Abstract
Chlamydomonas reinhardtii flagellar regeneration is accompanied by rapid induction of genes encoding a large set of flagellar structural components and provides a model system to study coordinate gene regulation and organelle assembly. After deflagellation, the abundance of a 70-kDa flagellar dynein intermediate chain (IC70, encoded by ODA6) mRNA increases approximately fourfold within 40 min and returns to predeflagellation levels by ∼90 min. We show by nuclear run-on that this increase results, in part, from increased rates of transcription. To localize cis induction elements, we created an IC70 minigene and measured accumulation, in C. reinhardtii, of transcripts from the endogenous gene and from introduced promoter deletion constructs. Clones containing 416 base pairs (bp) of 5′- and 2 kilobases (kb) of 3′-flanking region retained all sequences necessary for a normal pattern of mRNA abundance change after deflagellation. Extensive 5′- and 3′- flanking region deletions, which removed multiple copies of a proposed deflagellation-response element (the tub box), did not eliminate induction, and the IC70 5′-flanking region alone did not confer deflagellation responsiveness to a promoterless arylsulfatase (ARS) gene. Instead, an intron in the IC70 gene 5′-untranslated region was found to contain the deflagellation response element. These results suggest that the tub box does not play an essential role in deflagellation-induced transcriptional regulation of this dynein gene.
INTRODUCTION
Flagellar and ciliary regeneration is a common phenomenon among many organisms such as protozoan flagellates, ciliates, and ciliated invertebrate larvae (Lefebvre and Rosenbaum, 1986) and is accompanied by the rapid induction of genes encoding a large set of flagellar or ciliary structural components (Stephens, 1977; Guttman and Gorovsky, 1979; Lefebvre et al., 1980). Since these organelles contain hundreds of proteins that assemble in precise stoichiometric ratios, their regeneration provides a unique opportunity to study mechanisms of coordinate gene regulation during organelle assembly.
After deflagellation, the abundance of Chlamydomonas reinhardtii flagellar protein mRNAs rises rapidly and peaks after 20–50 min with a 2- to 15-fold increase over constitutive (nondeflagellated, ND) levels; transcript levels then gradually return to normal within 2 h (Lefebvre et al., 1980; Schloss et al., 1984; Williams et al., 1986). Nuclear run-on experiments have demonstrated that deflagellation-induced accumulation of α- and β-tubulin mRNA can be largely accounted for by increased transcription of new mRNAs (Keller et al., 1984), although changes in tubulin mRNA stability also contribute (Baker et al., 1984). The cis elements required for this response are located in 5′-flanking sequences, within 100 bp of the initiation site (Davies et al., 1992; Davies and Grossman, 1994). A 10-bp consensus motif (the tub box; Brunke et al., 1984) occurs in multiple copies upstream of all four C. reinhardtii tubulin genes and has been implicated in regulation of the tubB2 β-tubulin gene (Davies and Grossman, 1994). Similar sequences are found in other C. reinhardtii genes regulated by deflagellation, such as the radial spoke genes encoding RSP3, RSP4, and RSP6 (Williams et al., 1989; Curry et al., 1992), but the role of tub box sequences in transcriptional regulation of these genes has not been studied. Recent analysis of induction elements in an α1-tubulin gene concluded that two regions contributed to the deflagellation response and questioned the role of tub box elements in that promoter (Periz and Keller, 1997). The CBLP gene, which also has a tub box in the promoter region, is not up-regulated after deflagellation (Schloss, 1990). Because tubulins are far more abundant than other flagellar proteins and are used for cytoplasmic as well as flagellar microtubules, their regulation may not typify that of most flagellar protein genes. To identify promoter elements involved in transcriptional regulation of a typical flagellar protein gene, we have analyzed transcripts generated from introduced copies of a C. reinhardtii gene encoding the 70-kDa intermediate chain subunit of flagellar outer row dynein (ODA6 gene).
We previously found that IC70 mRNA abundance increases after deflagellation in a pattern similar to that of radial spoke gene transcripts (Williams et al., 1986). Since several copies of the tub box consensus were found within 1 kb upstream of the presumed IC70 transcription start site, this region was targeted for promoter deletions and for fusions to a reporter gene encoding C. reinhardtii arylsulfatase (ARS). Here we use nuclease protection to identify the IC70 transcription start site, show that the deflagellation-induced increase in IC70 mRNA abundance is regulated transcriptionally, and find that a cis promoter element located within an intron in the 5′-untranslated region (UTR) is both necessary and sufficient for this deflagellation response.
MATERIALS AND METHODS
Chlamydomonas Strains and Manipulations
Chlamydomonas stocks were obtained from the following sources: strain oda6–95 from R. Kamiya (University of Tokyo); wild-type strain 137c from J. Rosenbaum (Yale University); a cw15,nit1–305 strain and Nit+ strain A2 from P. A. LeFebvre (University of Minnesota). An oda6–95,nit1–305,cw15 strain was created by first crossing the oda6–95 mutation out of the 137c background into a Nit+ background and then in a second cross introducing the nit1–305 and cw15 mutations. All stocks were maintained by standard methods (Harris, 1989) on minimal medium I (M-I) or M-I supplemented with acetate (M-II) (Sager and Granick, 1953).
RNA Isolation and Northern Blot Analysis
Cells were grown in liquid M-I medium with a 12-h light/12-h dark cycle until cell density reached ∼1 × 107 cells per ml. Either before deflagellation, or at indicated times after pH shock deflagellation (Witman et al., 1972), cells were harvested and RNA was isolated by a guanidinium extraction procedure (Chomczynski and Sacchi, 1987) modified as previously described (Mitchell and Kang, 1991). For Northern blot analysis (Sambrook et al., 1989), 10–30 μg of total RNA were electrophoresed on a 1% agarose gel containing formaldehyde, transferred to nylon membranes (Micron Separations, Westboro, MA) by capillary blotting, baked at 80°C for 2 h, and hybridized with 32P-labeled RNA probes overnight at 58°C in an solution containing 50% formamide, 5× Denhardt’s solution, 6× SSC, and 100 μg/ml yeast tRNA. Membranes were then washed twice with 2× SSC, 0.1% SDS at 58°C and twice with 0.1× SSC, 0.1% SDS at 65°C. [32P]UTP-labeled RNA probes were synthesized using an in vitro transcription kit (Stratagene, La Jolla, CA). Autoradiographs were quantitated with a Shimadzu CS-9000 density scanner (Shimadzu Scientific Instruments, Columbia, MD). For comparing autoradiographic band intensities of different lanes, all densities were normalized with the RNA band density of a C. reinhardtii G protein β subunit-like polypeptide (CBLP), which is constitutively expressed after deflagellation (Schloss, 1990).
