
Having identified the putative leucine zipper as a probable protein–protein interaction site required for Rad3p function but not for kinase activity, we wished to further investigate the molecular mechanism of its function. Rad3p has been shown to self-associate (Bentley et al., 1996). Because leucine zipper motifs have been shown to be important in mediating both hetero- and homodimerization (Phizicky and Fields, 1995), the self-association capabilities of Rad3 proteins lacking the leucine zipper motif were examined. HA- and myc-tagged forms of wild-type Rad3p and Rad3-LZp were constructed and cotransformed in various combinations into wild-type cells (strain TE236; see Table 1). In parallel experiments, either HA-Rad3p or myc-Rad3p was immunoprecipitated with the appropriate monoclonal antibody. Immunoprecipitations were resolved by SDS-PAGE and probed with both the HA and myc antibodies. As has been shown by others (Bentley et al., 1996), Rad3p self-associates, because HA-Rad3p can be precipitated by anti-myc antibodies when coexpressed with myc-Rad3p (Figure 8A, lane 1) but was absent from immunoprecipitates from strains transformed with only the myc-tagging vector (Figure 8A, lane 6).
Using Rad3 proteins that contained deletions of the leucine zipper, we found that the leucine zipper is not required for Rad3p self-association. We were able to immunoprecipitate HA-Rad3-LZp with either myc-Rad3-LZp (Figure 8A, lane 2) or wild-type myc-Rad3p (Figure 8A, lane 3). We were also able to immunoprecipitate wild-type HA-Rad3p with myc-Rad3-LZp (Figure 8A, lane 4). Similar results were obtained in the reciprocal experiment, immunoprecipitating with an HA antibody and then blotting for myc-Rad3p protein (our unpublished results). Thus, the leucine zipper motif is not required for self-association as measured by this assay.
S. pombe Rad3p has recently been shown to coimmunoprecipitate with Chk1p (Martinho et al., 1998). We noticed a site in Chk1p that resembles a leucine zipper at amino acids 437–465 (Lx6Lx6Lx6Lx6K) and hypothesized that the putative leucine zipper in Rad3p may mediate its interaction with Chk1p. However, we were able to immunoprecipitate both wild-type HA-Rad3p and mutant HA-Rad3-LZp with myc-Chk1p (Figure 8B, lane 2), suggesting that the Rad3p leucine zipper motif is not required for the interaction observed in this assay.
DISCUSSION
Relatively little is known about the function of sequences outside the kinase domain in the PI3KR family of kinases. In this study, we have identified sites important for the normal cellular functions of S. pombe Rad3p, a member of this family. Our data indicate that sequences outside the kinase domain have at least two important functions. First, these sequences are required for catalytic activity. Second, we have identified two sites in the N terminus of Rad3p that may mediate interactions with regulators or substrates but are not required for catalytic activity.
The Isolated Rad3p Kinase Domain Is Catalytically Inactive and Does Not Complement rad3Δ Mutants
Our results demonstrate that the isolated Rad3p PI3-like-kinase domain is not sufficient for Rad3p function. Even the largest C-terminal fragment examined did not fully rescue the HU and UV sensitivities of rad3Δ cells (Figure 1). Furthermore, none of the C-terminal fragments exhibited kinase activity in our in vitro assay (Figure 2). The Rad3p kinase domain may require residues N-terminal to the kinase domain for catalytic activity for a variety of reasons. One possibility is that Rad3p kinase activity depends on Rad3p dimerization, mediated by sequences N-terminal to the kinase domain. Examples of this type of regulation include the receptor protein-tyrosine kinases, as well as some cytoplasmic protein-tyrosine kinases (for review, see Heldin, 1996). Indeed, Rad3p does self-associate, as shown by Bentley et al. (1996) and confirmed here (Figure 8), although it is not known whether this is necessary for catalytic activity. Alternatively, N-terminal sequences of Rad3p may function as an intramolecular positive regulator of the kinase domain. For example, they may be required to induce a catalytically active configuration or to stabilize interactions with substrates. Although we have not defined sequences outside the kinase domain required for catalytic activity, we have shown that the leucine zipper and the P-site are not required, as discussed below.
