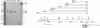Abstract
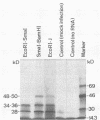
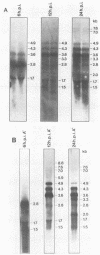
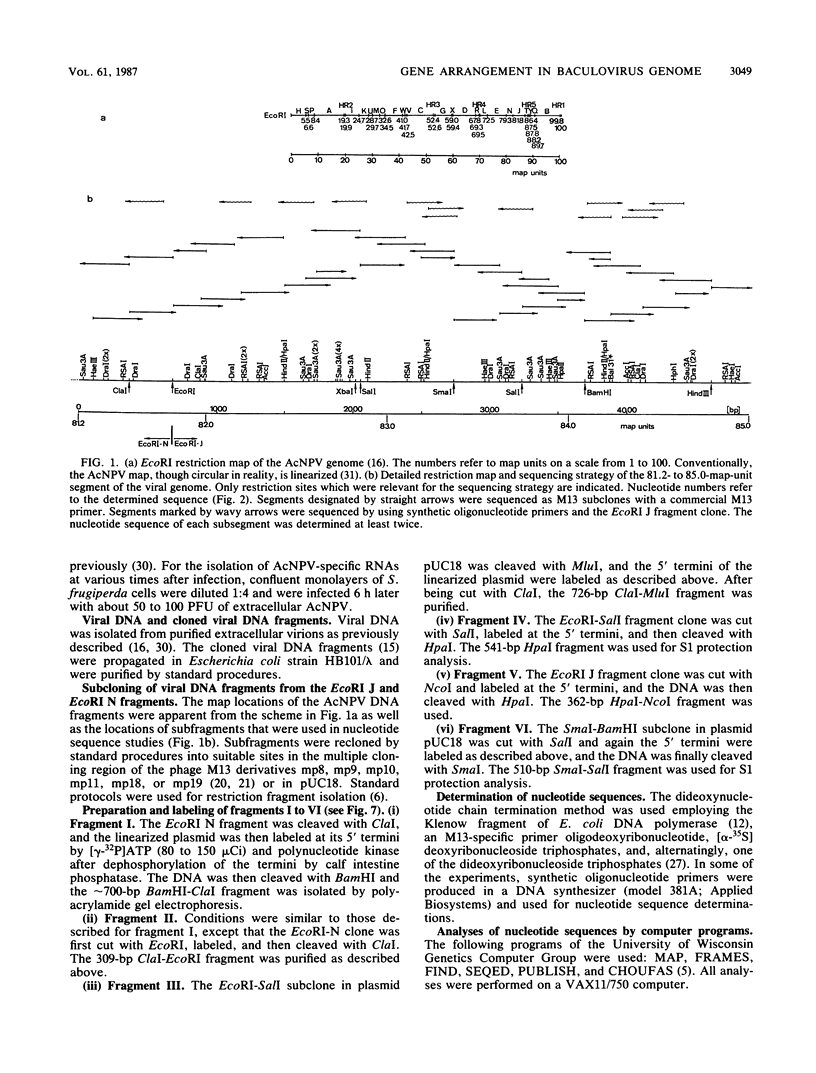
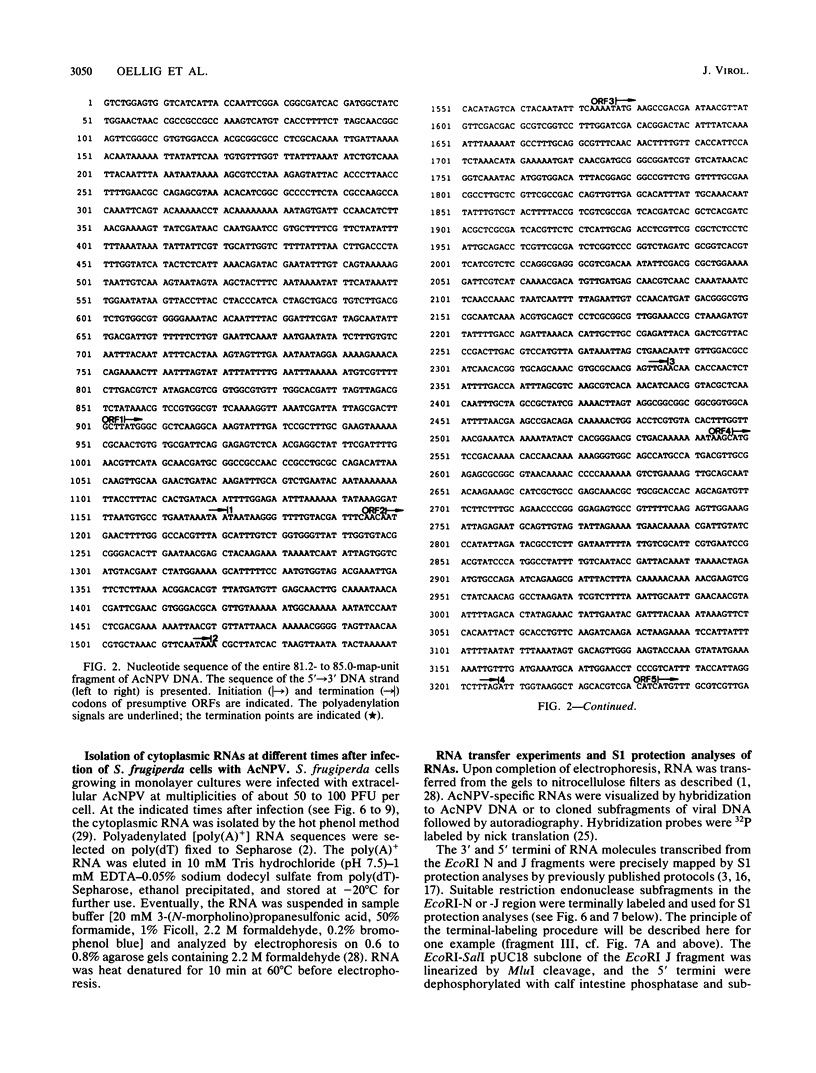
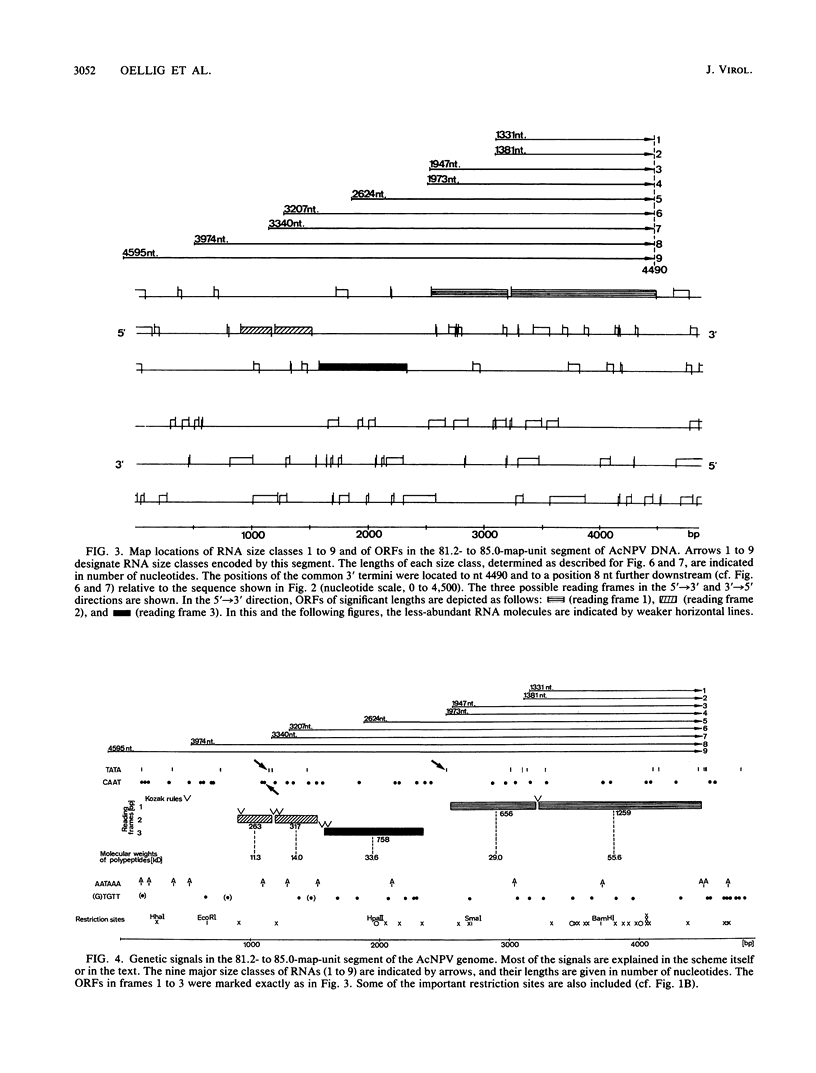
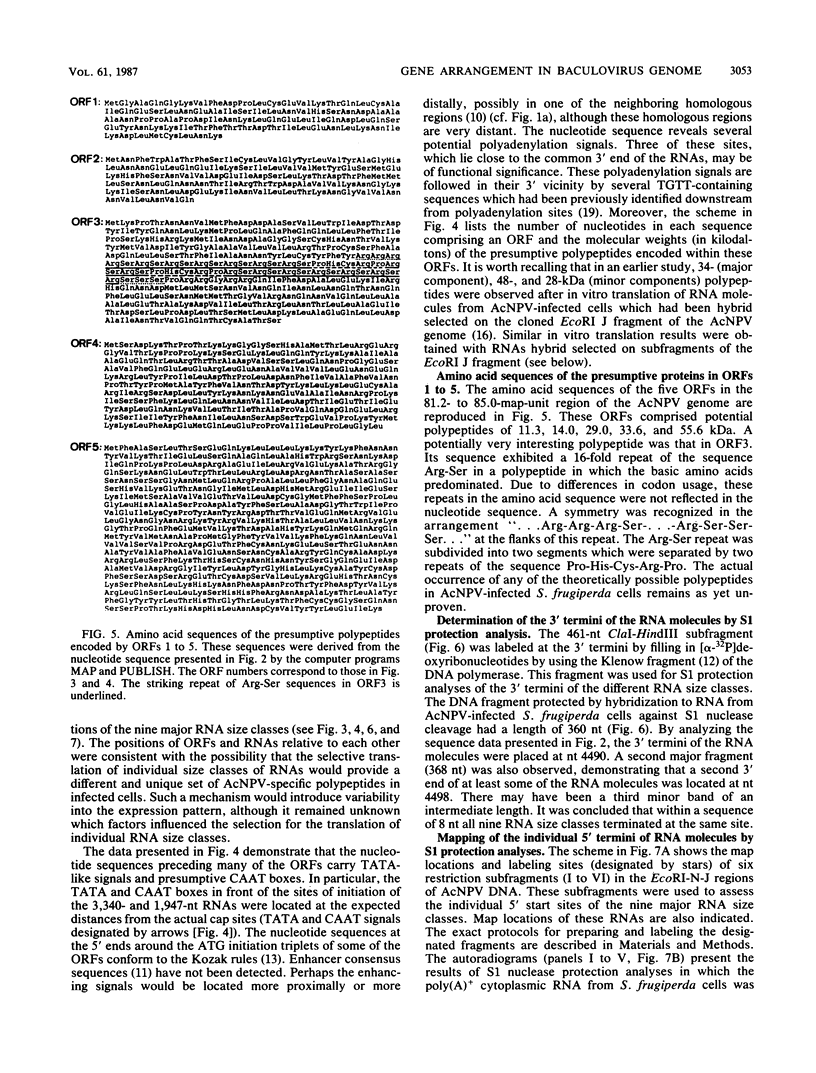
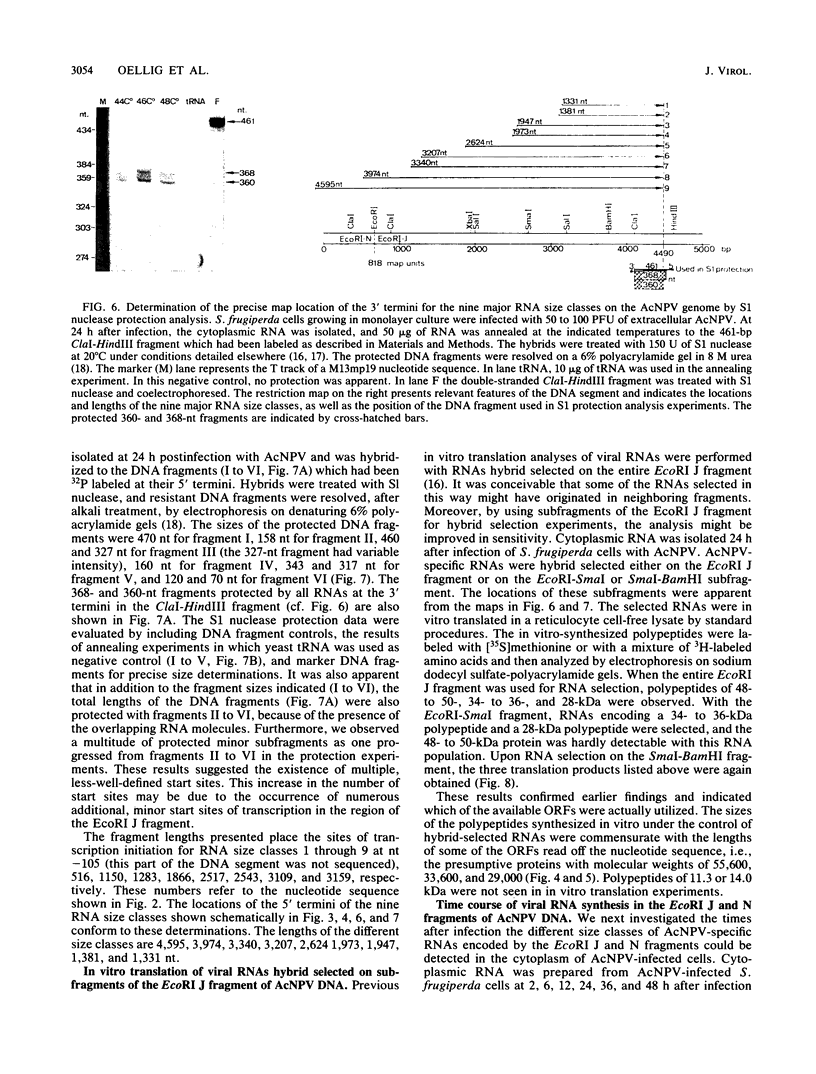
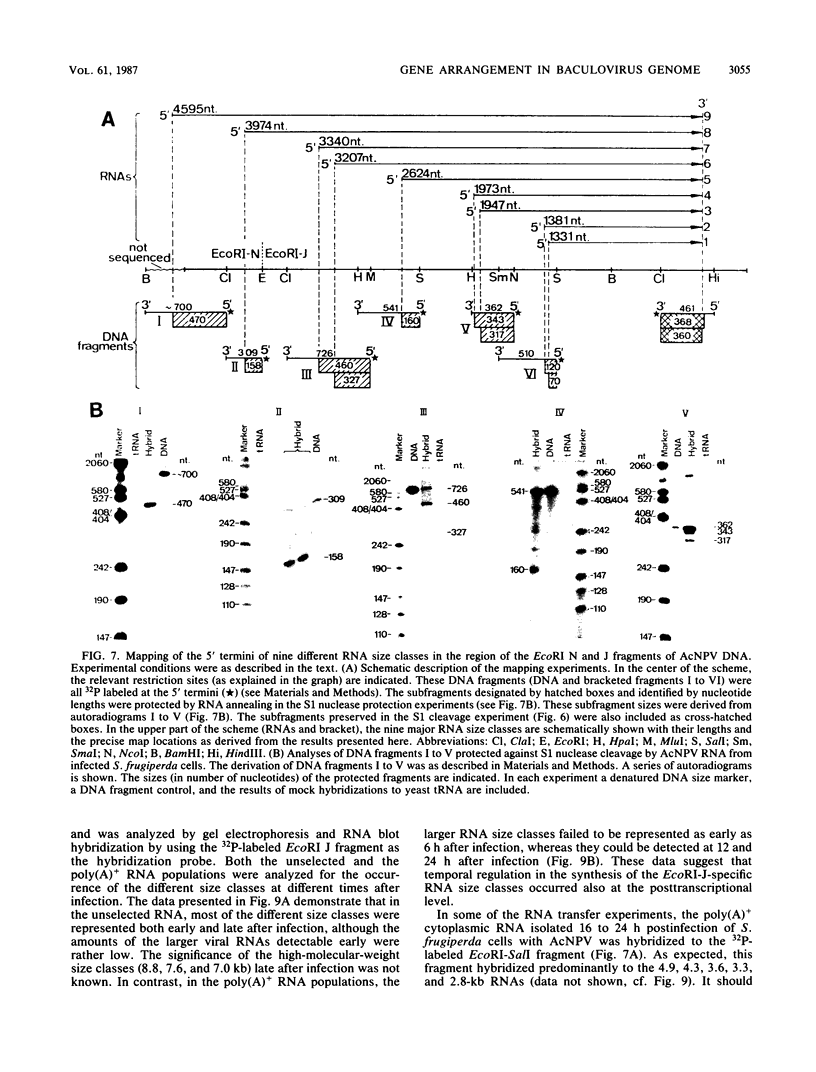
In several parts of the Autographa californica nuclear polyhedrosis virus (AcNPV) genome, nested sets of overlapping RNAs with common 3' or 5' termini have been recognized. In the present report, the pattern of viral transcription and the arrangement of viral gene products in the region of 81.2 to 85.0 map units were investigated. In this segment of the AcNPV genome, at least nine size classes of viral RNA were identified which ranged in size from 1.3 kilobases (kb) to 4.6 kb and exhibited common 3' termini. The detailed restriction map and the nucleotide sequence of this part of the AcNPV genome were determined. Computer analyses revealed several open reading frames (ORFs) on the rightward-transcribed strand with potential TATA and CAAT signals preceding many of the potential ORFs and the 5' termini of some of the mapped RNAs. The leftward-transcribed strand was devoid of major ORFs. The presumptive polypeptides encoded by the larger ORFs ranged in size from 11.3 to 55.6 kilodaltons (kDa). The amino acid sequence of the presumptive polypeptide