In Vitro Nuclear Run-on Transcription
A crude nuclear fraction was prepared from cw15,nit1 cells either before or 2 min after deflagellation by the method of Keller et al. (1984), mixed with 2× reaction buffer, quick-frozen in liquid nitrogen, and stored at −70°C. Buffers and procedures for freezing nuclei and for in vitro transcription reactions were taken from Nevins (1987). For labeling, 50 μl of nuclei from 6 × 107 cells were added to 50 μl of labeling mix containing 250 μCi α-32P UTP (3000 Ci/mmol). After 60 min at 26°C, 5 vol of DNase digestion mix were added (150 U DNase I, 10 mM Tris, 500 mM NaCl, 50 mM MgCl2, 2 mM CaCl2, pH 7.4), and incubation was continued tor 10 min at 30°C. Total nucleic acids were extracted, precipitated, and hybridized to duplicate dots of 2 μg denatured plasmid DNA. Blots were exposed for 38 h and were quantitated on a PhosphorImager (Molecular Dynamics, Sunnyvale, CA). Signal intensity readings were corrected for background, then deflagellated (DF) vs. ND signals were normalized for any difference in intensity of the CBLP signal (Schloss, 1990).
S1 Nuclease Mapping
S1 mapping followed standard procedures (Sambrook et al., 1989). A NheI–SalI restriction fragment from a 5′-deletion construct (see below), which covers from 114 bp downstream to 134 bp upstream of the 5′-end of IC70 cDNA c70–16 (Mitchell and Kang, 1991), was purified, dephosphorylated with calf intestinal alkaline phosphatase (New England Biolabs, Boston, MA), and labeled at the 5′-end with γ-32P ATP (Amersham, Arlington Heights, IL) and T4 polynucleotide kinase (New England Biolabs). The labeled double-stranded fragment was denatured in formamide at 90°C for 10 min and run on a 5% neutral polyacrylamide gel for 18 h, and separated strands were located by autoradiography and isolated by a crush and soak method. Each labeled single-stranded DNA (105 cpm) was denatured for 10 min at 90°C and hybridized with 1–3 μg mRNA overnight at 30°C in 80% formamide, 40 mM piperazine-N,N′-bis(2-ethanesulfonic acid) (pH 6.4), 400 mM NaCl, 1 mM EDTA. The annealed products were digested with S1 nuclease at 13°C for 1 h, the reaction was stopped by adding 0.25 vol of S1 stop buffer, and the products were precipitated with ethanol and separated on an 8% acrylamide gel. A phage M13 dideoxy sequence ladder was used as a size marker.
Cotransformation
Double-mutant strain oda6,nit1,cw15 was used for cotransformation with 2 μg of plasmid pMN24 containing the NIT1 gene and 10 μg of plasmid containing the IC70 promoter construct, as described by Kindle (1990). All plasmids were linearized within vector sequences before transformation. Cells grown in M-II to 1 × 107 cells/ml were harvested by centrifugation, and cell walls were removed by 30 min treatment with gametic cell wall lysin (Harris, 1989). Approximately 1 × 109 cells in 1 ml M-II lacking ammonia (NH4NO2 replaced with NaNO2) were used in each transformation, and cells were spread directly onto M-II plates lacking ammonia and grown under constant light for several days. Transformants were randomly selected and tested for cotransformation with promoter constructs by PCR amplification of plasmid sequences from genomic DNA as described below.
Genomic DNA was prepared from 107 cells by phenol/chloroform extraction and ethanol precipitation. DNA from 5 × 105 cells was amplified with 20 pmol of each primer in a total volume of 20 μl in a minicycler (MJ Research, Watertown, MA). IC70 minigene deletions were amplified with the pBluescript KS primer (CGAGGTCGACGGTATCG) as 5′-primer and a 3′-primer that covers the truncated IC70 gene from nucleotide +41 to +60 (GCACCGAATAGCTTGACCTG). If the size of an amplification product matched the expected length of the introduced promoter construct, we assumed that an intact promoter was present. For similar tests of 70F/ARS, 70FU/ARS, and 70FU I/ARS transformants, the 5′-primer covered the IC70 promoter from upstream −309 to −326 (TCGCATCCATCCTGTCTC); for 70+5′I/ARS and 70−5′I/ARS transformants the 5′-primers were the primers used to amplify the IC70 first intron (see below). A common 3′-primer (CGGGTTGAAGCACCACTA) was used that begins within the ARS gene 77 bp downstream of the translation start site.
Construction of Reporter Genes and Promoter Deletions
All IC70 constructs were derived from a 6-kb genomic SacI fragment (Mitchell and Kang, 1991) cloned into pBluescript (pB70S6). To create an IC70 minigene, two BglII fragments spanning 1411 bp, which includes the 400 bp second intron of the IC70 gene (Mitchell and Kang, 1993), were removed from pB70S6. The resulting truncated IC70 gene encodes a 1621-base mRNA instead of a full-length 2634 base transcript. An Erase-a-Base kit (Promega Biotec, Madison, WI) was used to create nested sets of 5′- and 3′- deletions that were sequenced to determine exact deletion endpoints. Small deletions within the 5′-flanking region were formed by joining pairs of 5′- and 3′-deletions that had been obtained as above. 5′- Deletions down to −32 and −13 were paired with 3′-deletions ending at −52 and −37 by ligating at pBluescript ClaI sites, so that deletion endpoints were joined by polylinker sequences (one ClaI and two half HindIII sites; AGCTTATCGATAAGCT).
