Abstract
To identify yeast cytosolic proteins that mediate targeting of precursor proteins to mitochondria, we developed an in vitro import system consisting of purified yeast mitochondria and a radiolabeled mitochondrial precursor protein whose C terminus was still attached to the ribosome. In this system, the N terminus of the nascent chain was translocated across both mitochondrial membranes, generating a translocation intermediate spanning both membranes. The nascent chain could then be completely chased into the mitochondrial matrix after release from the ribosome. Generation of this import intermediate was dependent on a mitochondrial membrane potential, mitochondrial surface proteins, and was stimulated by proteins that could be released from the ribosomes by high salt. The major salt-released stimulatory factor was yeast nascent polypeptide–associated complex (NAC). Purified NAC fully restored import of salt-washed ribosome-bound nascent chains by enhancing productive binding of the chains to mitochondria. We propose that ribosome-associated NAC facilitates recognition of nascent precursor chains by the mitochondrial import machinery.
INTRODUCTION
The great majority of mitochondrial proteins is encoded in the nucleus, synthesized on cytoplasmic ribosomes, and imported into mitochondria. Most matrix-targeted proteins carry an N-terminal, positively charged presequence that can form an amphiphatic α-helix and is usually cleaved by mitochondrial-processing peptidase (MPP) upon import into the matrix (Roise et al., 1986; von Heijne, 1986). Precursor proteins are recognized on the mitochondrial surface by “TOM” receptors (for “translocase of the outer membrane”) and translocated across the outer membrane through the TOM channel (Dekker et al., 1998; Hill et al., 1998; Künkele et al., 1998). According to the “acid chain” hypothesis, the precursor is guided through the TOM channel by acidic binding sites on Tom proteins of increasing affinity for the positively charged presequence (Bolliger et al., 1995; Dietmeier et al., 1997; Schatz, 1997; Komiya et al., 1998). The “TIM”-17/23 channel (for “translocase of the inner membrane”) transports presequence-carrying precursors across the inner membrane in a membrane potential–dependent way (Pfanner et al., 1997). All precursors that are imported completely into the matrix require ATP in the matrix for the proposed import motor, mitochondrial hsp70 (Pfanner et al., 1997). Some precursors also need ATP outside the mitochondria, probably for release from cytosolic chaperones (Wachter et al., 1994).
Genetic and biochemical experiments suggest that cytosolic proteins are involved in mitochondrial protein import. Several genetic screens identified candidate genes in yeast, but the phenotypes of the mutations are often subtle and pleiotropic. Mutations of members of the SSA subfamily of cytosolic hsp70 proteins or of the cytosolic DnaJ homologue Ydj1p impair protein import into mitochondria and the endoplasmic reticulum (ER) in vivo, indicating a general role for these chaperones in both translocation systems (Deshaies et al., 1988; Caplan et al., 1992). A mutation in the cytosolic protein Mft52p (Cartwright et al., 1997) or a deletion of the α-subunit of nascent polypeptide–associated complex (NAC) (George et al., 1998) inhibit delivery of artificial fusion proteins, but not of authentic precursors, to mitochondria in vivo.
Biochemical studies in mammalian systems identified the ATPase “mitochondrial import stimulation factor” and cytosolic hsp70 proteins as cytosolic factors that keep precursors in an import-competent state and deliver them to the TOM receptors in vitro, thereby stimulating protein import into isolated mitochondria (Komiya et al., 1996). Two additional cytosolic proteins, presequence binding factor (Murakami and Mori, 1990) and targeting factor (Ono and Tuboi, 1990), were reported to stimulate protein import into isolated mammalian mitochondria, but neither the amino acid sequence nor the mechanism of action of these proteins is known.
To identify yeast cytosolic “import stimulation factors” and to study the mechanism of the early steps of mitochondrial protein import, we set up a fully homologous in vitro import assay in that cytosol, mitochondria, and precursor were derived from yeast to avoid artifacts arising from heterologous combinations. In vivo, fully synthesized but unprocessed mitochondrial precursor proteins are almost undetectable (Fujiki and Verner, 1993), suggesting that their import is extremely fast and occurs either cotranslationally (Ades and Butow, 1980) or very soon after synthesis is complete. In both cases, import-stimulating factors are likely to associate with the nascent chain still bound to the ribosome. To identify such import-stimulating factors we have translated the precursor protein in vitro from an mRNA lacking the stop codon, thereby generating ribosome–nascent chain complexes (RNCs) in which the C terminus of the nascent chain remains bound to the ribosome (Mueckler and Lodish, 1986; Gilmore et al., 1991). RNCs have two major advantages over released precursor chains: 1) they can be separated from the bulk cytosol by sedimentation through a sucrose cushion, which allows supplementation with selected cytosolic factors during the import reaction; and 2) the precursor protein remains “trapped” in a “nascent conformation,” which prevents folding and might stabilize interactions with specific chaperones or targeting factors. An analogous approach has been a key to understanding the mechanisms of protein translocation into the ER (Gilmore et al., 1991).
Upon incubation of isolated RNCs with purified mitochondria, the N terminus of the nascent chain was imported into the matrix and processed by MPP, whereas the C terminus was still attached to the ribosome on the mitochondrial surface. Yeast NAC was identified as a factor that stimulated formation of this two-membrane spanning import intermediate by enhancing productive binding of the nascent chains to the mitochondria.
MATERIALS AND METHODS
Purification of NAC
A 24-l culture of the protease-deficient yeast strain C13 ABYS-86 (Heinemeyer et al., 1991) was grown at 30°C in YPD medium overnight to an OD600 of 1.2. Cells (120 g wet weight) were harvested and converted to spheroplasts by Zymolyase (Seikagaku America, Falmouth, MA) treatment. The spheroplasts were washed in 2.4 l of sorbitol buffer (1.4 M sorbitol, 50 mM KPi, pH 7.4, 5–10 mM DTT) and lysed in 200 ml of lysis buffer (20 mM HEPES, pH 7.4, 100 mM K-acetate, pH 7.4, 2 mM Mg-acetate, 2 mM DTT, 0.5 mM PMSF, 1.25 μg/ml leupeptin, 0.75 μg/ml antipain, 0.25 μg/ml chymostatin and elastinol, 5 μg/ml peptstatin) in an all-glass Dounce homogenizer. The homogenate was cleared by centrifugation for 15 min at 27,000 × g and then 30 min at 80,000 × g. The clear supernatant (140 ml) was layered on top of 2 vol of buffer S1 (25% sucrose, 20 mM HEPES, pH 7.4, 120 mM K-acetate, pH 7.4, 5 mM Mg-acetate, 2 mM DTT, 0.5 mM PMSF) and centrifuged for 2.5 h at 160,000 × g to sediment the ribosomes. The ribosomal pellet was resupended in buffer S1 containing 0.7 M K-acetate using a Dounce homogenizer with a Teflon piston, and the suspension was centrifuged for 2.5 h at 160,000 × g. The supernatant was saved and diluted with 3.5 vol of buffer A (buffer S1 lacking sucrose and K-acetate), cleared by centrifugation for 20 min at 1200 × g, and loaded onto a ResourceQ anion exchange column (Pharmacia, Piscataway, NJ). Bound proteins were eluted with a linear 80 ml K-acetate gradient (200–600 mM) and NAC eluted at ∼440 mM K-acetate. NAC-containing fractions were pooled, diluted with 0.33 vol of buffer A, and loaded onto a MonoQ anion exchange column (Pharmacia). Bound proteins were eluted with a linear 60-ml K-acetate gradient (500–700 mM) and NAC eluted at ∼620 mM K-acetate. NAC-containing fractions were pooled, diluted with 1 vol of buffer A, concentrated in a Centricon-30 device (Millipore, Bedford, MA), frozen in small aliquots in liquid nitrogen, and stored at −80°C.