encoded by ORF3, a 33.6-kDa molecule, exhibited an unusual, clustered 16-fold repeat of the dipeptide arginine-serine in a protein that showed an overall preponderance of basic amino acids. The results of in vitro translation experiments with hybrid-selected RNAs homologous to internal subfragments of the 81.2- to 85.0-map-unit region yielded polypeptides of approximately 28, 34 to 36, and 48 to 50 kDa, which were close in size to the lengths of the major ORFs derived from the nucleotide sequence. The localizations of individual size classes of RNAs in the 81.2- to 85.0-map-unit region of the viral genome were determined precisely at the 3' and 5' termini by S1 protection analyses. Within a sequence of eight nucleotides, all RNAs had the same 3' terminus, which lay close to multiple polyadenylation signals. The initiation sites of the nine different RNA size classes were precisely mapped. As the cap sites of the smaller RNAs (less than 1.8 kb) were determined by S1 protection analyses, a multitude of RNA initiation sites became apparent. It was also shown that the different RNA size classes in the 81.2- to 85.0-map-unit region were detectable as early as 2 h and at least until 36 to 48 h after infection. In unselected cytoplasmic RNA, the size classes of viral RNAs specific for the EcoRI J fragment were detectable early as well as late after infection, although at early times the larger RNAs were detectable in smaller amounts.(ABSTRACT TRUNCATED AT 400 WORDS)
