Abstract
Inoculation of diploid budding yeast onto nitrogen-poor agar media stimulates a MAPK pathway to promote filamentous growth. Characteristics of filamentous cells include a specific pattern of gene expression, elongated cell shape, polar budding pattern, persistent attachment to the mother cell, and a distinct cell cycle characterized by cell size control at G2/M. Although a requirement for MAPK signaling in filamentous gene expression is well established, the role of this pathway in the regulation of morphogenesis and the cell cycle remains obscure. We find that ectopic activation of the MAPK signal pathway induces a cell cycle shift to G2/M coordinately with other changes characteristic of filamentous growth. These effects are abrogated by overexpression of the yeast mitotic cyclins Clb1 and Clb2. In turn, yeast deficient for Clb2 or carrying cdc28-1N, an allele of CDK defective for mitotic functions, display enhanced filamentous differentiation and supersensitivity to the MAPK signal. Importantly, activation of Swe1-mediated inhibitory phosphorylation of Thr-18 and/or Tyr-19 of Cdc28 is not required for the MAPK pathway to affect the G2/M delay. Mutants expressing a nonphosphorylatable mutant Cdc28 or deficient for Swe1 exhibit low-nitrogen-dependent filamentous growth and are further induced by an ectopic MAPK signal. We infer that the MAPK pathway promotes filamentous growth by a novel mechanism that inhibits mitotic cyclin/CDK complexes and thereby modulates cell shape, budding pattern, and cell-cell connections.
INTRODUCTION
When diploid cells of dimorphic strains of the budding yeast Saccharomyces cerevisiae are inoculated onto agar media rich in a fermentable carbon source but poor in nitrogen, they can differentiate to enter filamentous growth. Previous studies (reviewed by Kron and Gow, 1995; Madhani and Fink, 1998a) have established that the filamentous cell represents a distinct cell type characterized by an elongated shape, a polar budding pattern, and persistent attachment of each daughter cell to the mother after septation. These features contribute to colony growth as branching filaments that invade the agar. Another characteristic of filamentous growth is that the cell division cycle is regulated by cell size control at mitosis (Kron et al., 1994). Filamentous cells remain budded until the cell doubles in size. Throughout most of this prolonged budded period, the cell exhibits phenotypes suggestive of G2 arrest (Surana et al., 1991), such as a short mitotic spindle, a single rounded nuclear mass, and an elongated, tubular bud. As a result of the long G2/M growth period, daughter cells are born larger than the critical size for progression to S phase at Start. Therefore, newly born daughter cells at the tip of filaments immediately bud and reenter the cell cycle. Thus, the G2/M cell cycle control likely contributes to the capacity of filamentous cells to continue mitotic growth even in the face of nitrogen limitation.
A comparison of the shape and cell division pattern of filamentous cells with the results of Lew and Reed (1993) regarding the effects of different cyclin/CDK complexes on cell morphogenesis suggests that the link between cell cycle and cell shape in filamentous growth may be direct. In the growth pattern of “well-fed” yeast (reviewed by Madden and Snyder, 1998), bud growth is initially polarized to the tip, leading to tube-like growth at S phase and into G2. During mitosis, bud growth becomes isotropic, leading to swelling growth. Finally, septation occurs upon completion of mitotic anaphase. Distinct cyclins combine with the Cdc28 CDK to mediate each of the nuclear transitions (reviewed by Mendenhall and Hodge, 1998). Altered expression of the cyclins or modulation of the activity of the cyclin/Cdc28 complexes has coordinate effects of delaying or accelerating both the nuclear transition and the related morphogenetic event (Lew and Reed, 1993). For example, increased expression of G1 cyclins or decreased expression of mitotic cyclins each lead to a delay before the onset of mitosis. This prolongs the period of tubular bud growth, leading to a “filamentous” appearance. It is reasonable to hypothesize that the cell cycle machinery may be a key target of the pathway regulating the morphogenetic events characteristic of filamentous differentiation. Recent data have shown that mutations in G1 cyclins can largely abrogate filamentous growth (Oehlen and Cross, 1998; Loeb et al., 1999), whereas deficiency of the Clb2 mitotic cyclin or induction of inhibitory tyrosine phosphorylation of the principal CDK Cdc28 or a particular point mutation of Cdc28 (Edgington et al., 1999) each significantly induces filamentous differentiation. What remains to be established is any specific link between the low-nitrogen signal and regulation of the cell cycle.