Import Assay
Mitochondria were isolated from the protease-deficient yeast strain C13 ABYS-86 (Heinemeyer et al., 1991) and purified on a Nycodenz gradient (Nycomed Pharma, Oslo, Norway) (Glick and Pon, 1995). For the study of the receptor mutants, the yeast strains deleted either in TOM70 (YVH1) (Hines and Schatz, 1993) or TOM20 (YTJB64) (Lithgow et al., 1994b) and a corresponding wild-type strain (JKR101) were grown on YPGal (1% yeast extract, 1% peptone, 2% galactose), because YTJB64 grows poorly on nonfermentable carbon sources. Crude mitochondria were prepared according to the method of Glick and Pon (1995), except that the last purification step on the Nycodenz gradient was omitted. Import reactions contained, unless stated otherwise, 0.8 mg/ml mitochondria in “import buffer” (0.32 mg/ml BSA, 20 mM HEPES, pH 7.4, 120 mM K-acetate, pH 7.4, 5 mM Mg-acetate, 0.6 M sorbitol, 0.05 U/μl RNase inhibitor [from human placenta], and 2 mM DTT) supplemented with 2 mM ATP, NADH, and KPi. The mitochondria were premixed with import buffer, and imports were started by the addition of 12.5% (vol/vol) RNCs (see below) and were carried out at 20°C for up to 20 min. Volumes of import varied from 100 to 500 μl. At the indicated time points, 100-μl samples were withdrawn for further analysis. Protease treatment to probe for import and folding was performed as described (Rospert and Schatz, 1998).
Construction of Truncated MDH1
The yeast mitochondrial MDH1 gene lacking the stop codon was amplified with Pfu polymerase from yeast genomic DNA using the primers MDH-F (ggccggtgcgggatccatgttgtcaagagtagct) and MDH-R2 (ctgcagtttactagcaacaaagttgacacc). The PCR product and the expression plasmid pSP65 (Promega, Madison, WI) were cut with BamHI and PstI, ligated, and transformed into the Escherichia coli strain DH10B. The resulting plasmid is termed pSP65-MDHt.
In Vitro Translation and Preparation of RNCs
Yeast translation extract was prepared as described (Garcia et al., 1991) from the protease-deficient strain C13-ABYS-86 (Heinemeyer et al., 1991). mRNA for truncated Mdh1p was generated by in vitro transcription using SP6 polymerase from plasmid pSP65-MDHt, phenol-chloroform extracted, ethanol precipitated, resuspended in water, and stored in aliquots at −80°C. Translations were performed as described (Garcia et al., 1991) at 20°C for 30 min and cleared by centrifugation for 10 min at 20,000 × g at 4°C. RNCs were pelleted by centrifugation through two volumes of buffer S2 (25% sucrose, 20 mM HEPES, pH 7.4, 5 mM Mg-acetate, 2 mM DTT, 0.005 U/μl RNase inhibitor [from human placenta]) containing either 120 mM K-acetate (untreated RNC) or 0.7 M K-acetate (salt-washed RNC) at 200,000 × g in a TLA-100 tabletop ultracentrifuge (Beckman Instruments, Palo Alto, CA) for 50 min at 2°C. The clear pellets were resupended in import buffer lacking BSA in a volume equal to the original translation reaction.
Solubilization of Mitochondria and Isolation of RNCs
Import was performed as described for 20 min at 20°C and stopped by transfer on ice, supplemented with protease inhibitors (1 mM PMSF, 1.25 μg/ml leupeptin, 0.75 μg/ml antipain, 0.25 μg/ml chymostatin and elastinol, 5 μg/ml peptstatin) and Triton X-100 to a final concentration of 1% in the presence or absence of 10 mM EDTA. Samples were left on ice for 10 min and cleared by centrifugation for 10 min at 14,000 × g at 4°C. The supernatants were layered on top of 2 vol of buffer S2 containing 120 mM K-acetate and 1% Triton X-100. RNCs were sedimented at 200,000 × g for 40 min at 4°C.
Preparation of Salt Wash Fluid and Immunodepletion
For the preparation of salt wash fluid, ribosomes from yeast translation extract were pelleted by centrifugation through 2 vol of buffer S2 containing 120 mM K-acetate for 50 min at 200,000 × g at 2°C. The ribosomes were resuspended in buffer S2 containing 0.7 M K-acetate and pelleted again for 50 min at 200,000 × g at 2°C. The resulting supernatant was the salt wash fluid. Immunoprecipitations were performed on ice for 2.5 h in 20 mM HEPES, pH 7.4, 120 mM K-acetate, pH 7.4, 5 mM Mg-acetate, 0.5 mM PMSF, 1.25 μg/ml leupeptin, 0.75 μg/ml antipain, 0.25 μg/ml chymostatin and elastinol, 5 μg/ml pepstatin; using 3 μl of polyclonal rabbit serum/1 μl of swollen protein A-Sepharose beads.
MPP Assay
Recombinant MPP was a gift from P. Luciano and V. Géli (LISM-CRNS, Marseille, France) (Luciano et al., 1998). RNCs were resuspended in import buffer and supplemented with 0.04 μg/μl NAC as indicated. MPP was added to 0.5 μM, and the samples were incubated at 20°C. After the indicated times, 30-μl aliquots were withdrawn and precipitated with trichloroacetic acid (TCA). Cleavage was assayed by SDS-PAGE and fluorography.
Miscellaneous
Protein concentrations were determined by the Bradford assay (from Bio-Rad, Hercules, CA) using BSA as a standard. 125I-labeled protein A was used to develop the immunoblots.
RESULTS
Ribosome-attached Nascent Chains Form Productive Import Intermediates
We chose the precursor of yeast mitochondrial malate dehydrogenase (Mdh1p) as a model protein, because it carries a typical N-terminal, cleavable presequence and folds into a protease-resistant conformation inside mitochondria (Dubaquiéet al., 1998). When the Mdh1p precursor was translated from an mRNA lacking a stop codon, the C terminus of the full-length precursor of 335 amino acids remained attached to the ribosome, forming a stable RNC (Figure 1B) (Mueckler and Lodish, 1986; Gilmore et al., 1991).
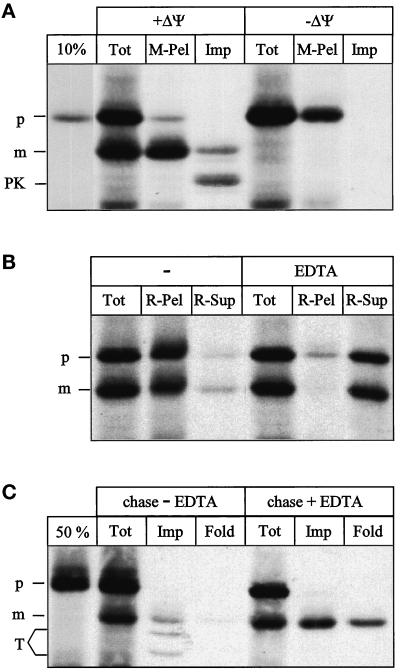
Figure 1.
Ribosome-associated nascent chains can generate a mitochondrial import intermediate spanning both membranes. (A) Generation of the import intermediate. RNCs carrying the radiolabeled Mdh1p precursor were isolated and incubated with yeast mitochondria as described in MATERIALS AND METHODS for 20 min at 20°C in the absence (+ΔΨ) or presence (−ΔΨ) of 1 μg/ml valinomycin. Equal aliquots were precipitated with TCA (Tot), centrifuged for 3 min at 14,000 × g to recover the mitochondria (M-Pel), or treated with 100 μg/μl proteinase K for 15 min on ice, followed by inactivation of the protease with 1 mM PMSF, reisolation of the mitochondria, and precipitation with TCA (Imp). All samples were analyzed by SDS-PAGE and fluorography. 10%, 10% of the RNCs added per lane. (B) The intermediate is ribosome attached. Import was performed as in A and stopped by transfer to 0°C. The sample was then split into two equal aliquots. The mitochondria in both aliquots were solubilized in 1% Triton X-100 either in the presence (EDTA) or absence (−) of 10 mM EDTA as described in MATERIALS AND METHODS. One-half of each aliquot was precipitated with TCA (Tot); the other half was separated into a ribosomal pellet (R-Pel) and a supernatant (R-Sup). All samples were analyzed by SDS-PAGE and fluorography. (C) Chase of the intermediate. Import was performed as in A, and further generation of the import intermediate was stopped after 20 min by the addition of 1 μg/ml valinomycin. The reaction was split into two equal aliquots. Onealiquot was left untreated at 20°C (chase − EDTA); the other aliquot was treated with 10 mM EDTA for 10 min at 20°C, followed by the addition of 10 mM MgAcetate (chase + EDTA). One-third of each aliquot was precipitated with TCA (Tot); the remaining two-thirds were treated with 100 μg/ml trypsin for 30 min on ice. After inactivation of the protease with 200 μg/ml soy bean trypsin inhibitor and reisolation of the mitochondria, the trypsin-treated mitochondria were subdivided in two equal samples. The first sample was precipitated with TCA (Imp), and the second sample was treated with 1% Triton X-100 plus 100 μg/ml proteinase K for 15 min on ice. After inactivation of the protease with 1 mM PMSF, the second sample was also precipitated with TCA (Fold). All samples were analyzed by SDS-PAGE and fluorography. 50%, 50% of the RNCs added per lane; p, Mdh1p precursor; m, mature Mdh1p; PK, proteinase K-protected fragment; T, trypsin-protected fragments.
