Abstract
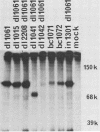
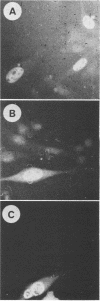
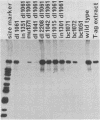
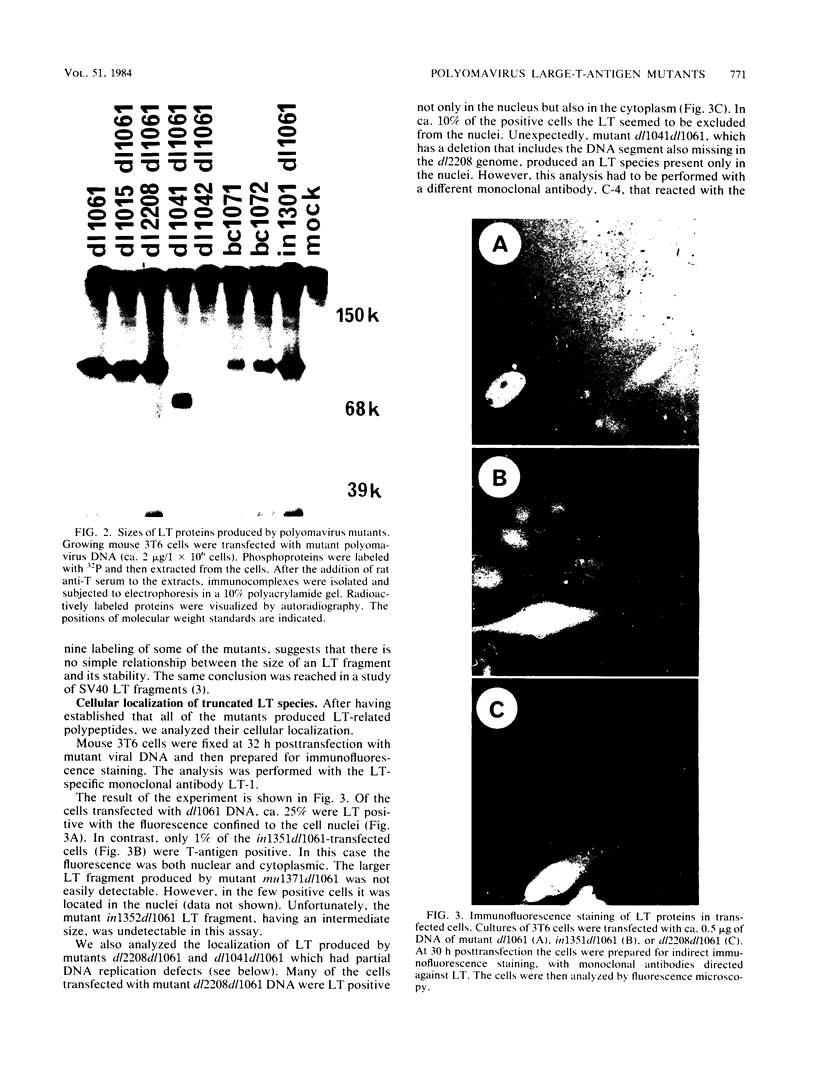
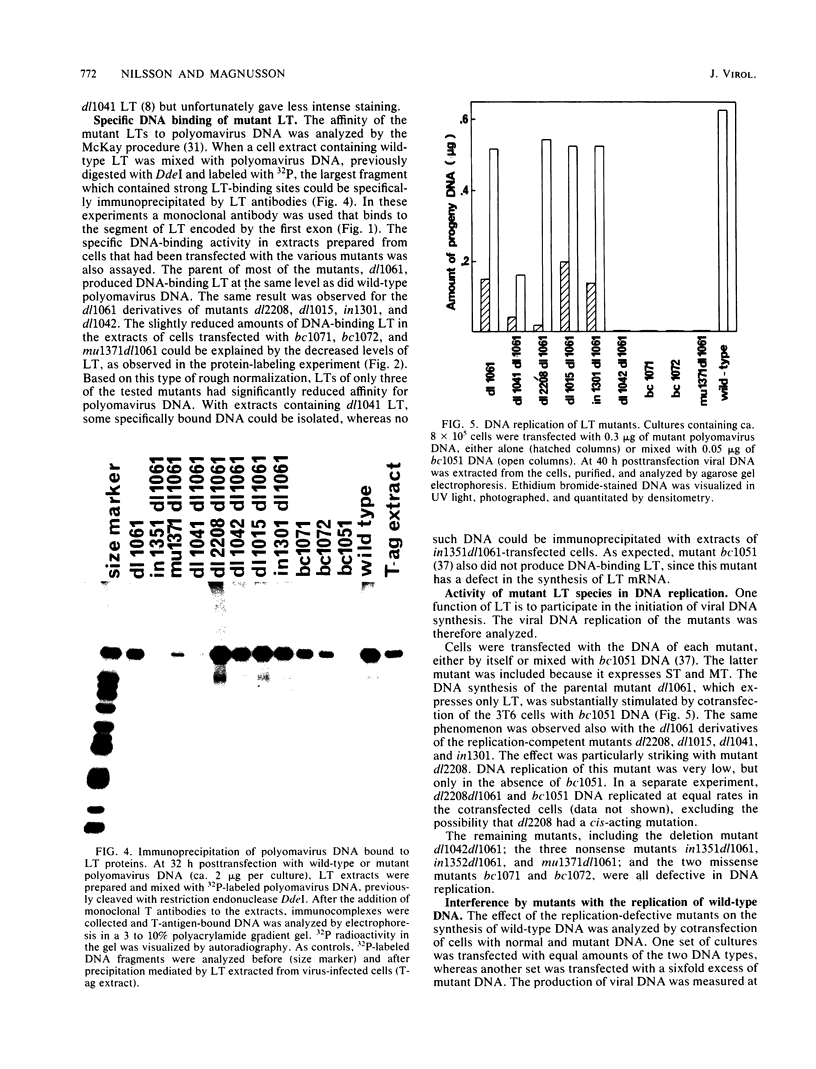
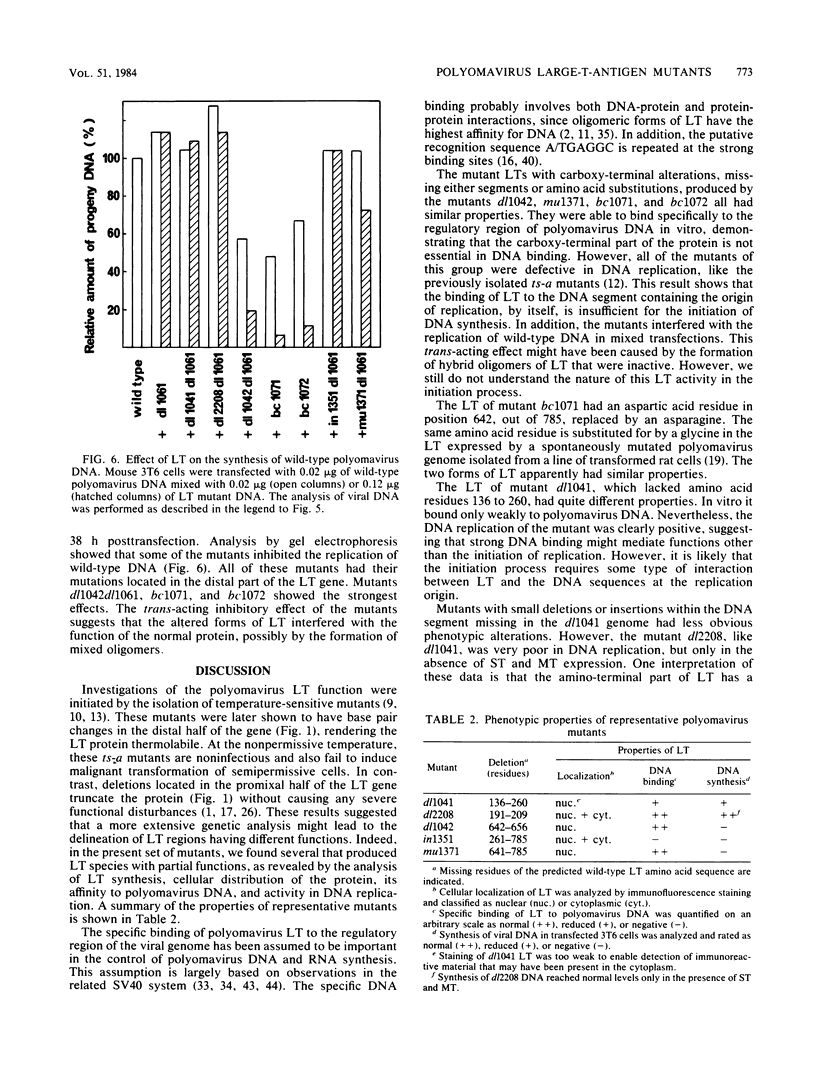
We constructed a set of polyomavirus mutants with alterations in the DNA sequences encoding large T-antigen. The mutant genomes were cloned and propagated as recombinants of plasmid pBR322, and the presence of the mutations was confirmed by nucleotide sequence analysis. To facilitate the analysis of defects in the function of large T-antigen, the dl1061 deletion was introduced into the mutant genomes. This deletion restricts the early gene expression to the synthesis of large T-antigen (Nilsson and Magnusson, EMBO J. 2:2095-2101, 1983). The mutant large T-antigens were identified after radioactive labeling. Their functional characterization was based on analysis of DNA binding, activity in the replication of viral DNA, and cellular localization. The native large T-antigen, which is 785 amino acid residues long, binds specifically to the regulatory region of polyomavirus DNA. This binding was significantly reduced by the deletion of amino acid residues 136 to 260. Nevertheless, this mutant large T-antigen was active in the initiation of viral DNA replication. Conversely, all of the mutants in this study that produced large T-antigens with alterations in the carboxy-terminal 146 amino acid residues had normal DNA-binding properties. However, these mutants were inactive in viral DNA synthesis and also inhibited the replication of wild-type DNA in cotransfected cells. The analysis of mutant dl2208 (Nilsson et al., J. Virol. 46:284-287, 1983) led to unexpected results. Its large T-antigen, missing amino acid residues 191 to 209, was overproduced. Although the protein had normal DNA-binding properties, it was not entering the cell nucleus normally. Furthermore, the dl2208 DNA replication was extremely low in the absence of small and middle T-antigens but was normal in the presence of these proteins.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bendig M. M., Thomas T., Folk W. R. Viable deletion mutant in the medium and large T-antigen-coding sequences of the polyoma virus genome. J Virol. 