Abstract
We purified from Dictyostelium lysates an 88-kDa protein that bound to a subset of small GTPases, including racE, racC, cdc42Hs, and TC4ran, but did not bind to R-ras or rabB. Cloning of the gene encoding this 88-kDa protein revealed that it contained multiple armadillo-like repeats most closely related to the mammalian GTP exchange factor smgGDS. We named this protein darlin (Dictyostelium armadillo-like protein). Disruption of the gene encoding darlin demonstrated that this protein is not essential for cytokinesis, pinocytosis, phagocytosis, or development. However, the ability of darlin null cells to aggregate in response to starvation is severely affected. When starved under liquid medium, the mutant cells were unable to form aggregation centers and streams, possibly because of a defect in cAMP relay signaling. This defect was not due to an inability of the darlin mutants to activate adenylate cyclase in response to G protein stimulation. These results suggest that the darlin protein is involved in a signaling pathway that may modulate the chemotactic response during early development.
INTRODUCTION
The ras superfamily of small GTPases are key regulatory proteins in a wide range of cellular processes, including membrane traffic, cell proliferation, gene regulation, and cytoskeletal organization. The activity of these proteins is determined by their guanine nucleotide content. Small GTPases are activated upon binding guanosine triphosphate (GTP)1 and inactivated when bound to guanosine diphosphate (GDP). Switching between these two states is modulated by regulatory proteins known as GTPase-activating proteins (GAPs) and guanine-nucleotide exchange factors (GEFs). In addition, small GTPases bind to a large number of effector proteins to exert their function. Thus, the identification of binding partners for small GTPases is essential to understanding their specific function.
In Dictyostelium discoideum ∼30 genes encoding different small GTPases have been found. Among these small GTPases, >5 belong to the ras family, 10 to the rab family, and 8 to the rac subfamily. In contrast to the large number of small GTPases identified in this organism, only a few GTPase-binding proteins have been discovered. A Dictyostelium ras-GEF protein was shown recently to be required for signal transduction during early development (Insall et al., 1996). Two proteins similar to the mammalian protein IQGAP were shown to be involved in cytokinesis (Adachi et al., 1996; Faix and Dittrich, 1996; Lee et al., 1997). Finally, a Dictyostelium gene encoding a rho GAP protein was isolated recently (Ludbrook et al., 1997).
We recently identified a novel member of the rac subfamily of small GTPases in a genetic screen designed to isolate cytokinesis mutants (Larochelle et al., 1996). This protein, racE, is essential for cytokinesis and maintenance of cortical tension but not for any other cytoskeletal function (Larochelle et al., 1997; Gerald et al., 1998). To better understand the role of this GTPase, we sought to identify Dictyostelium proteins that interact with racE. We report here the identification of darlin, a Dictyostelium armadillo-like protein that binds to racE and to other small GTPases. Sequence analysis of the gene encoding this protein (darA) revealed that it consists of 11 armadillo-like repeats and has significant homology to the mammalian GEF smgGDS (Yamamoto et al., 1990). Like smgGDS, darlin preferentially bound to the GDP-bound form of racE. Although darlin is not required for cytokinesis, it is necessary for the efficient streaming of cells early in development. This function is mediated by a pathway different from the one that activates adenylate cyclase in response to G protein stimulation. Thus, we postulate that darlin may be involved in the regulation of chemotaxis in early development.
MATERIALS AND METHODS
Materials
A randomly sheared Dictyostelium genomic library subcloned into the EcoRI site of Λ-Zap2 was a generous gift from Dr. Herb Ennis (Columbia University, New York, NY). A random-primed Dictyostelium cDNA library (prepared from cells at 6 h of development) in λ-gt11 was obtained from CLONTECH (Palo Alto, CA). Y1090 and XL1-MRF bacterial cells were obtained from Stratagene (La Jolla, CA). The pGEX-2T vector was obtained from Pharmacia (Piscataway, NJ). Glutathione beads and reduced glutathione were from Sigma (St. Louis, MO). The pMalc-2 vector, the amylose resin, and the polyclonal antibody against maltose-binding protein (MBP) were obtained from New England Biolabs (Beverly, MA). Other materials used were obtained from previously described sources (Larochelle et al., 1996; Vithalani et al., 1996).
Expression of racE and Other Small GTPases in Bacteria
Reverse transcription (RT)-PCR was used to obtain cDNAs coding for Dictyostelium racE and racC. Site-directed mutagenesis was used to create two independent mutations in the racE cDNA to create the constitutively active V20racE (glycine to valine at position 20) and constitutively inactive N25racE (threonine to asparagine at position 25). These cDNAs were then cloned into a Dictyostelium expression vector, pTIKL-Bsr-Exp (Larochelle et al., 1997). The cDNAs were removed from this vector using BglII and EcoRI and fused to the C-terminal end of the Schistosoma japonicum glutathione S-transferase (GST) gene by cloning them into the BamHI and EcoRI sites of pGEX-2T. Expression of the resulting racE–GST fusion protein is under the control of the tac promoter (Smith and Johnson, 1988). The pGEX-2T vector alone as well as pGEX-2T containing the respective cDNAs were each transformed into the Escherichia coli strain DH5-α and stored as glycerol stocks at −70°C. The bacterial strains expressing cdc42Hs-GST, TC4-ran-GST, and R-ras-GST were generous gifts from Drs. Daniel Lew and Sally Kornbluth (Duke University, Durham, NC), and Dr. Channing Der (University of North Carolina, Chapel Hill, NC), respectively.
To purify the fusion proteins, an overnight culture (100 ml) of the respective bacterial strain was diluted 1:10 into fresh L-broth containing 100 μg/ml ampicillin and incubated in 2-l flasks for 2 h at 37°C on an orbital shaker. Isopropyl-β-d-thiogalactopyranoside was added to 0.1 mM to induce expression and the culture was incubated overnight at room temperature with shaking. We found that if the induction was carried out at 37°C, it led to sequestration of racE into inclusion bodies. The cells were collected by centrifugation at 4000 rpm for 60 min at 4°C and resuspended to 20 ml in ice-cold lysis buffer (50 mM Tris-HCl, pH 7.5, 100 mM NaCl, 5 mM MgCl2, 0.5% Triton X-100, 1 mM dithiothreitol [DTT], 1 mg/ml lysozyme, 5 μg/ml leupeptin, 1.4 μg/ml pepstatin, 10 μg/ml phenylmethylsulfonyl fluoride [PMSF], and 2 mM sodium bisulfite). The resuspended bacteria were placed on ice for 30 min and lysed two times in a French press at 1200 psi. The lysate was centrifuged at 9000 × g for 10 min at 4°C, the supernatant was transferred to a chilled tube, and fresh protease inhibitors were added. Two milliliters of a prewashed 1:1 suspension of glutathione and agarose beads were added to the supernatant and incubated for 30 min on a rotating platform at 4°C. The beads were pelleted, the supernatant was discarded, and the beads were then washed three times with 20 ml lysis buffer without lysozyme, Triton X-100, or protease inhibitors. The bead-bound fusion proteins were stored as a 1:1 slurry on ice, or the proteins were eluted with 5 mM reduced glutathione in the same wash buffer.
