Abstract
To understand the factors specifically affecting tRNA nuclear export, we adapted in situ hybridization procedures to locate endogenous levels of individual tRNA families in wild-type and mutant yeast cells. Our studies of tRNAs encoded by genes lacking introns show that nucleoporin Nup116p affects both poly(A) RNA and tRNA export, whereas Nup159p affects only poly(A) RNA export. Los1p is similar to exportin-t, which facilitates vertebrate tRNA export. A los1 deletion mutation affects tRNA but not poly(A) RNA export. The data support the notion that Los1p and exportin-t are functional homologues. Because LOS1 is nonessential, tRNA export in vertebrate and yeast cells likely involves factors in addition to exportin-t. Mutation of RNA1, which encodes RanGAP, causes nuclear accumulation of tRNAs and poly(A) RNA. Many yeast mutants, including those with the rna1-1 mutation, affect both pre-tRNA splicing and RNA export. Our studies of the location of intron-containing pre-tRNAs in the rna1-1 mutant rule out the possibility that this results from tRNA export occurring before splicing. Our results also argue against inappropriate subnuclear compartmentalization causing defects in pre-tRNA splicing. Rather, the data support “feedback” of nucleus/cytosol exchange to the pre-tRNA splicing machinery.
INTRODUCTION
Nucleus/cytosol exchange of macromolecules is a complicated process requiring participation of “shared” gene products affecting exchange of many types of macromolecular cargo, as well as participation of “specific” gene products affecting a subset of the types of cargo. Generally, karyophilic proteins are imported into the nuclear interior, and newly synthesized RNAs are exported out to the cytosol, but some macromolecules pass in both directions. Entry and exit proceed through the same nuclear pores (Dworetzky and Feldherr, 1988). Nuclear pores are large supramolecular complexes comprising ∼50–100 separate proteins, called nucleoporins, that span the nuclear inner and outer membranes, creating an aqueous channel (for review see Fabre and Hurt, 1997; Ohno et al., 1998). Small molecules can diffuse through the pore, but most macromolecules are transported by an energy-requiring, signal-mediated process.
Nucleus/cytosol exchange seems to require a specific category of exchange components, called importin-β proteins, which are instrumental in docking of the cargo to the nuclear pore. The prototype of this family, importin-β, was identified as a cytoplasmic receptor for the nuclear localization signal containing karyophilic proteins (for review see Izaurralde and Adam, 1998; Ohno et al., 1998). Subsequent members of this family of receptors have been demonstrated to have different substrate specificity (Rout et al., 1997; Schlenstedt et al., 1997). RNA export, like protein import, requires the participation of receptors or exportins that are members of the importin-β family. Export of particular types of RNAs, i.e., tRNA, small nuclear RNA (snRNA), rRNA, and mRNA, is competed by an excess of that RNA type. However, the excess does not inhibit the export of other RNA types, indicating existence of limiting quantities of species-specific transit factors (Jarmolowski et al., 1994). Recent data on the roles of particular RNA binding proteins such as cap binding proteins, necessary for snRNA export, and hnRNP proteins, important for mRNA export, support the concept of RNA species-specific export pathways (Izaurralde and Adam, 1998). Some of the RNA binding proteins participating in the export process possess leucine-rich nuclear export sequences recognized by importin-β family member Crm1p/Xpo1p. Crm1p/Xpo1p has been shown to function as an exportin for the leucine-rich nuclear export sequence containing nucleus-localized proteins and to affect mRNA export to the cytosol (Fornerod et al., 1997; Fukuda et al., 1997; Kudo et al., 1997; Ossareh-Nazari et al., 1997; Stade et al., 1997; Ohno et al., 1998). Recently, another importin-β-like protein, exportin-t, has been proposed to serve as a tRNA-specific receptor for tRNA export in Xenopus and human cells by binding tRNA directly (Arts et al., 1998; Kutay et al., 1998).
Nucleus/cytosol exchange also requires participation of a small GTPase, Ran, and at least four proteins that regulate its GTP- or GDP-bound states. Although the role(s) of the Ran cycle in nucleus/cytosol exchange is not completely understood, several lines of evidence support the model that RanGDP/GTP exchange functions to release imported cargo from import receptors, and conversely, RanGTP hydrolysis functions to release exported cargo from export receptors (Izaurralde and Adam, 1998; Ohno et al., 1998). Although a functional Ran cycle is required for translocation of most RNAs (Izaurralde et al., 1997), export of the yeast heat shock mRNAs is apparently independent of the Ran cycle (Saavedra et al., 1996).
Our studies focus on tRNA biogenesis including those gene products necessary for pre-tRNA processing and export of the mature tRNAs to the cytosol. Yeast pre-tRNAs differ from their mature counterparts by possession of extra sequences located at the 5′ and 3′ extremities and, for ∼25% of tRNA families, by the presence of intron sequences located one nucleotide 3′ to the anticodon. Pre-tRNAs also lack numerous nucleoside modifications that are present on the mature tRNAs, posttranscriptionally added CCA nucleotides located at the 3′ end and sometimes a G located at the 5′ terminus (for review see Hopper and Martin, 1992). Although there appears to be a preferred order of processing steps, genetic studies and molecular analyses show that most of the steps are not in an obligatory order (Hopper et al., 1982; Martin and Hopper, 1982; O’Connor and Peebles, 1991; Hopper and Martin, 1992). Some order to the eukaryotic processing pathway may be imposed by the subcellular distribution of the processing activities, because particular processing activities such as m22G tRNA methyltransferase and splicing tRNA endonuclease appear to be located at the surface of the inner nuclear membrane (Peebles et al., 1983; Clark and Abelson, 1987; Rose et al., 1995), whereas other activities appear to be nucleolar (Bertrand et al., 1998; Hunter et al., 1998). Given what appears to be a nonobligatory order of processing steps, it is curious that those mutations that act indirectly in tRNA processing all affect the same step: excision of introns from the pre-tRNA.
Many of the genes that indirectly affect pre-tRNA splicing are known to play a role in nucleus/cytosol exchange, such as Rna1p and Prp20p (Amberg et al., 1992; Forrester et al., 1992; Kadowaki et al., 1993), and/or to be nucleoporins, such as Nsp1p, Nup49p, Nup116p, Nup133p, and Nup145p (Sharma et al, 1996). Why the first step of pre-tRNA splicing is often affected by gene products that act indirectly in pre-tRNA processing and how intervening sequence removal is coupled to nucleus/cytosol exchange are intriguing questions. In an effort to learn about gene products important for tRNA export and the coupling of nuclear export and pre-tRNA splicing, we successfully adapted in situ hybridization to locate endogenous levels of particular tRNAs in wild-type and mutant yeast cells. Here we describe the roles of particular yeast proteins in export of tRNAs to the cytosol.
MATERIALS AND METHODS
Strains and Media
The following yeast strains were used. EE1b-35 (MATa RNA1 rnh1::URA3 ura3-52 ade1 tyr1 his7 his4 Gal−) and EE1b-6 (MATa rna1-1 rnh1::URA3 ura3-52 ade1 tyr1 his7 his4 Gal−) were described in Traglia et al. (1989). Strains W303 α (MATα ade2-1 ura3-1 his3-11, 15 trp1-1 leu2-3, 112 NUP116) and SWY27 (MATα ade2-1 ura3-1 his3-11, 15 trp1-1 leu2-3, 112 nup116Δ::HIS) were obtained from S. Wente (Wente et al., 1993), and strains FY86 (MATα RAT7 his3Δ200 ura3-52 leu2Δ1) and LGY101 (MATα rat7-1 his3Δ200 ura3-52 leu2Δ1) were obtained from C.N. Cole (Gorsch et al., 1995). X2316-3C (MATα LOS1 SUP4 ade2-1 can1-100 lys1-1 his5-2 trp5-48 ura3-1) and IIIdIc-ΔV (los1-ΔV SUP4 ade2-1 can1-100 ura3-1) were described by Hurt et al. (1987). YEPD medium was used to grow yeast cells.