1980 Mar;33(3):1215–1220. doi: 10.1128/jvi.33.3.1215-1220.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradley M. K., Griffin J. D., Livingston D. M. Relationship of oligomerization to enzymatic and DNA-binding properties of the SV40 large T antigen. Cell. 1982 Jan;28(1):125–134. doi: 10.1016/0092-8674(82)90382-8. [DOI] [PubMed] [Google Scholar]
- Clark R., Peden K., Pipas J. M., Nathans D., Tjian R. Biochemical activities of T-antigen proteins encoded by simian virus 40 A gene deletion mutants. Mol Cell Biol. 1983 Feb;3(2):220–228. doi: 10.1128/mcb.3.2.220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clertant P., Cuzin F. Covalent affinity labeling by periodate-oxidized [alpha-32P]ATP of the large-T proteins of polyoma and SV40 viruses. J Biol Chem. 1982 Jun 10;257(11):6300–6305. [PubMed] [Google Scholar]
- Cogen B. Virus-specific early RNA in 3T6 cells infected by a tsA mutant of polyoma virus. Virology. 1978 Mar;85(1):222–230. doi: 10.1016/0042-6822(78)90426-9. [DOI] [PubMed] [Google Scholar]
- Collett M. S., Erikson R. L. Protein kinase activity associated with the avian sarcoma virus src gene product. Proc Natl Acad Sci U S A. 1978 Apr;75(4):2021–2024. doi: 10.1073/pnas.75.4.2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deininger P. L., LaPorte P., Friedmann T. Nucleotide sequence changes in polyoma ts-a mutants: correlation with protein structure. J Virol. 1981 Mar;37(3):871–875. doi: 10.1128/jvi.37.3.871-875.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Mayorca G., Callender J., Marin G., Giordano R. Temperature-sensitive mutants of polyoma virus. Virology. 1969 May;38(1):126–133. doi: 10.1016/0042-6822(69)90134-2. [DOI] [PubMed] [Google Scholar]
- Dilworth S. M., Griffin B. E. Monoclonal antibodies against polyoma virus tumor antigens. Proc Natl Acad Sci U S A. 1982 Feb;79(4):1059–1063. doi: 10.1073/pnas.79.4.1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckhart W. Complementation and transformation by temperature-sensitive mutants of polyoma virus. Virology. 1969 May;38(1):120–125. doi: 10.1016/0042-6822(69)90133-0. [DOI] [PubMed] [Google Scholar]