Plasmid pJD55 contains a promoterless ARS gene that lacks 5′-flanking sequences and retains only 67 bp of its 5′-UTR (Davies et al., 1992). To create promoter fusions, IC70 gene fragments extending from a KpnI site at −430 in the IC70 5′-flanking region to specific 3′-deletion sites were inserted into pJD55 polylinker sites KpnI and SalI. An IC70 fragment that had been deleted with ExoIII from the 3′-end to +1 (transcription initiation site) was cut at an adjacent pBluescript SalI site and ligated with pJD55 to create chimeric gene 70F/ARS. By inserting an IC70 KpnI–BstEII fragment spanning −430 to +430 into the promoterless ARS gene, 70FU/ARS was obtained. The IC70 first intron in 70FU/ARS was removed by two steps: first, a 288 bp NheI–BstEII fragment covering the first intron was removed from the IC70 minigene and replaced by a corresponding cDNA fragment, and then a 622-bp KpnI–BstEII fragment from this new construct was inserted into ARS plasmid pJD55. The resulting 70FUΔI/ARS chimeric gene has no IC70 first intron but retains the rest of the IC70 5′-sequence from −430 to +430.
Primers covering the 5′ (CTGGGTACCGCTGAACACAGTC) and 3′ (GACGTCCAGTGGGTACCGGT) end of the IC70 first intron were used to amplify the intron. The amplification product was cloned into plasmid pCRII by TA cloning (Invitrogen, San Diego, CA), and a KpnI fragment containing the intron was then inserted into the 5′-KpnI site of the 70FUΔI/ARS chimeric gene plasmid. The orientation of this fragment was determined by restriction mapping, and constructs with the intron in its original orientation (70+5′I/ARS), or the opposite orientation (70−5′I/ARS) were used for cotransformation and expression studies.
RESULTS
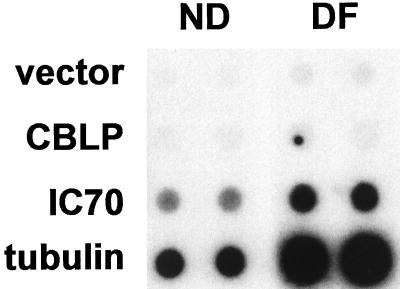
Since changes in tubulin mRNA levels in response to deflagellation are controlled by both changes in message stability and changes in transcription rate, we first tested whether changes in dynein IC70 abundance are controlled at the transcriptional level using nuclear run-on assays. Labeled RNA synthesized from an equal number of ND and DF cell nuclei were hybridized with dots of dynein cDNA clone pBc70–16 (Mitchell and Kang, 1991), β2 tubulin cDNA (Youngbloom et al., 1984), and of a cDNA-encoding CBLP, a constitutively expressed C. reinhardtii G protein β subunit-like protein (Schloss, 1990). As shown in Figure 1, IC70 expression increased dramatically in response to deflagellation. After subtraction of background and normalization to CBLP, transcription signals (measured by PhosphorImager) increased 2.2-fold for dynein and 6.3-fold for tubulin. Similar results were obtained using RNA synthesized from two independently prepared nuclear samples.
Figure 1.
Nuclear run-on shows that IC70 expression is regulated transcriptionally. Run-on transcripts generated in vitro from DF cell nuclei and ND cell nuclei were hybridized to dots of CBLP, IC70, and β2 tubulin cDNA. Transcription rates of IC70 and β2 tubulin were normalized to the CBLP signal. (Note that the dark spot in one CBLP dot is an artifact, and that dot was not included in quantitative normalization.)
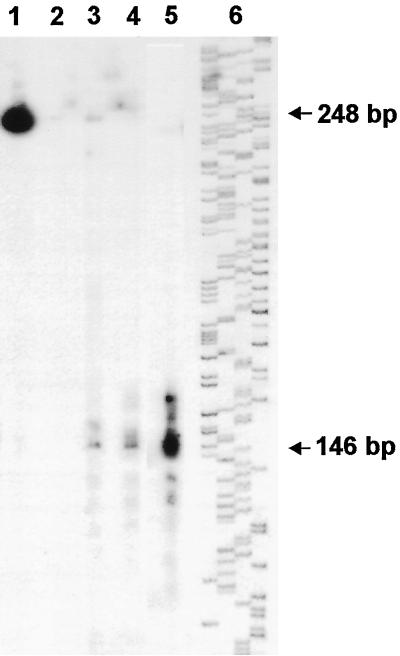
To map potential IC70 gene promoter elements, the probable transcription start site was first located through S1 nuclease mapping with a 248-base probe, which extends 134 bases beyond the 5′-end of cDNA clone pBc70–16 (Figure 2). The major protected length of the probe was 146 nucleotides (Figure 2A), which places the initiation site 32 bp beyond the 5′-end of cDNA clone c70–16 (Figure 2B). Primer extension analysis also demonstrated that a major stop site is present 32 bp beyond the c70–16 cDNA 5′-end.
Figure 2.