To test whether the ribosome-attached precursor chain could engage the mitochondrial import machinery, RNCs were isolated by centrifugation through a sucrose cushion and incubated with isolated yeast mitochondria (Figure 1A).
Approximately 45% of the chains were processed to the mature size (Tot), indicating that their N-terminus had reached the matrix. Import was dependent on a potential across the mitochondrial inner membrane (Tot, −ΔΨ). All processed chains, but very few unprocessed ones, were recovered in the mitochondrial pellet (M-Pel). After treatment of the mitochondria with proteinase K, only 14% of mature-sized chains was fully protected; the bulk was converted to a smaller protected fragment (Imp), indicating that a portion of the chain was accessible from outside. To test whether the processed chains were still attached to ribosomes, we isolated the RNCs from a total import reaction by solubilizing the mitochondria in 1% Triton X-100 and sedimentation through a sucrose cushion (Figure 1B). Triton X-100 solubilizes membranes but leaves ribosomes intact (Schneider et al., 1976). The majority of both precursor and mature Mdh1p was recovered in the ribosomal pellet (R-Pel), indicating that they were still attached to ribosomes. Solubilization in the presence of 10 mM EDTA dissociated the ribosomes and released most of the processed as well as unprocessed Mdh1p into the supernatant (Figure 1B, EDTA). Ribosome-attached nascent chains are thus not completely imported but accumulate as processed, two-membrane-spanning import intermediates.
To test whether these import intermediates could be chased into the matrix, we accumulated them as described in Figure 1A, stopped further accumulation by addition of the uncoupler valinomycin, and split the reaction in two. One-half was left untreated, and the other half was treated with EDTA to release the nascent chain from the ribosome. Both halves were then “chased” for 10 min (Figure 1C). When the chase was conducted in the absence of EDTA, one-third of the chains were processed to the mature size (Tot), and trypsin generated mature-sized chains as well as two smaller protected fragments (Imp). The pattern of protease-protected fragments depended on the type of protease used (Figure 1, compare A and C, Imp lanes). The protected fragments and most of the mature chains were not properly folded, because they were sensitive to proteinase K after solubilization of the mitochondria with Triton X-100 (Fold). When the chase was conducted in the presence of EDTA, 70% of the mature chains were protease-protected in intact mitochondria (Imp), and 34% had folded into a protease-resistant conformation (Fold). Ribosome-attached precursor chains can thus form productive import intermediates that can be completely imported into mitochondria upon release from the ribosome.
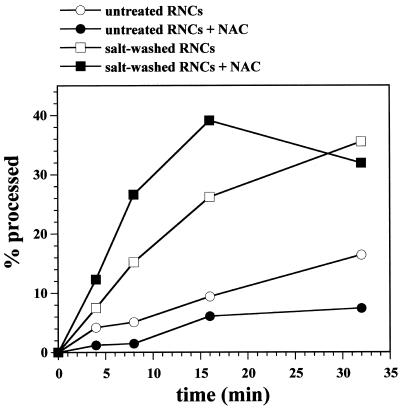
To test whether proteins peripherally associated with RNCs influenced the import of ribosome-attached chains, we prepared salt-washed RNCs by centrifugation through a sucrose cushion containing 0.7 M K-acetate. Salt washing of RNCs greatly reduced the import efficiency of the attached precursor chains (Figure 2).
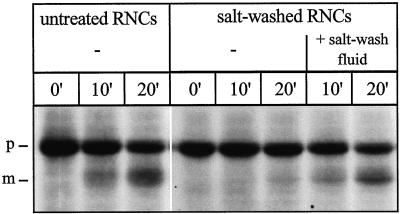
Figure 2.
Salt washing of RNCs reversibly reduces import of the attached nascent chains. Untreated RNCs, salt-washed RNCs, and a ribosomal salt wash fluid were prepared as described in MATERIALS AND METHODS. Salt wash fluid was prepared from an amount of ribosomes corresponding to the amount of ribosomes present as RNCs in the import reaction. Import reactions were performed with untreated RNCs, salt-washed RNCs, and salt-washed RNCs mixed with salt wash fluid (+ salt-wash fluid) as described in MATERIALS AND METHODS. After the indicated times, equal samples were precipitated with TCA and analyzed by SDS-PAGE and fluorography. p, Mdh1p precursor; m, mature Mdh1p.
This effect was reversible, readdition of a ribosomal salt wash fluid corresponding to the amount of proteins released from the RNCs, almost completely restored import. Heating of the salt wash fluid to 95°C for 5 min or protease treatment abolished the stimulatory activity, suggesting it is caused by a protein (our unpublished data).
Identification of the Stimulating Factor
We fractionated a ribosomal salt wash fluid by anion exchange chromatography and tested the fractions for import stimulation of ribosome-attached nascent chains. The active fractions contained equal amounts of two prominent proteins with apparent molecular masses of 28 and 21 kDa. By mass spectrometry of tryptic fragments, the two proteins were identified as the α- and β-subunits of yeast NAC. This complex has been described both in yeast and in mammals, although in a different context (Parthun et al., 1992; Wiedmann et al., 1994; Shi et al., 1995; George et al., 1998).
To verify that NAC acts indeed as an import stimulatory factor, a ribosomal salt wash fluid was either mock depleted with preimmuneserum or immunodepleted with immuneserum against yeast αNAC (George et al., 1998). After immunodepletion, αNAC was undetectable in the salt wash fluid and quantitatively recovered on the beads (Figure 3A).
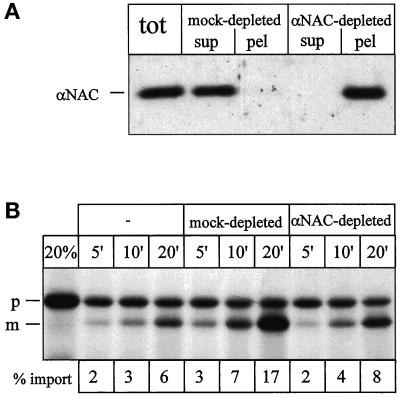
Figure 3.
Immunodepletion of NAC from salt wash fluid depletes most but not all import stimulatory activity. (A) Immunodepletion of salt wash fluid. Equal aliquots of salt wash fluid (see Figure 2) were either left untreated (tot), mock depleted with preimmuneserum (mock-depleted), or immunodepleted with immuneserum against αNAC (αNAC-depleted). The treated aliquots were separated into supernatant (sup) and beads (pel), and all samples were analyzed for αNAC by SDS-PAGE and immunoblotting. (B) Import stimulation by salt wash fluid. Import reactions were performed with salt-washed RNCs without addition (−), after addition of mock-depleted salt wash fluid (mock-depleted), or after addition of αNAC-depleted salt wash fluid (αNAC-depleted) as described in MATERIALS AND METHODS. After the indicated times, equal samples were centrifuged for 3 min at 14,000 × g to recover the mitochondria. All samples were analyzed by SDS-PAGE and fluorography and quantified by densitometry. % import, percentage of mature Mdh1p given as fraction of initially added Mdh1p; 20%, 20% of the RNCs added per lane; p, Mdh1p precursor; m, mature Mdh1p.