Our results are the first demonstration that sequences outside the kinase domain of any PI3KR kinase are required for catalytic or biological activity. In contrast to our findings, others have shown that a 390-amino-acid fragment of ATMp consisting of the isolated kinase domain is sufficient for complementation of the radioresistant DNA synthesis and radiosensitivity phenotypes of A-T cells (Morgan et al., 1997). In addition, the same ATMp fragment is capable of phosphorylating c-Abl without outside sequences (Baskaran et al., 1997). However, in another study, this fragment was unable to phosphorylate replication protein A when transfected into A-T cells (Morgan and Kastan, 1997), suggesting that sequences outside the domain may be required in certain circumstances or for interactions with some substrates.
Because the isolated Rad3p kinase domain is not catalytically active, our results suggest that residues outside the kinase domain of Rad3p are more important for function than equivalent regions in ATMp. An interesting possibility is that the ATR and ATM PI3KR subgroups are structurally different and have distinct requirements for catalytic activity. Because the ATR subgroup exhibits regions of homology in the middle of the proteins, which are not present in the ATM subgroup (Savitsky et al., 1995b; Bentley et al., 1996), it is possible that these regions are required for the catalytic activity of proteins in the ATR subgroup, including Rad3p.
Another difference between the ATR and ATM-like PI3KR proteins is their sensitivity to the drug wortmannin. Our results indicate that Rad3p catalytic activity is relatively insensitive to wortmannin (Figure 2). The kinase activity of human ATRp is also relatively insensitive to the drug wortmannin (Sarkaria et al., 1998). In contrast, the catalytic activity of human ATMp is inhibited by low concentrations of wortmannin (Banin et al., 1998; Sarkaria et al., 1998). Thus, these data further support the idea that Rad3p is more similar to human ATRp than ATMp.
It is also possible that differences in our experimental systems account for the requirement for non-kinase domain residues that we have observed. We have assayed Rad3p activity in strains harboring a large internal deletion in the rad3 coding sequence (Bentley et al., 1996). In contrast, the nature of the mutation in the ATM gene in the A-T cell line used by Morgan et al. (1997) is not known, leaving open the possibility of intragenic complementation, although the authors were careful to point out that they could not detect endogenous protein or message. Furthermore, functional complementation of A-T cells has proved problematic in the past; a number of rescuing clones were initially identified that did not map to the A-T locus, suggesting that suppression can occur by indirect mechanisms (Jongmans et al., 1995; Shiloh, 1995; Ziv et al., 1995). In contrast, no extragenic suppressors were isolated during the cloning of rad3+ (Carr, unpublished observations), arguing that complementation of yeast mutants is a highly specific assay for rad3 function. In light of our results, it will be interesting to examine the biological and catalytic activity of the isolated kinase domain of the newly identified fission yeast Tel1p (Naito et al., 1998), which is more closely related to ATMp that Rad3p.
Use of Rad3p Dominant Negative Alleles to Identify Sequences Required for the Checkpoint Response
Sequences outside the kinase domain are required for Rad3p checkpoint function. To begin to understand the functions of nonkinase regions of Rad3p, we focused on the N terminus of Rad3p. Overexpression of N-terminal sequences of Rad3p causes wild-type cells to become checkpoint deficient (Figures 3 and 4), indicating that these sequences may be capable of sequestering proteins into nonfunctional complexes. Similarly, Morgan et al. (1997) demonstrated that expression of fragments of human ATMp, which contain a putative leucine zipper motif, caused dominant negative checkpoint phenotypes in a human tumor cell line. However, this study did not address what sites are required for these activities. To determine what sites are required for the dominant negative activity exhibited by overexpression of the Rad3-N775p, we performed deletion analysis (Figure 5). We found that the leucine zipper is necessary for the dominant negative activity of the N terminus. Interestingly, another site we call the P-site, located between amino acids 541 and 690 of the protein, is also necessary for dominant negative activity of the N terminus. Moreover, in the context of the full-length dominant negative allele rad3-KD, mutating both sites reduced dominant negative activity more than mutating either site alone (Figure 6), indicating that these sites are likely to be affecting two separate functions.