Oligonucleotide Probes
Probe 02 contains 50 residues of deoxythymidine. The sequences for probes 03, 04, and 05 are 5′-CGTTGCTTTTAAAGGCCTGTTTGAAAGGTCTTTGGCACAGAAACTTCGGAAACCGAATGTTGCTAT-3′, 5′GTGGGGATTGAACCCACGACGGTCGCGTTATAAGCACGAAGCTCTAACCACTGAGCTACA-3′, and 5′-GCGGGATCGAACCGCTGATCCCCGCGTTATTAGCACGGTGCCTTAACCAACTGGGCCAAG-3′, respectively. All oligonucleotides used as probes for Northern analysis and in the subsequent fluorescence in situ hybridization analyses were synthesized by the Pennsylvania State University College of Medicine Macromolecular Core Facility.
Preparation of RNA and Northern Analysis
RNA was isolated by phenol extraction from log phase yeast cells as described by Hopper et al. (1980). Approximately 15 μg of each RNA sample were used for Northern analysis, which was done according to Wang et al. (1988).
Fluorescence In Situ Hybridization
This procedure has been adapted from the previously published procedure of Kadowaki et al. (1992). Strains were grown at 23°C to log phase in YEPD and were either maintained at 23°C or shifted to 37°C for the indicated periods of time. Cells were prefixed in the culture by the addition of 0.1 volume of 37% formaldehyde. After 15 min, 5 ml cells were harvested by centrifugation and resuspended in 6 ml 4% paraformaldehyde, 0.1 M KPO4 (pH 6.5), and 5 mM MgCl2. After 3 h, cells were washed twice with solution B (1.2 M sorbitol and 0.1 M KPO4, pH 6.5) and resuspended in 2.8 ml solution B containing 0.05% β-mercaptoethanol and 50 μl 2 mg/ml freshly prepared 100T Zymolyase (ICN Biochemicals, Cosa Mesa, CA). Spheroplasting was conducted at 37°C for 20 min. Spheroplasts were washed three times in solution B and resuspended in 0.3 ml solution B. Cells were adhered to the wells of Teflon-faced slides (Cel-Line/Erie Scientific, Portsmouth, NH) that had been pretreated with a 0.1% (wt/vol) poly-l-lysine-containing solution (Sigma Chemical, St. Louis, MO). Nonadhered cells were removed by aspiration. Cells were treated with 70, 90, and 100% ethanol successively for a duration of 5 min each. The slides were placed in a dessicator for 5 min. Cells were then incubated in prehybridization buffer containing 10% dextran sulfate, 0.2% BSA, 2× SSC (1× SSC is 0.15 M NaCl and 0.015 M Na-citrate), 125 μg Escherichia coli tRNA/ml, and 500 μg denatured sonicated salmon sperm DNA/ml for 2 h at 37°C in a humid chamber. Hybridization buffer had the same composition with 450 pg/ml digoxigenin-labeled probes. All probes were labeled at their 3′ end using terminal transferase (Life Technologies, Gaithersburg, MD) and digoxigenin-11-UTP (Boehringer Mannheim, Indianapolis, IN) according to the previously described procedure of Amberg et al. (1992). Both the prehybridization and hybridization buffers contained RNasin (Promega, Madison, WI) at a concentration of 1 U/μl. Hybridization was carried out at 37°C overnight. Cells were washed three times with 2× SSC at 45°C for tRNA probes and at 37°C for the oligo(dT)50 probe. Cells were then washed three times for 10 min with 1× SSC at room temperature. Cells were briefly washed with 4× SSC containing 1% Triton X-100 and then blocked for 2 h using 1% BSA containing 4× SSC. Fluoresceinated anti-digoxigenin Fab fragment (Boehringer Mannheim) was diluted according to the manufacturer’s recommendation in solution containing 1% BSA and 4× SSC, and the cells were incubated with the diluted antibody for 2 h. Cells were washed twice with 4× SSC followed by two more washes with 4× SSC containing 1% Triton X-100, each wash lasting for 10 min. After two more rapid washes with 4× SSC, cell nuclei were counterstained with 0.1 μg/ml 4′,6-diamidino-2-phenylindole dihydrochloride (DAPI). After two rapid washes with water, the slides were mounted under 90% glycerol and 1× PBS containing 1 mg/ml p-phenylenediamine and stored at −20°C.
Fluorescence In Situ Hybridization and Indirect Immunofluorescence
Fluorescence in situ hybridization was carried out as described above, but after incubation with fluoresceinated anti-digoxigenin Fab fragment all subsequent washes were of only 5 min duration each. Cells were incubated with 1:20,000 dilution of mouse monoclonal antibody 32D6 (anti-Nsp1p) (Hunter et al., 1998) for 2 h followed by five rapid washes with 1× SSC and then incubated for 1 h with 1:400 dilution of CY3-conjugated goat-anti-mouse antibody (Jackson ImmunoResearch Laboratories, West Grove, PA). Cells were washed five times with 1× SSC and stained with DAPI as described above.
Microscopic Imaging
Fluorescence images were obtained by using a Nikon Microphot-FX microscope (Nikon Instrument Group, Melville, NY) equipped with a SenSys charged-coupled device camera (Photometrics, Tucson, AZ). Image processing was done with QED software (Pittsburgh, PA), NIH Image (http://rsb.info.nih.gov/nih-image), and Adobe Photoshop (Adobe Systems, Mountain View, CA).
RESULTS
In Situ Hybridization Analysis of the Subcellular Distributions of Endogenous Levels of tRNAs in Wild-Type Cells
In situ hybridization is a powerful method to study gene products important for the export of RNAs from their nuclear site of synthesis to their final cytoplasmic destination. Using oligo(dT) to detect total poly(A)-containing mRNA populations, numerous yeast mutants altered in mRNA transport have been identified (Amberg et al, 1992; Kadowaki et al., 1992). It has also been possible to study the location of particular mRNAs via in situ hybridization. Usually single mRNA species are detected by overexpression of the gene in question and by using multiple probes complementary to the mRNA species (Saavedra et al., 1996; Long et al., 1997); however, there is one report of successful use of in situ hybridization to locate endogenous levels of a mRNA (Takizawa et al., 1997). Because tRNAs have no sequences such as poly(A) in common with each other, it is not possible to study the cellular distribution of the total tRNA population. However, tRNAs are rather abundant molecules. Only ∼2.5 × 104 of the 1.2 × 107 nucleotide yeast genome is devoted to tRNA genes (for review see Hani and Feldmann, 1998), yet tRNAs constitute ∼20% of the total RNA population. One reason for tRNA abundance is long half-life. Given 42 tRNA species (Hani and Feldmann, 1998), an individual species should constitute ∼0.5% of the total cellular RNA, within an order of magnitude of the sum of the total mRNA population (∼1–5% of total RNA). Therefore, we anticipated that in situ hybridization would be sufficiently sensitive to locate a single species of mature tRNA within yeast cells. In contrast, tRNA precursors (pre-tRNAs) have shorter half-lives than their mature counterparts, and the ability to detect individual pre-tRNAs by in situ techniques could be problematic.
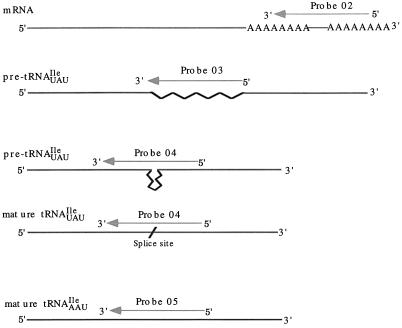
To determine whether it is possible to use in situ hybridization to locate individual tRNAs within yeast, we chose to study tRNAIle and designed oligonucleotide probes to detect particular tRNAIle species (Figure 1). Probe 05 contains 60 nucleotides and is complementary to tRNAIleAAU, which is encoded by 13 identical genes that do not contain introns (Hani and Feldmann, 1998). The specificity of the probe was determined by RNA blot analysis. Using this assay, a single RNA species that migrates at the expected molecular weight for mature tRNAIleAAU was detected (Figure 2). tRNAIleUAU is encoded by two identical genes that contain 60-nucleotide-long introns, the longest of the yeast tRNA introns (Hopper and Martin, 1992). To detect intron-containing pre-tRNAIleUAU, we used a 66-nucleotide probe complementary to the entire intron plus 3 nucleotides 5′ and 3′ to the intron (Figure 1, Probe 03). To detect the entire tRNAIleUAU population, we used an oligonucleotide containing 60 nucleotides complementary to a region of the mature tRNA sequence that spans the anticodon loop (Figure 1, Probe 04). By Northern analysis probe 04 detected three RNAs migrating at the expected molecular weights for mature tRNAIleUAU, end-matured intron-containing pre-tRNA, and 5′ and 3′ end-extended intron-containing pre-tRNA species. Probe 03, as expected, detected only the two intron-containing pre-tRNAs. Therefore, the probes detect only the anticipated tRNA species.