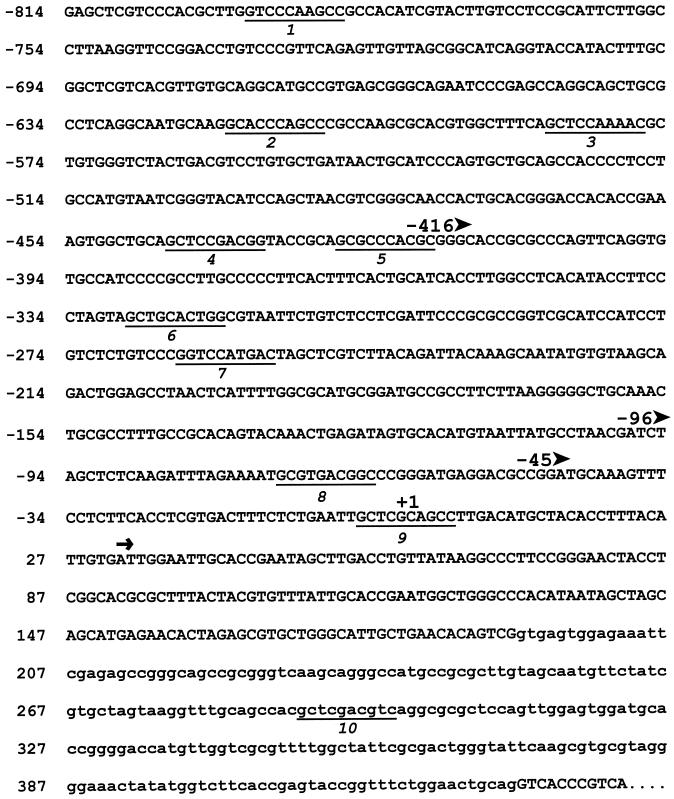
Mapping the IC70 5′-end. (A) S1 nuclease mapping to identify the transcriptional initiation site. Lane 1, the 5′-end–labeled DNA fragment alone; lane 2, 2 μg of total RNA were hybridized with the 5′-end–labeled sense strand DNA and digested with 50 U S1 nuclease; lane 3, 1 μg of total RNA was hybridized with a 5′- end–labeled antisense DNA and digested with 10 U S1 nuclease; lane 4, 1 μg of total RNA was hybridized with a 5′-end–labeled antisense DNA and digested with 50 U S1 nuclease; lane 5, 2 μg of total RNA was hybridized with a 5′-end–labeled antisense DNA and digested with 100 U S1 nuclease; lane 6, phage M13 sequencing ladder was used as a size marker. (B) Sequence of the IC70 promoter region indicating the major transcriptional start site (+1), the 10 tub box sequences (underlined), and the beginning of cDNA c70–16 (arrow). Arrowheads at −416, −96 and −45 mark the ends of deletion constructs described in the text and in Figure 3; an intron in the 5′-UTR is shown in lowercase.
Promoter Deletion Analysis of the Truncated IC70 Gene
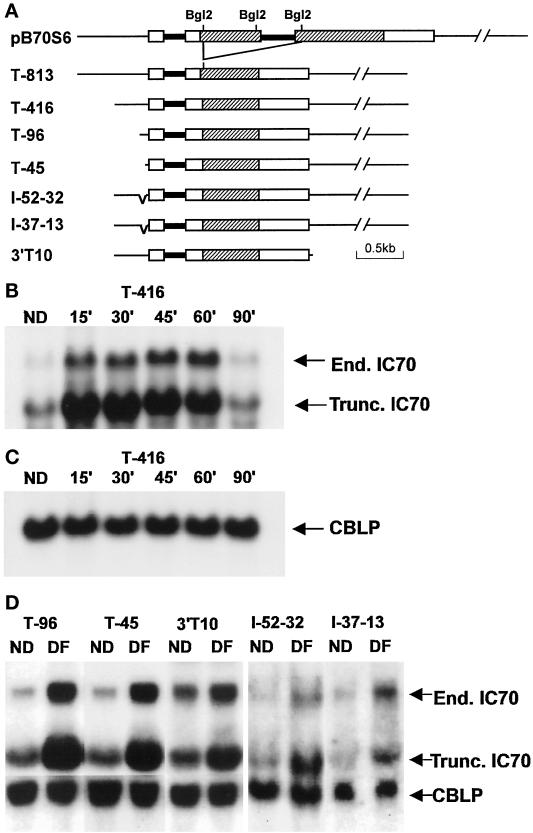
Because no enzymatic reporter gene assay system has been adapted successfully for analysis of rapid changes in transcript abundance in Chlamydomonas, transcripts of introduced genes were measured directly by Northern blot hybridization. A truncated IC70 clone was created in which an internal 1411-bp BglII fragment was deleted so that an mRNA smaller than the endogenous IC70 mRNA would be encoded (Figure 3). When this truncated gene was cotransformed into oda6,nit1,cw15 cells, the truncated message was expressed, and transformants showed increased levels of this mRNA after deflagellation, suggesting that the deflagellation response element(s) was contained in the 813 bp of upstream sequences present in this construct. Examination of this 813-bp upstream region revealed the presence of eight sequences (underlined in Figure 2B) with at least 70% identity to the 10-bp tub box consensus GCTC(G/C)AAGGC (a ninth copy spans the initiation site, and a tenth resides within an intron in the 5′-UTR). To determine whether these or other regulatory elements control deflagellation responsiveness, a series of 5′-flanking region deletions were made from the truncated gene by exonuclease digestion, and the constructs diagrammed in Figure 3A were selected for further use. Deletion constructs T-416, T-96, and T-45, which result in deletion of five, seven, or all eight upstream tub boxes (see arrowheads in Figure 2B), were introduced, and RNA samples were isolated before and at various times after deflagellation. The constitutively expressed CBLP mRNA was used as a normalizing control for the amount of RNA loaded in each lane. When RNA that had been isolated from a T-416 transformant was probed with an IC70 cDNA antisense transcript, bands of 1.6 kb (truncated gene) and 2.6 kb (endogenous gene) were detected (Figure 3B). Both bands show a similar time course of abundance changes, indicating that T-416 contains most of the element(s) required for normal regulation in response to a deflagellation signal. Peak abundance of the endogenous transcript was 7.9-fold above control (ND) levels in this experiment, while peak abundance of the truncated transcript was 5.7-fold above control. Absolute levels of truncated gene mRNA were 2.1 ± 0.4 (n = 8) fold higher than those of the endogenous IC70 message (averaged over multiple time points from two separate deflagellations). Southern blots used to analyze T-416 copy number indicated that one partial and two complete copies of the minigene had been integrated in this transformant. Two T-96 and one T-45 transformant were similarly tested and in each case expressed the truncated gene at levels that were detectable in ND cells. Both constructs supported induction 30′ after deflagellation similar to that of the endogenous IC70 gene (Figure 3D). To determine whether a regulatory element is located 3′ to the IC70 gene, a deletion was created in which all sequences beyond 10 bp 3′ from the polyadenylation site were removed (Figure 3A, 3′T10). When this deleted gene was introduced into oda6,nit1,cw15 cells, it still responded to deflagellation. The ratio of the fold-induction of truncated:endogenous transcripts was close to 1 in all cases (0.81 ± 0.18 (n = 3) for T-96, 1.24 ± 0.63 (n = 3) for T-45, and 0.96 ± 0.22 (n = 5) for 3′T10).