- FRIED M. CELL-TRANSFORMING ABILITY OF A TEMPERATURE-SENSITIVE MUTANT OF POLYOMA VIRUS. Proc Natl Acad Sci U S A. 1965 Mar;53:486–491. doi: 10.1073/pnas.53.3.486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fanning E., Westphal K. H., Brauer D., Cörlin D. Subclasses of simian virus 40 large T antigen: differential binding of two subclasses of T antigen from productively infected cells to viral and cellular DNA. EMBO J. 1982;1(9):1023–1028. doi: 10.1002/j.1460-2075.1982.tb01290.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francke B., Eckhart W. Polyoma gene function required for viral DNA synthesis. Virology. 1973 Sep;55(1):127–135. doi: 10.1016/s0042-6822(73)81014-1. [DOI] [PubMed] [Google Scholar]
- Friedmann T., Esty A., LaPorte P., Deininger P. The nucleotide sequence and genome organization of the polyoma early region: extensive nucleotide and amino acid homology with SV40. Cell. 1979 Jul;17(3):715–724. doi: 10.1016/0092-8674(79)90278-2. [DOI] [PubMed] [Google Scholar]
- Gaudray P., Clertant P., Cuzin F. ATP phosphohydrolase (ATPase) activity of a polyoma virus T antigen. Eur J Biochem. 1980 Aug;109(2):553–560. doi: 10.1111/j.1432-1033.1980.tb04827.x. [DOI] [PubMed] [Google Scholar]
- Gaudray P., Tyndall C., Kamen R., Cuzin F. The high affinity binding site on polyoma virus DNA for the viral large-T protein. Nucleic Acids Res. 1981 Nov 11;9(21):5697–5710. doi: 10.1093/nar/9.21.5697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffin B. E., Maddock C. New classes of viable deletion mutants in the early region of polyoma virus. J Virol. 1979 Sep;31(3):645–656. doi: 10.1128/jvi.31.3.645-656.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayday A. C., Chaudry F., Fried M. Loss of polyoma virus infectivity as a result of a single amino acid change in a region of polyoma virus large T-antigen which has extensive amino acid homology with simian virus 40 large T-antigen. J Virol. 1983 Feb;45(2):693–699. doi: 10.1128/jvi.45.2.693-699.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchinson M. A., Hunter T., Eckhart W. Characterization of T antigens in polyoma-infected and transformed cells. Cell. 1978 Sep;15(1):65–77. doi: 10.1016/0092-8674(78)90083-1. [DOI] [PubMed] [Google Scholar]