Figure 1.
Diagram of the regions of RNAs complementary to oligonucleotide probes. The straight line depicts mature RNA regions; the wavy line represents the IVS sequence, and the interrupted line marks the position from where an intron has been removed. Lines with arrows indicate the complementary regions of each probe.
Figure 2.
Northern blot detection of pre-tRNAIleUAU, mature tRNAIleUAU, and mature tRNAIleAAU. tRNAs were isolated from EE1b-35 (lanes 1, 2, 5, 6, 9, and 10) and EE1b-6 (lanes 3, 4, 7, 8, 11, and 12) cells grown either at 23°C (lanes 1, 3, 5, 7, 9, and 11) or shifted to 37°C for 1 h (lanes 2, 4, 6, 8, 10, and 12). Lanes 1–4 were incubated with probe 04, lanes 5–8 were incubated with probe 05, and lanes 9–11 were incubated first with probe 05 and then stripped and reprobed with probe 03. In lanes 1–4, the top band corresponds to 5′ and 3′ extended intron-containing pre-tRNA species, the next band corresponds to end-matured intron-containing pre-tRNA, and the bottom band corresponds to mature tRNAIleUAU.
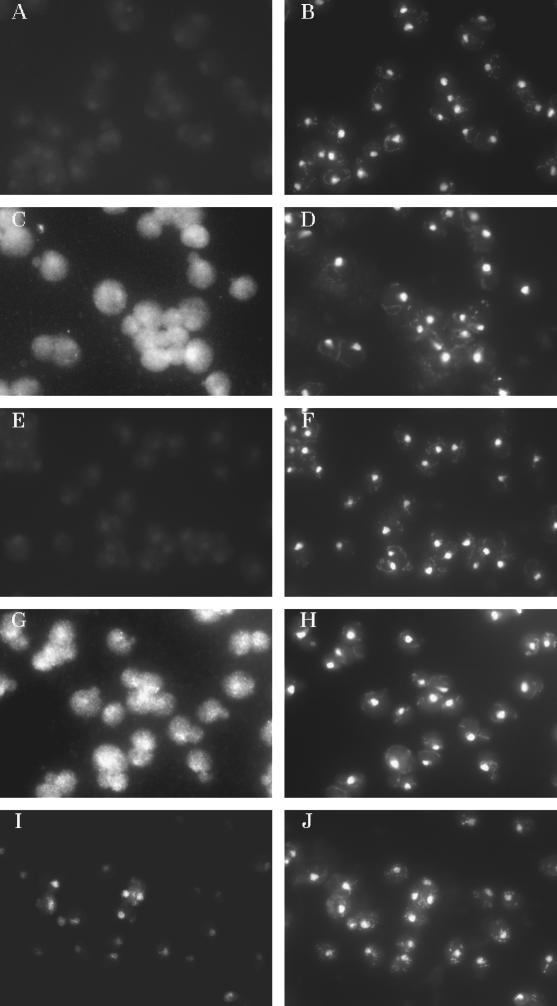
In situ hybridization studies using probes 03, 04, and 05 were conducted to locate the tRNAs within wild-type yeast strain EE1b-35 (Figure 3; see MATERIALS AND METHODS). The positions of nuclei were assessed by costaining cells with the DNA-specific dye DAPI (Figure 3) or by combining in situ hybridization with immunofluorescence techniques and using an antibody specific for nuclear pores (see below). Control cells treated identically to experimental cells except for the absence of probe had little or no fluorescent signal (Figure 3, A and B). Using probe 05, which is specific for mature tRNAIleAAU, wild-type cells grown at 23°C had signal throughout the cells, with some accumulation in nuclei (Figure 3, C and D). When the same cells were incubated for 1 or 3 h at 37°C, the signal was primarily cytoplasmic, and the nucleus appeared to be rather depleted for tRNAIleAAU (see below). Controls in which 1000-fold excess of unlabeled probe 05 was included during hybridization resulted in loss of the fluorescent signal (Figure 3, E and F), whereas including 1000-fold excess of probes 03 or 04 during hybridization did not affect the intensity or location of the probe 05 signal (our unpublished results). The results show that the hybridization signal detected is specific for tRNAIleAAU and indicate, as expected, that mature tRNAs are located primarily in the cytosol. Detection of a nuclear pool at 23°C but not at 37°C probably reflects a lower rate of nuclear export of tRNAIleAAU at the lower temperature growth conditions.
Figure 3.
Detection of endogenous levels of single species of tRNA is possible by fluorescence in situ hybridization. EE1b-35 cells were grown at 23°C and then subjected to fluorescence in situ hybridization. (A) In situ hybridization done in the absence of digoxigenin-labeled probe. (C) In situ hybridization detection of mature tRNAIleAAU using digoxigenin-labeled probe 05. (E) In situ hybridization detection of mature tRNAIleAAU using digoxigenin-labeled probe 05 in the presence of 1000-fold excess of unlabeled probe 05. (G) In situ hybridization detection of mature tRNAIleUAU using digoxigenin-labeled probe 04. (I) In situ hybridization detection of pre-tRNAIleUAU using digoxigenin-labeled probe 03. (B, D, F, H, and J) DAPI staining of cells shown in A, C, E, G, and I, respectively.
In situ hybridization using probe 04 complementary to the two forms of precursor and the mature tRNAIleUAU gave a cytosolic signal along with evidence of nuclear staining when the cells were grown at 23°C (Figure 3, G and H) or when incubated at 37°C for 1 or 3 h (our unpublished results). This signal was completely competed by the addition of 1000-fold excess of unlabeled probe 04 in the hybridization mix but not by heterologous probe 05 (our unpublished results), again documenting specificity. Because probe 04 detects pre-tRNAIleUAU as well as mature tRNAIleUAU, whereas probe 05 detects only mature tRNAAAU, the nuclear signal detected for wild-type cells incubated at 37°C using probe 04 but not 05 could be indicative of a nuclear pool of pre-tRNAIleUAU at both low and high temperatures. To test this, we used probe 03, which detects only pre-tRNAIleUAU as documented by Northern analysis for in situ hybridization. These studies revealed only a somewhat uniformly distributed nuclear signal whether the cells were grown at 23°C (Figure 3, I and J) or incubated at 37°C for 1 or 3 h (see below).
Absence of signal in the absence of probe, competition of signal for each probe when an excess of unlabeled probe was included during hybridization, and lack of competition when the same molar excess of either heterologous unlabeled probe was used, provide strong evidence that in situ hybridization can be used to detect endogenous levels of specific tRNAs in yeast. Moreover, the nuclear location of pre-tRNA and primarily cytosolic location of mature tRNAs substantiate the efficacy of the in situ hybridization procedure and further indicate the usefulness of this method for studies of subcellular distributions of tRNA species and/or pre-tRNA processing.
The Effects of Nucleoporins, Los1p, and RanGAP on Nuclear Export of tRNAs Encoded by Genes Lacking Introns
Nucleoporins.
Our goal is to identify genes important for the export of tRNAs to the cytosol. As described above, tRNA export is coupled to pre-tRNA splicing. To study export of tRNAs independent of pre-tRNA splicing, we chose to assess the location of tRNAIleAAU, which is encoded by genes lacking introns. In wild-type yeast cells tRNAIleAAU is located primarily in the cytosol (Figure 3, C and D). Because tRNAs are very stable molecules, cells will have high levels of cytosolic tRNAIleAAU whether or not they are blocked in tRNA nuclear export when incubated at nonpermissive conditions. Therefore, it was not clear whether it would be possible to detect by in situ hybridization increased nuclear pools over high cytosolic signals in cells with tRNA export defects. To determine this, we chose to compare the location of tRNAIleAAU in the nup116Δ mutant to its location in the parent strain. NUP116 encodes a nucleoporin, and a deletion of this gene causes a temperature-sensitive growth defect resulting from aberrant, sealed nucleopores and subsequent defects in nucleus/cytosol exchange (Wente and Blobel, 1993). Because of the aberrant nucleopores, we anticipated that export of tRNA would also be defective and that this strain would be useful to test whether in situ hybridization could be used to study tRNA nuclear retention.