Importantly, both sites identified through dominant negative analysis are required for the normal cellular functions of Rad3p, because constructs deleted for either site fail to rescue rad3Δ cells (Figure 7). The failure of the mutants to complement rad3Δ cells is not due to destabilization of Rad3p, because these mutant proteins are expressed at levels comparable with wild type and retain full catalytic activity. This indicates that the gross structure of the Rad3 protein is likely to be intact, particularly because catalytic activity requires sequences outside the kinase domain. However, it is possible that the deletions create more localized disruptions of Rad3p structure.
We initially focused on the P-site because of its similarity to the PCNA binding motif found in a number of proteins from diverse organisms and considered the possibility that the P-site might mediate Rad3p binding to PCNA, the processivity factor for DNA polymerases. However, we have been unable to identify a P-site-dependent interaction between Rad3p and PCNA in preliminary studies (Chapman and Enoch, unpublished observations) and, moreover, have not identified PCNA binding sites in other proteins in the PI3KR family. Interestingly, S. cerevisiae Mec1p has a related site at an equivalent position, amino acids 537–545, ESLLSGILF (conserved residues with P-site are bold; similar amino acid is underlined). Because this sequence is conserved in budding yeast but even less related to the PCNA binding site consensus, we believe that the P-site in Rad3p could be required for interactions with another protein that has yet to be identified. Possible candidates could include other fission yeast checkpoint proteins (see INTRODUCTION), particularly those which have S. cerevisiae homologues, such as Rad1p and Rad17p.
The putative leucine zipper domain of Rad3p is conserved in other PI3KR family members, but this is the first demonstration that the motif is functionally important. Leucine zippers frequently mediate homodimerization or heterodimerization with closely related proteins (Phizicky and Fields, 1995). However, surprisingly, we found that the putative leucine zipper is not required for self-association of Rad3p. Furthermore, it is not required to interact with Chk1p (Figure 8). It is possible that the coimmunoprecipitation assay we used (Bentley et al., 1996), which uses overexpressed proteins, may not be sufficiently sensitive to detect differences in binding between the wild-type and mutant proteins. Alternatively, other sequences that have yet to be identified may mediate Rad3p self-association and association with Chk1p, and the leucine zipper could be required for interactions with other proteins that have leucine zipper motifs. One interesting candidate for an interaction partner is the newly identified fission yeast Tel1p (Naito et al., 1998), which has a leucine zipper-like site. Because putative leucine zippers are found in other members of the PI3KR family, interactions directed by this motif are likely to be an important aspect of regulation of this important class of kinases.
ACKNOWLEDGMENTS
We thank Juanita Campos Torres for FACS analysis, Merl Hoekstra for supplying PHAS-1, Gary Rathbun for helpful discussions and for providing wortmannin, and Ed Harlow for supplying anti-HA (12CA5) antibodies. Thanks to Stuart MacNeill for pointing out the site with similarity to the PCNA-binding motif. Elspeth Stewart, Matt Endrizzi, Sidong Huang, and Aimee Dudley provided technical advice and help. We are grateful to F. Winston, G. Rathbun, K. Forbes, C. Kaplan, C. Kostrub, M. Leung, A. Martin, E. Moynihan, and T. Wolkow for critical reading of the manuscript. Gladys Reimundo provided excellent lab support. Work in T.E.’s laboratory is supported by National Institutes of Health grant GM50015. C.R.C. was supported by a grant from the Massachusetts Department of Public Health Breast Cancer Research Program, followed by Department of Defense Breast Cancer Program grant DAMD17-96-1-6100.
