Abstract
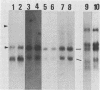
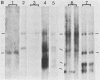
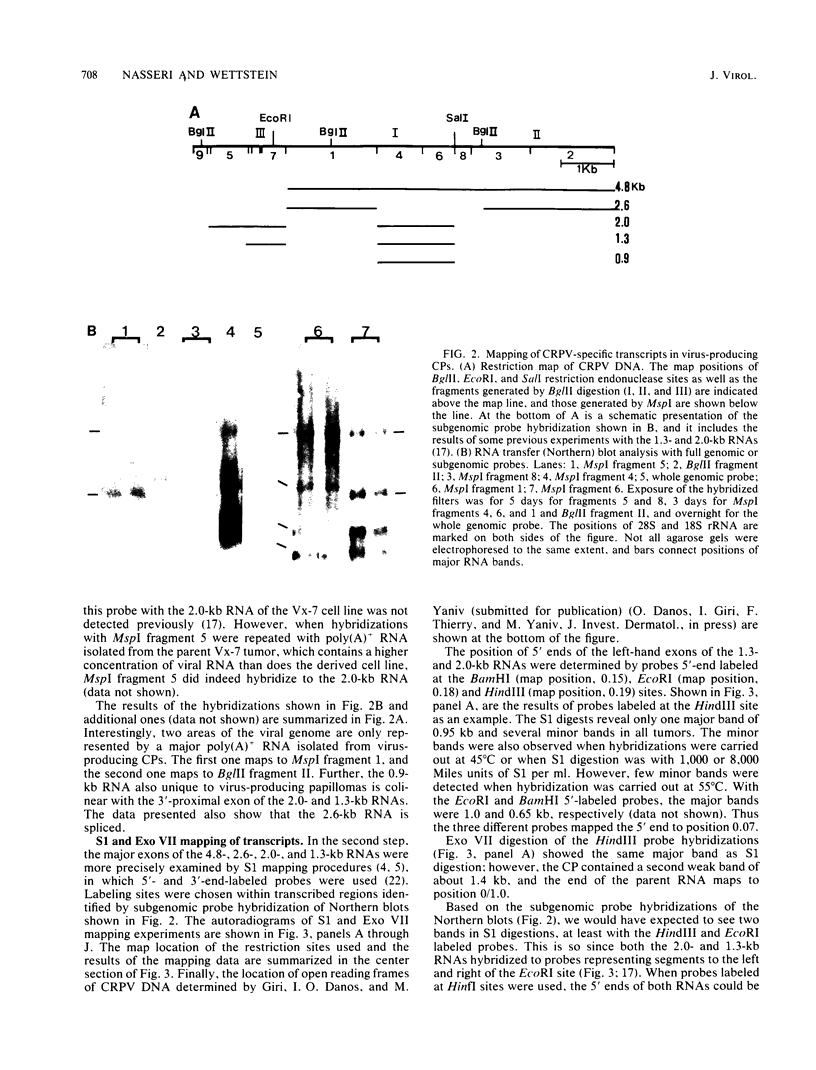
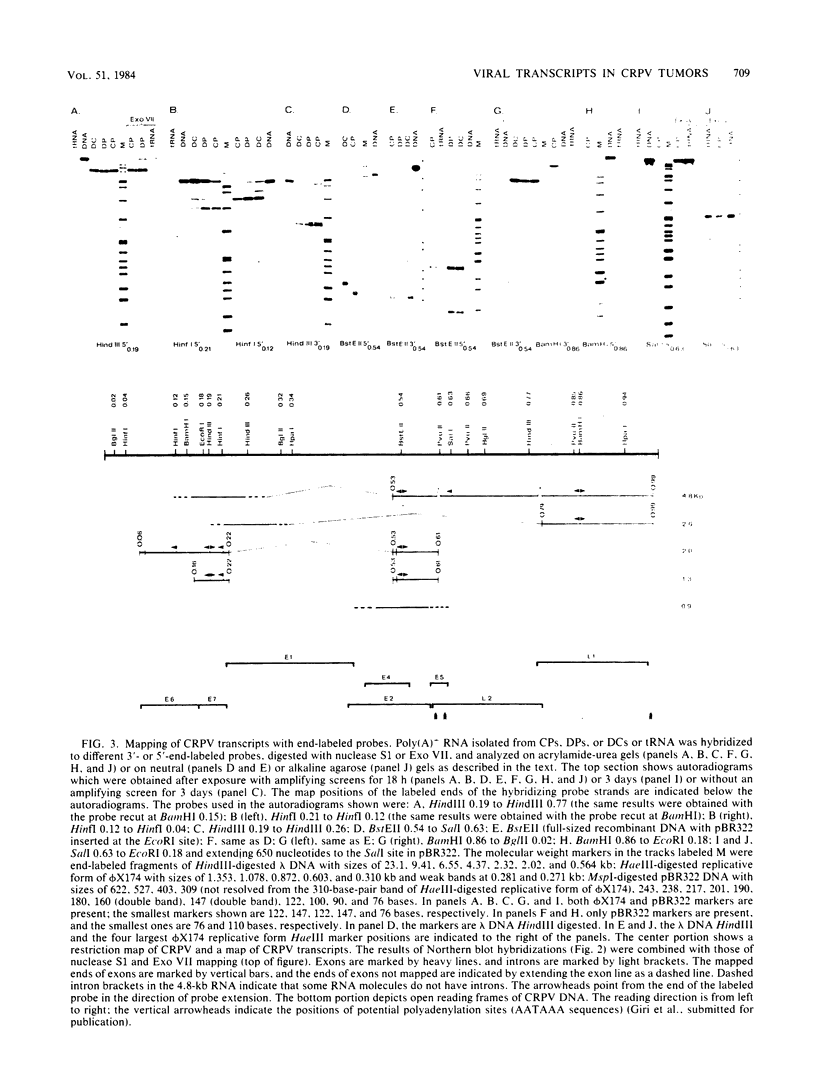
Five major cottontail rabbit papillomavirus-specific polyadenylated RNA species with sizes of 4.8, 2.6, 2.0, 1.3, and 0.9 kilobases (kb) were found in virus-producing tumors of cottontail rabbits (the natural host for the virus). Two of the RNA species (sizes, 2.0 and 1.3 kb) are indistinguishable with respect to size and map position from the RNA species detected previously in non-virus-producing benign and malignant tumors (Nasseri et al., J. Virol. 44:263-268, 1982). The 2.0-kb RNA in virus-producing benign tumors is more abundant than the 1.3-kb RNA. This, together with similar observations of benign non-virus-producing tumors, suggests that the predominance of the 2.0-kb RNA is a general feature of benign tumors. The change to a preferential synthesis of the 1.3-kb RNA appears to be a phenomenon of tumor progression from papillomas to carcinomas. Three transcripts of 4.8, 2.6, and 0.9 kb are unique to virus-producing tumors. The RNA molecules were mapped in two steps. First, hybridization of Northern blots with subgenomic probes revealed the approximate map position of the transcripts. Second, with nuclease S1 and exonuclease VII mapping procedures and end-labeled probes, the major exons of the 4.8-, 2.6-, 2.0-, and 1.3-kb RNAs were mapped precisely, and it is shown that all RNAs are transcribed from the same DNA strand. Both 1.3- and 2.0- kb RNAs consist of two exons which are separated by an identical 2.45-kb intron. The 5' ends of the 5'-proximal exons of the 2.0- and 1.3-kb RNAs map to positions 0.07 and 0.16, respectively. Some of the 2.0-kb RNA molecules, especially in the carcinoma, have an alternative 5' end at position 0.06. The 3' ends of both exons map to position 0.22, where two ends were found about seven nucleotides apart. The sizes of the 5'-proximal exons of the 2.0- and 1.3-kb RNAs are 1.23 and 0.48 kb, respectively. The 1.3- and 2.0-kb RNAs share a common 3'-proximal exon of 0.66 (0.61) kb. This exon has two 5' ends 50 nucleotides apart at map position 0.53 and a 3' end at map position 0.61. Only the 3'-proximal part of the 4.8- and 2.6-kb RNAs have been mapped precisely. Both RNAs share a common 3' end at position 0.99. The 2.6-kb RNA part consists of a single 1.59-kb exon which extends to map position 0.79.(ABSTRACT TRUNCATED AT 400 WORDS)