Parent W303 strain and the nup116Δ strain were grown at a permissive temperature and incubated at 37°C for 1 h, and the locations of RNAs were determined by in situ hybridization (Figure 4). To be certain that the cells showed the appropriate defect in nucleus/cytosol exchange under these conditions and in the in situ procedures we use, we assessed the location of poly(A)-containing RNA in these cells using a 50-nucleotide oligo(d)T probe. Appropriate controls to confirm the specificity of the oligo(dT)50 probe were conducted (our unpublished results). For the parent grown at 23°C (our unpublished results) or incubated for 1 h at 37°C (Figure 4, A and B), poly(A)-containing RNA was distributed throughout the cells. For nup116Δ cells grown at the permissive temperature, the signal was indistinguishable from the parent strain (our unpublished results). In contrast, for nup116Δ cells incubated for 1 h at 37°C (Figure 4, C and D), poly(A)-containing RNA was predominately nuclear. Thus, nup116Δ cells display the previously reported defect in mRNA nuclear export (Wente and Blobel, 1993). Mature tRNAIleAAU was distributed throughout parental cells when grown at 23°C (our unpublished results). When the parent strain was incubated for 1 h at 37°C (Figure 4, E and F), the tRNAIleAAU nuclear signal was less prominent than at 23°C (our unpublished results). The signal for the nup116Δ mutants grown at 23°C was indistinguishable from its parent (our unpublished results). However, upon incubation of the nup116Δ cells for 1 h (Figure 4, G and H) at 37°C, prominent nuclear accumulation of tRNAIleAAU resulted. Even though there is considerable tRNAIleAAU in the cytosol of nup116Δ cells incubated at 37°C, within 1 h there is clear nuclear accumulation of tRNAIleAAU above this background. We conclude that it is possible to detect RNA export defects by in situ hybridization even for a stable molecule such as tRNA and that Nup116p is important for the movement of tRNA from the nucleus to the cytosol.
Figure 4.
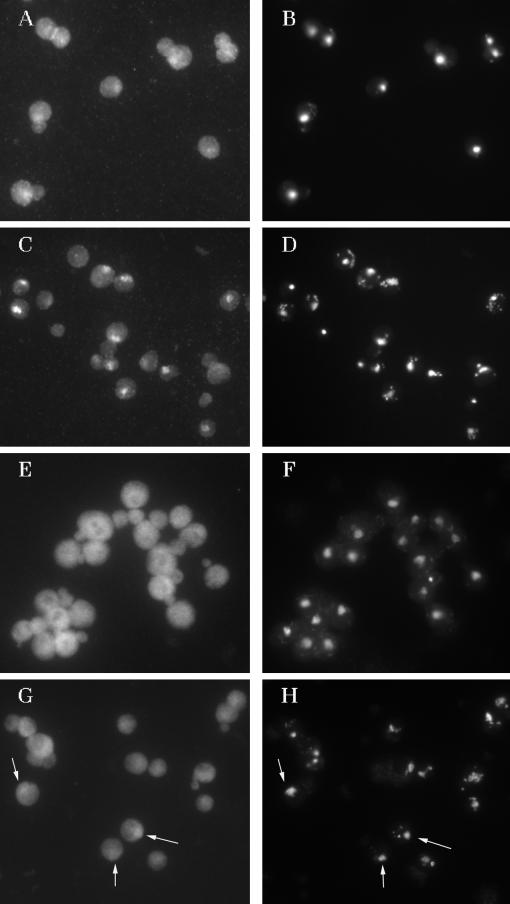
Identification of tRNA transport mutants is possible using fluorescence in situ hybridization. Parent W303 and nup116Δ strain SWY27 were grown at 23°C, and log phase cells were shifted to 37°C for 1 h. (A) In situ hybridization detection of Poly(A) RNA with digoxigenin-labeled probe 02 in W303 cells. (C) Detection of Poly(A) RNA with digoxigenin-labeled probe 02 in SWY27 cells. (E) Detection of mature tRNAIleAAU with digoxigenin-labeled probe 05 in W303 cells. (G) Mature tRNAIleAAU detection with digoxigenin-labeled probe 05 in SWY27 cells. (B, D, F, and H) DAPI staining of cells shown in A, C, E, and G, respectively.
Mutations of certain nucleoporins appear to affect nucleus/cytosol exchange in a single direction only. RAT7 encodes an essential nucleoporin, Nup159p, which contains GLFG and XFXFG repeats found in numerous other nucleoporins (Gorsch et al., 1995). The rat7-1 mutation causes temperature-sensitive growth defects at 37°C and defects in mRNA nuclear export, as assessed by in situ hybridization using oligo(dT) probes, but does not appear to affect nuclear import of at least some particular protein cargoes (Gorsch et al., 1995). To address the possibility that Nup159p could have substrate specificity for exported cargo, we assessed the locations of poly(A) RNA and tRNAIleAAU in rat7-1 cells and the parent to this mutant, strain FY86. As anticipated, rat7-1 cells accumulate substantially more poly(A) RNA in the nucleus when incubated for 1 h at 37°C than do the parental cells (Figure 5, A–D). As for the parent strain of nup116Δ, tRNAIleAAU was distributed throughout the FY86 cells when they were grown at 23°C, and there was a less prominent nuclear signal when these cells were incubated for 1 h at 37°C (Figure 5, E and F). However, in contrast to the results obtained for nup116Δ, tRNAIleAAU distribution in rat7-1 cells was indistinguishable from the isogenic FY86 parent cells. No tRNAIleAAU nuclear accumulation was evident when the rat7-1 cells were incubated for 1 h at 37°C (Figure 5, G and H). We conclude that not all nucleoporins that are important for the distribution of mRNA to the cytosol are important for the distribution of tRNA to the cytosol.
Figure 5.
Nucleoporins that are required for Poly(A) RNA export may not be required for mature tRNA export out of the nucleus. Parent FY86 and rat7-1 mutant strain LGY101 were grown at 23°C, and log phase cells were shifted to 37°C for 1 h. In situ hybridization was performed using the following probes and cells: (A) poly(A) RNA probe 02, FY86 cells; (C) poly(A) RNA probe 02, LGY101 cells; (E) mature tRNAIleAAU probe 05, FY86 cells; (G) mature tRNAIleAAU probe 05, LGY101 cells. (B, D, F, and H) DAPI staining of cells shown in A, C, E, and G, respectively.
Los1p.
Yeast Los1p bears similarity to the importin-β family of proteins (Görlich et al., 1997), specifically to vertebrate exportin-t, which has been shown to facilitate nuclear tRNA export (Arts et al., 1998; Kutay et al., 1998). LOS1 is an unessential yeast gene. Mutations of the LOS1 gene cause accumulation of intron-containing pre-tRNAs (Hopper et al., 1980, Simos et al., 1996) but do not appear to affect production of rRNA or most mRNAs (Hopper et al., 1980; Shen et al., 1996). If yeast Los1p is the functional homologue of exportin-t, then one might expect that in addition to the defects in pre-tRNA splicing, los1 mutants might show defects in the distribution of tRNA to the cytosol. To study the effects of Los1p on nuclear export independent of the affects on pre-tRNA splicing, we used in situ hybridization to locate tRNAIleAAU that is encoded by intronless genes.
The locations of poly(A) RNA and tRNAIleAAU (Figure 6A) were determined for wild-type strain X2316-3C and the related strain IIId1c-ΔV, which possesses a deletion allele, los1ΔV (Hurt et al., 1987). As expected, poly(A)-containing RNA and tRNAIleAAU were distributed throughout the X2316-3C cells when they were grown at 23°C (our unpublished results). When X2316-3C cells were incubated for 1 h (Figure 6) or 3 h at 37°C (our unpublished results), tRNAIleAAU was substantially depleted from nuclei (Figure 6, A, panels E and F, and B). In contrast, los1ΔV mutant cells showed significant tRNAIleAAU nuclear accumulation when the cells were incubated for 1 h (Figure 6, A, panels G and H, and B) or 3 h at 37°C (our unpublished results). The accumulation of nucleus-located RNA appeared to be tRNA specific, because the same cells showed no accumulation of nucleus-located poly(A) RNA (Figure 6A, panels C and D). Thus, los1 mutations affect export of tRNA but not mRNA at the nonpermissive temperature. Although it is difficult to obtain quantitative information regarding the amounts of nuclear signal to cytosolic signal by these methods, the nup116Δ strain appears to accumulate tRNAIleAAU in the nucleus more rapidly and to a higher level than do los1ΔV cells.