The import stimulatory activity of αNAC-depleted salt wash fluid was reduced to 20% compared with the mock-depletion (Figure 3B). Consistent with published data (Parthun et al., 1992; Wiedmann et al., 1994; Reimann et al., 1999), we identified α- and βNAC as a stable complex and the purified complex could be quantitatively immunoprecipitated with immuneserum against αNAC (Figure 4 and our unpublished data). The complex of α- and βNAC thus constitutes the major stimulatory activity in the salt wash fluid.
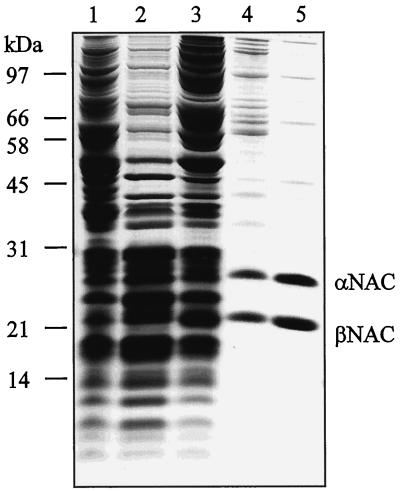
Figure 4.
Purification of NAC from yeast cytosol. Yeast NAC was purified as described in MATERIALS AND METHODS, and the following fractions were analyzed by SDS-PAGE and staining with Coomassie blue: lane 1, cytosol; lane 2, ribosomal pellet; lane 3, salt wash fluid; lane 4, pooled fractions after ResourceQ chromatography; lane 5, pooled fractions after MonoQ chromatography. Lanes 1–3 contain 100 μg of protein; lanes 4 and 5 contain 10 μg of protein. Molecular mass standards are indicated on the left.
After refinement of the fractionation scheme used to identify NAC, we purified NAC by a factor of 900 from total yeast cytosol (Figure 4 and Table 1).
Table 1.
Purification of NAC and the import-stimulatory activity from yeast cytosol
| Fraction | Total protein (mg) | NAC (mg)a | Yield of NAC (%) | Purification of NAC (fold) | Total activity (U)b | Yield of activity (%) | Specific activity (U/mg) | Purification of activity (fold) |
|---|---|---|---|---|---|---|---|---|
| Cytosol | 2,200 | 2.5 | 100 | 1 | 121,000 | 100 | 55 | 1 |
| Ribosomal pellet | 380 | 2.0 | 83 | 5 | 21,600 | 18 | 57 | 1 |
| Salt wash fluid | 38 | 0.63 | 25 | 15 | 20,000 | 17 | 530 | 10 |
| ResourceQ column eluate | 1.70 | 0.51 | 20 | 272 | 4,688 | 4 | 2,800 | 50 |
| MonoQ column eluate | 0.62 | 0.62 | 25 | 892 | 2,530 | 2 | 4,000 | 74 |
Estimated by quantitative immunoblotting using purified NAC as a standard.
One unit is defined as the activity that doubles import of Mdh1p bound to salt-washed RNCs into isolated mitochondria under the import conditions specified in MATERIALS AND METHODS.
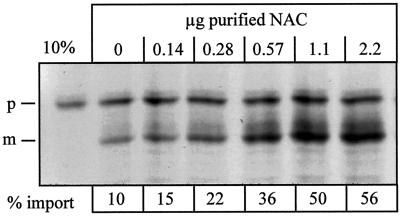
Purified NAC stimulated mitochondrial import of precursor chains on salt-washed RNCs in a dose-dependent manner from 10 up to 56% (Figure 5).
Figure 5.
Purified NAC stimulates import of ribosome-associated nascent chains into mitochondria. Import reactions were performed with salt-washed RNCs in the presence of the indicated amounts of purified NAC in a total volume of 100 μl as described in MATERIALS AND METHODS. After 15 min of import, all samples were centrifuged for 3 min at 14,000 × g to recover the mitochondria and were analyzed by SDS-PAGE and fluorography. The resulting bands on the fluorogram were quantified by densitometry, and the percentage of processed chains was determined, with the amount of initially added Mdh1p set to 100% (% import). 10%, 10% of the RNCs added per lane; p, Mdh1p precursor; m, mature Mdh1p.
Untreated RNCs containing endogenous NAC at concentrations of 0.06 μg/100 μl of import reaction were translocated with an efficiency of 20%. To restore import efficiency of salt-washed RNCs to 20%, 0.24 μg of our purified NAC preparation was required per 100-μl import reaction. A larger excess of purified NAC stimulated import to a significantly greater extent than the pool of factors bound to untreated RNCs (compare DISCUSSION).
Accessibility of the Precursor to Added MPP
A possible explanation for the low import efficiency of salt-washed RNCs would be that the positively charged presequence folds back onto the negatively charged ribosome and cannot be recognized by the TOM receptors on the mitochondrial surface. To probe the accessibility of the presequence, untreated and salt-washed RNCs were incubated with purified yeast MPP (Luciano et al., 1998) in the presence or absence of NAC (Figure 6).
Figure 6.
Salt washing of RNCs increases the accessibility of the attached nascent chains to MPP in vitro. Untreated and salt-washed RNCs were incubated in import buffer with 0.5 μM purified MPP in the presence or absence of 40 μg/ml purified NAC at 20°C for the indicated times. Cleavage of the Mdh1p precursor was analyzed by SDS-PAGE and fluorography. The percentage of cleaved chains was determined by densitometry of the fluorogram, with the amount of initially added Mdh1p set to 100%.
The precursor on untreated RNCs was cleaved only slowly, suggesting that it is relatively inaccessible. Addition of concentrations of purified NAC that fully stimulated translocation, decreased the cleavage rate even further. In contrast, the precursor on salt-washed RNCs was more accessible to MPP. Addition of purified NAC did not restore protection but increased the cleavage rate even further. We conclude that the MPP cleavage site on ribosome-associated nascent chains is protected by salt-extractable factors other than NAC, and that the presequence on salt-washed RNCs is accessible to surface receptors on the mitochondria.
NAC Stimulates Productive Binding of RNCs to Protease-sensitive Sites on Mitochondria
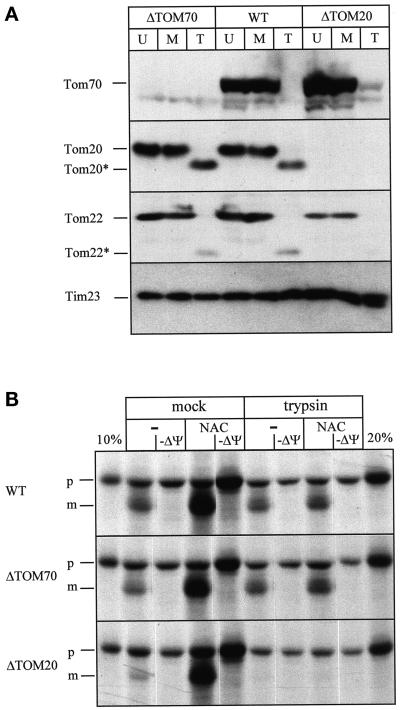
To test the influence of mitochondrial surface proteins on import stimulation, we compared import of nascent chains into trypsin-shaved or mock-treated mitochondria (Figure 7). Pretreatment of the mitochondria with 100 μg/ml trypsin for 5 min on ice completely removed the protease-sensitive cytosolic domains of the receptors Tom70p, Tom20p, and Tom22p, whereas the protease-sensitive inner membrane protein Tim23p remained intact (Figure 7A, WT). Salt-washed RNCs were incubated with trypsin or mock treated mitochondria in the presence or absence of purified NAC (Figure 7B, WT).
Figure 7.
