Abstract
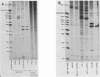
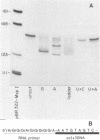
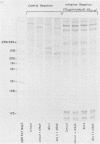
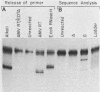
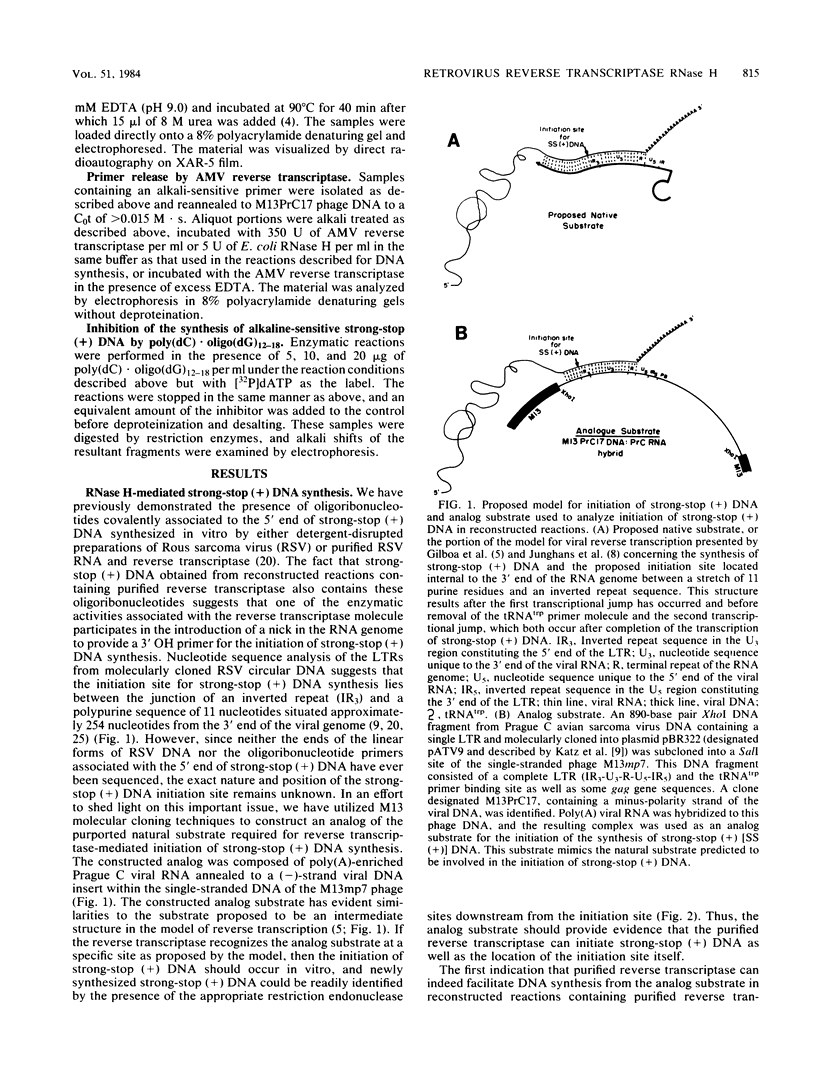
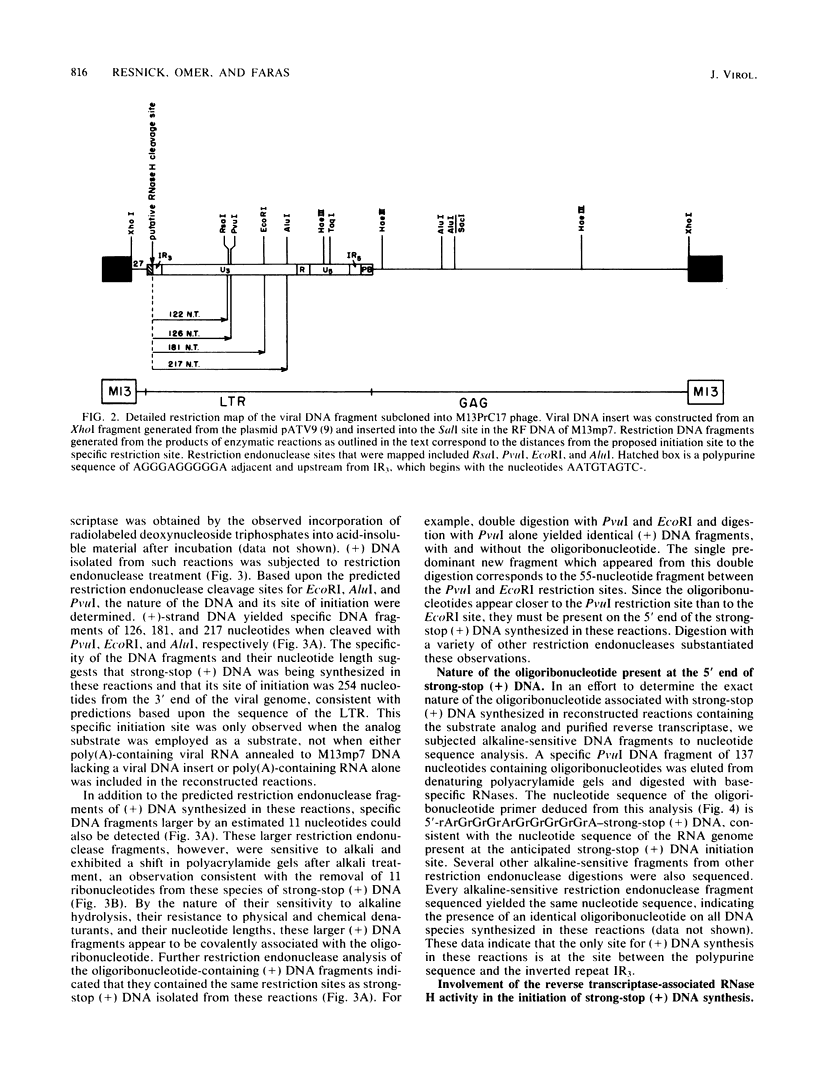
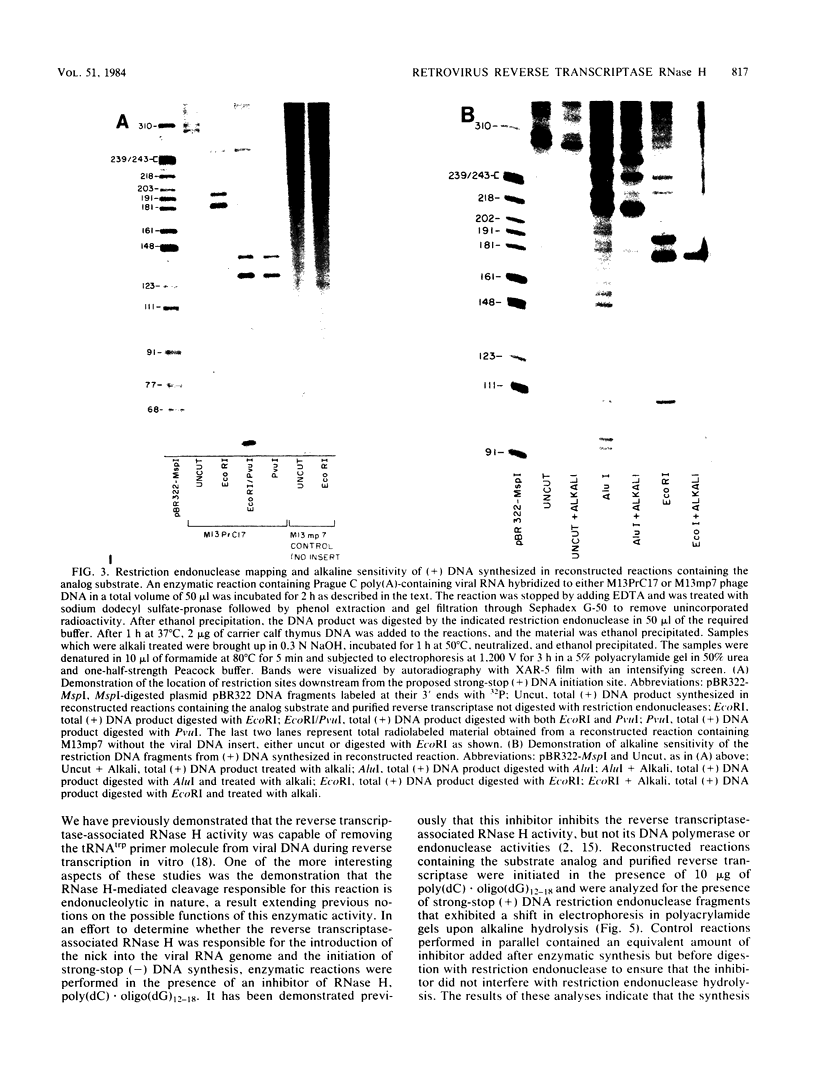
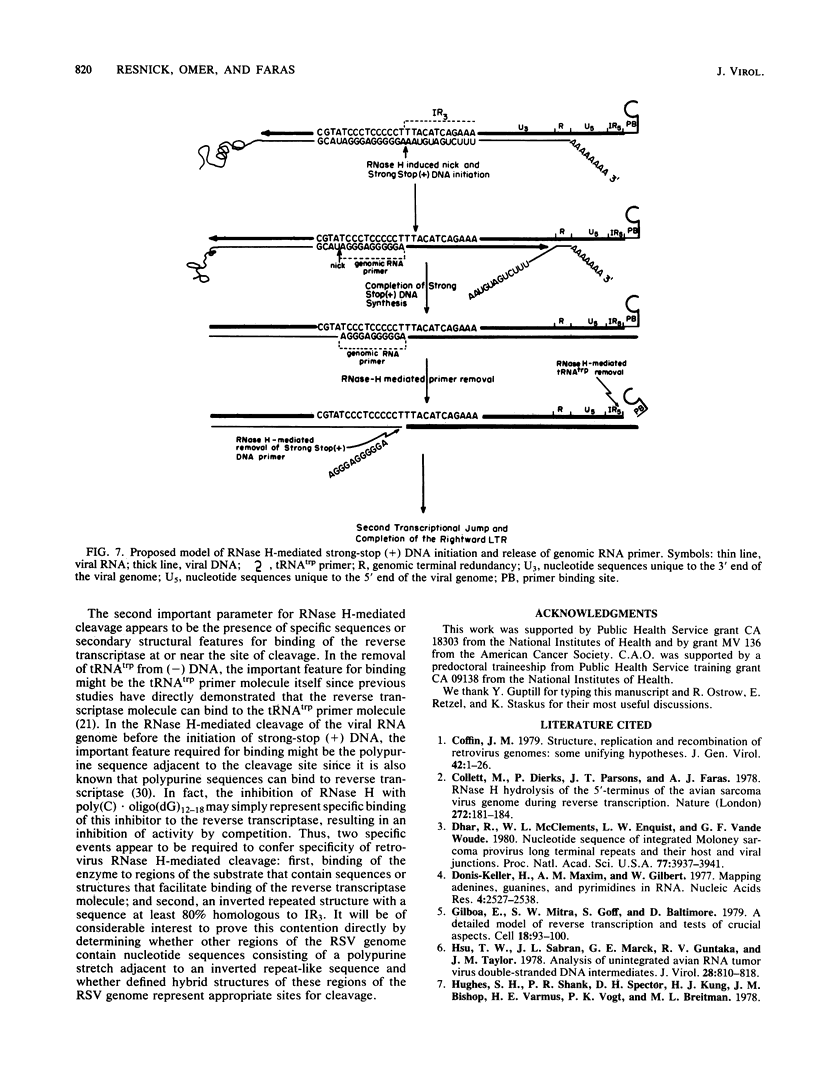
Reconstructed enzymatic reactions containing purified reverse transcriptase and defined analog substrates which mimic those purported to be natural substances for reverse transcription in vivo were employed to delineate the mechanism of strong-stop (+) DNA synthesis. Our analysis of this system has indicated that strong-stop (+) DNA synthesis is initiated after the introduction of a nick in the viral RNA genome between a polypurine sequence and an inverted repeat that represents the end of the long terminal repeat. Since inhibitors of the reverse transcriptase-associated RNase H activity prevent the introduction of the nick and the synthesis of strong-stop (+) DNA synthesis, it appears that this particular reverse transcriptase-associated enzymatic activity is responsible for the initiation of strong-stop (+) DNA. Our data also indicated that the RNase H activity creates a second nick in the viral RNA genome 11 nucleotides upstream from the strong-stop (+) DNA initiation site since the strong-stop (+) DNA synthesized in these reactions is covalently linked to an oligoribonucleotide 11 residues in length. Nucleotide sequence analysis of the oligoribonucleotide primer molecule indicated that a single homogenous oligomer was associated with strong-stop (+) DNA exhibiting the sequence rArGrGrGrArGrGrGrGrGrA. The oligoribonucleotide primer can be removed from strong-stop (+) DNA by the purified reverse transcriptase, which creates a nick at the junction between the primer and strong-stop (+) DNA. These data demonstrate that the initiation of strong-stop (+) DNA synthesis is mediated by RNase H and that the site of initiation is exactly at the end of the long terminal repeat, providing evidence for yet another function of this reverse transcriptase-associated enzymatic activity in the synthesis of retrovirus DNA.