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Alwine J. C., Kemp D. J., Stark G. R. Method for detection of specific RNAs in agarose gels by transfer to diazobenzyloxymethyl-paper and hybridization with DNA probes. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5350–5354. doi: 10.1073/pnas.74.12.5350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aviv H., Leder P. Purification of biologically active globin messenger RNA by chromatography on oligothymidylic acid-cellulose. Proc Natl Acad Sci U S A. 1972 Jun;69(6):1408–1412. doi: 10.1073/pnas.69.6.1408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berk A. J., Sharp P. A. Sizing and mapping of early adenovirus mRNAs by gel electrophoresis of S1 endonuclease-digested hybrids. Cell. 1977 Nov;12(3):721–732. doi: 10.1016/0092-8674(77)90272-0. [DOI] [PubMed] [Google Scholar]
- Bonner W. M., Laskey R. A. A film detection method for tritium-labelled proteins and nucleic acids in polyacrylamide gels. Eur J Biochem. 1974 Jul 1;46(1):83–88. doi: 10.1111/j.1432-1033.1974.tb03599.x. [DOI] [PubMed] [Google Scholar]
- Chou P. Y., Fasman G. D. Prediction of the secondary structure of proteins from their amino acid sequence. Adv Enzymol Relat Areas Mol Biol. 1978;47:45–148. doi: 10.1002/9780470122921.ch2. [DOI] [PubMed] [Google Scholar]