Stimulation of import by NAC depends on mitochondrial surface proteins. (A) Trypsin treatment of mitochondria. Three aliquots of crude mitochondria from yeast strains YVH1 (ΔTOM70), JKR101 (WT), or YVH1 (ΔTOM20) each were treated as follows. The first aliquot was left untreated on ice (U); the second aliquot was treated with 100 μg/ml trypsin for 5min on ice, followed by inactivation of trypsin with 200 μg/ml soybean trypsin inhibitor for 5 min and reisolation of the mitochondria (T); and the third aliquot was mock-treated with 100 μg/ml trypsin that had been inactivated by preincubation with 200 μg/ml trypsin inhibitor for 5 min on ice followed by reisolation of the mitochondria (M). Equal amounts of mitochondrial protein (75 μg) were analyzed by SDS-PAGE and immunoblotting for Tom70p, Tom20p, Tom22p, and Tim23p as indicated on the side. Tom20* and Tom22* denote a trypsin-generated fragment of Tom20p and Tom22p, respectively. (B) Mitochondrial surface proteins are required for import stimulation by NAC. Imports were performed with the same trypsin-treated (trypsin) or mock-treated (mock) mitochondria as shown in A with salt-treated RNCs in the presence (NAC) or absence (−) of 10 μg/ml NAC. The strain is indicated on the left. After 7.5 min, mitochondria were recovered by centrifugation for 3 min at 14,000 × g, and analyzed by SDS-PAGE and fluorography. 10 and 20%, 10 and 20% of the RNCs added per lane, respectivey; −ΔΨ, import in the presence of 1 μg/ml valinomycin and 25 μM carbonyl cyanide p-trifluoromethoxyphenyl-hydrazone; p, Mdh1p precursor; m, mature Mdh1p.
In mock-treated wild-type mitochondria, NAC stimulated import 2.5-fold. After pretreatment of the mitochondria with trypsin, import stimulation by NAC was almost abolished, indicating that it depends on trypsin-sensitive mitochondrial surface proteins (Figure 7B, WT). The two best studied receptors on the outer mitochondrial membrane, Tom70p and Tom20p, show differential specificities in posttranslational precursor recognition (Lithgow et al., 1995; Lill and Neupert, 1996). To test the influence of Tom70p and Tom20p on NAC-dependent import of nascent chains, we compared import into mitochondria prepared from a wild-type strain (WT), a strain carrying a deletion of TOM70 (ΔTOM70), and a strain carrying a deletion in TOM20 (ΔTOM20) (Figure 7B) (Hines and Schatz, 1993; Lithgow et al., 1994b). The deletion mutations were confirmed by immunoblotting for Tom70p and Tom20p (Figure 7A). Stimulation of translocation in the presence of NAC was independent of Tom20p or Tom70p, indicating that these two proteins are not responsible for the trypsin sensitivity of NAC stimulation (Figure 7B).
In wild-type and ΔTOM70 mitochondria, residual NAC-independent import was not affected by trypsin-treatment. However, it was almost abolished in ΔTOM20 mitochondria. Possibly, the protease-resistant fragment of Tom20p (Figure 7A, Tom20*) is sufficient for NAC-independent import in the other two strains. Alternatively, other components of the TOM complex, e.g., Tom22p, might be destabilized and more easily degraded in ΔTOM20 mitochondria (Figure 7A, Tom22*) (Lithgow et al., 1994a).
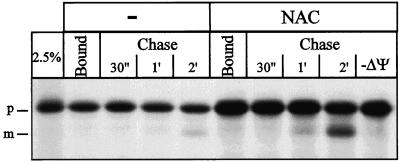
To determine the stage of import that was stimulated by NAC, we performed a binding and chase experiment. Salt-washed RNCs were first incubated with nonenergized mitochondria on ice in the presence or absence of NAC (Figure 8) to allow binding of RNCs.
Figure 8.
NAC promotes productive binding of RNCs to mitochondria. Salt-washed RNCs were incubated with 0.15 mg/ml mitochondria in import buffer for 5 min on ice in the presence (NAC) or absence (−) of 11 μg/ml NAC. The mitochondria were reisolated, resuspended in import buffer, and divided in two aliquots. One aliquot was precipitated with TCA (bound). The other aliquot was supplemented with 2 mM ATP, NADH, and KPi and shifted to 20°C. After the indicated chase times, samples were withdrawn and precipitated with TCA and analyzed by SDS-PAGE and fluorography. −ΔΨ, chase for 2 min in the presence of 1 μg/ml valinomycin; 2.5%, 2.5% of the RNCs added per lane; p, Mdh1p precursor; m, mature Mdh1p.
Reisolation of the mitochondria revealed that NAC stimulated binding of RNCs to mitochondria twofold (Bound). Bound precursor chains were chased into the mitochondria by energizing the mitochondria and raising the temperature to 20°C. After 2 min, only 6% of the chains that had bound to mitochondria in the absence of NAC were imported, whereas 23% were imported after binding in the presence of NAC (Chase). Addition of NAC during the chase did not influence import (our unpublished data). NAC thus stimulates productive binding of RNCs to mitochondria when bound to RNCs before their contact with the mitochondrial surface. If formation of a membrane potential was prevented by the addition of valinomycin after the binding step, but before the chase, import was completely inhibited (Figure 8; Chase, −ΔΨ). Incubation of RNCs with nonenergized mitochondria on ice thus only allows binding but not import of the attached precursor chains.
DISCUSSION
Purification of NAC as an Import Stimulatory Factor
Using a novel in vitro assay for the study of the initial steps of mitochondrial protein import in vitro, we have identified yeast NAC, a complex of α- and β-subunits, as a factor that stimulates import of ribosome-associated nascent chains into mitochondria. Yeast NAC was originally described in an entirely different context as a complex of two nonessential proteins that stabilizes the binding of the transcriptional activator Gal4p to DNA (Parthun et al., 1992; Shi et al., 1995), suggesting a function in the nucleus. The individual subunits of mammalian NAC have also been described as factors involved in transcription (Zheng et al., 1990; Moreau et al., 1998; Yotov et al., 1998). In addition, and consistent with our findings, Wiedmann et al. (1994) identified mammalian NAC as a ribosome-associated complex that can be cross-linked to short nascent chains destined to various compartments of the cell. NAC was proposed to enhance targeting fidelity by preventing promiscuous interaction of signal recognition particle (SRP) with nascent chains not destined for the ER (Wiedmann et al., 1994). Alternatively NAC might modulate the interaction of SRP with the ribosome (Powers and Walter, 1996) or the interaction of RNCs with the ER (Lauring et al., 1995a,b; Möller et al., 1998). The latter finding has been challenged (Neuhof et al., 1998; Raden and Gilmore, 1998). Recent experiments describe yeast NAC as a ribosome-associated complex involved in mitochondrial protein import and possibly targeting to the ER (George et al., 1998). As will be discussed below, NAC shows a stimulatory effect on mitochondrial protein import, whereas it acts as a negative control element during import into the ER.
NAC Alone Does Not Shield the Nascent Chain against Purified MPP In Vitro
Mammalian MDH is cleaved in a two-step process involving both MPP and intermediate processing peptidase (Shimokata et al., 1997). The same was proposed for yeast Mdh1p (Branda and Isaya, 1995). We observed that Mdh1p was indeed cleaved to an intermediate form by recombinant MPP (our unpublished data), suggesting that MPP cleaves off the first 9 amino acids of the 17-amino acid-long presequence.
Salt washing of RNCs enhanced presequence cleavage by purified MPP (Figure 6), and concentrations of NAC that stimulated import efficiently did not restore protection against MPP. This result suggests that additional factors are bound to untreated RNCs and protect the presequence part of the nascent chain. These factors, however, are dispensable for import in our system, because purified NAC fully restores import of salt-washed RNCs. Wang et al. (1995) have shown that NAC was both necessary and sufficient to protect short nascent chains (up to 44 amino acids) against proteolysis. This is in good agreement with the finding that NAC can be cross-linked to very short nascent chains and probably is in close contact to the exit site of the nascent chain on the ribosome (Wiedmann et al., 1994). We used a nascent chain of 335 amino acids, which was too long for the N-terminal presequence to be protected by NAC. The accessibility of the nascent chains to MPP, as well as the reversibility of the import competence, suggest that the nascent chain on salt-washed RNCs does not irreversibly aggregate or adhere to the ribosomal surface.
Mode of Action of NAC
NAC stimulated productive binding of RNCs to mitochondria. The first recognition of precursor proteins on the outer mitochondrial membrane is thought to be mediated by Tom20p and by Tom70p, two receptors that exhibit different specificities for mitochondrial precursor proteins. Tom20p binds preferentially to the charged presequence of the precursor protein, whereas Tom70p binds more tightly to internal sequences (Moczko et al., 1994; Komiya et al., 1998; Brix et al., 1999). As a consequence, productive binding of presequence-containing precursor proteins, such as Mdh1p, to the mitochondrial surface is predominantly mediated by Tom20p (Haucke et al., 1995). Our results indicated that translocation of ribosome-bound Mdh1p in the presence of NAC was unaffected by the absence of either Tom20p or Tom70p. Although NAC-mediated import of nascent chains seems to bypass these two receptor components, trypsin-sensitive proteins on the mitochondrial surface were strictly required.