Figure 6.
(A) Deletion of LOS1 affects mature tRNA nuclear export. Parent X2316-3C and los1Δ mutant strain IIId1c-ΔV were grown at 23°C, and log phase cells were shifted to 37°C for 1 h. In situ hybridization was as follows: (panel A) detection of Poly(A) RNA with probe 02 in X2316-3C cells; (panel C) detection of Poly(A) RNA with probe 02 in IIId1c-ΔV cells; (panel E) detection of mature tRNAIleAAU with probe 05 in X2316-3C cells; (panel G) in situ hybridization detection of mature tRNAIleAAU with digoxigenin-labeled probe 05 in IIId1c-ΔV cells. (panels B, D, F, and H) DAPI staining of cells shown in panels A, C, E, and G, respectively. (B) Simultaneous in situ hybridization analysis of tRNAIleAAU and immunofluorescent location of Nsp1. Parent X2316-3C and los1Δ mutant strain IIId1c-ΔV were grown at 23°C and log phase cells were shifted to 37°C for 1 h. (Panel A) Detection of mature tRNAIleAAU with probe 05 in X2316-3C cells. (Panel B) Indirect immunofluorescence detection of nucleoporin Nsp1p with 32D6 antibody in X2316-3C cells. (Panel C) Detection of mature tRNAIleAAU with probe 05 in IIId1c-ΔV cells. (Panel D) Indirect immunofluorescence detection of nucleoporin Nsp1p with 32D6 antibody in IIId1c-ΔV cells. Arrows point to cells displaying nuclear accumulations.
The tRNAIleAAU nuclear signal in nup116Δ (Figure 4) or los1ΔV (Figure 6A) strains appeared more diffuse and to extend beyond the DAPI signal. To confirm that accumulated tRNAIleAAU was within the confines of the nuclear border, we probed for tRNAIleAAU and simultaneously stained the nuclear membrane using monoclonal antibody 32D6, which is specific for nucleoporin Nsp1p (Hunter et al., 1998; see MATERIALS AND METHODS). Despite the diffuse signal, the vast majority of the accumulated tRNAIleAAU is indeed located within the nuclear interior in nup116Δ (our unpublished results) and los1ΔV mutant cells (Figure 6B, panels A–D).
RanGAP.
Alteration of components of the RanGTPase cycle causes nuclear accumulation of mRNA (Amberg et al., 1992; Forrester et al., 1992; Kadowaki et al., 1993; Schlenstedt et al., 1995) and defects in nuclear protein import (Corbett et al., 1995). The yeast RNA1 gene encodes the RanGTPase-activating protein RanGAP, which is necessary for GTP hydrolysis of RanGTP to RanGDP (Becker et al., 1995; Bischoff and Ponstingl, 1995; Corbett et al., 1995). If alteration of Ran components also affects tRNA export, then one might expect mutations in RNA1 to cause nuclear accumulation of mature tRNAs. We determined the location of mature tRNAIleAAU encoded by intronless genes in rna1-1 cells. As for the studies of nup116Δ and rat7-1 mutants, we compared the cellular distributions of poly(A)-containing RNA and tRNAIleAAU in EE1b-6 rna1-1 mutant cells with the distributions in EE1b-35, the isogenic wild-type strain. Poly(A)-containing RNA distributions were, as anticipated, largely cytoplasmic in the wild-type strain and nuclear in the rna1-1 mutant strain when the cells were incubated at 37°C for 1 h (our unpublished results) or 3 h (Figure 7, A–D). Also as anticipated, tRNAIleAAU was predominantly cytosolic when EE1b-35 cells were incubated at 37°C for 1 h (our unpublished results) or 3 h (Figure 7, E–F). Interestingly, rna1-1 mutant cells showed nuclear accumulation of tRNAIleAAU when exposed for 1 h (our unpublished results) or 3 h (Figure 7, G–H) at the nonpermissive temperature. As for the studies of los1ΔV, it appears that the nup116Δ cells accumulate tRNAIleAAU in the nucleus more rapidly and to a higher level than rna1-1 cells. Our results are in agreement with the studies of Izaurralde et al. (1997), who demonstrated a role for a functional RanGTPase cycle for the export of all tested RNAs.
Figure 7.
Mutation in RNA1 causes accumulation of mature tRNA in the nucleus. The same cultures of parent EE1b-35 and rna1-1 mutant strain EE1b-6 that were grown at 23°C and used for Figure 3 were shifted to 37°C for 3 h. (A) Detection of Poly(A) RNA with probe 02 in EE1b-35 cells. (C) Detection of Poly(A) RNA with probe 02 in EE1b-6 cells. (E) Detection of mature tRNAIleAAU with probe 05 in EE1b-35 cells. (G) Detection of mature tRNAIleAAU with probe 05 in EE1b-6 cells. (B, D, F, and H) DAPI staining of cells shown in A, C, E, and G, respectively. Arrows point to cells displaying nuclear accumulations.
Defects in RanGAP Cause Nuclear Accumulation of Intron-Containing Pre-tRNA
As pre-tRNA splicing precedes export of mature tRNA from the nucleus, it is remarkable that mutations of genes involved in nucleus/cytosol exchange—nuclear pore structural components and the RanGTPase pathway—affect pre-tRNA intron removal (Hopper et al., 1978; Kadowaki et al., 1993; Sharma et al., 1996). At least four different scenarios, or combinations thereof, could account for this conundrum. First, there could be “feedback” of information from the exchange process to the splicing endonuclease machinery thereby indirectly causing intron-containing species to accumulate within the nucleus. Second, pre-tRNAs could fail to be delivered from their site of synthesis to the nuclear membrane where the tRNA splicing machinery is located (Peebles et al., 1983; Clark and Abelson, 1987). Third, alteration of the nuclear pores and/or the Ran pathway could lead to structural changes in the nuclear membrane which, in turn, could alter the topology of the nuclear membrane-located tRNA splicing endonuclease or cause leakage of nuclear components. Previous studies have implicated a role for the RanGTPase cycle in nuclear membrane integrity in mammalian and Schizosaccharomyces pombe cells (for review see Sazer, 1996). Fourth, alteration of nuclear transport components could affect the regulation of the ordered path of pre-tRNA splicing preceding nuclear export causing export of intervening sequence (IVS)-containing RNAs. There is precedence for alterations in the nucleus/cytosol exchange machinery affecting the ordered steps of RNA processing and export. For example, when the HIV Rev gene is expressed in yeast, unspliced RRE-containing mRNAs are detected in the cytosol and the levels of these cytoplasmic pre-mRNAs are modulated by overexpression or disruption of RIP1/NUP42, a gene encoding a yeast nucleoporin (Stutz et al., 1995). Consequences of either the third or fourth scenarios could generate cytoplasmic pools of intron-containing pre-tRNAs that could not be spliced because they would be physically separated from the tRNA splicing endonuclease located at the inner surface of the nuclear membrane.
Temperature-sensitive rna1-1 mutants are defective in nucleus/cytosol exchange at the nonpermissive temperature (Amberg et al., 1992; Corbett et al., 1995), and they accumulate intron-containing pre-tRNAs (Hopper et al., 1978; Knapp et al., 1978). As assessed by Northern analysis, using probe 03 complementary to the entire pre-tRNAIleUAU intron, there was no difference in the amount of intron containing pretRNAIleUAU when the wild-type, EE1b-35 or rna1-1 mutant, EE1b-6, cells were grown at the permissive temperature (our unpublished results). However, after an exposure to the elevated temperature of 37°C for 1 h, rna1-1 cells had an increased level pre-tRNAIleUAU compared with the isogenic wild-type cells (Figure 2, compare lane 4 with lanes 1–3). To test whether the increased levels of pre-tRNAIleUAU in the rna1-1 cells were due to precocious movement of pre-tRNAs to the cytosol in cells with an altered RanGTPase pathway, we used in situ hybridization to locate intron-containing pre-tRNAs in wild-type cells and rna1-1 cells incubated at the nonpermissive temperature. In agreement with the Northern analysis, the in situ hybridization signal using probe 03 was substantially more intense in rna1-1 cells when they were incubated at the nonpermissive temperature in comparison with the control cells (Figure 8, compare A with C). This intron-specific signal is restricted to the nucleus in both parental and rna1-1 mutant cells, and the signal is somewhat uniformly distributed throughout the nucleus in both. Thus, the data indicate that pre-tRNA accumulation in rna1-1 cells does not result from precocious pre-tRNA nuclear export or accumulation in an inappropriate nuclear subcompartment. The nuclear location of intron-containing pre-tRNAs supports the notion that the tRNA splicing pathway is tightly coupled to nucleus/cytosol exchange (Hopper et al., 1978; Kadowaki et al., 1993; Sharma et al., 1996).