Abbreviations used:
- A-T
ataxia-telangiectasia
- FACS
fluorescence-activated cell-sorting
- HA
hemagglutinin
- HU
hydroxyurea
- LZ
leucine zipper
- PCNA
proliferating cell nuclear antigen
- PI3KR
phosphatidylinositol 3-kinase-related
- TBS
Tris-buffered saline
- TBST
TBS and Tween 20
REFERENCES
- Al-Khodairy F, Carr AM. DNA repair mutants defining G2 checkpoint pathways in Schizosaccharomyces pombe. EMBO J. 1992;11:1343–1350. doi: 10.1002/j.1460-2075.1992.tb05179.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Khodairy F, Fotou E, Sheldrick KS, Griffiths DJF, Lehmann AR, Carr AM. Identification and characterization of new elements involved in checkpoint and feedback controls in fission yeast. Mol Biol Cell. 1994;5:147–160. doi: 10.1091/mbc.5.2.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banin S, et al. Enhanced phosphorylation of p53 by ATM in response to DNA damage. Science. 1998;281:1674–1677. doi: 10.1126/science.281.5383.1674. [DOI] [PubMed] [Google Scholar]
- Barlow C, et al. Atm-deficient mice: a paradigm of ataxia telangiectasia. Cell. 1996;86:159–171. doi: 10.1016/s0092-8674(00)80086-0. [DOI] [PubMed] [Google Scholar]
- Basi G, Schmid E, Maundrell K. TATA box mutations in the Schizosaccharomyces pombe nmt1 promoter affect transcription efficiency but not the transcription start point or thiamine repressibility. Gene. 1993;123:131–136. doi: 10.1016/0378-1119(93)90552-e. [DOI] [PubMed] [Google Scholar]
- Baskaran R, et al. Ataxia telangiectasia mutant protein activates c-Abl tyrosine kinase in response to ionizing radiation. Nature. 1997;387:516–519. doi: 10.1038/387516a0. [DOI] [PubMed] [Google Scholar]
- Beamish H, Lavin MF. Radiosensitivity in ataxia-telangiectasia: anomalies in radiation-induced cell cycle delay. Int J Radiat Biol. 1994;65:175–184. doi: 10.1080/09553009414550211. [DOI] [PubMed] [Google Scholar]
- Bentley NJ, Holtzman DA, Flaggs G, Keegan KS, DeMaggio A, Ford JC, Hoekstra M, Carr AM. The Schizosaccharomyces pombe rad3 checkpoint gene. EMBO J. 1996;15:6641–6651. [PMC free article] [PubMed] [Google Scholar]
- Brown EJ, Albers MW, Shin TB, Ichikawa K, Keith CT, Lane WS, Schreiber SL. A mammalian protein targeted by G1-arresting rapamycin-receptor complex. Nature. 1994;369:756–758. doi: 10.1038/369756a0. [DOI] [PubMed] [Google Scholar]
- Canman CE, Lim DS, Cimprich KA, Taya Y, Tamai K, Sakaguchi K, Appella E, Kastan MB, Siliciano JD. Activation of the ATM kinase by ionizing radiation and phosphorylation of p53. Science. 1998;281:1677–1679. doi: 10.1126/science.281.5383.1677. [DOI] [PubMed] [Google Scholar]
- Carr AM, Moudjou M, Bentley NJ, Hagan IM. The chk1 pathway is required to prevent mitosis following cell-cycle arrest at “start.”. Curr Biol. 1995;5:1179–1190. doi: 10.1016/s0960-9822(95)00234-x. [DOI] [PubMed] [Google Scholar]
- Chan DW, Lees-Miller SP. The DNA-dependent protein kinase is inactivated by autophosphorylation of the catalytic subunit. J Biol Chem. 1996;271:8936–8941. doi: 10.1074/jbc.271.15.8936. [DOI] [PubMed] [Google Scholar]
- Chen BP, Hai T. Expression vectors for affinity purification and radiolabeling of proteins using Escherichia coli as host. Gene. 1994;139:73–75. doi: 10.1016/0378-1119(94)90525-8. [DOI] [PubMed] [Google Scholar]