Figure 3.
Expression of truncated minigene deletion constructs. (A) Diagrams depicting deletion constructs. Thin lines indicate untranscribed flanking regions, thick lines indicate introns, and boxes indicate exons (open boxes represent UTRs and shaded boxes translated regions of the exons). A truncated IC70 gene was created by removing two BglII fragments from 6 kb genomic clone pB70S6. 5′- And 3′-deletions were then created by unidirectional exonuclease III digestion, and internal deletions were obtained by ligating together selected 5′- and 3′-deletions. (B) Expression of truncated gene T-416. RNA isolated before deflagellation (ND) and at indicated times after deflagellation was probed with an IC70 cDNA. The truncated gene expressed a smaller mRNA (1.6 kb) distinguishable from that of endogenous IC70 mRNA (2.6 kb). (C) The blot from panel B was stripped and reprobed with CBLP to confirm that equal amounts of mRNA were loaded. (D) Northern blots of RNA isolated before deflagellation (ND) and 30 min after deflagellation (DF) from cells transformed with the constructs shown in panel A. Blots were hybridized separately (T-96, T-45) or simultaneously (I-52–32, I-37–13) with IC70 and CBLP probes.
To further test the function of flanking sequences between −45 and the transcriptional start site, we tested a 5′-deletion to −13 (T−13) and two internal deletions in which bp −52 to −32 (I−52−32) and −37 to −13 (I−37−13) were replaced by 16 bp of plasmid sequence. None of 17 colonies transformed with T-13 expressed the truncated gene at a detectable level, either before or after deflagellation, suggesting that some elements between −45 and −13 might be necessary for basal gene expression. However, both I-52–32 and I-37–13 transformed cells expressed the truncated gene, and in both cases transcript levels had increased several fold by 30 min after deflagellation (Figure 3D).
Use of the Promoterless ARS Gene as a Reporter Gene at the Transcriptional Level
Because extensive 5′ and 3′-deletions did not eliminate the response of this truncated gene to deflagellation, we reasoned that either plasmid sequences were functioning as deflagellation response elements, or an essential element was located inside the gene. To test these possibilities, a promoterless ARS gene was used as a reporter gene. Endogenous ARS expression is completely repressed by the presence of low concentrations of SO42− in the medium (Davies et al., 1992), which allows the activity of introduced ARS gene constructs to be monitored. Although ARS enzyme activity was detectable in cells transformed with an ARS gene under the control of the β-2 tubulin promoter, this activity did not parallel the rapid increase in ARS mRNA observed when cells were deflagellated (Davies et al., 1992), perhaps reflecting slow processing of the translation product to its mature, catalytically active form. We therefore used transcript abundance rather than ARS activity to measure transcription of ARS under the control of a modified IC70 promoter. Transformants of chimeric IC70/ARS genes used for tests of transcriptional regulation were all selected based upon a PCR assay that ensured that ARS had not fortuitously recombined with genomic DNA so as to lose IC70 sequences and fall under the influence of another promoter.
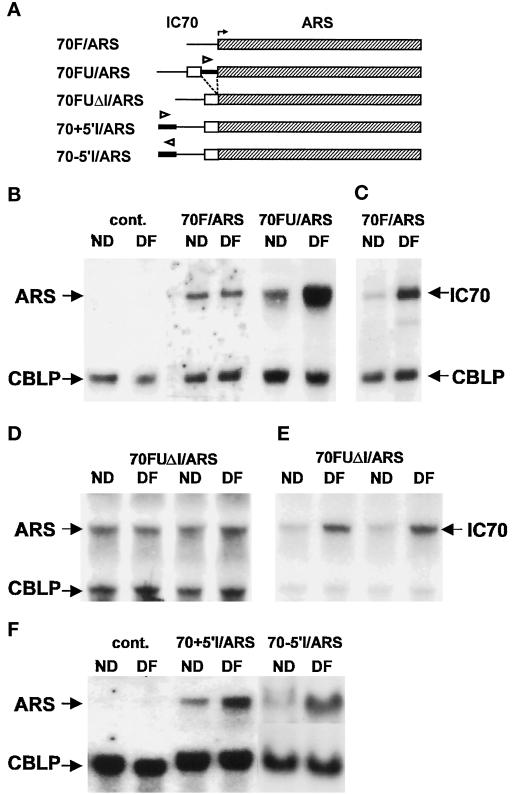
Several IC70/ARS transcriptional fusions were generated to locate the deflagellation response element (Figure 4A). In construct 70F/ARS, the IC70 5′-flanking region was ligated to a promoterless ARS gene that extends 67 bp 5′ of its translation start site. Additional constructs contain both the 5′-flanking region and portions of the 5′-UTR of IC70. We first confirmed that endogenous ARS expression is repressed in untransformed cells grown in the presence of SO42− (cont. lanes in Figure 4B). 70F/ARS transformants expressed the chimeric gene but no mRNA abundance increase was seen after deflagellation, suggesting that neither the IC70 5′-flanking region nor adjacent plasmid sequences can support deflagellation induction (Figure 4B, 70F/ARS). To exclude the possibility that the promoter fusion failed to respond because the pH shock induction signal was weak, this blot was stripped and reprobed with IC70 cRNA (Figure 4C), and a 4.8-fold increase in the endogenous IC70 mRNA abundance was revealed (both probes could not be used at the same time because the ARS and IC70 transcripts are of similar size).
Figure 4.