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Coffin J. M. Structure, replication, and recombination of retrovirus genomes: some unifying hypotheses. J Gen Virol. 1979 Jan;42(1):1–26. doi: 10.1099/0022-1317-42-1-1. [DOI] [PubMed] [Google Scholar]
- Collett M. S., Dierks P., Parsons J. T., Faras A. J. RNase H hydrolysis of the 5' terminus of the avian sarcoma virus genome during reverse transcription. Nature. 1978 Mar 9;272(5649):181–184. doi: 10.1038/272181a0. [DOI] [PubMed] [Google Scholar]
- Dhar R., McClements W. L., Enquist L. W., Vande Woude G. F. Nucleotide sequences of integrated Moloney sarcoma provirus long terminal repeats and their host and viral junctions. Proc Natl Acad Sci U S A. 1980 Jul;77(7):3937–3941. doi: 10.1073/pnas.77.7.3937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donis-Keller H., Maxam A. M., Gilbert W. Mapping adenines, guanines, and pyrimidines in RNA. Nucleic Acids Res. 1977 Aug;4(8):2527–2538. doi: 10.1093/nar/4.8.2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilboa E., Mitra S. W., Goff S., Baltimore D. A detailed model of reverse transcription and tests of crucial aspects. Cell. 1979 Sep;18(1):93–100. doi: 10.1016/0092-8674(79)90357-x. [DOI] [PubMed] [Google Scholar]
- Hsu T. W., Sabran J. L., Mark G. E., Guntaka R. V., Taylor J. M. Analysis of unintegrated avian RNA tumor virus double-stranded DNA intermediates. J Virol. 1978 Dec;28(3):810–818. doi: 10.1128/jvi.28.3.810-818.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes S. H., Shank P. R., Spector D. H., Kung H. J., Bishop J. M., Varmus H. E., Vogt P. K., Breitman M. L. Proviruses of avian sarcoma virus are terminally redundant, co-extensive with unintegrated linear DNA and integrated at many sites. Cell. 1978 Dec;15(4):1397–1410. doi: 10.1016/0092-8674(78)90064-8. [DOI] [PubMed] [Google Scholar]