- Ito Y. Polyoma virus-specific 55K protein isolated from plasma membrane of productively infected cells is virus-coded and important for cell transformation. Virology. 1979 Oct 15;98(1):261–266. doi: 10.1016/0042-6822(79)90545-2. [DOI] [PubMed] [Google Scholar]
- Kamen R., Favaloro J., Parker J., Treisman R., Lania L., Fried M., Mellor A. Comparison of polyoma virus transcription in productively infected mouse cells and transformed rodent cell lines. Cold Spring Harb Symp Quant Biol. 1980;44(Pt 1):63–75. doi: 10.1101/sqb.1980.044.01.009. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Luthman H., Magnusson G. High efficiency polyoma DNA transfection of chloroquine treated cells. Nucleic Acids Res. 1983 Mar 11;11(5):1295–1308. doi: 10.1093/nar/11.5.1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luthman H., Nilsson M. G., Magnusson G. Non-contiguous segments of the polyoma genome required in cis for DNA replication. J Mol Biol. 1982 Nov 15;161(4):533–550. doi: 10.1016/0022-2836(82)90406-5. [DOI] [PubMed] [Google Scholar]
- Magnusson G., Berg P. Construction and analysis of viable deletion mutants of polyoma virus. J Virol. 1979 Nov;32(2):523–529. doi: 10.1128/jvi.32.2.523-529.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magnusson G., Nilsson M. G., Dilworth S. M., Smolar N. Characterization of polyoma mutants with altered middle and large T-antigens. J Virol. 1981 Sep;39(3):673–683. doi: 10.1128/jvi.39.3.673-683.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandel M., Higa A. Calcium-dependent bacteriophage DNA infection. J Mol Biol. 1970 Oct 14;53(1):159–162. doi: 10.1016/0022-2836(70)90051-3. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- McCutchan J. H., Pagano J. S. Enchancement of the infectivity of simian virus 40 deoxyribonucleic acid with diethylaminoethyl-dextran. J Natl Cancer Inst. 1968 Aug;41(2):351–357. [PubMed] [Google Scholar]
- McKay R. D. Binding of a simian virus 40 T antigen-related protein to DNA. J Mol Biol. 1981 Jan 25;145(3):471–488. doi: 10.1016/0022-2836(81)90540-4. [DOI] [PubMed] [Google Scholar]
- Morrison B., Kress M., Khoury G., Jay G. Simian virus 40 tumor antigen: isolation of the origin-specific DNA-binding domain. J Virol. 1983 Jul;47(1):106–114. doi: 10.1128/jvi.47.1.106-114.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers R. M., Rio D. C., Robbins A. K., Tjian R. SV40 gene expression is modulated by the cooperative binding of T antigen to DNA. Cell. 1981 Aug;25(2):373–384. doi: 10.1016/0092-8674(81)90056-8. [DOI] [PubMed] [Google Scholar]