- Dretzen G., Bellard M., Sassone-Corsi P., Chambon P. A reliable method for the recovery of DNA fragments from agarose and acrylamide gels. Anal Biochem. 1981 Apr;112(2):295–298. doi: 10.1016/0003-2697(81)90296-7. [DOI] [PubMed] [Google Scholar]
- Esche H., Schilling R., Doerfler W. In vitro translation of adenovirus type 12-specific mRNA isolated from infected and transformed cells. J Virol. 1979 Apr;30(1):21–31. doi: 10.1128/jvi.30.1.21-31.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friesen P. D., Miller L. K. Temporal regulation of baculovirus RNA: overlapping early and late transcripts. J Virol. 1985 May;54(2):392–400. doi: 10.1128/jvi.54.2.392-400.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guarino L. A., Gonzalez M. A., Summers M. D. Complete Sequence and Enhancer Function of the Homologous DNA Regions of Autographa californica Nuclear Polyhedrosis Virus. J Virol. 1986 Oct;60(1):224–229. doi: 10.1128/jvi.60.1.224-229.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hearing P., Shenk T. The adenovirus type 5 E1A transcriptional control region contains a duplicated enhancer element. Cell. 1983 Jul;33(3):695–703. doi: 10.1016/0092-8674(83)90012-0. [DOI] [PubMed] [Google Scholar]
- Klenow H., Overgaard-Hansen K., Patkar S. A. Proteolytic cleavage fo native DNA polymerase into two different catalytic fragments. Influence of assay condtions on the change of exonuclease activity and polymerase activity accompanying cleavage. Eur J Biochem. 1971 Oct 14;22(3):371–381. doi: 10.1111/j.1432-1033.1971.tb01554.x. [DOI] [PubMed] [Google Scholar]