- Chiu MI, Katz H, Berlin V. RAPT1, a mammalian homolog of yeast Tor, interacts with the FKBP12/rapamycin complex. Proc Natl Acad Sci USA. 1994;91:12574–12578. doi: 10.1073/pnas.91.26.12574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cimprich KA, Shin TB, Keith CT, Schreiber SL. cDNA cloning and gene mapping of a candidate human cell cycle checkpoint protein. Proc Natl Acad Sci USA. 1996;93:2850–2855. doi: 10.1073/pnas.93.7.2850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cliby WA, Roberts CJ, Cimprich KA, Stringer CM, Lamb JR, Schreiber SL, Friend SH. Overexpression of a kinase-inactive ATR protein causes sensitivity to DNA-damaging agents and defects in cell cycle checkpoints. EMBO J. 1998;17:159–169. doi: 10.1093/emboj/17.1.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enoch T, Carr AM, Nurse P. Fission yeast genes involved in coupling mitosis to completion of DNA replication. Genes & Dev. 1992;6:2035–2046. doi: 10.1101/gad.6.11.2035. [DOI] [PubMed] [Google Scholar]
- Forsburg SL. Comparison of Schizosaccharomyces pombe expression systems. Nucleic Acids Res. 1993;21:2955–2956. doi: 10.1093/nar/21.12.2955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenwell PW, Kronmal SL, Porter SE, Gassenhuber J, Obermaier B, Petes TD. TEL1, a gene involved in controlling telomere length in S. cerevisiae, is homologous to the human ataxia telangiectasia gene. Cell. 1995;82:823–829. doi: 10.1016/0092-8674(95)90479-4. [DOI] [PubMed] [Google Scholar]
- Hari KL, Santerre A, Sekelsky JJ, McKim KS, Boyd JB, Hawley RS. The mei-41 gene of D. melanogaster is a structural and functional homolog of the human ataxia telangiectasia gene. Cell. 1995;82:815–821. doi: 10.1016/0092-8674(95)90478-6. [DOI] [PubMed] [Google Scholar]
- Harnden DG. The nature of ataxia-telangiectasia: problems and perspectives. Int J Radiat Biol. 1994;66:S13–S19. [PubMed] [Google Scholar]
- Hartley KO, Gell D, Smith GC, Zhang H, Divecha N, Connelly MA, Admon A, Lees-Miller SP, Anderson CW, Jackson SP. DNA-dependent protein kinase catalytic subunit: a relative of phosphatidylinositol 3-kinase and the ataxia telangiectasia gene product. Cell. 1995;82:849–856. doi: 10.1016/0092-8674(95)90482-4. [DOI] [PubMed] [Google Scholar]
- Hartwell LH, Weinert TA. Checkpoints: controls that ensure the order of cell cycle events. Science. 1989;246:629–634. doi: 10.1126/science.2683079. [DOI] [PubMed] [Google Scholar]
- Heitman J, Movva NR, Hall MN. Targets for cell cycle arrest by the immunosuppressant rapamycin in yeast. Science. 1991;253:905–909. doi: 10.1126/science.1715094. [DOI] [PubMed] [Google Scholar]
- Heldin CH. Protein tyrosine kinase receptors. Cancer Surv. 1996;27:7–24. [PubMed] [Google Scholar]
- Jeggo PA, Taccioli GE, Jackson SP. Menage a trois: double strand break repair, V(D)J recombination and DNA-PK. Bioessays. 1995;11:949–957. doi: 10.1002/bies.950171108. [DOI] [PubMed] [Google Scholar]