- Myers R. M., Tjian R. Construction and analysis of simian virus 40 origins defective in tumor antigen binding and DNA replication. Proc Natl Acad Sci U S A. 1980 Nov;77(11):6491–6495. doi: 10.1073/pnas.77.11.6491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers R. M., Williams R. C., Tjian R. Oligomeric structure of a simian virus 40 T antigen in free form and bound to DNA. J Mol Biol. 1981 Jun 5;148(4):347–353. doi: 10.1016/0022-2836(81)90180-7. [DOI] [PubMed] [Google Scholar]
- Nilsson S. V., Magnusson G. Sealing of gaps in duplex DNA by T4 DNA ligase. Nucleic Acids Res. 1982 Mar 11;10(5):1425–1437. doi: 10.1093/nar/10.5.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson S. V., Magnusson G. T-antigen expression by polyoma mutants with modified RNA splicing. EMBO J. 1983;2(12):2095–2101. doi: 10.1002/j.1460-2075.1983.tb01708.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson S. V., Tyndall C., Magnusson G. Deletion mapping of a short polyoma virus middle T antigen segment important for transformation. J Virol. 1983 Apr;46(1):284–287. doi: 10.1128/jvi.46.1.284-287.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pipas J. M., Peden K. W., Nathans D. Mutational analysis of simian virus 40 T antigen: isolation and characterization of mutants with deletions in the T-antigen gene. Mol Cell Biol. 1983 Feb;3(2):203–213. doi: 10.1128/mcb.3.2.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pomerantz B. J., Mueller C. R., Hassell J. A. Polyomavirus large T antigen binds independently to multiple, unique regions on the viral genome. J Virol. 1983 Sep;47(3):600–610. doi: 10.1128/jvi.47.3.600-610.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prives C., Barnet B., Scheller A., Khoury G., Jay G. Discrete regions of simian virus 40 large T antigen are required for nonspecific and viral origin-specific DNA binding. J Virol. 1982 Jul;43(1):73–82. doi: 10.1128/jvi.43.1.73-82.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rassoulzadegan M., Cowie A., Carr A., Glaichenhaus N., Kamen R., Cuzin F. The roles of individual polyoma virus early proteins in oncogenic transformation. Nature. 1982 Dec 23;300(5894):713–718. doi: 10.1038/300713a0. [DOI] [PubMed] [Google Scholar]