Expression of chimeric IC70/ARS gene constructs. (A) A promoterless ARS gene (shaded) was joined to IC70 flanking sequences (−430 to +1) to form 70F/ARS, or to a larger fragment spanning the IC70 5′-UTR as well (−430 to +430) to form 70FU/ARS. By removing the first intron from 70FU/ARS, 70FUΔI/ARS was obtained. 70+5′I/ARS and 70–5′I/ARS resulted from the insertion of the first intron upstream of 70FUΔI/ARS. Arrowheads indicate the orientation of intron sequences. (B) RNA isolated before deflagellation (ND) and 30 min after deflagellation (DF) from untransformed cells (C) and from selected transformants of 70F/ARS and 70FU/ARS was probed with ARS and CBLP. Endogenous ARS gene expression was totally repressed by the presence of SO42− in the medium (cont. lanes), 70F/ARS was expressed but not induced, whereas 70FU/ARS transcripts increased after deflagellation. (C) The 70F/ARS blot from panel B was stripped and reprobed with IC70 and CBLP to reveal a normal induction of the endogenous IC70 gene. (D) RNA from two 70FUΔI/ARS transformants probed with ARS and CBLP. Loss of the intron eliminates deflagellation inducibility. (E) The same blot as panel D stripped and reprobed with IC70. (F) RNA from a control strain and from strains transformed with 70+5′I/ARS and 70−5′I/ARS probed with ARS and CBLP. Insertion of the intron restores deflagellation inducibility.
To determine whether sequences needed for induction were present in the IC70 5′-UTR, a restriction fragment spanning the IC70 5′-flanking region plus most of the 5′-UTR (from −430 to +430) was ligated to the promoterless ARS gene to create construct 70FU/ARS (Figure 4A). When this construct was introduced into oda6/nit1 cells, two of nine randomly selected Nit+ colonies expressed the chimeric gene, and in both cases the gene was inducible. The Northern blot results of one such transformant are shown in Figure 4B. The strong induction seen (3.3-fold) suggests that a deflagellation-responsive element is located within the first 430 bp of the IC70 5′-UTR, which is interrupted by a 239-bp intron. To remove the intron from 70FU/ARS, an IC70 restriction fragment containing the intron was removed and a corresponding IC70 cDNA fragment substituted to create 70FUΔI/ARS (Figure 4A). After transformation of pMN24 and 70FUΔI/ARS into oda6/nit1 cells, two of nine Nit+ colonies expressed the chimeric gene, but neither of them responded to deflagellation (Figure 4D). As in Figure 4C, the blot was stripped and reprobed with IC70 (Figure 4E), and densitometry showed that the endogenous IC70 mRNA abundance increased by 3.6-fold and 3.3-fold after deflagellation.
Most mammalian enhancer and yeast upstream elements have the ability to function in both orientations, and at long and variable distances with respect to the transcription initiation site. The IC70 first intron was therefore tested for position and orientation dependence. The intron was amplified and inserted in both orientations into an upstream (−430) KpnI site of 70FUΔI/ARS (Figure 4A). When introduced into oda6/nit1 cells, reporter plasmids containing the intron in either orientation (70+5′I/ARS and 70−5′I/ARS) supported ARS expression and responded to deflagellation (3.2-fold and 3.7-fold, respectively), indicating that the enhancer activity of the intron is position and orientation independent (Figure 4F).
DISCUSSION
Our analysis of the deflagellation-induced increase in IC70 mRNA abundance indicates that a primary mode of regulation for this gene is transcriptional, as first shown by nuclear run-on assays and subsequently confirmed by localizing a necessary regulatory region in an intron. Because basal promoter elements have not been extensively characterized in Chlamydomonas, and the IC70 gene lacks an obvious TATA homology domain, its promoter could not be identified by sequence inspection. S1 nuclease protection assays were used to reveal one major transcription initiation site that defines the approximate 3′-end of the promoter. Multiple transcription initiation sites have been observed for other Chlamydomonas genes that lack an obvious TATA homology, such as those for radial spoke proteins 3 and 4 (Curry et al., 1992) and CBLP (Schloss, 1990), but promoter deletion studies that might define important basal promoter elements in these genes have not been reported.
In several transformants the IC70 minigenes were expressed at levels equal to or higher than the endogenous IC70 gene, as seen also for α1-tubulin gene constructs (Periz and Keller, 1997). In contrast, transformants containing chimeric CABII-1/NIT1 genes (Blankenship and Kindle, 1992) and tubulin/ARS genes (Davies et al., 1992) express these genes at levels much lower than those of endogenous CABII-1 or tubulin genes. Whether the high levels of expression we observed result from high levels of mRNA stability or from high levels of transcription, which in turn might be due to fortuitous integration into favorable genomic sites, to integration of multiple gene copies, or to an intrinsic property of the IC70 promoter sequences used, was not determined. Although several of these transformants contain more than one introduced gene copy, we did not determine how many of these copies were transcribed actively. Approximately 25% of the cotransformants (identified by PCR) failed to express the truncated gene at a detectable level. Although promoter regions of these genes were intact (PCR primers were chosen to only amplify complete recombinant promoters), absence of a detectable expression product could have resulted from rearrangement of coding sequences during the integration event or, alternatively, to integration into an unfavorable genomic region.