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ahola H., Stenlund A., Moreno-Lopez J., Pettersson U. Sequences of bovine papillomavirus type 1 DNA--functional and evolutionary implications. Nucleic Acids Res. 1983 May 11;11(9):2639–2650. doi: 10.1093/nar/11.9.2639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amtmann E., Sauer G. Bovine papilloma virus transcription: polyadenylated RNA species and assessment of the direction of transcription. J Virol. 1982 Jul;43(1):59–66. doi: 10.1128/jvi.43.1.59-66.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bantle J. A., Maxwell I. H., Hahn W. E. Specificity of oligo (dT)-cellulose chromatography in the isolation of polyadenylated RNA. Anal Biochem. 1976 May 7;72:413–427. doi: 10.1016/0003-2697(76)90549-2. [DOI] [PubMed] [Google Scholar]
- Berk A. J., Sharp P. A. Sizing and mapping of early adenovirus mRNAs by gel electrophoresis of S1 endonuclease-digested hybrids. Cell. 1977 Nov;12(3):721–732. doi: 10.1016/0092-8674(77)90272-0. [DOI] [PubMed] [Google Scholar]
- Berk A. J., Sharp P. A. Spliced early mRNAs of simian virus 40. Proc Natl Acad Sci U S A. 1978 Mar;75(3):1274–1278. doi: 10.1073/pnas.75.3.1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen E. Y., Howley P. M., Levinson A. D., Seeburg P. H. The primary structure and genetic organization of the bovine papillomavirus type 1 genome. Nature. 1982 Oct 7;299(5883):529–534. doi: 10.1038/299529a0. [DOI] [PubMed] [Google Scholar]
- Danos O., Katinka M., Yaniv M. Human papillomavirus 1a complete DNA sequence: a novel type of genome organization among papovaviridae. EMBO J. 1982;1(2):231–236. doi: 10.1002/j.1460-2075.1982.tb01152.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engel L. W., Heilman C. A., Howley P. M. Transcriptional organization of bovine papillomavirus type 1. J Virol. 1983 Sep;47(3):516–528. doi: 10.1128/jvi.47.3.516-528.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Favre M. Structural polypeptides of rabbit, bovine, and human papillomaviruses. J Virol. 1975 May;15(5):1239–1247. doi: 10.1128/jvi.15.5.1239-1247.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freese U. K., Schulte P., Pfister H. Papilloma virus-induced tumors contain a virus-specific transcript. Virology. 1982 Feb;117(1):257–261. doi: 10.1016/0042-6822(82)90525-6. [DOI] [PubMed] [Google Scholar]
- Heilman C. A., Engel L., Lowy D. R., Howley P. M. Virus-specific transcription in bovine papillomavirus-transformed mouse cells. Virology. 1982 May;119(1):22–34. doi: 10.1016/0042-6822(82)90061-7. [DOI] [PubMed] [Google Scholar]
- Lusky M., Botchan M. R. Characterization of the bovine papilloma virus plasmid maintenance sequences. Cell. 1984 Feb;36(2):391–401. doi: 10.1016/0092-8674(84)90232-0. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- McMaster G. K., Carmichael G. G. Analysis of single- and double-stranded nucleic acids on polyacrylamide and agarose gels by using glyoxal and acridine orange. Proc Natl Acad Sci U S A. 1977 Nov;74(11):4835–4838. doi: 10.1073/pnas.74.11.4835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meinke W., Meinke G. C. Isolation and characterization of the major capsid protein of bovine papilloma virus type 1. J Gen Virol. 1981 Jan;52(Pt 1):15–24. doi: 10.1099/0022-1317-52-1-15. [DOI] [PubMed] [Google Scholar]