Figure 8.
Accumulation of pre-tRNAs in rna1-1 cells is not due to precocious movement of pre-tRNAs out of the nucleus. Parent EE1b-35 and rna1-1 mutant strain EE1b-6 were grown at 23°C, and log phase cells were shifted to 37°C for 1 h. (A) Detection of pre-tRNAIleUAU with probe 03 in EE1b-35 cells. (C) Detection of pre-tRNAIleUAU with probe 03 in EE1b-6 cells. (B and D) DAPI staining of cells shown in A and C, respectively.
DISCUSSION
Three lines of evidence document successful adaptation of in situ hybridization to assess intracellular locations of endogenous levels of individual tRNA species: 1) competition studies showing specificity of the signals for particular tRNA probes used, 2) location of mature tRNAs in the cytosol and IVS-containing pre-tRNAs in the nucleus, and (3) nuclear accumulation of mature tRNA in yeast cells with sealed nuclear pores. Detection of endogenous levels of individual species of IVS-containing tRNA processing intermediates is due, in part, to pre-tRNA splicing being a slow step in the biogenesis pathway. Our ability to detect nuclear accumulation of mature tRNAs above the cytosolic pool has allowed analyses of particular yeast mutants for defects in tRNA export. We intend to extend this approach to characterize roles of other known nucleoporins in tRNA export. In principle, we should be able to adapt this type of analysis to screen among collections of yeast temperature-sensitive mutants to uncover novel essential genes important to the tRNA export pathway.
Studies of yeast mutants with lesions in genes encoding nucleoporins have demonstrated that many of the nucleoporins are important for both nuclear import and export. However, other nucleoporins have been found to affect transit in a single direction (Fabre and Hurt, 1997). Yeast RAT7 encoding nucleoporin Nup159p has been reported to affect only outward-bound nuclear traffic (Gorsch et al., 1995). Here we show that even though rat7-1 cells are defective in poly(A) RNA export, they appear not to be defective in nuclear export of mature tRNAIleAAU. Thus, Rat7p/Nup159p appears to have a species-specific role in RNA export. Yeast NUP42/RIP1 encoding nucleoporin Nup42p provides another example of an RNA species-specific nucleoporin. NUP42/RIP1 is an unessential gene and rip1 mutants show no defect in RNA nuclear export when the location of poly(A) mRNA is analyzed using oligo(d)T probes for in situ hybridization. However, the mutant cells are unable to export heat shock mRNAs (Saavedra et al., 1997). These two examples, the role of Nup159p in poly(A) export but not in tRNA export, and the role of Nup42p in heat shock mRNA export but not in general poly(A) export, indicate that the roles of other nucleoporins in the exchange processes need to be evaluated for multiple types of RNA cargo.
Pre-tRNA splicing is highly coupled to nuclear export. Perhaps the most compelling evidence for this is accumulation of IVS-containing pre-tRNAs in yeast strains with mutations in any of several genes encoding nucleoporins (Nsp1p, Nup49p, Nup116p, Nup133p, and Nup145p; Sharma et al., 1996). We originally identified the RNA1 and LOS1 genes in searches for yeast mutants defective in pre-tRNA processing, and we and others showed that rna1-1 and los1 mutants accumulate intron-containing end-matured pre-tRNAs (Hopper et al., 1978, 1980; Knapp et al., 1978; Simos et al., 1996). In this study we demonstrate that the rna1-1 and los1ΔV mutations cause nuclear accumulation of tRNAs encoded by genes lacking introns. Thus, RNA1 and LOS1 functions are also important for tRNA export independent of the effects on pre-tRNA splicing.
Los1p is located primarily in nuclei and it is a member of the importin-β family of proteins (Shen et al., 1993; Görlich et al., 1997). Recently, a human Ran binding protein, exportin-t, was shown to interact with tRNA and, when overexpressed, to facilitate export of tRNA from the nucleus to the cytosol in Xenopus oocytes and HeLa cells (Arts et al., 1998; Kutay et al., 1998). Human exportin-t is 19% identical to yeast Los1p (Kutay et al., 1998). Here we demonstrate that a disruption of the LOS1 gene causes nuclear accumulation of mature tRNA. Thus, an excess of human exportin-t facilitates tRNA nuclear export and yeast los1 deletion inhibits tRNA nuclear export. Hence our data is consistent with the model that Los1p and human exportin-t are functionally homologous because both affect tRNA export. Because the yeast genome contains a single LOS1 gene and it is unessential (Hurt et al., 1987; Goffeau et al., 1997), Los1p cannot be absolutely required for tRNA export. Moreover, our results indicate that los1 mutant cells accumulate nuclear tRNA more slowly and to a lesser extent than do nup116 mutant cells, which have nucleopore structural defects. If Los1p is indeed the yeast exportin-t homologue, then there must be other factors, at least in yeast, that also play a role in tRNA export.
In our efforts to identify other proteins that may function like Los1p, we found the SOL family of genes as multicopy suppressors of los1 mutations (Shen et al., 1996). However, the SOL genes do not appear to be involved in tRNA nuclear export (Sarkar, Stanford, and Hopper, unpublished results) or pre-tRNA splicing, because we do not see any accumulation of pre-tRNAs in the sol mutants by in situ hybridization (Sarkar, Stanford, and Hopper, unpublished results), and accumulation of intron-containing pre-tRNAs, observed in los1 mutants, is not reversed by overexpressing the SOL genes (Shen et al., 1996). Using the strategy of synthetic lethality, Simos et al. (1996) uncovered three-way genetic interactions between LOS1 and PUS1, which encodes tRNA pseudouridine synthase, and NSP1. NSP1 encodes an essential member of the FXFG family of nucleoporins and nsp1 mutants show defects in nuclear protein import but not nuclear export of poly(A) RNAs. A testable possibility is that NSP1 affects tRNA nuclear export without affecting poly(A) export. It would also be valuable to determine the phenotypes of yeast strains that have lesions in LOS1 in addition to lesions in genes encoding other members of the importin-β family.
Although the RanGTPase path has been shown to be required for nuclear exit of most RNAs, yeast heat shock mRNAs exit the nucleus by a Ran-independent path (Saavedra et al., 1996). The role of the RanGTPase pathway in tRNA nuclear export has been somewhat controversial. Studying cells with a mutant RanGDP/ GTP exchange factor, Cheng et al. (1995) found that tRNA export was independent of the RanGTPase pathway. In contrast, injection of excess RanGAP into nuclei to deplete nuclei of RanGTP, led Izaurralde et al. (1997) to conclude that the RanGTPase pathway is necessary for tRNA nuclear export. However, even for the studies using excess nuclear RanGAP, tRNA export was affected to a lesser extent than were other RNAs such as snRNAs. Although not conclusive from our work here, it appears that rna1-1 cells, like los1ΔV cells, accumulate nuclear tRNA slower and to lesser extent than do nup116 mutant cells. Thus, in yeast as in higher eukaryotic cells, it may be that nuclear export of tRNAs is less dependent on the RanGTPase cycle than are other RNAs. Why the RanGTPase pathway has different effects on particular RNA substrates is an intriguing unresolved question.