Affinity Chromatography
Wild-type Dictyostelium AX2 cells were seeded in HL5 medium at 1 × 105 cells/ml, grown at 21°C at 240 rpm, and harvested while still in the logarithmic phase. The cells were resuspended to 5 × 107 cells/ml in ice-cold binding buffer (20 mM piperazine-N,N′-bis[2-ethanesulfonic acid], pH 6.8, 1.5 mM EDTA, 15 mM MgCl2, 1 mM DTT, 5 μg/ml leupeptin, 1.4 μg/ml pepstatin, 10 μg/ml PMSF, and 2 mM sodium bisulfite with or without 400 mM NaCl) and lysed in an ice-water slurry by sonication (Virsonic 50, Virtis, Gardiner, NY; 50% output power) for 3 × 30 sec. Lysis was checked at 40× magnification, and if incomplete, sonication was repeated as needed. The lysate was aliquoted, 1 ml per Eppendorf tube (Eppendorf North America, Madison, WI), and cleared by centrifugation at 10,000 rpm (table-top Eppendorf microcentrifuge) for 10 min at 4°C. The supernatant was transferred to clean chilled tubes, and fresh protease inhibitors were added. Approximately 20–30 μg of the appropriate bead-bound GST fusion protein were added to 1 ml lysate. The beads were incubated with the lysate for 1 h at 4°C on a rotating platform and pelleted at 5000 rpm for 2 min. The supernatant was aspirated, and the beads were washed three times in binding buffer. The GST fusion proteins were then eluted along with any interacting proteins from the agarose beads with 15 mM reduced glutathione. The eluate was mixed 1:1 with 2× SDS sample buffer, heated to 100°C for 2 min, and loaded onto 12% reducing polyacrylamide gels. Some gels were fixed overnight in 20% trichloroacetic acid and subsequently silver stained (Dunn, 1990). Other gels were transferred to nitrocellulose membranes and processed for Western blot analysis using a polyclonal anti-darlin antiserum.
Peptide Sequencing of Darlin
Darlin protein was purified as described above from 2–3 × 1010 AX2 cells. The eluted proteins were loaded on a 12% polyacrylamide-SDS preparative gel and transferred overnight at 4 W at 4°C in CAPS buffer (10 mM 3-[cyclohexylamino]-1-propanesulfonic acid buffer, pH 11.0, and 10% methanol) onto a polyvinylidene difluoride protein-sequencing membrane (Bio-Rad, Hercules, CA). The polyvinylidene difluoride membrane was then stained with freshly made Ponceau S and washed in distilled H2O. The major protein band, with an apparent molecular mass of 88 kDa, was cut out, transferred to an Eppendorf tube, washed five times with distilled H2O, and frozen at −70°C. The membrane-bound protein was then submitted to the Harvard Microsequencing Facility (Cambridge, MA) for microsequencing. Three peptides, peptides 1–3, were sequenced. The sequences were as follows: peptide 1, EGYYENSFANDLVSSLSTLSLN; peptide 2, VEDNRETIIRSPSNVIEK; and peptide 3, DTEHYSEEAVELLI.
Molecular Cloning of Darlin
The Dictyostelium gene encoding the darlin protein is designated darA. Using the Dictyostelium codon bias (Sharp and Devine, 1989), we designed six primers based on the sequence of each peptide obtained: AO-171 (ggt tat tat gaa aat tca ttt gc), AO-172 (taa tga tga aac taa atc att agc), AO-173 (gtg aaa cta tta ttc gtt caa c), AO-174 (caa taa cat ttg atg gtg aac), AO-175 (gaa cat tat tca gaa gaa gct gtt g), and AO-176 (caa cag ctt ctt ctg aat aat gtt c). Dictyostelium mRNA was made by incubating biotinylated oligo(dT) with total RNA prepared by the diethyl-pyrocarbonate method (Nellen et al., 1987) followed by affinity chromatography with streptavidin-coupled magnetic beads (PolyATract mRNA Isolation Systems; Promega, Madison, WI). The mRNA thus obtained was used as a template with oligo(dT) for RT (Superscript RNase H− reverse transcriptase, Life Technologies, Gaithersburg, MD) to obtain cDNA (Ausubel et al., 1994). The appropriate primer pairs were used for PCR (AmpliTaq; Perkin Elmer-Cetus, Norwalk, CT). The only primers that generated a legitimate PCR product were AO-175 and AO-172. This 700-bp product was cloned into the pT7-Blue vector (Novagen, Milwaukee, WI) and subsequently used to probe genomic and cDNA libraries using standard protocols (Ausubel et al., 1994). Comparison of the sequence of genomic and cDNA clones allowed the mapping of three introns. Clone PCR3 was obtained by PCR using Dictyostelium DNA as a template to complete the sequence of the third intron. The cDNA clones did not include the 3′ portion of the gene; thus, we used 3′ rapid amplification of cDNA ends–PCR to determine the end of the coding region. The product of this PCR also contained the polyadenylation site for the darlin mRNA as shown in Figure 5. We designed two primers to obtain the contiguous full-length darA coding region. One was complementary to the 5′ end (AO-212: aag gat cca tgg aag aga tac aaa aat taa tta atg aat tag gtg gtt cac) and another to the 3′ end (AO-210: tat aag ctt aaa ttg tta att gaa cta aaa ttt ttt gaa tta aat ttg tta atg att gtg gtg c). These primers contained a BamHI site and a HindIII site, respectively, and were used to amplify the full-length cDNA by PCR.
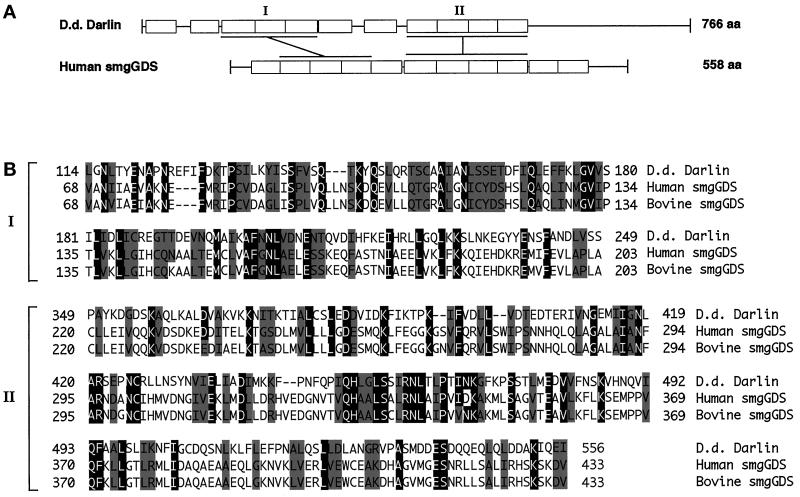
Figure 5.
Darlin is similar to mammalian smgGDS and has 11 armadillo-like repeats. (A) Diagram indicating the position of armadillo-like repeats in darlin and smgGDS. Each rectangle represents a single repeat. Almost the entire sequence of human smgGDS protein consists of 11 armadillo repeats. The Dictyostelium (D.d.) darlin protein also has 11 armadillo-like repeats. The shaded regions (I–II) indicate regions of homology between the two proteins as shown in B. (B) The darlin sequence was compared with the bovine and human smgGDS sequences from the GenBank database using BlastP. Residues that are identical between the darlin protein and either mammalian smgGDS protein are shown in white letters on a black background. Residues that represent conservative substitutions are shown in black letters on a gray background. The amino acid sequence is numbered on both sides of these regions of similarity.