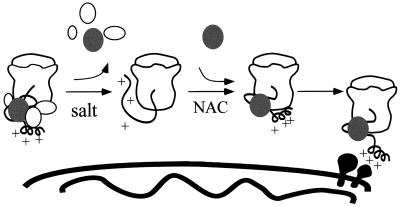
The acid chain hypothesis suggests that the presequence of a precursor binds to components of the components of the TOM complex in successive steps determined by its binding affinity (Bolliger et al., 1995; Dietmeier et al., 1997; Schatz, 1997; Komiya et al., 1998). Our results would be in agreement with a model in which NAC directs RNCs to more central parts of the TOM complex than posttranslationally imported precursors use to enter the TOM complex. The TOM channel is composed of the two essential proteins Tom22p and Tom40p and the small Tom proteins Tom5p, Tom6p, and Tom7p (Dekker et al., 1998). Tom22p, Tom5p, and Tom40p have been implicated in binding to precursor proteins at later stages than Tom20p and Tom70p (Lithgow et al., 1994a; Dietmeier et al., 1997; Kanamori et al., 1997; Rapaport et al., 1997). Our results would also be in agreement with the existence of as yet unidentified proteins in the outer membrane that specifically interact with the precursor–NAC complex. However, NAC is in contact with nascent chains destined for many cellular compartments (Wiedmann et al., 1994), thus a specific “NAC receptor” on the mitochondrial surface might not exist. In fact, we were unable to detect significant binding of purified NAC to trypsin-sensitive sites on the mitochondrial surface in vitro (our unpublished data). NAC resembles chaperones in that it unspecificaly binds to unfolded proteins. Similar to many chaperone-like proteins, NAC might serve different functions in the cell. NAC positively affects translocation of RNCs into mitochondria, yet at the same time, it prevents unspecific targeting to the ER. This apparent contradiction might reflect the fundamental differences between import to these two cellular compartments. In both cases, NAC initially interacts with the nascent chain. In case of import into the ER, NAC is displaced by SRP, which is essential for specific translocation into this organelle. Specificity of mitochondrial targeting is achieved by properties that reside in the precursor protein itself. We favor the model that binding of NAC to mitochondrially targeted nascent chains affects the structure of the precursor, presenting it in a conformation that is efficiently recognized by the mitochondrial import machinery (Figure 9).
Figure 9.
Proposed role of NAC in mitochondrial protein import. From left to right: untreated RNCs contain several salt-extractable proteins, including NAC (black ellipse), which keep the nascent chain import competent (depicted by the helical conformation of the positively charged presequence). Salt washing removes these proteins and exposes the nascent chain to purified MPP and lowers its import competence. Readdition of purified NAC does not shield the nascent chain against purified MPP but restores import competence of the nascent chain by presenting it in a conformation recognized by mitochondrial surface receptors.
NAC probably acts on nascent chains only in the context of the ribosome (Wiedmann et al., 1994). Released chains might therefore require other cytosolic factors. In our assay, import of released chains was more efficient than import of ribosome-attached nascent chains (our unpublished data). However, the import of a complex mixture of full-length mitochondrial precursor proteins (Dubaquiéet al., 1998) into mitochondria is significantly reduced when the translation is performed in the absence of functional NAC (our unpublished data).
Purification of NAC
We purified NAC from yeast cytosol ∼900-fold. In contrast, the total import stimulatory activity was purified by a factor of 74 (Table 1).
This difference reflects a combination of at least two effects: 1) NAC might be inactivated during purification. Because immunodepletion showed that NAC constitutes a major import stimulatory activity in the salt wash fluid (Figure 3), loss of activity during fractionation of the salt wash fluid without concomitant loss of NAC protein suggests that NAC was either partially inactivated, or that NAC is part of a larger complex that was coimmunodepleted by the immuneserum against αNAC but was disrupted during the purification. 2) Several proteins might contribute to the total activity independently. This possibility is likely as the immunodepletion experiment showed that 20% of the stimulatory activity in the ribosomal salt wash fluid is not caused by NAC (Figure 3). This is furthermore in good agreement with the fact that NAC is not essential (Parthun et al., 1992; Shi et al., 1995; Reimann et al., 1999).
Co- versus Post-translational Import
The question whether mitochondrial protein import in vivo occurs co- or post-translationally is open. Under normal growth conditions fully synthesized, but unprocessed precursor proteins are essentially not detected, suggesting that they are translocated into mitochondria either very soon after synthesis or co-translationally (Fujiki and Verner, 1993). When translation is slowed by addition of cycloheximide, yeast mitochondria are covered with ribosomes (Kellems et al., 1974; Kellems et al., 1975), suggesting that the ribosome-bound precursors accumulate on the surface of mitochondria. On the other hand, import can occur post-translationally in vivo: mitochondrial precursor proteins, accumulated in vivo by treatment of yeast cells with the uncoupler carbonyl cyanide m-chlorophenyl-hydrazone, can be chased subsequently into mitochondria by removal of carbonyl cyanide m-chlorophenyl-hydrazone (Reid and Schatz, 1982). Chimeric precursor proteins with the potential to form a stably folded C-terminal domain can accumulate in vivo as two-membrane-spanning intermediates with a folded domain outside mitochondria (Wienhues et al., 1991; Schulke et al., 1997). This indicates that protein synthesis is at least not coupled tightly enough with import to prevent folding of the C-terminal domain. It was proposed that co-translational import might be involved in sorting of proteins that are found in dual locations in both mitochondria and the cytosol (Stein et al., 1994; Nobumoto et al., 1998).
For the first time, we show that ribosome-attached nascent chains can initiate import into mitochondria in a homologous in vitro system. This import system now enables us to combine mutant mitochondria (e.g., in the Tom proteins) with cytosolic extracts from mutant strains (e.g., NAC and cytosolic hsp70s). We hope to develop a more detailed understanding of the early steps of mitochondrial protein import that can facilitate in vivo investigation of the process.
ACKNOWLEDGMENTS
We are indebted to Prof. Gottfried Schatz for generous support throughout the project and for critical reading of the manuscript. We thank Vincent Géli and Pierre Luciano for purified MPP, Trevor Lithgow for the αNAC antibody, Paul Jenö for mass spectroscopy, Renate Looser for technical assistance, and members of the Schatz group for support and fruitful discussions. This study was supported by grants from the Swiss National Science Foundation (to S.R. and G. Schatz), the European Economic Community (to G. Schatz), the Human Frontiers Science Program (to G. Schatz).