- Junghans R. P., Boone L. R., Skalka A. M. Products of reverse transcription in avian retrovirus analyzed by electron microscopy. J Virol. 1982 Aug;43(2):544–554. doi: 10.1128/jvi.43.2.544-554.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz R. A., Omer C. A., Weis J. H., Mitsialis S. A., Faras A. J., Guntaka R. V. Restriction endonuclease and nucleotide sequence analyses of molecularly cloned unintegrated avian tumor virus DNA: structure of large terminal repeats in circle junctions. J Virol. 1982 Apr;42(1):346–351. doi: 10.1128/jvi.42.1.346-351.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller W., Crouch R. Degradation of DNA RNA hybrids by ribonuclease H and DNA polymerases of cellular and viral origin. Proc Natl Acad Sci U S A. 1972 Nov;69(11):3360–3364. doi: 10.1073/pnas.69.11.3360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leis J. P., Berkower I., Hurwitz J. Mechanism of action of ribonuclease H isolated from avian myeloblastosis virus and Escherichia coli. Proc Natl Acad Sci U S A. 1973 Feb;70(2):466–470. doi: 10.1073/pnas.70.2.466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majors J. E., Varmus H. E. Nucleotide sequences at host-proviral junctions for mouse mammary tumour virus. Nature. 1981 Jan 22;289(5795):253–258. doi: 10.1038/289253a0. [DOI] [PubMed] [Google Scholar]
- Messing J., Crea R., Seeburg P. H. A system for shotgun DNA sequencing. Nucleic Acids Res. 1981 Jan 24;9(2):309–321. doi: 10.1093/nar/9.2.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitra S. W., Chow M., Champoux J., Baltimore D. Synthesis of murine leukemia virus plus strong stop DNA initiates at a unique site. J Biol Chem. 1982 Jun 10;257(11):5983–5986. [PubMed] [Google Scholar]
- Modak M. J., Marcus S. L. Specific inhibition of DNA polymerase-associated RNase H by DNA. J Virol. 1977 Apr;22(1):243–246. doi: 10.1128/jvi.22.1.243-246.