Expression of Darlin in Bacteria
The full-length darA cDNA was cloned into the BamHI and HindIII sites of pMAL-C2 (New England Biolabs, Beverly, MA). The darA gene was inserted downstream of the malE gene, which encodes MBP and results in the expression of an MBP fusion protein that is expressed under the control of the tac promoter. The resulting vector was then transformed into DH5-α cells, and miniprep DNA was made from these clones. After restriction-digest analysis, those clones containing the entire coding region of darA were screened for optimal expression levels of the darlin–MBP fusion protein. Two clones were chosen for large-scale protein preparations.
To purify MBP or the darlin–MBP fusion protein, 100 ml L-broth containing 100 μg/ml ampicillin were inoculated with the respective bacterial strain and incubated at 37°C overnight. The culture was then diluted 1:10 into fresh L-broth and ampicillin in 2-l flasks and incubated for 2 h at room temperature on an orbital shaker. Isopropyl-β-d-thiogalactopyranoside was added to 0.1 mM to induce expression, and the culture was incubated overnight at room temperature at 250 rpm on an orbital shaker. The cells were then collected by centrifugation at 4000 rpm for 60 min at 4°C and resuspended to 20 ml in ice-cold lysis buffer (20 mM Tris-HCl, pH 7.5, 200 mM NaCl, 1 mM EDTA, 1 mM DTT, 5 μg/ml leupeptin, 1.4 μg/ml pepstatin, 10 μg/ml PMSF, and 2 mM sodium bisulfite). The resuspended bacteria were lysed two times in a French press at 1200 psi. The lysate was centrifuged at 9000 × g for 10 min at 4°C, the supernatant was transferred to a chilled tube, and fresh protease inhibitors were added. Two milliliters of a prewashed 1:1 suspension of amylose resin were added to the supernatant and incubated for 30 min on a rotating platform at 4°C. The beads were centrifuged at 1000 × g for 5 min, the supernatant was discarded, and the beads were then washed three times with 20 ml lysis buffer without protease inhibitors. The bead-bound fusion proteins were stored as a 1:1 slurry on ice, or the proteins were eluted with 10 mM maltose in the same buffer.
The soluble darlin–MBP fusion protein was used to raise polyclonal anti-darlin antibodies in rabbits (Cocalico Biologicals, Reamstown, PA).
Construction of Darlin Knockout Mutants
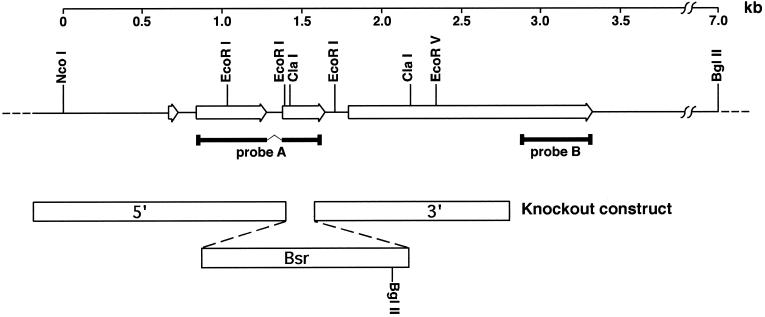
A construct was designed to disrupt the darA locus using the blasticidin resistance marker from pBsrΔBam (Adachi et al., 1994) flanked by 1.5 kb of 5′ flanking and 1.4 kb of 3′ flanking sequences from the darA gene as diagramed in Figure 2. Recombination of this construct into the darA locus leads to the insertion of a BglII site that is 5.5 kb from a BglII site in the 3′ flanking region of the darA gene.
Figure 2.
Genomic organization of the darA gene locus. Probe A was obtained by RT-PCR using oligonucleotides designed from peptide sequences obtained from purified darlin protein. This probe was used to obtain genomic and cDNA clones that encompassed the entire locus. Comparison of the sequence of these clones revealed that the darA open reading frame (open arrows) is interrupted by three introns as indicated. A knockout construct was designed using the indicated portions of the darA gene (5′ and 3′ open bars) and a blasticidin-resistance selectable marker (Bsr). When this construct recombined into the darA locus by a double crossover, it introduced a BglII site 5.5 kb away from the BglII site in the 3′ flanking region of the darA gene (see Figure 7A). Probe B is a 450-bp PCR product from the 3′ end of the open reading frame. It does not overlap with portions included in the knockout construct.
The disruption construct was introduced as a linear fragment into Dictyostelium AX2 cells by electroporation (Adachi et al., 1994), and transformed cell lines were selected in 96-well dishes with HL5 medium supplemented with 10 μg/ml blasticidin S. Individual clones were expanded and analyzed by Southern and Western blots to determine those that contained a disruption of the darA locus.
Phenotypic Analysis of Darlin Mutants
The development phenotype of the darlin null cells was assessed under three conditions. First, cells were seeded on a bacterial lawn on an SM/5 agar plate as described (Vithalani et al., 1996). Second, cells were plated on nonnutrient agar at 22°C, as previously described (Devreotes et al., 1987). Briefly, cells were washed in developmental buffer (5 mM Na2HPO4, 5 mM NaH2PO4, pH 6.2, 2 mM MgSO4, and 200 μM CaCl2), resuspended at 1 × 107 cells/ml, and deposited on the agar plate. Typically, 1 × 107 cells were placed on a 35-mm plate. Once the cells adhered to the surface, the excess buffer was aspirated, and the plates were placed in a humidified chamber. In wild-type cells, development was complete within 24 h. Cells then were starved under a layer of buffer as described previously (De Lozanne and Spudich, 1987).
Adenylyl Cyclase Assays
Enzyme activity was measured on differentiated cells. Briefly, cells were harvested by centrifugation, resuspended at 2 × 107 cells/ml in developmental buffer, and shaken for 5 h at 100 rpm with repeated exogenous pluses of cAMP (final concentration, 50 nM). This starvation period induces the maximal expression of the chemoattractant receptor, heterotrimeric G protein subunits, and adenylyl cyclase along with its cytosolic regulators. After a 30-min treatment with 2 mM caffeine (to bring the cells to a basal state), the cells were washed twice in development buffer without CaCl2 and resuspended at 8 × 107 cells/ml. Adenylyl cyclase activity was measured in filter-lysed cells for 2 min at room temperature in the presence of 5 mM MnSO4 (intrinsic activity), 2 mM MgSO4 (basal activity), or 40 μM GTPγS and 1 μM cAMP (G protein-mediated activity) as previously described (Parent and Devreotes, 1995).
RESULTS
Identification of a Dictyostelium Protein that Binds to Small GTPases
To identify Dictyostelium proteins that interact with members of the rho family of small GTPases, we used a bacterially expressed racE–GST fusion protein as an affinity ligand. The racE–GST fusion protein was bound to glutathione and agarose beads and incubated with a Dictyostelium lysate. We subsequently eluted the racE–GST fusion protein from the beads and analyzed them by SDS-PAGE. Using this approach, we identified a single protein with an apparent molecular mass of ∼88 kDa (p88) that bound to racE but not to GST alone (Figure 1A). Moreover, p88 did not bind to a GST fusion protein containing only the C-terminal 40 amino acids of racE or to a GST fusion protein of the tail portion of a Caenorhabditis elegans myosin I protein (our unpublished results). These results suggested that the p88 protein bound specifically to racE–GST but not to other unrelated GST fusion proteins.