Abbreviations used:
- ER
endoplasmic reticulum
- MPP
mitochondrial processing peptidase
- NAC
nascent polypeptide–associated complex
- RNC
ribosome–nascent chain complex
- SRP
signal recognition particle
- TCA
trichloroacetic acid
- TIM
translocase of the inner membrane
- TOM
translocase of the outer membrane
REFERENCES
- Ades IZ, Butow RA. The products of mitochondria-bound cytoplasmic polysomes in yeast. J Biol Chem. 1980;255:9918–9924. [PubMed] [Google Scholar]
- Bolliger L, Junne T, Schatz G, Lithgow T. Acidic receptor domains on both sides of the outer membrane mediate translocation of precursor proteins into yeast mitochondria. EMBO J. 1995;14:6318–6326. doi: 10.1002/j.1460-2075.1995.tb00322.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branda SS, Isaya G. Prediction and identification of new natural substrates of the yeast mitochondrial intermediate peptidase. J Biol Chem. 1995;270:27366–27373. doi: 10.1074/jbc.270.45.27366. [DOI] [PubMed] [Google Scholar]
- Brix J, Rüdiger S, Bukau B, Schneider-Mergener J, Pfanner N. Distribution of binding sequences for the mitochondrial import receptors Tom20, Tom22, and Tom70 in a presequence-carrying preprotein and a noncleavable preprotein. J Biol Chem. 1999;274:16522–16530. doi: 10.1074/jbc.274.23.16522. [DOI] [PubMed] [Google Scholar]
- Caplan AJ, Cyr DM, Douglas MG. YDJ1p facilitates polypeptide translocation across different intracellular membranes by a conserved mechanism. Cell. 1992;71:1143–1155. doi: 10.1016/s0092-8674(05)80063-7. [DOI] [PubMed] [Google Scholar]
- Cartwright P, Beilharz T, Hansen P, Garrett J, Lithgow T. Mft52, an acid-bristle protein in the cytosol that delivers precursor proteins to yeast mitochondria. J Biol Chem. 1997;272:5320–5325. doi: 10.1074/jbc.272.8.5320. [DOI] [PubMed] [Google Scholar]
- Dekker PJT, Ryan MT, Brix J, Müller H, Hönlinger A, Pfanner N. Preprotein translocase of the outer mitochondrial membrane: molecular dissection and assembly of the general import pore complex. Mol Cell Biol. 1998;18:6515–6524. doi: 10.1128/mcb.18.11.6515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshaies RJ, Koch BD, Werner-Washburne M, Craig EA, Schekman R. A subfamily of stress proteins facilitates translocation of secretory and mitochondrial precursor polypeptides. Nature. 1988;332:800–805. doi: 10.1038/332800a0. [DOI] [PubMed] [Google Scholar]
- Dietmeier K, Hönlinger A, Bömer U, Dekker PJ, Eckerskorn C, Lottspeich F, Kubrich M, Pfanner N. Tom5 functionally links mitochondrial preprotein receptors to the general import pore [see comments] Nature. 1997;388:195–200. doi: 10.1038/40663. [DOI] [PubMed] [Google Scholar]
- Dubaquié Y, Looser R, Fünfschilling U, Jenö P, Rospert S. Identification of in vivo substrates of the yeast mitochondrial chaperonins reveals overlapping but nonidentical requirement for hsp60 and hsp10. EMBO J. 1998;17:5868–5876. doi: 10.1093/emboj/17.20.5868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiki M, Verner K. Coupling of cytosolic protein synthesis and mitochondrial protein import in yeast. Evidence for cotranslational import in vivo. J Biol Chem. 1993;268:1914–1920. [PubMed] [Google Scholar]
- Garcia PD, Hansen W, Walter P. In vitro protein translocation across microsomal membranes of Saccharomyces cerevisiae. Methods Enzymol. 1991;194:675–682. doi: 10.1016/0076-6879(91)94049-i. [DOI] [PubMed] [Google Scholar]
- George R, Beddoe T, Landl K, Lithgow T. The yeast nascent polypeptide-associated complex initiates protein targeting to mitochondria in vivo. Proc Natl Acad Sci USA. 1998;95:2296–2301. doi: 10.1073/pnas.95.5.2296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilmore R, Collins P, Johnson J, Kellaris K, Rapiejko P. Transcription of full-length and truncated mRNA transcripts to study protein translocation across the endoplasmic reticulum. Methods Cell Biol. 1991;34:223–239. doi: 10.1016/s0091-679x(08)61683-0. [DOI] [PubMed] [Google Scholar]
- Glick BS, Pon LA. Isolation of highly purified mitochondria from Saccharomyces cerevisiae. Methods Enzymol. 1995;260:213–223. doi: 10.1016/0076-6879(95)60139-2. [DOI] [PubMed] [Google Scholar]
- Haucke V, Lithgow T, Rospert S, Hahne K, Schatz G. The yeast mitochondrial protein import receptor Mas20p binds precursor proteins through electrostatic interaction with the positively charged presequence. J Biol Chem. 1995;270:5565–5570. doi: 10.1074/jbc.270.10.5565. [DOI] [PubMed] [Google Scholar]
- Heinemeyer W, Kleinschmidt JA, Saidowsky J, Escher C, Wolf DH. Proteinase yscE, the yeast proteasome/multicatalytic-multifunctional proteinase: mutants unravel its function in stress induced proteolysis and uncover its necessity for cell survival. EMBO J. 1991;10:555–562. doi: 10.1002/j.1460-2075.1991.tb07982.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill K, Model K, Ryan MT, Dietmeier K, Martin F, Wagner R, Pfanner N. Tom40 forms the hydrophilic channel of the mitochondrial import pore for preproteins. Nature. 1998;395:516–521. doi: 10.1038/26780. [DOI] [PubMed] [Google Scholar]
- Hines V, Schatz G. Precursor binding to yeast mitochondria. A general role for the outer membrane protein Mas70p. J Biol Chem. 1993;268:449–454. [PubMed] [Google Scholar]
- Kanamori T, Nishikawa S, Shin I, Schultz PG, Endo T. Probing the environment along the protein import pathways in yeast mitochondria by site-specific photocrosslinking. Proc Natl Acad Sci USA. 1997;94:485–490. doi: 10.1073/pnas.94.2.485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellems RE, Allison VF, Butow RA. Cytoplasmic type 80 S ribosomes associated with yeast mitochondria. II. Evidence for the association of cytoplasmic ribosomes with the outer mitochondrial membrane in situ. J Biol Chem. 1974;249:3297–3303. [PubMed] [Google Scholar]
- Kellems RE, Allison VF, Butow RA. Cytoplasmic type 80S ribosomes associated with yeast mitochondria. IV. Attachment of ribosomes to the outer membrane of isolated mitochondria. J Cell Biol. 1975;65:1–14. doi: 10.1083/jcb.65.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komiya T, Rospert S, Koehler C, Looser R, Schatz G, Mihara K. Interaction of mitochondrial targeting signals with acidic receptor domains along the protein import pathway: evidence for the “acid chain” hypothesis. EMBO J. 1998;17:3886–3898. doi: 10.1093/emboj/17.14.3886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komiya T, Sakaguchi M, Mihara K. Cytoplasmic chaperones determine the targeting pathway of precursor proteins to mitochondria. EMBO J. 1996;15:399–407. [PMC free article] [PubMed] [Google Scholar]
- Künkele KP, Heins S, Dembowski M, Nargang FE, Benz R, Thieffry M, Walz J, Lill R, Nussberger S, Neupert W. The preprotein translocation channel of the outer membrane of mitochondria. Cell. 1998;93:1009–1019. doi: 10.1016/s0092-8674(00)81206-4. [DOI] [PubMed] [Google Scholar]
- Lauring B, Kreibich G, Weidmann M. The intrinsic ability of ribosomes to bind to endoplasmic reticulum membranes is regulated by signal recognition particle and nascent-polypeptide-associated complex [see comments] Proc Natl Acad Sci USA. 1995a;92:9435–9439. doi: 10.1073/pnas.92.21.9435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauring B, Sakai H, Kreibich G, Wiedmann M. Nascent polypeptide-associated complex protein prevents mistargeting of nascent chains to the endoplasmic reticulum. Proc Natl Acad Sci USA. 1995b;92:5411–5415. doi: 10.1073/pnas.92.12.5411. (erratum 92, 8088). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lill R, Neupert W. Mechanisms of protein import across the mitochondrial outer membrane. Trends Cell Biol. 1996;6:56–61. doi: 10.1016/0962-8924(96)81015-4. [DOI] [PubMed] [Google Scholar]
- Lithgow T, Glick BS, Schatz G. The protein import receptor of mitochondria. Trends Biochem Sci. 1995;20:98–101. doi: 10.1016/s0968-0004(00)88972-0. [DOI] [PubMed] [Google Scholar]
- Lithgow T, Junne T, Suda K, Gratzer S, Schatz G. The mitochondrial outer membrane protein Mas22p is essential for protein import and viability of yeast. Proc Natl Acad Sci USA. 1994a;91:11973–11981. doi: 10.1073/pnas.91.25.11973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lithgow T, Junne T, Wachter C, Schatz G. Yeast mitochondria lacking the two import receptors Mas20p and Mas70p can efficiently and specifically import precursor proteins. J Biol Chem. 1994b;269:15325–15330. [PubMed] [Google Scholar]