Less than 100 bp of Chlamydomonas β2 tubulin gene 5′-flanking region is sufficient for deflagellation induction (Davies and Grossman, 1994); deletions of this tubulin promoter to −95 retained inducibility while deletions to −65 or −36 removed deflagellation control without affecting basal transcription. A Chlamydomonas α-tubulin gene promoter has been similarly tested and loses full inducibility between −176 and −122, but retains weak inducibility down to −85 and basal transcription down to −16 (Periz and Keller, 1997). Here we have demonstrated continued transcription of the IC70 gene after deletion of 5′-flanking regions down to −45 from the major initiation site, and after replacing sequences from −52 to −32 and −37 to −13 with approximately equal lengths of plasmid sequence, suggesting that, as with tubulin, all essential promoter elements are found in a small region close to the initiation site. Unlike tubulin genes, the regulatory element responsible for deflagellation induction of the IC70 gene is not found in the 5′-flanking region but instead is an enhancer located in a 239-bp intron. Intronic enhancers have been identified in many other genes (Chung and Perry, 1989; Franklin et al., 1991; Bai et al., 1993), including β-tubulin genes of Drosophila (Buttgereit and Renkawitz-Pohl, 1993), and the RBCS2 gene in Chlamydomonas (Lumbreras et al., 1998). Sequence comparisons between this intron and either the 5′-proximal promoter region or the introns of several other C. reinhardtii flagellar genes including α1, α2, β1, and β2 tubulin genes (Brunke et al., 1984; Schloss et al., 1984; Silflow et al., 1985) and radial spoke protein 3, 4, and 6 genes (Williams et al., 1989; Curry et al., 1992) did not reveal any flagellar gene-exclusive consensus sequence. Although one region with an 80% match to the tub box consensus is seen in the IC70 intron, deletion of nine other tub box consensus sequences between −813 and +1 (and four additional tub box consensus sequences in reverse orientation on the opposite strand) did not affect induction. The presence of multiple matches to the tub box consensus may be a consequence of the high G + C content of the C. reinhardtii genome with no direct relevance to transcriptional regulation. Lack of a highly conserved sequence motif among all genes induced by deflagellation suggests that different genes may use different regulatory element(s) for deflagellation-induced transcriptional regulation, or that the element(s) is degenerate enough to be missed by sequence alignments.
Although α- and β-tubulin mRNA accumulation can be largely accounted for by new transcription (Keller et al., 1984), comparison of the synthesis rates with accumulation fold change indicated that an elongated mRNA half-life contributes as well (Baker et al., 1984). Whether dynein mRNA stability likewise increases after deflagellation has not been directly determined, but a similar fold change in mRNA abundance was observed for the IC70 minigene, which has intact 5′- and 3′-UTRs, and the IC70/ARS chimeric gene constructs, in which the IC70 3′-UTR was replaced by the ARS 3′-UTR. Gene-specific 3′-UTR sequences do not appear to contribute to deflagellation-induced mRNA abundance increases. Further, when the intronic transcriptional control region is absent, no change in IC70/ARS mRNA abundance is seen after deflagellation (Figure 4B, 70F/ARS; Figure 4D). The contribution of 5′-UTR–dependent mRNA stability changes to increases in mRNA abundance must therefore be below the level of detection in these Northern blot experiments.
The signal transduction pathway that mediates deflagellation-induced transcriptional activation is not yet well characterized. Although mRNA abundance changes are usually coupled with flagellar outgrowth and synthesis of new flagellar proteins, several lines of evidence indicate that protein synthesis is not required for deflagellation-induced increases in mRNA synthesis (Baker et al., 1986) and that induction is actually independent of flagellar excision and regeneration (Cheshire and Keller, 1991; Quarmby et al., 1992). The calmodulin antagonist W-7, the protein kinase inhibitor staurosporine, and conditions that reduce calcium concentrations each partially inhibit flagellar gene induction, implicating several second messengers in the deflagellation induction cascade (Cheshire and Keller, 1991). This pathway must ultimately lead to cis elements important to the deflagellation response such as the enhancer identified here. A fuller understanding of the coordinate regulation of flagellar assembly will require a more detailed analysis of transcriptional regulation with multiple genes, preferably genes encoding flagellar components that ultimately assemble in similar stoichiometric ratios.
ACKNOWLEDGMENTS
We thank Pete Lefebvre for providing the NIT1 gene, John Davies for ARS constructs, Jeff Schloss for the CBLP gene, Mike Lane for generous use of his scanner, and Bob West for reading the manuscript critically. Kim Brown provided valuable technical assistance. This work was supported by grant GM-44228 from the General Medical Sciences division of the National Institutes of Health (to D.R.M.).
REFERENCES
- Bai G, Stuebing EW, Parker HR, Harlow P, Nemer M. Combinatorial regulation by promoter and intron 1 regions of the metallothionein SpMTA gene in the sea urchin embryo. Mol Cell Biol. 1993;13:993–1001. doi: 10.1128/mcb.13.2.993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker EJ, Keller LR, Schloss JA, Rosenbaum JL. Protein synthesis is required for rapid degradation of tubulin mRNA and other deflagellation-induced RNAs in Chlamydomonas reinhardtii. Mol Cell Biol. 1986;6:54–61. doi: 10.1128/mcb.6.1.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker EJ, Schloss JA, Rosenbaum JL. Rapid changes in tubulin RNA synthesis and stability induced by deflagellation in Chlamydomonas. J Cell Biol. 1984;99:2074–2081. doi: 10.1083/jcb.99.6.2074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blankenship JE, Kindle KL. Expression of chimeric genes by the light-regulated cabII-1 promoter in Chlamydomonas reinhardtii: a cabII-1/nit1 gene functions as a dominant selectable marker in a nit1−nit2− strain. Mol Cell Biol. 1992;12:5268–5279. doi: 10.1128/mcb.12.11.5268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunke KJ, Anthony JG, Sternberg EJ, Weeks DP. Repeated consensus and pseudopromoters in the four coordinately regulated tubulin genes of Chlamydomonas reinhardtii. Mol Cell Biol. 1984;4:1115–1124. doi: 10.1128/mcb.4.6.1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buttgereit D, Renkawitz-Pohl R. Expression of 1 tubulin (Tub56D) in Drosophila testis stem cells is regulated by a short upstream sequence while intron elements guide expression in somatic cells. Mol Gen Genet. 1993;241:263–270. doi: 10.1007/BF00284677. [DOI] [PubMed] [Google Scholar]