Previous studies of filamentous differentiation have established a branching signal transduction pathway involving elements of the RAS GTPase/cAMP-dependent kinase cascade and the STE MAPK pathway (reviewed by Banuett, 1998). The STE MAPK cascade and gene expression pathway (reviewed by Leberer et al., 1997) is best known for its role in mediating yeast mating response. Pheromones induce a sequence of phosphorylations in the STE MAPK cascade, consisting of a PAK kinase homolog Ste20, a MEK kinase Ste11, a MEK Ste7, and the MAPKs Fus3 and Kss1. The downstream target is a transcriptional activator, Ste12, that binds to pheromone response elements in promoters of effector genes such as Far1 (Hagen et al., 1991). Far1 promotes G1 arrest in pheromone-stimulated cells (Chang and Hersko-witz, 1990), acting as a stoichiometric inhibitor that sequesters Cln1,2 G1 cyclin/Cdc28 complexes (Peter and Herskowitz, 1994).
Filamentous differentiation requires only a subset of the STE pathway genes required for mating responses (Madhani and Fink, 1998b). Mutations in the Ste20, Ste11, or Ste7 kinases or the Ste12 transcription factor abrogate filamentous differentiation (Liu et al., 1993; Roberts and Fink, 1994). The MAPK Kss1 serves a dual role in filamentous differentiation as both inhibitor and activator of Ste12-dependent gene expression (Cook et al., 1997; Madhani et al., 1997). Ectopic activation of the STE pathway by dominant active alleles of STE11 (Liu et al., 1993), STE7 (Madhani et al., 1997), and KSS1 (Madhani et al., 1997) or by overexpression of STE12 (Liu et al., 1993) all promote enhanced filamentous growth and can partially bypass upstream blockades in the pathway.
Ectopic activation of RAS/cAMP signaling can also promote filamentous growth. The dominant active allele RAS2-Val19 markedly enhances filamentous growth in nitrogen-starved cells (Gimeno et al., 1992; Mosch et al., 1996) via activation of both cAMP-dependent and MAPK responses (Mosch et al., 1996, 1999). In turn, activation of the STE pathway alone by dominant active alleles such as STE11-4 (Stevenson et al., 1992; Liu et al., 1993) apparently enhances filamentous growth by cAMP-independent mechanisms. Recent data suggest that RAS signaling induces its cAMP-mediated effects and activates the STE MAPK cascade independently (Mosch et al., 1999; Pan and Heitman, 1999) but that the signals transmitted via these parallel pathways converge on similar targets (Robertson and Fink, 1998; Pan and Heitman, 1999; Rupp et al., 1999).
Thus, elements of the RAS/cAMP and STE MAPK pathways constitute a single “RAS/STE” filamentous response pathway. This compound signaling pathway mediates its effects predominantly via regulation of gene expression. RAS/STE signals induce cell type–specific transcription (Mosch et al., 1996) via a distinct UAS, the filamentous response element (FRE) (Madhani and Fink, 1997), that consists of adjacent Ste12 and Tec1 binding sites. Cells lacking either factor lack FRE-dependent gene expression and are defective in filamentous differentiation (Gavrias et al., 1996; Lo et al., 1997; Madhani and Fink, 1997; Mosch and Fink, 1997). To date, only one effector essential for filamentous differentiation, the MUC1/FLO11 flocculin (Lambrechts et al., 1996; Lo and Dranginis, 1998), has been shown to be expressed under RAS/STE control via a FRE (Lo and Dranginis, 1998; Rupp et al., 1999). Cells deficient in Flo11 do not remain linked in chains and form only disorganized colonies that fail to invade the substratum. However, these cells remain responsive to RAS/STE signals, retaining the elongated cell shape, polar budding, and G2/M delay characteristic of filamentous cells (Kron and Dranginis, unpublished results). As yet, the genes that mediate these aspects of filamentous development remain to be described.