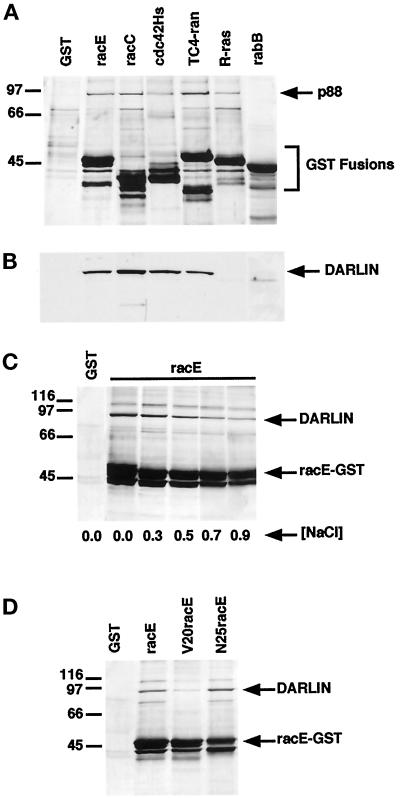
Figure 1.
Identification of darlin as a small GTPase-binding protein from Dictyostelium. (A) Isolation of Dictyostelium proteins that bind to small GTPases. In bacteria we expressed GST and GST fusion proteins of Dictyostelium racE, racC, and rabB, and human cdc42Hs, TC4-ran, and R-ras. The different GST fusion proteins were incubated with Dictyostelium lysates and washed, and the proteins were eluted off the beads. The eluted proteins were subsequently analyzed by SDS-PAGE and visualized by silver staining. A protein of ∼88 kDa bound to all GST fusion proteins except that for rabB or to GST alone. GST migrates below the area shown. (B) Darlin binds to a subset of small GTPases. Western blot analysis of the same samples shown in A using a polyclonal antibody against the purified darlin protein. Darlin bound to the GST fusion proteins for racE, racC, cdc42Hs, and TC4-ran but not to those for R-ras or rabB. Note that the 88-kDa protein that bound to R-ras (A) did not react with the anti-darlin antibody. (C) Darlin binds to racE under high ionic strength. The racE–GST fusion protein was incubated with Dictyostelium lysates in a range of NaCl concentrations and analyzed as in A. Darlin binds to racE even under high ionic strength buffer conditions. (D) Darlin binds preferentially to the GDP-bound form of racE. GST and GST fusion proteins of racE, the constitutively GTP-bound V20RacE, and the constitutively GDP-bound N25racE were incubated with Dictyostelium lysates in 0.4 M NaCl and analyzed as in A.
To determine the specificity of binding of p88 to racE, we tested the ability of various other small GTPases to bind to p88 from Dictyostelium lysates. We expressed in bacteria GST fusion proteins of Dictyostelium racC and rabB, and human cdc42Hs, TC4-ran, and R-ras (Boguski and McCormick, 1993; Hall, 1993). The different GST fusion proteins were incubated with Dictyostelium lysates and analyzed as before. As shown in Figure 1A, Dictyostelium proteins of ∼88 kDa also bound to the GST fusions of racC, cdc42Hs, TC4-ran, and R-ras, but not to that of rabB. However, it was possible that different proteins of ∼88 kDa bound to the different small GTPases. Therefore, we raised a polyclonal antibody against the protein bound by racE (p88) and used it to process a similar gel by Western blot analysis (Figure 1B). This experiment demonstrated that the GST fusion proteins of racE, racC, cdc42Hs, and TC4-ran were bound by the same 88-kDa protein that reacted with our antiserum. On the other hand, the 88-kDa protein that bound to the R-ras–GST fusion protein did not react with our antiserum and therefore is distinct from p88. Finally, the rabB-GST fusion protein did not bind to p88 or any other protein of similar size.
Once we had determined that p88 bound to a subset of small GTPases, we explored its binding properties in more detail. We first examined the binding of p88 to racE in a buffer containing a range of NaCl concentrations from 0 to 0.9 M. Although the amount of p88 protein bound to racE–GST decreased at high salt concentrations, substantial p88 protein remained bound at 0.5 M NaCl (Figure 1C). This result suggests that the binding of p88 to racE–GST was not the result of low-affinity ionic interactions. Because the binding of many proteins to small GTPases is nucleotide dependent (Boguski and McCormick, 1993), we determined whether the binding of p88 was influenced by the nucleotide bound to racE. We used the constitutively active (GTP-bound) and inactive (GDP-bound) forms of racE. Proteins corresponding to the GTP-bound V20racE (valine for a glycine) and the GDP-bound N25racE (asparagine for a threonine) were expressed as GST fusion proteins, purified, and assayed for their ability to bind to p88 from Dictyostelium cytosolic extracts. In the absence of salt, the p88 protein bound equally well to V20racE, N25racE, and wild-type racE. However, when the NaCl concentration was increased to 0.4 M, p88 bound preferentially to the GDP-bound N25racE (Figure 1D).
Cloning of the Dictyostelium Small GTPase-binding Protein
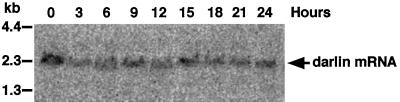
The affinity chromatography approach used to identify p88 was scaled up to purify ∼50 pmol of protein for amino acid microsequencing. Three peptides were isolated and microsequenced. Using the Dictyostelium codon bias (Sharp and Devine, 1989), we designed PCR primers from the sequence of these peptides and amplified a specific 700-bp cDNA product (Figure 2, probe A). The sequence of the PCR product revealed that peptide 2 was in fact nested between peptides 1 and 3 (Figure 3), confirming that the three peptides were derived from the same protein. The 700-bp PCR product was then used in Northern blot analysis to probe total RNA prepared from Dictyostelium vegetative cells. This probe detected an mRNA of ∼2.3 kb, a size consistent with the molecular mass of the p88 protein (Figure 4, 0 h). The same PCR product was used to probe RNA isolated from Dictyostelium cells at different stages of the developmental program (3–24 h poststarvation). Figure 4 shows that the mRNA encoding p88 was expressed throughout the Dictyostelium developmental program, suggesting that this protein may be needed throughout the entire life cycle.
Figure 3.
Sequence of the darA gene. The open reading frame of the darA gene is divided into four exons (bold capital letters) that extend over 2301 bp and encode a protein of 766 amino acids with a predicted molecular mass of 87.7 kDa. The darA coding region is interrupted by three introns (underlined bold lowercase letters) of 110, 100, and 144 bp. The 5′ and 3′ flanking sequences are shown in lower case letters. The 5′ flanking portion of the gene has two GC-rich clusters (boxed letters) that are often associated with promoter elements in Dictyostelium (Driscoll et al., 1988). After the stop codon at the 3′ end of the gene, there are four poly(A) signals (AATAAA) immediately preceding the site of polyadenylation at residue 3158. The predicted amino acid sequence is shown below the coding sequence. The three peptides that were derived from microsequencing are shown in white letters on a black background. The amino acid sequence is numbered on the left and the nucleic acid sequence on the right. This sequence has the GenBank accession number AF006055.
Figure 4.