How are pre-tRNA splicing and nuclear export coupled in rna1-1 and los1 mutants? Unless Los1p and Rna1p function at more than one step in the tRNA biogenesis pathway, the simplest explanation for their functions is that they play direct roles in tRNA nucleus/cytosol exchange and affect pre-tRNA splicing indirectly. Nuclear accumulation of intron-containing pre-tRNAs in rna1-1 (Figure 8) and los1ΔV (our unpublished results) cells rule out one model wherein the IVS-containing pre-tRNAs accumulate because they are physically separated from the splicing machinery due to precocious movement to the cytosol. Our studies showing similar intranuclear distribution of IVS-containing pre-tRNAs in wild-type and mutant strains argue against yet another model wherein pre-tRNA accumulation in the mutant strains is caused by inappropriate subnuclear compartmentalization. A model (Sharma et al., 1996) that could account for the phenotypes of nup mutants posits that the tRNA splicing machinery is located within nucleopore channels and is aberrant in cells with lesions of genes encoding nucleoporins. However, by this model it is more difficult to account for the coupling of nuclear export and pre-tRNA splicing evidenced by los1 and rna1 strains. It is possible that each alters nuclear pore structure, but there is no evidence to support this. Alternatively, it is possible that faulty export blocks upstream pre-tRNA splicing. It is not likely that splicing defects are caused by inappropriately high concentrations of mature tRNAs in nuclear export-deficient mutants because it has been demonstrated that high concentrations of mature tRNA failed to act as a competitor for endonuclease activity (Peebles et al., 1979). An attractive and testable model for feedback inhibition is the possible shuttling of splicing endonuclease subunits between the nucleus and the cytosol (Trotta and Abelson, personal communication). This would result in pre-tRNA splicing dependence on continuous appropriate nucleus/cytosol exchange.
ACKNOWLEDGMENTS
We thank Dr. A.M. Tartakoff for technical advice, Dr. D. Engelke for information before publication, and Dr. E. Phizicky for stimulating conversations. We thank Dr. J.E. Hopper, Dr. T. Zoladek, and the members of Dr. Hopper’s laboratory for comments on the manuscript. This work was supported by a Public Health Services grant from the National Institutes of Health to A.K.H.
REFERENCES
- Amberg DC, Goldstein AL, Cole CN. Isolation and characterization of RAT1: an essential gene of Saccharomyces cerevisiae required for the efficient nucleocytoplasmic trafficking of mRNA. Genes Dev. 1992;6:1173–1189. doi: 10.1101/gad.6.7.1173. [DOI] [PubMed] [Google Scholar]
- Amberg DC, Fleischmann M, Stagljar I, Cole CN, Abei M. Nuclear PRP20 protein is required for mRNA export. EMBO J. 1993;12:233–241. doi: 10.1002/j.1460-2075.1993.tb05649.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arts G, Fornerod M, Mattaj IW. Identification of a nuclear export receptor for tRNA. Curr Biol. 1998;8:305–314. doi: 10.1016/s0960-9822(98)70130-7. [DOI] [PubMed] [Google Scholar]
- Becker J, Melchior F, Gerke V, Bischoff FR, Ponstingl H, Wittinghofer A. RNA1 encodes a GTPase-activating protein specific for Gsp1p, the Ran/TC4 homologue of Saccharomyces cerevisiae. J Biol Chem. 1995;270:11860–11865. doi: 10.1074/jbc.270.20.11860. [DOI] [PubMed] [Google Scholar]
- Bertrand E, Houser-Scott F, Kendall A, Singer RH, Engelke DR. Nucleolar localization of early tRNA processing. Genes Dev. 1998;12:2463–2468. doi: 10.1101/gad.12.16.2463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bischoff FR, Krebber H, Kempf T, Hermes I, Ponstingl H. Human RanGTPase-activating protein RanGAP1 is a homologue of yeast Rna1p involved in mRNA processing and transport. Proc Natl Acad Sci USA. 1995;92:1749–1753. doi: 10.1073/pnas.92.5.1749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Y, Dahlberg JE, Lund E. Diverse effects of the guanine nucleotide exchange factor RCC1 on RNA transport. Science. 1995;267:1807–1810. doi: 10.1126/science.7534442. [DOI] [PubMed] [Google Scholar]
- Clark MW, Abelson J. The subnuclear localization tRNA ligase in yeast. J Cell Biol. 1987;105:1515–1526. doi: 10.1083/jcb.105.4.1515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbett AH, Koepp DM, Schlenstedt G, Lee MS, Hopper AK, Silver PA. Rna1p, a Ran/TC4 GTPase activating protein, is required for nuclear import. J Cell Biol. 1995;130:1017–1026. doi: 10.1083/jcb.130.5.1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dworetzky SI, Feldherr CM. Translocation of RNA-coated gold particles through the nuclear pores of oocytes. J Cell Biol. 1988;106:575–584. doi: 10.1083/jcb.106.3.575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabre E, Hurt E. Yeast genetics to dissect the nuclear pore complex and nucleocytoplasmic trafficking. Annu Rev Genet. 1997;31:277–313. doi: 10.1146/annurev.genet.31.1.277. [DOI] [PubMed] [Google Scholar]
- Fornerod M, Ohno M, Yoshida M, Mattaj IW. CRM1 is an export receptor for leucine-rich nuclear export signals. Cell. 1997;90:1051–1060. doi: 10.1016/s0092-8674(00)80371-2. [DOI] [PubMed] [Google Scholar]
- Forrester W, Stutz F, Roshbash M, Wickens M. Defects in mRNA 3′-end formation, transcription initiation, and mRNA transport associated with the yeast mutation prp20: possible coupling of mRNA processing and chromatin structure. Genes Dev. 1992;6:1914–1926. doi: 10.1101/gad.6.10.1914. [DOI] [PubMed] [Google Scholar]
- Fukuda M, Asano S, Nakamura T, Adachi M, Yoshida M, Yanagida M, Nishida E. CRM1 is responsible for intracellular transport mediated by the nuclear export signal. Nature. 1997;390:308–311. doi: 10.1038/36894. [DOI] [PubMed] [Google Scholar]
- Goffeau A, et al. The yeast genome directory. Nature. 1997;387:5–105. [PubMed] [Google Scholar]
- Görlich D, Dabrowski M, Bischoff FR, Kutay U, Bork P, Hartmann E, Prehn S, Izaurralde E. A novel class of RanGTP binding proteins. J Cell Biol. 1997;138:65–80. doi: 10.1083/jcb.138.1.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorsch LC, Dockendorff TC, Cole CN. A conditional allele of a novel repeat containing yeast nucleoporin RAT7/NUP159 causes both rapid cessation of mRNA export and reversible clustering of nuclear pore complexes. J Cell Biol. 1995;129:939–955. doi: 10.1083/jcb.129.4.939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hani J, Feldmann H. tRNA genes and retroelements in the yeast genome. Nucleic Acids Res. 1998;26:689–696. doi: 10.1093/nar/26.3.689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopper AK, Banks F, Evangelides V. A yeast mutant which accumulates precursor tRNAs. Cell. 1978;14:211–219. doi: 10.1016/0092-8674(78)90108-3. [DOI] [PubMed] [Google Scholar]
- Hopper AK, Furukawa AH, Pham HD, Martin NC. Defects in modification of cytoplasmic and mitochondrial transfer RNA are caused by single nuclear mutation. Cell. 1982;28:543–550. doi: 10.1016/0092-8674(82)90209-4. [DOI] [PubMed] [Google Scholar]
- Hopper AK, Martin NC. Processing of yeast cytoplasmic and mitochondrial precursor tRNAs. In: Jones EW, Pringle JR, Broach JR, editors. The Molecular and Cellular Biology of the Yeast Saccharomyces: Gene Expression. Vol. 2. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1992. pp. 99–141. [Google Scholar]
- Hopper AK, Schultz LD, Shapiro RA. Processing of intervening sequences: a new yeast mutant which fails to excise intervening sequences from precursor tRNAs. Cell. 1980;19:741–751. doi: 10.1016/s0092-8674(80)80050-x. [DOI] [PubMed] [Google Scholar]
- Hunter, L.A., Benko, A.L., Aris, J.P., Stanford, D.R., Martin, N.C., and Hopper, A.K. (1998). S. cerevisiae Mod5p-II contains sequences antagonistic for nuclear and cytosolic locations. Genetics (in press). [DOI] [PMC free article] [PubMed]