- Jimenez G, Yucel J, Rowley R, Subramani S. The rad3+ gene of Schizosaccharomyces pombe is involved in multiple checkpoint functions and in DNA repair. Proc Natl Acad Sci USA. 1992;89:4952–4956. doi: 10.1073/pnas.89.11.4952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jongmans W, Verhaegh GW, Jaspers NG, Oshimura M, Stanbridge EJ, Lohman PH, Zdzienicka MZ. Studies on phenotypic complementation of ataxia-telangiectasia cells by chromosome transfer. Am J Hum Genet. 1995;56:438–443. [PMC free article] [PubMed] [Google Scholar]
- Kato R, Ogawa H. An essential gene, ESR1, is required for mitotic cell growth, DNA repair and meiotic recombination in Saccharomyces cerevisiae. Nucleic Acids Res. 1994;22:3104–3112. doi: 10.1093/nar/22.15.3104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keith CT, Schreiber SL. PIK-related kinases: DNA repair, recombination, and cell cycle checkpoints. Science. 1995;270:50–51. doi: 10.1126/science.270.5233.50. [DOI] [PubMed] [Google Scholar]
- Kostrub CF, Knudsen K, Subramani S, Enoch T. Hus1p, a conserved fission yeast checkpoint protein, interacts with Rad1p and is phosphorylated in response to DNA damage. EMBO J. 1998;17:2055–2066. doi: 10.1093/emboj/17.7.2055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunz J, Henriquez R, Schneider U, Deuter-Reinhard M, Movva NR, Hall MN. Target of rapamycin in yeast, TOR2, is an essential phosphatidylinositol kinase homolog required for G1 progression. Cell. 1993;73:585–596. doi: 10.1016/0092-8674(93)90144-f. [DOI] [PubMed] [Google Scholar]
- Lees-Miller S, Sakaguchi K, Ullrich S, Appella E, Anderson C. Human DNA-activated protein kinase phosphorylates serine 15 and 37 in the amino-terminal transactivation domain of human p53. Mol Cell Biol. 1992;12:5041–5049. doi: 10.1128/mcb.12.11.5041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieber MR, Grawunder U, Wu X, Yaneva M. Tying loose ends: roles of Ku and DNA-dependent protein kinase in the repair of double-strand breaks. Curr Opin Genet Dev. 1997;7:99–104. doi: 10.1016/s0959-437x(97)80116-5. [DOI] [PubMed] [Google Scholar]
- Lindsay HD, Griffiths DJ, Edwards RJ, Christensen PU, Murray JM, Osman F, Walworth N, Carr AM. S-phase-specific activation of Cds1 kinase defines a subpathway of the checkpoint response in Schizosaccharomyces pombe. Genes & Dev. 1998;12:382–395. doi: 10.1101/gad.12.3.382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinho RG, Linsay HD, Flaggs G, DeMaggio AJ, Hoekstra MF, Carr AM, Bentley NJ. Analysis of Rad3 and Chk1 protein kinases defines different checkpoint responses. EMBO J. 1998;17:7239–7249. doi: 10.1093/emboj/17.24.7239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maundrell K. nmt1 of fission yeast. A highly transcribed gene completely repressed by thiamine. J Biol Chem. 1990;265:10857–10864. [PubMed] [Google Scholar]
- McFarlane RJ, Carr AM, Price C. Characterization of the Schizosaccharomyces pombe rad4/cut5 mutant phenotypes: dissection of DNA replication and G2 checkpoint control function. Mol Gen Genet. 1997;255:332–340. doi: 10.1007/s004380050504. [DOI] [PubMed] [Google Scholar]
- Montecucco A, Rossi R, Levin DS, Gary R, Park MS, Motycka TA, Ciarrocchi G, Villa A, Biamonti G, Tomkinson AE. DNA ligase I is recruited to sites of DNA replication by an interaction with proliferating cell nuclear antigen: identification of a common targeting mechanism for the assembly of replication factories. EMBO J. 1998;17:3786–3795. doi: 10.1093/emboj/17.13.3786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno SA, Klar A, Nurse P. Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol. 1991;194:795–823. doi: 10.1016/0076-6879(91)94059-l. [DOI] [PubMed] [Google Scholar]