The darA gene is expressed throughout the Dictyostelium developmental program. Northern blot analysis of total RNA harvested from vegetative wild-type cells (time 0) and every 3 h during development (times 3–24). The blot was probed with a darA PCR probe (Figure 2, probe A). Molecular weights in kilobases are indicated on the left.
We then used probe A to screen Dictyostelium genomic and cDNA libraries and obtained multiple overlapping clones. The sequences of the different genomic and cDNA clones were used to compile the sequence of the entire gene encoding the p88 protein (Figure 3). The open reading frame of this gene extends over 2301 bp and encodes a protein of 766 amino acids with a predicted molecular mass of 87.7 kDa. The coding region is interrupted by three small introns, 110, 100, and 144 bp in size. As is typical of Dictyostelium genes, these introns and the 5′ and 3′ flanking portions of this gene are extremely A-T rich. The 5′ flanking portion of the gene has two clusters of GC base pairs that are often associated with promoter elements in Dictyostelium (Driscoll et al., 1988). At the 3′ end of the gene there are four consensus poly(A) signals (AATAAA) immediately preceding the site of polyadenylation.
Darlin: a Dictyostelium Protein that Contains Armadillo-like Repeats
When we compared the sequence of the gene encoding the p88 protein to the GenBank database, we discovered that two regions exhibited significant homology to the mammalian GEF, smgGDS (Yamamoto et al., 1990) (Figure 5, A and B). The first region, amino acids 114–249 of p88, bears 22% identity and 53% similarity to amino acids 68–203 of smgGDS. Likewise, the second region, p88 residues 349–556, shows 21% identity and 43% similarity to smgGDS residues 220–433. Both regions were aligned by BlastP (National Center for Biotechnology Information, Bethesda, MD) with a probability of 3 × 10−6.
Mammalian smgGDS is known to contain 11 armadillo-like repeats that comprise nearly the entire protein. Originally identified in the Drosophila armadillo protein (Peifer and Wieschaus, 1990; McCrea et al., 1991), multiple copies of these 42 amino acid motifs are also found in proteins with diverse functions, such as β-catenin, plakoglobin, the adenomatous polyposis coli gene product, and p120 (Peifer et al., 1994; Shimizu et al., 1996). The crystal structure of β-catenin revealed recently that the armadillo repeat consists of three α-helices folded tightly against each other and against those from other repeats (Huber et al., 1997). Thus, the multiple repeats form an elongated superhelix with a long positively charged groove. Careful analysis of the p88 protein sequence indicated the presence of 11 regions (illustrated schematically in Figure 5A) that resemble the armadillo repeat. The alignment of these regions (Figure 6) shows that the motifs present in p88 are distantly related to the armadillo repeat but possess many of the characteristic residues that are important for folding. Therefore, we named this protein darlin, which is encoded by the darA gene.
Figure 6.
Alignment of the armadillo-like repeats of darlin with the universal armadillo consensus motif. The 11 recognizable armadillo-like repeats of darlin were aligned with the smgGDS consensus and the universal armadillo consensus sequences according to Peifer et al., 1994. Residues that are identical between the darlin repeats and either the smgGDS or the universal armadillo consensus sequences are shown in white letters on a black background. Individual consensus matches within darlin are shown in white letters on a gray background. Dashes in the repeats indicate gaps inserted in the sequence for best alignment. The amino acid sequence is numbered on both sides of the darlin repeats.
Darlin Is Not an Essential Protein
To test the function of darlin in vivo, we generated Dictyostelium mutant cell lines defective in this protein. We constructed a gene disruption vector for the darA gene using the blasticidin-resistance selectable marker (Figure 2). This vector integrated into the darA locus at a frequency of 15%. Of 61 blasticidin resistant cell lines obtained, 9 had the disruption vector integrated at the darA gene. Southern blot analysis of these cell lines demonstrated that in seven of those mutant cell lines the disruption vector integrated by a double crossover event. In these cell lines, a probe not included in the disruption vector (probe B) hybridized to a 5.5-kb BglII genomic fragment (Figure 7A). The same probe hybridized to a 7-kb NcoI–BglII fragment in wild-type or nonrecombinant cell lines (Figure 7A). The other two recombinant cell lines contained the entire disruption vector inserted into the darA locus by a single crossover event (our unpublished results).
Figure 7.
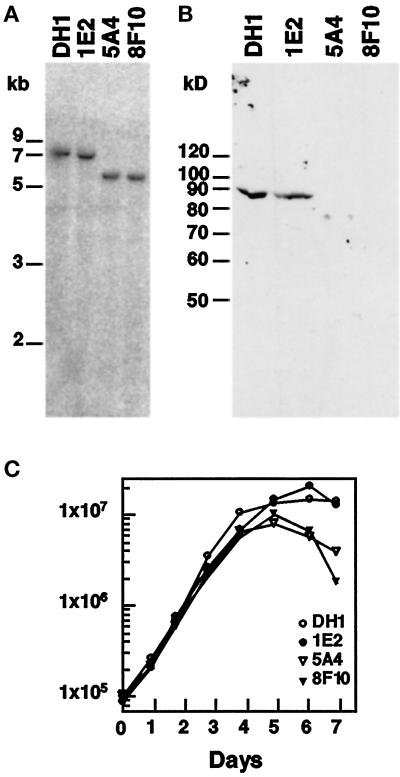
The darA gene is not essential for growth. (A) Disruption of the darA gene. Dictyostelium wild-type cells (DH1 strain) were transformed with a darlin knockout construct (Figure 2). Genomic DNA from individual clones was digested with NcoI and BglII and analyzed on Southern blots probed with probe B (Figure 2). This probe hybridizes to a 7-kb band in both wild-type and nonrecombinant clones (1E2). Integration of the knockout construct at the darA locus results in the formation of a 5.5-kb BglII band that hybridizes to probe B (clones 5A4 and 8F10). (B) Disruption of the darA gene abolishes darlin expression. Western blot analysis of cell extracts of wild-type (DH1), nonrecombinant strains (1E2), or darA knockout strains (5A4 and 8F10) probed with an anti-darlin polyclonal antibody. The knockout strains do not have any detectable darlin protein. (C) darlin− strains grow at normal rates. Cells were placed in suspension culture and monitored for growth for several days. The darlin− strains increased in cell density at the same rate as wild-type and control cells. After reaching saturation, the darlin− strains decreased in cell density at a faster rate than wild-type and control cells.
To determine whether the disruption of the darA gene resulted in the loss of darlin protein, we analyzed the different cell lines by Western blot analysis using our anti-darlin antibody. This antibody recognizes specifically the 88-kDa darlin protein in wild-type or nonrecombinant cell lines (Figure 7B). As expected, the knockout cell lines did not contain any traces of darlin protein (Figure 7B).
Because the darlin protein binds to racE, we tested the possibility that darlin plays an important role during cytokinesis. Darlin− and control cells were placed in suspension cultures and monitored for growth. Under these conditions, darlin− cells grew at the same rate as control cells (Figure 7C) and remained uninucleate (our unpublished results). This is in contrast with racE null cells or other cytokinesis mutants, which fail to grow in suspension and become large multinucleate cells (Larochelle et al., 1996). Therefore, darlin is not essential for cytokinesis. The darlin− cultures did have the tendency to reach saturation slightly sooner than wild-type cultures. The titers on the saturated darlin− cultures also decreased faster than those of wild-type cultures, but we did not observe a significant number of multinucleate cells in these cultures.