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers J. C., Dobkin C., Spiegelman S. RNA primer used in synthesis of anticomplementary DNA by reverse transcriptase of avian myeloblastosis virus. Proc Natl Acad Sci U S A. 1980 Mar;77(3):1316–1320. doi: 10.1073/pnas.77.3.1316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen J. C., Watson K. F. Avian retrovirus RNA-directed DNA synthesis by purified reverse transcriptase. Covalent linkage of RNA to plus strand DNA. Biochem Biophys Res Commun. 1980 Dec 31;97(4):1376–1383. doi: 10.1016/s0006-291x(80)80019-2. [DOI] [PubMed] [Google Scholar]
- Omer C. A., Faras A. J. Mechanism of release of the avian rotavirus tRNATrp primer molecule from viral DNA by ribonuclease H during reverse transcription. Cell. 1982 Oct;30(3):797–805. doi: 10.1016/0092-8674(82)90284-7. [DOI] [PubMed] [Google Scholar]
- Omer C. A., Parsons J. T., Faras A. J. Direct proof of the 5' to 3' transcriptional jump during reverse transcription of the avian retrovirus genome by DNA sequencing. J Virol. 1981 Apr;38(1):398–402. doi: 10.1128/jvi.38.1.398-402.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omer C. A., Resnick R., Faras A. J. Evidence for involvement of an RNA primer in initiation of strong-stop plus DNA synthesis during reverse transcription in vitro. J Virol. 1984 May;50(2):465–470. doi: 10.1128/jvi.50.2.465-470.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panet A., Haseltine W. A., Baltimore D., Peters G., Harada F., Dahlberg J. E. Specific binding of tryptophan transfer RNA to avian myeloblastosis virus RNA-dependent DNA polymerase (reverse transcriptase). Proc Natl Acad Sci U S A. 1975 Jul;72(7):2535–2539. doi: 10.1073/pnas.72.7.2535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabran J. L., Hsu T. W., Yeater C., Kaji A., Mason W. S., Taylor J. M. Analysis of integrated avian RNA tumor virus DNA in transformed chicken, duck and quail fibroblasts. J Virol. 1979 Jan;29(1):170–178. doi: 10.1128/jvi.29.1.170-178.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Coulson A. R. The use of thin acrylamide gels for DNA sequencing. FEBS Lett. 1978 Mar 1;87(1):107–110. doi: 10.1016/0014-5793(78)80145-8. [DOI] [PubMed] [Google Scholar]