- Morgan SE, Kastan MB. Dissociation of radiation-induced phosphorylation of replication protein A from the S-phase checkpoint. Cancer Res. 1997;57:3386–3389. [PubMed] [Google Scholar]
- Morgan SE, Lovly C, Pandita TK, Shiloh Y, Kastan MB. Fragments of ATM which have dominant-negative or complementing activity. Mol Cell Biol. 1997;17:2020–2029. doi: 10.1128/mcb.17.4.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrow DM, Tagle DA, Shiloh Y, Collins FS, Hieter P. TEL1, an S. cerevisiae homolog of the human gene mutated in ataxia telangiectasia, is functionally related to the yeast checkpoint gene MEC1. Cell. 1995;82:831–840. doi: 10.1016/0092-8674(95)90480-8. [DOI] [PubMed] [Google Scholar]
- Naito T, Matsuura A, Ishikawa F. Circular chromosome formation in a fission yeast mutant defective in two ATM homologues. Nat Genet. 1998;20:203–206. doi: 10.1038/2517. [DOI] [PubMed] [Google Scholar]
- Painter RB, Young BR. Radiosensitivity in ataxia-telangiectasia: a new explanation. Proc Natl Acad Sci USA. 1980;77:7315–7317. doi: 10.1073/pnas.77.12.7315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phizicky EM, Fields S. Protein-protein interactions: methods for detection and analysis. Microbiol Rev. 1995;59:94–123. doi: 10.1128/mr.59.1.94-123.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prentice HL. High efficiency transformation of Schizosaccharomyces pombe by electroporation. Nucleic Acids Res. 1992;20:621. doi: 10.1093/nar/20.3.621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowley R, Subramani S, Young PG. Checkpoint controls in Schizosaccharomyces pombe: rad1. EMBO J. 1992;11:1335–1342. doi: 10.1002/j.1460-2075.1992.tb05178.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabatini DM, Erdjument-Bromage H, Lui M, Tempst P, Snyder SH. RAFT1: a mammalian protein that binds to FKBP12 in a rapamycin-dependent manner and is homologous to yeast TORs. Cell. 1994;78:35–43. doi: 10.1016/0092-8674(94)90570-3. [DOI] [PubMed] [Google Scholar]
- Saka Y, Esashi F, Matsusaka T, Mochida S, Yanagida M. Damage and replication checkpoint control in fission yeast is ensured by interactions of Crb2, a protein with BRCT motif, with Cut5 and Chk1. Genes & Dev. 1997;11:3387–3400. doi: 10.1101/gad.11.24.3387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkaria JN, Tibbetts RS, Busby EC, Kennedy AP, Hill DE, Abraham RT. Inhibition of phosphoinositide 3-kinase related kinases by the radiosensitizing agent wortmannin. Cancer Res. 1998;58:4375–4382. [PubMed] [Google Scholar]
- Savitsky K, et al. A single ataxia telangiectasia gene with a product similar to PI-3 kinase. Science. 1995a;268:1749–1753. doi: 10.1126/science.7792600. [DOI] [PubMed] [Google Scholar]
- Savitsky K, Sfez S, Ziv Y, Tagle DA, Sartiel A, Collins FS, Shiloh Y, Rotman G. A complete ATM gene product is similar to cell cycle regulators in different species. Hum Mol Genet. 1995b;4:2025–2032. doi: 10.1093/hmg/4.11.2025. [DOI] [PubMed] [Google Scholar]