- Kozak M. Comparison of initiation of protein synthesis in procaryotes, eucaryotes, and organelles. Microbiol Rev. 1983 Mar;47(1):1–45. doi: 10.1128/mr.47.1.1-45.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lübbert H., Doerfler W. Mapping of Early and Late Transcripts Encoded by the Autographa californica Nuclear Polyhedrosis Virus Genome: Is Viral RNA Spliced? J Virol. 1984 May;50(2):497–506. doi: 10.1128/jvi.50.2.497-506.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lübbert H., Doerfler W. Transcription of overlapping sets of RNAs from the genome of Autographa californica nuclear polyhedrosis virus: a novel method for mapping RNAs. J Virol. 1984 Oct;52(1):255–265. doi: 10.1128/jvi.52.1.255-265.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lübbert H., Kruczek I., Tjia S., Doerfler W. The cloned EcoRI fragments of Autographa californica nuclear polyhedrosis virus DNA. Gene. 1981 Dec;16(1-3):343–345. doi: 10.1016/0378-1119(81)90092-5. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- McLauchlan J., Gaffney D., Whitton J. L., Clements J. B. The consensus sequence YGTGTTYY located downstream from the AATAAA signal is required for efficient formation of mRNA 3' termini. Nucleic Acids Res. 1985 Feb 25;13(4):1347–1368. doi: 10.1093/nar/13.4.1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messing J., Vieira J. A new pair of M13 vectors for selecting either DNA strand of double-digest restriction fragments. Gene. 1982 Oct;19(3):269–276. doi: 10.1016/0378-1119(82)90016-6. [DOI] [PubMed] [Google Scholar]