- Hurt DJ, Wang SS, Li Y, Hopper AK. Cloning and characterization of LOS1, a Saccharomyces cerevisiae gene that affects tRNA splicing. Mol Cell Biol. 1987;7:1208–1216. doi: 10.1128/mcb.7.3.1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izaurralde E, Adam S. Transport of macromolecules between the nucleus and the cytoplasm. RNA. 1998;4:351–364. [PMC free article] [PubMed] [Google Scholar]
- Izaurralde E, Kutay U, von Kobbe C, Mattaj IW, Görlich D. The asymmetric distribution of the constituents of the Ran system is essential for transport into and out of the nucleus. EMBO J. 1997;16:6535–6547. doi: 10.1093/emboj/16.21.6535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarmolowski A, Boelens WC, Izaurralde E, Mattaj IW. Nuclear export of different classes of RNA is mediated by specific factors. J Cell Biol. 1994;124:627–635. doi: 10.1083/jcb.124.5.627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadowaki T, Goldfarb D, Spitz LM, Tartakoff AM, Ohno M. Regulation of RNA processing and transport by a nuclear guanine nucleotide release protein and members of the Ras superfamily. EMBO J. 1993;12:2929–2937. doi: 10.1002/j.1460-2075.1993.tb05955.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadowaki T, Zhao Y, Tartakoff AM. A conditional yeast mutant deficient in mRNA transport from the nucleus to cytoplasm. Proc Natl Acad Sci USA. 1992;89:2312–2316. doi: 10.1073/pnas.89.6.2312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knapp G, Beckmann JS, Johnson PF, Fuhrman SA, Abelson J. Transcription and processing of intervening sequences in yeast tRNA genes. Cell. 1978;14:221–236. doi: 10.1016/0092-8674(78)90109-5. [DOI] [PubMed] [Google Scholar]
- Kudo N, Khochbin S, Nishi K, Kitano K, Yanagida M, Yoshida M, Horinouchi S. Molecular cloning and cell cycle dependent expression of mammalian CRM1, a protein involved in nuclear export of proteins. J Biol Chem. 1997;272:29742–29751. doi: 10.1074/jbc.272.47.29742. [DOI] [PubMed] [Google Scholar]
- Kutay U, Lipowsky G, Izaurralde E, Bischoff FR, Schwarzmaier P, Hartmann E, Görlich D. Identification of a tRNA-specific nuclear export receptor. Mol Cell. 1998;1:359–369. doi: 10.1016/s1097-2765(00)80036-2. [DOI] [PubMed] [Google Scholar]
- Long RM, Singer RH, Meng X, Gonzalez I, Nasmyth K, Jansen RP. Mating type switching in yeast controlled by asymmetric localization of ASH1 mRNA. Science. 1997;277:383–387. doi: 10.1126/science.277.5324.383. [DOI] [PubMed] [Google Scholar]
- Martin NC, Hopper AK. Isopentenylation of both cytoplasmic and mitochondrial tRNA is affected by a single nuclear mutation. J Biol Chem. 1982;257:10562–10565. [PubMed] [Google Scholar]
- O’Connor JP, Peebles CL. In vivo tRNA processing in Saccharomyces cerevisiae. Mol Cell Biol. 1991;11:425–439. doi: 10.1128/mcb.11.1.425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohno M, Fornerod M, Mattaj IW. Nucleocytoplasmic transport: the last 200 nanometers. Cell. 1998;92:327–336. doi: 10.1016/s0092-8674(00)80926-5. [DOI] [PubMed] [Google Scholar]
- Ossareh-Nazari B, Bachelerie F, Dargemont C. Evidence of the role of CRM1 in signal-mediated nuclear protein export. Science. 1997;278:141–144. doi: 10.1126/science.278.5335.141. [DOI] [PubMed] [Google Scholar]
- Peebles CL, Gegenheimer P, Abelson J. Precise excision of intervening sequences from precursor tRNAs by a membrane-associated yeast endonuclease. Cell. 1983;32:525–536. doi: 10.1016/0092-8674(83)90472-5. [DOI] [PubMed] [Google Scholar]
- Peebles CL, Ogden RC, Knapp G, Abelson J. Splicing of yeast tRNA precursors: a two stage reaction. Cell. 1979;18:27–35. doi: 10.1016/0092-8674(79)90350-7. [DOI] [PubMed] [Google Scholar]
- Rose AM, Belford HG, Shen WC, Greer CL, Hopper AK, Martin NC. Location of N2, N2-dimethylguanosine-specific tRNA methyltransferase. Biochimie. 1995;77:45–53. doi: 10.1016/0300-9084(96)88103-x. [DOI] [PubMed] [Google Scholar]
- Rout MP, Blobel G, Aitchison JD. A distinct nuclear import pathway used by ribosomal proteins. Cell. 1997;89:715–725. doi: 10.1016/s0092-8674(00)80254-8. [DOI] [PubMed] [Google Scholar]
- Saavedra CA, Hummell CM, Heath CV, Cole CN. Yeast heat shock mRNAs are exported through a distinct pathway defined by Rip1p. Genes Dev. 1997;11:2845–2856. doi: 10.1101/gad.11.21.2845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saavedra CA, Tung KS, Amberg DC, Hopper AK, Cole CN. Regulation of mRNA export in response to stress in Saccharomyces cerevisiae. Genes Dev. 1996;10:1608–1620. doi: 10.1101/gad.10.13.1608. [DOI] [PubMed] [Google Scholar]
- Sazer S. The search for the primary function of the Ran GTPase continues. Trends Biochem Sci. 1996;6:81–84. doi: 10.1016/0962-8924(96)80992-5. [DOI] [PubMed] [Google Scholar]
- Schlenstedt G, Smirnova E, Deane R, Solsbacher J, Kutay U, Görlich D, Ponstingl H, Bischoff FR. Yrb4p, a yeast Ran-GTP-binding protein involved in import of ribosomal protein L25 into the nucleus. EMBO J. 1997;16:6237–6249. doi: 10.1093/emboj/16.20.6237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlenstedt G, Wong DH, Koepp DM, Silver PA. Mutants in a yeast Ran binding protein are defective in nuclear transport. EMBO J. 1995;14:5367–5378. doi: 10.1002/j.1460-2075.1995.tb00221.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma K, Fabre E, Tekotte H, Hurt EC, Tollervey D. Yeast nucleoporin mutants are defective in pre-tRNA splicing. Mol Cell Biol. 1996;16:294–301. doi: 10.1128/mcb.16.1.294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen WC, Selvakumar D, Stanford DR, Hopper AK. The Saccharomyces cerevisiae LOS1 gene involved in pre-tRNA splicing encodes a nuclear protein that behaves as a component of the nuclear matrix. J Biol Chem. 1993;268:19436–19444. [PubMed] [Google Scholar]
- Shen WC, Stanford DR, Hopper AK. Los1p, involved in yeast pre-tRNA splicing, positively regulates members of the SOL gene family. Genetics. 1996;143:699–712. doi: 10.1093/genetics/143.2.699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simos G, Tekotte H, Grosjean H, Segref A, Sharma K, Tollervey D, Hurt EC. Nuclear pore proteins are involved in the biogenesis of functional tRNA. EMBO J. 1996;15:2270–2284. [PMC free article] [PubMed] [Google Scholar]
- Stade K, Ford CS, Guthrie C, Weis K. Exportin 1 (Crm1p) is an essential nuclear export factor. Cell. 1997;90:1041–1050. doi: 10.1016/s0092-8674(00)80370-0. [DOI] [PubMed] [Google Scholar]
- Stutz F, Neville M, Rosbash M. Identification of a novel nuclear pore-associated protein as a functional target for the HIV-1 Rev protein in yeast. Cell. 1995;82:495–506. doi: 10.1016/0092-8674(95)90438-7. [DOI] [PubMed] [Google Scholar]
- Takizawa PA, Sil A, Swedlow JR, Herskowitz I, Vale RD. Actin-dependent localization of an RNA encoding a cell-fate determinant in yeast. Nature. 1997;389:90–93. doi: 10.1038/38015. [DOI] [PubMed] [Google Scholar]
- Traglia HM, Atkinson NS, Hopper AK. Structural and functional analyses of Saccharomyces cerevisiae wild-type and mutant RNA1 genes. Mol Cell Biol. 1989;9:2989–2999. doi: 10.1128/mcb.9.7.2989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang SS, Hopper AK. Isolation of a yeast gene involved in species-specific pre-tRNA processing. Mol Cell Biol. 1988;8:5140–5149. doi: 10.1128/mcb.8.12.5140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wente SR, Blobel G. A temperature sensitive NUP116 null mutant forms a nuclear envelope seal over the yeast nuclear pore complex thereby blocking nucleocytoplasmic traffic. J Cell Biol. 1993;123:275–284. doi: 10.1083/jcb.123.2.275. [DOI] [PMC free article] [PubMed] [Google Scholar]