Potential links between the RAS/STE signal transduction pathway and the cell cycle machinery in the control of filamentous differentiation remain poorly defined. This is surprising, considering the well-established roles for cyclins and CDK in determining yeast cell shape (Lew et al., 1997; Madden and Snyder, 1998; Mendenhall and Hodge, 1998). In pheromone response, ectopic activation of the Cln G1 cyclins can antagonize both gene expression (Oehlen and Cross, 1994) and cell cycle arrest (Edwards et al., 1997; Kron, unpublished results). Recent data have suggested that Cln2 cyclin complexes with Cdc28 CDK mediate this effect by regulation of Ste20 (Oehlen and Cross, 1998; Wu et al., 1998; Leza and Elion, 1999) and/or regulation of Ste11 (Wassmann and Ammerer, 1997). Importantly, Cln1,2/Cdc28 phosphorylation of Ste20 has been proposed to underlie the requirement for Cln G1 cyclins in filamentous differentiation (Oehlen and Cross, 1998). A pathway linking Clb1 and Clb2 mitotic cyclins to morphogenesis via Cdc28 phosphorylation of a Ste20 homolog, Cla4, has been demonstrated (Tjandra et al., 1998), but any relationship to signal transduction remains uncharacterized. A broad interpretation of these studies would place the cell cycle machinery as a regulator acting upstream in the RAS/STE signaling pathway.
An alternative hypothesis based on the role of the CDK inhibitor Far1 in pheromone-induced cell cycle arrest is that the cell cycle effect of RAS/STE signaling may be mediated by a FRE-regulated effector, such as a mitotic inhibitor. Such a factor might inhibit Clb1- and Clb2-dependent forms of the Cdc28 CDK and function to delay both depolarization of the bud and completion of mitosis. To test this hypothesis, we set out to examine the relationship between elements of the cell cycle machinery and the signaling pathways modulating filamentous growth. We observed that 1) ectopic activation of the RAS/STE pathway slows mitotic progression, and 2) modulation of mitotic cyclin expression dramatically affects filamentous differentiation. Cells deficient in mitotic cyclins are activated for filamentous growth, whereas cells overexpressing mitotic cyclins cannot form filamentous colonies, even in the presence of an inducing signal. Epistasis analysis places the activity of the cyclins downstream of the RAS/STE signaling pathway. Our results establish a pathway for filamentous differentiation in which the cell cycle engine is a critical target of the RAS/STE pathway involved in promoting the elongated cell shape, polarized cell division, and invasive growth characteristic of the differentiated state.
MATERIALS AND METHODS
Yeast Media
Media were prepared as previously described (Kron et al., 1994) with yeast extract, peptone, and yeast nitrogen base from United States Biochemical (Cleveland, OH). Other reagents were obtained from Fisher Scientific (Pittsburgh, PA) and Sigma (St. Louis, MO). All low-nitrogen media contained 50 μM ammonium sulfate and 6.8 g/l yeast nitrogen base without amino acids or ammonium sulfate and were filter sterilized through a 0.45-μm filter. Synthetic low-ammonia dextrose (SLAD) medium also contained 2% dextrose. Agar (bacteriological, United States Biochemical) was prepared by autoclaving at 4% wt/vol in deionized water and diluting to 2% final concentration with 2× liquid media. Low-ammonia agar media were made using agar washed four times in deionized water before sterilization.
Yeast Strains and Plasmids
All yeast strains were derived in the Σ1278b background (Grenson et al., 1966; Gimeno et al., 1992) using conventional genetic methods. Marker segregation, PCR assay, and/or Southern blotting determined genotypes. Yeast strains are described in Table 1. Transformations were performed by a lithium acetate procedure based on that of Schiestl and Gietz (1989). Plasmids are described in Table 2.
Table 1.