- Shank P. R., Hughes S. H., Kung H. J., Majors J. E., Quintrell N., Guntaka R. V., Bishop J. M., Varmus H. E. Mapping unintegrated avian sarcoma virus DNA: termini of linear DNA bear 300 nucleotides present once or twice in two species of circular DNA. Cell. 1978 Dec;15(4):1383–1395. doi: 10.1016/0092-8674(78)90063-6. [DOI] [PubMed] [Google Scholar]
- Swanstrom R., DeLorbe W. J., Bishop J. M., Varmus H. E. Nucleotide sequence of cloned unintegrated avian sarcoma virus DNA: viral DNA contains direct and inverted repeats similar to those in transposable elements. Proc Natl Acad Sci U S A. 1981 Jan;78(1):124–128. doi: 10.1073/pnas.78.1.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanstrom R., Varmus H. E., Bishop J. M. The terminal redundancy of the retrovirus genome facilitates chain elongation by reverse transcriptase. J Biol Chem. 1981 Feb 10;256(3):1115–1121. [PubMed] [Google Scholar]
- Taylor J. M., Hsu T. W., Yeater C., Mason W. S. Synthesis and integration of avian sarcoma virus DNA. Cold Spring Harb Symp Quant Biol. 1980;44(Pt 2):1091–1096. doi: 10.1101/sqb.1980.044.01.117. [DOI] [PubMed] [Google Scholar]
- Temin H. M. Structure, variation and synthesis of retrovirus long terminal repeat. Cell. 1981 Nov;27(1 Pt 2):1–3. doi: 10.1016/0092-8674(81)90353-6. [DOI] [PubMed] [Google Scholar]
- Varmus H. E., Heasley S., Kung H. J., Oppermann H., Smith V. C., Bishop J. M., Shank P. R. Kinetics of synthesis, structure and purification of avian sarcoma virus-specific DNA made in the cytoplasm of acutely infected cells. J Mol Biol. 1978 Mar 25;120(1):55–82. doi: 10.1016/0022-2836(78)90295-4. [DOI] [PubMed] [Google Scholar]
- Verma I. M. The reverse transcriptase. Biochim Biophys Acta. 1977 Mar 21;473(1):1–38. doi: 10.1016/0304-419x(77)90005-1. [DOI] [PubMed] [Google Scholar]
- Yamamoto K. R., Alberts B. M., Benzinger R., Lawhorne L., Treiber G. Rapid bacteriophage sedimentation in the presence of polyethylene glycol and its application to large-scale virus purification. Virology. 1970 Mar;40(3):734–744. doi: 10.1016/0042-6822(70)90218-7. [DOI] [PubMed] [Google Scholar]