- Norrander J., Kempe T., Messing J. Construction of improved M13 vectors using oligodeoxynucleotide-directed mutagenesis. Gene. 1983 Dec;26(1):101–106. doi: 10.1016/0378-1119(83)90040-9. [DOI] [PubMed] [Google Scholar]
- Pelham H. R., Jackson R. J. An efficient mRNA-dependent translation system from reticulocyte lysates. Eur J Biochem. 1976 Aug 1;67(1):247–256. doi: 10.1111/j.1432-1033.1976.tb10656.x. [DOI] [PubMed] [Google Scholar]
- Rankin C., Ladin B. F., Weaver R. F. Physical mapping of temporally regulated, overlapping transcripts in the region of the 10K protein gene in Autographa californica nuclear polyhedrosis virus. J Virol. 1986 Jan;57(1):18–27. doi: 10.1128/jvi.57.1.18-27.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricciardi R. P., Miller J. S., Roberts B. E. Purification and mapping of specific mRNAs by hybridization-selection and cell-free translation. Proc Natl Acad Sci U S A. 1979 Oct;76(10):4927–4931. doi: 10.1073/pnas.76.10.4927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigby P. W., Dieckmann M., Rhodes C., Berg P. Labeling deoxyribonucleic acid to high specific activity in vitro by nick translation with DNA polymerase I. J Mol Biol. 1977 Jun 15;113(1):237–251. doi: 10.1016/0022-2836(77)90052-3. [DOI] [PubMed] [Google Scholar]