Yeast strain list
| Strain | Genotype | Source |
|---|---|---|
| L5683 | MATα ura3–52 | G.R. Fink |
| SKY754 | MATa ura3–52 | This work |
| SKY757 | MATa/MATα ura3–52/ura3–52 | This work |
| L5791 | MATa/MATα ura3–52/ura3–52 leu2::hisg/leu2::hisg | G.R. Fink |
| L5624 | MATa/MATα ura3–52/ura3–52 trp1::hisg/trp1::hisg ste20::TRP1/ste20::TRP1 | G.R. Fink |
| L5535 | MATa/MATα ura3–52/ura3–52 leu2::hisg/leu2::hisg ste7::LEU2/ste7::LEU2 | G.R. Fink |
| L5540 | MATa/MATα ura3–52/ura3–52 leu2::hisg/leu2::hisg ste11::LEU2/ste11::LEU2 | G.R. Fink |
| L5537 | MATa/MATα ura3–52/ura3–52 leu2::hisg/leu2::hisg ste12::LEU2/ste12::LEU2 | G.R. Fink |
| SKY2183 | MATa/MATα ura3–52/ura3–52 leu2::hisg/leu2::hisg STE11::STE11–4::LEU2/STE11 | This work |
| SKY1056 | MATa/MATα ura3–52/ura3–52 CDC28–T18A, Y19F/CDC28–T18A, Y19F | This work |
| SKY1057 | MATa/MATα ura3–52/ura3–52 leu2::hisg/leu2::hisg mih1::LEU2/mih1::LEU2 | This work |
| SKY1058 | MATa/MATα ura3–52/ura3–52 leu2::hisg/leu2::hisg swe1::LEU2/swe1::LEU2 | This work |
| SKY776 | MATa/MATα ura3–52/ura3–52 trp1::hisg/trp1::hisg clb1::TRP1/clb1::TRP1 | This work |
| SKY778 | MATa/MATα ura3–52/ura3–52 leu2::hisg/leu2::hisg clb2::LEU2/clb2::LEU2 | This work |
| SKY2187 | MATa/MATα ura3–52/ura3–52 cdc28–1N/cdc28–1N | This work |
| SKY2184 | MATa/MATα ura3–52/ura3–52 leu2::hisg::GAL1–CLB1::LEU2/leu2::hisg | This work |
| SKY2185 | MATa/MATα ura3–52/ura3–52 leu2::hisg::GAL1–CLB2::LEU2/leu2::hisg | This work |
| SKY2186 | MATa/MATα ura3–52/ura3–52 leu2::hisg/leu2::hisg ste7::LEU2/ste7::LEU2 clb2::LEU2/clb2::LEU2 | This work |
| SKY2188 | MATa/MATα ura3–52/ura3–52 trp1::hisg/trp1::hisg leu2::hisg/leu2::hisg ste20::TRP1/ste20::TRP1 | This work |
| clb2::LEU2/clb2::LEU2 | ||
| SKY2189 | MATa/MATα ura3–52/ura3–52 leu2::hisg/leu2::hisg ste12::LEU2/ste12::LEU2 clb2::LEU2/clb2::LEU2 | This work |
| SKY2225 | MATa/MATα ura3–52/ura3–52 leu2::hisg/leu2::hisg STE11::STE11–4::LEU2/STE11::STE11–4 | This work |
| CDC28–T18A, Y19F/CDC28–T18A, Y19F | ||
| SKY2226 | MATa/MATα ura3–52/ura3–52 leu2::hisg/leu2::hisg STE11::STE11–4::LEU2/STE11::STE11–4 | This work |
All strains are Σ1287b (Grenson et al., 1966) derivatives.
Table 2.