- Rio D. C., Tjian R. SV40 T antigen binding site mutations that affect autoregulation. Cell. 1983 Apr;32(4):1227–1240. doi: 10.1016/0092-8674(83)90305-7. [DOI] [PubMed] [Google Scholar]
- Rio D., Robbins A., Myers R., Tjian R. Regulation of simian virus 40 early transcription in vitro by a purified tumor antigen. Proc Natl Acad Sci U S A. 1980 Oct;77(10):5706–5710. doi: 10.1073/pnas.77.10.5706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaffhausen B. S., Benjamin T. L. Phosphorylation of polyoma T antigens. Cell. 1979 Dec;18(4):935–946. doi: 10.1016/0092-8674(79)90206-x. [DOI] [PubMed] [Google Scholar]
- Schaffhausen B. S., Silver J. E., Benjamin T. L. Tumor antigen(s) in cell productively infected by wild-type polyoma virus and mutant NG-18. Proc Natl Acad Sci U S A. 1978 Jan;75(1):79–83. doi: 10.1073/pnas.75.1.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seif R., Cuzin F. Temperature-sensitive growth regulation in one type of transformed rat cells induced by the tsa mutant of polyoma virus. J Virol. 1977 Dec;24(3):721–728. doi: 10.1128/jvi.24.3.721-728.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shortle D., Nathans D. Local mutagenesis: a method for generating viral mutants with base substitutions in preselected regions of the viral genome. Proc Natl Acad Sci U S A. 1978 May;75(5):2170–2174. doi: 10.1073/pnas.75.5.2170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smart J. E., Ito Y. Three species of polyoma virus tumor antigens share common peptides probably near the amino termini of the proteins. Cell. 1978 Dec;15(4):1427–1437. doi: 10.1016/0092-8674(78)90066-1. [DOI] [PubMed] [Google Scholar]
- Smolar N., Griffin B. E. DNA sequences of polyoma virus early deletion mutants. J Virol. 1981 Jun;38(3):958–967. doi: 10.1128/jvi.38.3.958-967.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas T., Vollmer P., Folk W. R. Nucleotide sequence changes in polyoma virus A gene mutants. J Virol. 1981 Mar;37(3):1094–1098. doi: 10.1128/jvi.37.3.1094-1098.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]