- Rohel D. Z., Faulkner P. Time Course Analysis and Mapping of Autographa californica Nuclear Polyhedrosis Virus Transcripts. J Virol. 1984 Jun;50(3):739–747. doi: 10.1128/jvi.50.3.739-747.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schirm S., Doerfler W. Expression of viral DNA in adenovirus type 12-transformed cells, in tumor cells, and in revertants. J Virol. 1981 Sep;39(3):694–702. doi: 10.1128/jvi.39.3.694-702.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott M. R., Westphal K. H., Rigby P. W. Activation of mouse genes in transformed cells. Cell. 1983 Sep;34(2):557–567. doi: 10.1016/0092-8674(83)90388-4. [DOI] [PubMed] [Google Scholar]
- Tjia S. T., Carstens E. B., Doerfler W. Infection of Spodoptera frugiperda cells with Autographa californica nuclear polyhedrosis virus II. The viral DNA and the kinetics of its replication. Virology. 1979 Dec;99(2):399–409. doi: 10.1016/0042-6822(79)90018-7. [DOI] [PubMed] [Google Scholar]
- Vlak J. M., Smith G. E. Orientation of the Genome of Autographa californica Nuclear Polyhedrosis Virus: a Proposal. J Virol. 1982 Mar;41(3):1118–1121. doi: 10.1128/jvi.41.3.1118-1121.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogt V. M. Purification and further properties of single-strand-specific nuclease from Aspergillus oryzae. Eur J Biochem. 1973 Feb 15;33(1):192–200. doi: 10.1111/j.1432-1033.1973.tb02669.x. [DOI] [PubMed] [Google Scholar]