We also tested the ability of darlin− mutants to grow on agar plates with bacteria as a food source and found that they grew to the same extent as control cells (our unpublished results). Therefore, darlin is not essential for phagocytosis or for the digestion of bacteria under these conditions.
Darlin Plays an Important Role During Early Development in Dictyostelium
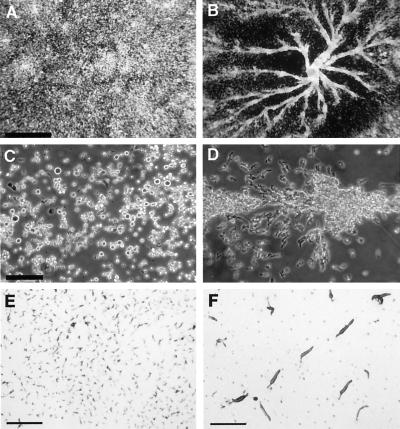
When a Dictyostelium culture depletes the nutrients in the surrounding medium, the starving cells initiate a complex developmental program that culminates with the formation of a fruiting body. To determine whether darlin plays any role during the early stages of development, we placed wild-type and darlin-null cells under starvation buffer on plastic petri dishes. Under these conditions, monolayers of Dictyostelium wild-type cells began development and formed large streams of elongated cells that congregated into very large aggregation centers (Figure 8, B and D). In contrast, monolayers of darlin-null cells completely failed to initiate aggregation and did not form any streams or aggregation centers (Figure 8, A and C). To further explore the developmental properties of darlin-null cells, we plated them on nonnutrient agar plates. Under these conditions, the deposited cells are not submerged under liquid and can complete the developmental program, forming fruiting bodies within 24 h. Although both wild-type (Figure 8F) and darlin-null cells were able to form slugs and fruiting bodies of normal size, the darlin-null mutants formed small aggregating centers that lead to the formation of very small slugs (Figure 8E) and fruiting bodies.
Figure 8.
Darlin− mutants have a defect in early development. Dictyostelium wild-type (B and D) and darlin− (A and C) cells were allowed to grow to confluency on plastic Petri dishes. The media were then replaced by starvation buffer, and the cultures were photographed after 12 h. Wild-type cells formed large aggregation centers that congregated into organized streams of elongated cells. Darlin− cells failed to form aggregation centers and streams. In addition these cells did not elongate to the same extent as wild-type cells. Bars: (A and B), 1 mm; (C and D), 0.2 mm. (E and F) Cells were plated on nonnutrient agar and allowed to undergo development at room temperature. The pictures were taken 24 h after plating. Wild-type (F) and control cells formed normal size slugs that differentiated into normal fruiting bodies (our unpublished observations). The darlin-null cells (E) formed very small aggregating centers that lead to the formation of very small slugs and fruiting bodies. Bar, 0.3 cm.
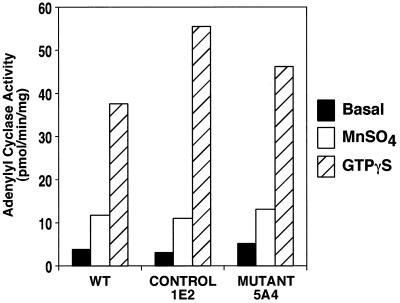
The inability of darlin-null cells to initiate development under buffer contrasts with their ability to form normal aggregates on agar. This difference could potentially be caused by a defect in the signaling pathway mediated by cAMP (Parent and Devreotes, 1996). To address this possibility, we first performed Western analysis to assess the presence of key proteins participating in the activation of adenylyl cyclase in Dictyostelium. In both wild-type and darlin-null cells, we observed equivalent levels of the cAMP receptor, cAR1, the adenylyl cyclase, and one of its cytosolic regulators, pianissimo (our unpublished results). We then measured the ability of darlin-null lysates to activate adenylyl cyclase in response to G protein stimulation. This response is known to be mediated by the G protein βγ subunits and at least two cytosolic regulators (Chen et al., 1997). As shown in Figure 9, the darlin-null cells displayed robust GTPγS-mediated adenylyl cyclase activity that was comparable with that from wild-type cells or control cells. These results demonstrate that the signaling pathway leading to the activation of adenylyl cyclase in the darlin-null cells is intact.
Figure 9.
Darlin− cells have normal adenylyl cyclase activation in response to G protein activation. Adenylyl cyclase activity was measured in cell lysates under basal conditions or in the presence of 40 μM GTPγS and 1 μM cAMP at room temperature for 2 min. The unregulated intrinsic enzyme activity was measured by adding 5 mM MnSO4. The G protein-mediated activation was as robust in the darlin-null cells as in either wild-type or nonrecombinant control cells. In addition, the levels of MnSO4 activity were similar in each cell line, suggesting that they were expressing comparable levels of adenylate cyclase. Results are presented in picomoles per minute per milligrams protein and represent the average of experiments performed in duplicate.
Finally, we explored the developmental properties of the darlin-null cells by plating them on agar plates in the presence of bacteria as a food source. The mutant cells were able to ingest bacteria and form colonies at the same rate as wild-type controls. Furthermore, when the bacteria were depleted, the darlin− cells initiated development and formed aggregates and fruiting bodies indistinguishable from those formed by wild-type cells under these conditions (our unpublished observations).
DISCUSSION
We described the isolation and characterization of darlin, a Dictyostelium small GTPase-binding protein. Cloning of the gene encoding darlin revealed that this protein contains multiple repeats similar to those found in armadillo and related proteins (Peifer et al., 1994). The three-dimensional structure of the β-catenin region composed of armadillo repeats was determined recently (Huber et al., 1997). These studies revealed that each armadillo repeat is composed of three α-helices that interact extensively with each other and with helices from adjacent repeats to form an elongated superhelical structure. Given the similarity of the darlin and armadillo repeats, it seems likely that this region of darlin folds into a similar elongated structure. Furthermore, because the repeat region of armadillo has been shown to be involved in interactions with multiple binding partners, we postulate that the repeat region of darlin will be implicated in a similar function. However, unlike armadillo, the repeat region of darlin is mildly acidic (pI 4.7 vs. 8.3). Therefore, it is possible that darlin may interact with proteins that are more basic than those that bind to armadillo.
Among those proteins that contain armadillo repeats, darlin displays the most similarity to the mammalian GEF smgGDS (Yamamoto et al., 1990). The repeats of smgGDS are the most divergent among those of other proteins. Thus, it seems that the repeats of darlin and smgGDS may represent a divergent subclass of armadillo repeats.
Originally identified in bovine brain as a regulator of Rap1B (Yamamoto et al., 1990), smgGDS has subsequently been shown to modulate nucleotide exchange on the rho family proteins as well as other ras-related proteins (Kawamura et al., 1991; Hiraoka et al., 1992). Mammalian smgGDS binds to a series of small GTPases including rhoA, cdc42Hs, rac1, rap1A, rap1B, and the ras splice variant K-ras4B (Takai et al., 1993). SmgGDS does not bind to K-ras4A, H-ras, N-ras, or rab3A (Takai et al., 1992). Because darlin shares structural homology with smgGDS, we tested the binding of darlin with various small GTP-binding proteins. We found that darlin bound to GST fusion proteins of racE, racC, cdc42Hs, and TC4-ran. Furthermore, darlin did not bind to GST–R-ras or GST–rabB. Thus, similar to smgGDS, darlin is also a GTPase binding protein that binds to a broad spectrum of GTPases.