- Sazer S, Nurse P. A fission yeast RCC1-related protein is required for the mitosis to interphase transition. EMBO J. 1994;13:606–615. doi: 10.1002/j.1460-2075.1994.tb06298.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sazer S, Sherwood SW. Mitochondrial growth and DNA synthesis occur in the absence of nuclear DNA replication in fission yeast. J Cell Sci. 1990;97:509–516. doi: 10.1242/jcs.97.3.509. [DOI] [PubMed] [Google Scholar]
- Scott SP, Zhang N, Khanna KK, Khromykh A, Hobson K, Watters D, Lavin MF. Cloning and expression of the ataxia-telangiectasia gene in baculovirus. Biochem Biophys Res Commun. 1998;245:144–148. doi: 10.1006/bbrc.1998.8137. [DOI] [PubMed] [Google Scholar]
- Scully R, Chen J, Ochs RL, Keegan K, Hoekstra M, Feunteun J, Livingston DM. Dynamic changes of BRCA1 subnuclear location and phosphorylation state are initiated by DNA damage. Cell. 1997;90:425–435. doi: 10.1016/s0092-8674(00)80503-6. [DOI] [PubMed] [Google Scholar]
- Seaton BL, Yucel J, Sunnerhagen P, Subramani S. Isolation and characterization of the Schizosaccharomyces pombe rad3 gene, involved in the DNA damage and DNA synthesis checkpoints. Gene. 1992;119:83–89. doi: 10.1016/0378-1119(92)90069-2. [DOI] [PubMed] [Google Scholar]
- Shiloh Y. Ataxia-telangiectasia: closer to unraveling the mystery. Eur J Hum Genet. 1995;3:116–138. doi: 10.1159/000472285. [DOI] [PubMed] [Google Scholar]
- Sunnerhagen P, Seaton BL, Nasim A, Subramani S. Cloning and analysis of a gene involved in DNA repair and recombination, the rad1 gene of Schizosaccharomyces pombe. Mol Cell Biol. 1990;10:3750–3760. doi: 10.1128/mcb.10.7.3750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walworth NC, Bernards R. rad-dependent response of the chk1-encoded protein kinase at the DNA damage checkpoint. Science. 1996;271:353–356. doi: 10.1126/science.271.5247.353. [DOI] [PubMed] [Google Scholar]
- Warbrick E, Lane DP, Glover DM, Cox LS. A small peptide inhibitor of DNA replication defines the site of interaction between the cyclin-dependent kinase inhibitor p21WAF1 and proliferating cell nuclear antigen. Curr Biol. 1995;5:275–282. doi: 10.1016/s0960-9822(95)00058-3. [DOI] [PubMed] [Google Scholar]
- Weinert TA. Dual cell cycle checkpoints sensitive to chromosome replication and DNA damage in the budding yeast Saccharomyces cerevisiae. Radiat Res. 1992;132:141–143. [PubMed] [Google Scholar]
- Willson J, Wilson S, Warr N, Watts FZ. Isolation and characterization of the Schizosaccharomyces pombe rhp9 gene: a gene required for the DNA damage checkpoint but not the replication checkpoint. Nucleic Acids Res. 1997;25:2138–2146. doi: 10.1093/nar/25.11.2138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Ashley T, Brainerd EE, Bronson RT, Meyn MS, Baltimore D. Targeted disruption of ATM leads to growth retardation, chromosomal fragmentation during meiosis, immune defects, and thymic lymphoma. Genes & Dev. 1996;10:2411–2422. doi: 10.1101/gad.10.19.2411. [DOI] [PubMed] [Google Scholar]
- Ziv Y, et al. Human cDNA clones that modify radiomimetic sensitivity of ataxia-telangiectasia (group A) cells. Somat Cell Mol Genet. 1995;21:99–111. doi: 10.1007/BF02255785. [DOI] [PubMed] [Google Scholar]