- Luciano P, Tokatlidis K, Chambre I, Germanique JC, Geli V. The mitochondrial processing peptidase behaves as a zinc-metallopeptidase. J Mol Biol. 1998;280:193–199. doi: 10.1006/jmbi.1998.1858. [DOI] [PubMed] [Google Scholar]
- Moczko M, Ehmann B, Gartner F, Honlinger A, Schafer E, Pfanner N. Deletion of the receptor MOM19 strongly impairs import of cleavable preproteins into Saccharomyces cerevisiae mitochondria. J Biol Chem. 1994;269:9045–9051. [PubMed] [Google Scholar]
- Möller I, Jung M, Beatrix B, Levy R, Kreibich G, Zimmermann R, Wiedmann M, Lauring B. A general mechanism for regulation of access to the translocon: competition for a membrane attachment site on ribosomes. Proc Natl Acad Sci USA. 1998;95:13425–13430. doi: 10.1073/pnas.95.23.13425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreau A, Yotov WV, Glorieux FH, St-Arnaud R. Bone-specific expression of the alpha chain of the nascent polypeptide-associated complex, a coactivator potentiating c-Jun-mediated transcription. Mol Cell Biol. 1998;18:1312–1321. doi: 10.1128/mcb.18.3.1312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueckler M, Lodish HF. The human glucose transporter can insert posttranslationally into microsomes. Cell. 1986;44:629–637. doi: 10.1016/0092-8674(86)90272-2. [DOI] [PubMed] [Google Scholar]
- Murakami K, Mori M. Purified presequence binding factor (PBF) forms an import-competent complex with a purified mitochondrial precursor protein. EMBO J. 1990;9:3201–3208. doi: 10.1002/j.1460-2075.1990.tb07518.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuhof A, Rolls MM, Jungnickel B, Kalies KU, Rapoport TA. Binding of signal recognition particle gives ribosome/nascent chain complexes a competitive advantage in endoplasmic reticulum membrane interaction. Mol Biol Cell. 1998;9:103–115. doi: 10.1091/mbc.9.1.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nobumoto M, Yamada M, Song S, Inouye S, Nakazawa A. Mechanism of mitochondrial import of adenylate kinase isozymes. J Biochem. 1998;123:128–135. doi: 10.1093/oxfordjournals.jbchem.a021899. [DOI] [PubMed] [Google Scholar]
- Ono H, Tuboi S. Purification and identification of a cytosolic factor required for import of precursors of mitochondrial proteins into mitochondria. Arch Biochem Biophys. 1990;280:299–304. doi: 10.1016/0003-9861(90)90333-t. [DOI] [PubMed] [Google Scholar]
- Parthun MR, Mangus DA, Jaehning JA. The EGD1 product, a yeast homolog of human BTF3, may be involved in GAL4 DNA binding. Mol Cell Biol. 1992;12:5683–5689. doi: 10.1128/mcb.12.12.5683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfanner N, Craig EA, Hönlinger A. Mitochondrial preprotein translocase. Annu Rev Cell Dev Biol. 1997;13:25–51. doi: 10.1146/annurev.cellbio.13.1.25. [DOI] [PubMed] [Google Scholar]
- Powers T, Walter P. The nascent polypeptide-associated complex modulates interactions between the signal recognition particle and the ribosome. Curr Biol. 1996;6:331–338. doi: 10.1016/s0960-9822(02)00484-0. [DOI] [PubMed] [Google Scholar]
- Raden D, Gilmore R. Signal recognition particle-dependent targeting of ribosomes to the rough endoplasmic reticulum in the absence and presence of the nascent polypeptide-associated complex. Mol Biol Cell. 1998;9:117–130. doi: 10.1091/mbc.9.1.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rapaport D, Neupert W, Lill R. Mitochondrial protein import. Tom40 plays a major role in targeting and translocation of preproteins by forming a specific binding site for the presequence. J Biol Chem. 1997;272:18725–18731. doi: 10.1074/jbc.272.30.18725. [DOI] [PubMed] [Google Scholar]
- Reid GA, Schatz G. Import of proteins into mitochondria. Yeast cells grown in the presence of carbonyl cyanide m-chlorophenylhydrazone accumulate massive amounts of some mitochondrial precursor polypeptides. J Biol Chem. 1982;257:13056–13061. [PubMed] [Google Scholar]
- Reimann B, Bradsher J, Franke J, Hartmann E, Wiedmann M, Prehn S, Wiedmann B. Initial characterization of the nascent polypeptide-associated complex in yeast. Yeast. 1999;15:397–407. doi: 10.1002/(SICI)1097-0061(19990330)15:5<397::AID-YEA384>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- Roise D, Horvath SJ, Tomich JM, Richards JH, Schatz G. A chemically synthesized presequence of an imported mitochondrial protein can form an amphiphilic helix and perturb natural and artificial phospholipid bilayers. EMBO J. 1986;5:1327–1334. doi: 10.1002/j.1460-2075.1986.tb04363.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rospert S, Schatz G. Protein translocation into mitochondria. In: Celis JE, editor. Cell Biology, a Laboratory Handbook. Vol. 2. San Diego: Academic Press; 1998. pp. 277–285. [Google Scholar]
- Schatz G. Just follow the acid chain [news; comment] Nature. 1997;388:121–122. doi: 10.1038/40510. [DOI] [PubMed] [Google Scholar]
- Schneider E, Lochmann ER, Lother H. Distribution of membrane-bound and free ribosomes in growing yeast. Biochim Biophys Acta. 1976;432:92–97. doi: 10.1016/0005-2787(76)90044-7. [DOI] [PubMed] [Google Scholar]
- Schulke N, Sepuri NB, Pain D. In vivo zippering of inner and outer mitochondrial membranes by a stable translocation intermediate. Proc Natl Acad Sci USA. 1997;94:7314–7319. doi: 10.1073/pnas.94.14.7314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi X, Parthun MR, Jaehning JA. The yeast EGD2 gene encodes a homologue of the alpha NAC subunit of the human nascent-polypeptide-associated complex. Gene. 1995;165:199–202. doi: 10.1016/0378-1119(95)00577-s. [DOI] [PubMed] [Google Scholar]
- Shimokata K, Nishio T, Song MC, Kitada S, Ogishima T, Ito A. Substrate recognition by mitochondrial processing peptidase toward the malate dehydrogenase precursor. J Biochem. 1997;122:1019–1023. doi: 10.1093/oxfordjournals.jbchem.a021841. [DOI] [PubMed] [Google Scholar]
- Stein I, Peleg Y, Even-Ram S, Pines O. The single translation product of the FUM1 gene (fumarase) is processed in mitochondria before being distributed between the cytosol and mitochondria in Saccharomyces cerevisiae. Mol Cell Biol. 1994;14:4770–4778. doi: 10.1128/mcb.14.7.4770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Heijne G. Mitochondrial targeting sequences may form amphiphilic helices. EMBO J. 1986;5:1335–1342. doi: 10.1002/j.1460-2075.1986.tb04364.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wachter C, Schatz G, Glick BS. Protein import into mitochondria: the requirement for external ATP is precursor-specific whereas intramitochondrial ATP is universally needed for translocation into the matrix. Mol Biol Cell. 1994;5:465–474. doi: 10.1091/mbc.5.4.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Sakai H, Wiedmann M. NAC covers ribosome-associated nascent chains thereby forming a protective environment for regions of nascent chains just emerging from the peptidyl transferase center. J Cell Biol. 1995;130:519–528. doi: 10.1083/jcb.130.3.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiedmann B, Sakai H, Davis TA, Wiedmann M. A protein complex required for signal-sequence-specific sorting and translocation. Nature. 1994;370:434–440. doi: 10.1038/370434a0. [DOI] [PubMed] [Google Scholar]
- Wienhues U, Becker K, Schleyer M, Guiard B, Tropschug M, Horwich AL, Pfanner N, Neupert W. Protein folding causes an arrest of preprotein translocation into mitochondria in vivo. J Cell Biol. 1991;115:1601–1609. doi: 10.1083/jcb.115.6.1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yotov WV, Moreau A, St-Arnaud R. The alpha chain of the nascent polypeptide-associated complex functions as a transcriptional coactivator. Mol Cell Biol. 1998;18:1303–1311. doi: 10.1128/mcb.18.3.1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng XM, Black D, Chambon P, Egly JM. Sequencing and expression of complementary DNA for the general transcription factor BTF3. Nature. 1990;344:556–559. doi: 10.1038/344556a0. [DOI] [PubMed] [Google Scholar]