The similarity of darlin and smgGDS suggests that darlin may also be a GEF. In support of this possibility, we found that the binding of darlin to racE was nucleotide dependent. Darlin bound preferentially to the GDP-loaded N25racE–GST compared with the GTP-loaded V20racE–GST. This is consistent with the relative affinities of GEFs to GDP- and GTP-bound forms of GTPases (Chuang et al., 1994; Hart et al., 1994; Hart and Powers, 1995). However, we have not been able to demonstrate an exchange activity in assays using bacterially expressed darlin and racE (Vithalani, unpublished observations). Further studies are aimed at purifying both proteins from Dictyostelium extracts to determine whether darlin does possess a GEF activity.
Alternatively, it is possible that darlin is not a GEF. In fact, smgGDS, unlike other exchange factors such as rasGEF or cdc24, is a poor exchanger. A large molar excess of smgGDS over small G protein is required to observe an appreciable exchange activity (Hiraoka et al., 1992). This may indicate that the primary function of smgGDS and darlin is not to exchange nucleotides but to serve as linkers or adaptors, bringing the small GTPase in contact with a particular effector. In this regard, it is interesting that smgGDS has been shown recently to also bind to the armadillo repeat-containing protein KAP3, a protein associated with the kinesin KIF3A/B molecule (Shimizu et al., 1998). Finally, although darlin is most similar to smgGDS, they are quite divergent and could certainly have very different functions in vivo.
What then is the role of darlin in vivo? We isolated this protein in our search for binding partners of racE, a small GTPase required for cytokinesis. However, as we have demonstrated here, darlin-null mutant cells are not defective in cytokinesis. Clearly, the interaction of racE and darlin is not essential for cell division. A potential explanation for this result is that other proteins may exist that can substitute for darlin during cytokinesis. In this regard it should be noted that in addition to darlin we also found a Dictyostelium protein that bound to GST–R-ras. This protein, which is the same size as darlin, may be similar enough to darlin that it could potentially replace darlin during cytokinesis.
The analysis of darlin-null mutants also demonstrated that darlin is not required for phagocytosis, pinocytosis, or cell movement. Interestingly, the mutants were not able to initiate development when placed under a layer of starvation buffer. Furthermore, under less stringent plating conditions, the cells gave rise to very small aggregating centers, suggesting that the darlin-null cells were somehow impaired in cell–cell signaling. However, the darlin-null cells showed no alterations in the signal transduction pathway that activates adenylyl cyclase in response to G protein activation. Thus, darlin might be involved in an alternate pathway that couples receptor activation and the motility apparatus. For example, aimless, a RasGEF that was recently cloned in Dictyostelium, was also found to be involved in the regulation of chemotaxis (Insall et al., 1996). Aimless and darlin thus seem to be members of a class of proteins that regulates the activity of small G proteins that are involved in the control of directed cell movement, and depending on their target, these modifying proteins present distinct phenotypes. Additional studies will reveal the link between chemoattractant receptors, these small G protein regulators, and the cell motility apparatus.
ACKNOWLEDGMENTS
We gratefully acknowledge Drs. Daniel Lew, Sally Kornbluth, and Channing Der for their generous contributions of cdc42Hs, TC4-ran, and R-ras expression vectors. Special thanks to Melinda Maready for excellent technical support. We also thank Drs. Daniel Kiehart, Steven Garrett, Terry O’Halloran, Katherine Swenson, and Margaret Titus, and the members of the De Lozanne and O’Halloran laboratories for useful comments throughout this work. This work was supported by National Institutes of Health grants GM-48745 (to A.D.) and GM-28007 (to P.N.D.).
Abbreviations used:
- DTT
dithiothreitol
- GAP
GTPase-activating protein
- GDP
guanosine diphosphate
- GEF
guanine nucleotide exchange factor
- GST
glutathione S-transferase
- GTP
guanosine triphosphate
- MBP
maltose-binding protein
- PMSF
phenylmethylsulfonyl fluoride
- RT
reverse transcription
REFERENCES
- Adachi H, Hasebe T, Yoshinaga K, Ohta T, Sutoh K. Isolation of Dictyostelium discoideum cytokinesis mutants by restriction enzyme-mediated integration of the blasticidin S resistance marker. Biochem Biophys Res Commun. 1994;205:1808–1814. doi: 10.1006/bbrc.1994.2880. [DOI] [PubMed] [Google Scholar]
- Adachi H, Takahashi Y, Hasebe T, Shirouzu M, Yokoyama S, Sutoh K. Dictyostelium IQGAP-related protein specifically involved in the completion of cytokinesis. J Cell Biol. 1997;137:891–898. doi: 10.1083/jcb.137.4.891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ausubel FM, Brent R, Kingston RE, Moore DA, Seidman JG, Smith JA, Struhl K. Current Protocols. K. Janssen, New York: John Wiley & Sons; 1994. Current protocols in molecular biology. [Google Scholar]
- Boguski MS, McCormick F. Proteins regulating Ras and its relatives. Nature. 1993;366:643–654. doi: 10.1038/366643a0. [DOI] [PubMed] [Google Scholar]
- Chen MY, Long Y, Devreotes PN. A novel cytosolic regulator, pianissimo, is required for chemoattractant receptor and G protein-mediated activation of the twelve transmembrane domain adenylyl cyclase in Dictyostelium. Genes Dev. 1997;11:3218–3231. doi: 10.1101/gad.11.23.3218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuang T-H, Xu X, Quilliam LA, Bokoch GM. SmgGDS stabilizes nucleotide-bound and -free forms of the Rac1 GTP-binding protein and stimulates GTP/GDP exchange through a substituted enzyme mechanism. Biochem J. 1994;303:761–767. doi: 10.1042/bj3030761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Lozanne A, Spudich JA. Disruption of the Dictyostelium myosin heavy chain gene by homologous recombination. Science. 1987;236:1086–1091. doi: 10.1126/science.3576222. [DOI] [PubMed] [Google Scholar]
- Devreotes PN, Fontana D, Klein P, Sherring J, Theibert A. Transmembrane signaling in Dictyostelium. Methods Cell Biol. 1987;28:299–331. doi: 10.1016/s0091-679x(08)61653-2. [DOI] [PubMed] [Google Scholar]
- Driscoll DM, Pears CJ, Williams JG. Characterization of two divergently transcribed Dictyostelium gene pairs and identification of G-rich sequence element lying between them with the characteristics of a basal promoter element. Dev Genet. 1988;9:455–468. doi: 10.1002/dvg.1020090423. [DOI] [PubMed] [Google Scholar]