- Nakabayashi Y., Chattopadhyay S. K., Lowy D. R. The transforming function of bovine papillomavirus DNA. Proc Natl Acad Sci U S A. 1983 Oct;80(19):5832–5836. doi: 10.1073/pnas.80.19.5832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasseri M., Wettstein F. O., Stevens J. G. Two colinear and spliced viral transcripts are present in non-virus-producing benign and malignant neoplasms induced by the shope (rabbit) papilloma virus. J Virol. 1982 Oct;44(1):263–268. doi: 10.1128/jvi.44.1.263-268.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigby P. W., Dieckmann M., Rhodes C., Berg P. Labeling deoxyribonucleic acid to high specific activity in vitro by nick translation with DNA polymerase I. J Mol Biol. 1977 Jun 15;113(1):237–251. doi: 10.1016/0022-2836(77)90052-3. [DOI] [PubMed] [Google Scholar]
- Schwarz E., Dürst M., Demankowski C., Lattermann O., Zech R., Wolfsperger E., Suhai S., zur Hausen H. DNA sequence and genome organization of genital human papillomavirus type 6b. EMBO J. 1983;2(12):2341–2348. doi: 10.1002/j.1460-2075.1983.tb01744.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp P. A., Sugden B., Sambrook J. Detection of two restriction endonuclease activities in Haemophilus parainfluenzae using analytical agarose--ethidium bromide electrophoresis. Biochemistry. 1973 Jul 31;12(16):3055–3063. doi: 10.1021/bi00740a018. [DOI] [PubMed] [Google Scholar]
- Stevens J. G., Wettstein F. O. Multiple copies of Shope virus DNA are present in cells of benign and malignant non-virus-producing neoplasms. J Virol. 1979 Jun;30(3):891–898. doi: 10.1128/jvi.30.3.891-898.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver R. F., Weissmann C. Mapping of RNA by a modification of the Berk-Sharp procedure: the 5' termini of 15 S beta-globin mRNA precursor and mature 10 s beta-globin mRNA have identical map coordinates. Nucleic Acids Res. 1979 Nov 10;7(5):1175–1193. doi: 10.1093/nar/7.5.1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wettstein F. O., Stevens J. G. Shope papilloma virus DNA is extensively methylated in non-virus-producing neoplasms. Virology. 1983 Apr 30;126(2):493–504. doi: 10.1016/s0042-6822(83)80007-5. [DOI] [PubMed] [Google Scholar]
- Wettstein F. O., Stevens J. G. Transcription of the viral genome in papillomas and carcinomas induced by the Shope virus. Virology. 1981 Mar;109(2):448–451. doi: 10.1016/0042-6822(81)90517-1. [DOI] [PubMed] [Google Scholar]
- Wettstein F. O., Stevens J. G. Variable-sized free episomes of Shope papilloma virus DNA are present in all non-virus-producing neoplasms and integrated episomes are detected in some. Proc Natl Acad Sci U S A. 1982 Feb;79(3):790–794. doi: 10.1073/pnas.79.3.790. [DOI] [PMC free article] [PubMed] [Google Scholar]