- Cheshire JL, Keller LR. Uncoupling of Chlamydomonas flagellar gene expression and outgrowth from flagellar excision by manipulation of Ca2+ J Cell Biol. 1991;115:1651–1659. doi: 10.1083/jcb.115.6.1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- Chung S, Perry RP. Importance of intron for expression of mouse ribosomal protein gene rpL32. Mol Cell Biol. 1989;9:2075–2082. doi: 10.1128/mcb.9.5.2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curry AM, Williams BD, Rosenbaum JL. Sequence analysis reveals homology between two proteins of the flagellar radial spoke. Mol Cell Biol. 1992;12:3967–3977. doi: 10.1128/mcb.12.9.3967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies JP, Grossman AR. Sequences controlling transcription of the Chlamydomonas reinhardtii β2-tubulin gene after deflagellation and during the cell cycle. Mol Cell Biol. 1994;14:5165–5174. doi: 10.1128/mcb.14.8.5165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies JP, Weeks DP, Grossman AR. Expression of the arylsulfatase gene from the β2-tubulin promoter in Chlamydomonas reinhardtii. Nucleic Acids Res. 1992;20:2959–2965. doi: 10.1093/nar/20.12.2959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franklin GC, Donovan M, Adams GIA, Holmgren L, Pfieffer-Ohlsson S, Ohlsson R. Expression of the human PDGF-beta gene is regulated by both positively and negatively acting cell type-specific regulatory elements isolated in the first intron. EMBO J. 1991;10:1365–1373. doi: 10.1002/j.1460-2075.1991.tb07656.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttman SD, Gorovsky MA. Cilia regeneration in starved Tetrahymena: an inducuble system for studying gene expression and organelle biogenesis. Cell. 1979;17:307–317. doi: 10.1016/0092-8674(79)90156-9. [DOI] [PubMed] [Google Scholar]
- Harris EH. The Chlamydomonas Sourcebook. San Diego, CA: Academic Press; 1989. [Google Scholar]
- Keller LR, Schloss JA, Silflow CD, Rosenbaum JL. Transcription of alpha and beta tubulin genes in vitro in isolated Chlamydomonas reinhardtii nuclei. J Cell Biol. 1984;98:1138–1143. doi: 10.1083/jcb.98.3.1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kindle KL. High-frequency nuclear transformation of Chlamydomonas reinhardtii. Proc Natl Acad Sci USA. 1990;87:1228–1232. doi: 10.1073/pnas.87.3.1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefebvre PA, Rosenbaum J. Regulation of the synthesis and assembly of ciliary and flagellar proteins during regeneration. Annu Rev Cell Biol. 1986;2:517–546. doi: 10.1146/annurev.cb.02.110186.002505. [DOI] [PubMed] [Google Scholar]
- Lefebvre PA, Silflow CD, Wieben ED, Rosenbaum JL. Increased levels of mRNAs for tubulin and other flagellar proteins after amputation or shortening of Chlamydomonas flagella. Cell. 1980;20:469–477. doi: 10.1016/0092-8674(80)90633-9. [DOI] [PubMed] [Google Scholar]
- Lumbreras V, Stevens DR, Purton S. Efficient foreign gene expression in Chlamydomonas reinhardtii mediated by an endogenous intron. Plant J. 1998;14:441–447. [Google Scholar]
- Mitchell DR, Kang Y. Identification of oda6 as a Chlamydomonas dynein mutant by rescue with the wild-type gene. J Cell Biol. 1991;113:835–842. doi: 10.1083/jcb.113.4.835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell DR, Kang Y. Reversion analysis of dynein intermediate chain function. J Cell Sci. 1993;105:1069–1078. doi: 10.1242/jcs.105.4.1069. [DOI] [PubMed] [Google Scholar]
- Nevins JR. Isolation and analysis of nuclear RNA. Methods Enzymol. 1987;152:234–241. doi: 10.1016/0076-6879(87)52025-0. [DOI] [PubMed] [Google Scholar]
- Periz G, Keller LR. DNA elements regulating α1-tubulin gene inductionn during regeneration of eukaryotic flagella. Mol Cell Biol. 1997;17:3858–3866. doi: 10.1128/mcb.17.7.3858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quarmby LM, Yueh YG, Cheshire JL, Keller LR, Snell WJ, Crain RC. Inositol phospholipid metabolism may trigger flagellar excision in Chlamydomonas reinhardtii. J Cell Biol. 1992;116:737–744. doi: 10.1083/jcb.116.3.737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sager R, Granick S. Nutritional studies with Chlamydomonas reinhardtii. Ann NY Acad Sci. 1953;466:18–30. doi: 10.1111/j.1749-6632.1953.tb30261.x. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Schloss JA. A Chlamydomonas gene encodes a G protein subunit-like polypeptide. Mol Gen Genet. 1990;221:443–452. doi: 10.1007/BF00259410. [DOI] [PubMed] [Google Scholar]
- Schloss JA, Silflow CD, Rosenbaum JL. mRNA abundance changes during flagellar regeneration in Chlamydomonas reinhardtii. Mol Cell Biol. 1984;4:424–434. doi: 10.1128/mcb.4.3.424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silflow CD, Chisholm RL, Conner TW, Ranum LPW. The two alpha-tubulin genes of Chlamydomonas reinhardtii code for slightly different proteins. Mol Cell Biol. 1985;5:2389–2398. doi: 10.1128/mcb.5.9.2389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens RE. Differential protein synthesis and utilization during cilia formation in sea urchin embryos. Dev Biol. 1977;61:311–329. doi: 10.1016/0012-1606(77)90301-3. [DOI] [PubMed] [Google Scholar]
- Williams BD, Mitchell DR, Rosenbaum JL. Molecular cloning and expression of flagellar radial spoke and dynein genes of Chlamydomonas. J Cell Biol. 1986;103:1–11. doi: 10.1083/jcb.103.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams BD, Velleca MA, Curry AM, Rosenbaum JL. Molecular cloning and sequence analysis of the Chlamydomonas gene coding for radial spoke protein 3: flagellar mutation pf-14 is an ochre allele. J Cell Biol. 1989;109:235–245. doi: 10.1083/jcb.109.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witman GB, Carlson K, Berliner J, Rosenbaum JL. Chalmydomonas flagella. I. Isolation and electrophoretic analysis of microtubules, matrix, membranes, and mastigonemes. J Cell Biol. 1972;54:507–539. doi: 10.1083/jcb.54.3.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youngbloom J, Schloss JA, Silflow CD. The two β-tubulin genes of Chlamydomonas reinhardtii code for identical proteins. Mol Cell Biol. 1984;4:2686–2696. doi: 10.1128/mcb.4.12.2686. [DOI] [PMC free article] [PubMed] [Google Scholar]