- Dunn MJ. Protein Purification Methods. E.L.V. Harris and S. Angal, New York: Oxford University Press; 1990. Determination of total protein concentration; pp. 10–66. [Google Scholar]
- Faix J, Dittrich W. DGAP1, a homologue of ras GTPase activating proteins that controls growth, cytokinesis, and development in Dictyostelium discoideum. FEBS Lett. 1996;394:251–257. doi: 10.1016/0014-5793(96)00963-5. [DOI] [PubMed] [Google Scholar]
- Gerald N, Dai J, Ting-Beall HP, De Lozanne A. A role for Dictyostelium racE in cortical tension and cleavage furrow progression. J Cell Biol. 1998;141:483–492. doi: 10.1083/jcb.141.2.483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall A. Ras-related proteins. Curr Opin Cell Biol. 1993;5:265–268. doi: 10.1016/0955-0674(93)90114-6. [DOI] [PubMed] [Google Scholar]
- Hart M, Powers S. Ras-Cdc25 and Rho-Dbl binding assays: complex formation in vitro. Methods Enzymol. 1995;255:129–135. doi: 10.1016/s0076-6879(95)55016-x. [DOI] [PubMed] [Google Scholar]
- Hart MJ, Eva A, Zangrilli D, Aaronson SA, Evans T, Cerione RA, Zheng Y. Cellular transformation and guanine nucleotide exchange activity are catalyzed by a common domain on the dbl oncogene product. J Biol Chem. 1994;269:62–65. [PubMed] [Google Scholar]
- Hiraoka K, et al. Both stimulatory and inhibitory GDP/GTP exchange proteins, smgGDS and rho GDI, are active on multiple small GTP-binding proteins. Biochem Biophys Res Commun. 1992;182:921–930. doi: 10.1016/0006-291x(92)91820-g. [DOI] [PubMed] [Google Scholar]
- Huber AH, Nelson WJ, Weis WI. Three-dimensional structure of the armadillo repeat region of beta-catenin. Cell. 1997;90:871–882. doi: 10.1016/s0092-8674(00)80352-9. [DOI] [PubMed] [Google Scholar]
- Insall RH, Borleis J, Devreotes PN. The aimless RasGEF is required for processing of chemotactic signals through G-protein-coupled receptors in Dictyostelium. Curr Biol. 1996;6:719–729. doi: 10.1016/s0960-9822(09)00453-9. [DOI] [PubMed] [Google Scholar]
- Kawamura S, Kaibuchi K, Hiroyoshi M, Hata Y, Takai Y. Stoichiometric interaction of smg p21 with its GDP/GTP exchange protein and its novel action to regulate the translocation of smg p21 between membrane and cytoplasm. Biochem Biophys Res Commun. 1991;174:1095–1102. doi: 10.1016/0006-291x(91)91533-i. [DOI] [PubMed] [Google Scholar]
- Larochelle DA, Vithalani K, De Lozanne A. A novel member of the rho family of small GTP-binding proteins is specifically required for cytokinesis. J Cell Biol. 1996;133:1321–1330. doi: 10.1083/jcb.133.6.1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larochelle DA, Vithalani KK, De Lozanne A. The role of Dictyostelium racE in cytokinesis: mutational analysis and localization studies by use of green fluorescent protein. Mol Biol Cell. 1997;8:935–944. doi: 10.1091/mbc.8.5.935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S, Escalante R, Firtel RA. A ras GAP is essential for cytokinesis and spatial patterning in Dictyostelium. Development. 1997;124:983–996. doi: 10.1242/dev.124.5.983. [DOI] [PubMed] [Google Scholar]
- Ludbrook SB, Eccleston JF, Strom M. Cloning and characterization of a rhoGAP homolog from Dictyostelium discoideum. J Biol Chem. 1997;272:15682–15686. doi: 10.1074/jbc.272.25.15682. [DOI] [PubMed] [Google Scholar]
- McCrea PD, Turck CW, Gumbiner B. A homolog of the armadillo protein in Drosophila (plakoglobin) associated with E-cadherin. Science. 1991;254:1359–1361. doi: 10.1126/science.1962194. [DOI] [PubMed] [Google Scholar]
- Nellen W, Datta S, Reymond C, Siversen A, Mann S, Crowley T, Firtel RA. Dictyostelium discoideum: Molecular Approaches to Cell Biology. J.A. Spudich, New York: Academic Press; 1987. Molecular biology in Dictyostelium: tools and applications; pp. 67–100. [DOI] [PubMed] [Google Scholar]
- Parent CA, Devreotes PN. Isolation of inactive and G protein-resistant adenylyl cyclase mutants using random mutagenesis. J Biol Chem. 1995;270:22693–22696. doi: 10.1074/jbc.270.39.22693. [DOI] [PubMed] [Google Scholar]
- Parent CA, Devreotes PN. Molecular genetics of signal transduction in Dictyostelium. Annu Rev Biochem. 1996;65:411–440. doi: 10.1146/annurev.bi.65.070196.002211. [DOI] [PubMed] [Google Scholar]
- Peifer M, Berg S, Reynolds AB. A repeating amino acid motif shared by proteins with diverse cellular roles. Cell. 1994;76:789–791. doi: 10.1016/0092-8674(94)90353-0. [DOI] [PubMed] [Google Scholar]
- Peifer M, Wieschaus E. The segment polarity gene armadillo encodes a functionally modular protein that is the Drosophila homolog of human plakoglobin. Cell. 1990;63:1167–1176. doi: 10.1016/0092-8674(90)90413-9. [DOI] [PubMed] [Google Scholar]
- Sharp PM, Devine KM. Codon usage and gene expression level in Dictyostelium discoideum: highly expressed genes do “prefer” optimal codons. Nucleic Acids Res. 1989;17:5029–5039. doi: 10.1093/nar/17.13.5029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu K, Kawabe H, Minami S, Honda T, Takaishi K, Shirataki H, Takai Y. SMAP, an smgGDS-associating protein having arm repeats and phosphorylated by src tyrosine kinase. J Biol Chem. 1996;271:27013–27017. doi: 10.1074/jbc.271.43.27013. [DOI] [PubMed] [Google Scholar]
- Shimizu K, Shirataki H, Honda T, Minami S, Takai Y. Complex formation of SMAP/KAP3, a KIF3A/B ATPase motor-associated protein, with a human chromosome-associated polypeptide. J Biol Chem. 1998;273:6591–6594. doi: 10.1074/jbc.273.12.6591. [DOI] [PubMed] [Google Scholar]
- Smith DB, Johnson KS. Single-step purification of polypeptides expressed in Escherichia coli as fusions with glutathione S-transferase. Gene. 1988;67:31–40. doi: 10.1016/0378-1119(88)90005-4. [DOI] [PubMed] [Google Scholar]
- Takai Y, Kaibuchi K, Kikuchi A, Kawata M. Small GTP-binding proteins. Int Rev Cytol. 1992;133:187–230. doi: 10.1016/s0074-7696(08)61861-6. [DOI] [PubMed] [Google Scholar]
- Takai Y, Kaibuchi K, Kikuchi A, Sasaki T, Shirataki H. Regulators of small GTPases. Ciba Found Symp. 1993;176:128–138. doi: 10.1002/9780470514450.ch9. [DOI] [PubMed] [Google Scholar]
- Vithalani KK, Shoffner JD, De Lozanne A. Isolation and characterization of a novel cytokinesis-deficient mutant in Dictyostelium discoideum. J Cell Biochem. 1996;62:290–301. doi: 10.1002/(sici)1097-4644(199608)62:2<290::aid-jcb16>3.0.co;2-p. [DOI] [PubMed] [Google Scholar]
- Yamamoto T, Kaibuchi K, Mizuno T, Hiroyoshi M, Shirataki H, Takai Y. Purification and characterization from bovine brain cytosol of proteins that regulate the GDP/GTP exchange reaction of smg p21s, ras p21-like GTP-binding proteins. J Biol Chem. 1990;265:16626–16634. [PubMed] [Google Scholar]