Plasmid list
| Plasmid | Markers | Source |
|---|---|---|
| pIL30 | CEN URA3 Ty1::TDH3::lacZ | Laloux et al. (1994) |
| pSL1509 | YCp50 CEN URA3 STE11–4 | Stevenson et al. (1992) |
| SKB4166 | pRS305 LEU2 STE11–4 | This work |
| p3303 | plac22 CEN TRP1 CDC28–A18F19 | Amon et al. (1992) |
| SKB4167 | pRS316 CEN URA3 CDC28–A18F19 | This work |
| SKB4168 | pRS413 CEN HIS3 cdc28–1N | This work |
| SKB4169 | pRS306 URA3 cdc28–1N | This work |
| B2255 | Ycp50 CEN URA3 RAS2–Val19 | Fink, personal communication |
| B2553 | 2 μm URA3 STE12 | Fink, personal communication |
| YIpG2::CLB1 | GAL1::CLB1 LEU2 | Stueland et al. (1993) |
| YIpG2::CLB2 | GAL1::CLB2 LEU2 | Stueland et al. (1993) |
| pSWE1–15 | 2 μm LEU2 SWE1 | Booher et al. (1993) |
| pWAS5–1 | 2 μm LEU2 MIH1 | Booher, personal communication |
| pRS305 | LEU2 | Sikorski and Hieter (1989) |
| pRS413 | CEN HIS3 | Sikorski and Hieter (1989) |
To derive strains that stably express STE11-4 from the STE11 chromosomal locus, a 3.6-kilobase XbaI fragment from pSL1509 (yCP50-STE11-4) (Stevenson et al., 1992) was subcloned into pRS30 (Sikorski and Hieter, 1989) to construct SKB4166, an integrating plasmid carrying the STE11-4 gene and a LEU2 marker. A diploid LEU− strain, L5791, was transformed with SKB4166 guided to one STE11 locus by digestion at a unique NcoI site in the STE11 ORF. We chose a transformant, SKY2183, that exhibited slow growth in rich media and striking filamentous growth on SLAD plus 1 mM uracil. When transformed with the Ty1-lacZ reporter pIL30 (Laloux et al., 1994; Mosch et al., 1996), a high constitutive level of expression was observed. Sporulation of SKY2183 yielded 2:0 LEU+ segregants that when crossed to wild-type haploids dominantly conferred enhanced filamentous growth to the resulting diploids.
A URA3-marked centromeric plasmid carrying the dominant CDC28-T18A, Y19F allele, SKB4167, was constructed by subcloning a 2.3-kilobase XhoI/PvuII fragment from p3303 (Ycplac22 CDC28-T18A, Y19F) (Amon et al., 1992) into XhoI/SmaI-digested pRS316 (Sikorski and Hieter, 1989). To construct a double mutant diploid carrying both STE11-4 and CDC28-T18A, Y19F alleles on both pairs of chromosomes, SKY2183 and SKY1056 were sporulated and dissected. Segregants were then mated and the heterozygous diploid was then sporulated and dissected to yield double mutant STE11-4, CDC28-T18A, Y19F haploid segregants. These were then mated to each other to create the STE11-4/STE11-4, CDC28-T18A, Y19F/CDC28-T18A, Y19F diploid strain SKY2225. SKY2226 (STE11-4/STE11-4) was formed from LEU+ segregants of SKY2183.
The cdc28-1N allele of K406 (Surana et al., 1991) was amplified by high-fidelity PCR and the XhoI/MunI fragment was subcloned into pRS413 (Sikorski and Hieter, 1989) to make plasmid SKB4168. The complete ORF of this clone was sequenced and the mutation confirmed. A URA3-marked integrating plasmid carrying the cdc28-1N allele, SKB4169, was constructed by subcloning a 1.3-kilobase BamHI/XhoI fragment from SKB4168 into pRS306 (Sikorski and Hieter, 1989). To create a pair of haploid strains carrying only the cdc28-1N allele, SKB4169 was digested with HindIII and transformed into SKY754 and L5683 to integrate at the CDC28 locus. Transformants were selected on synthetic media lacking uracil, colony purified, and grown nonselectively for several generations at 22°C. Clones that had evicted the URA3 plasmid, leaving behind the cdc28-1N allele, were selected on media containing 1 mg/ml 5-fluoro-orotic acid and then screened for lack of growth at 36°C. A pair of cells of opposite mating type were then mated to form a ts diploid, SKY2187, with the genotype cdc28-1N/cdc28-1N.
Strains overexpressing Clb1 and Clb2 were constructed by transforming L5791 with the plasmids YIpG2::CLB1 and YIpG2::CLB2 (Stueland et al., 1993), respectively, each digested with BstEII to direct integration to the LEU2 locus.
Microscopy Methods
Microcolonies were imaged through the agar and plastic Petri dish with a Zeiss (Thornwood, NY) Axiovert 25 with bright-field illumination and a 32× LD Achroplan objective. Cells from suspension cultures were spread gently onto agar plates and imaged similarly. Pixera VCS image-acquisition software and a Pixera charge-coupled device camera were used to capture images at 1280 ×1024 resolution. Images were converted to gray scale, filtered to remove noise and enhance contrast, cropped, and assembled in Photoshop (Adobe, Mountain View, CA).
Staining of fixed cells with DAPI was carried out essentially as described (Pringle et al., 1991). DAPI-stained experimental and control cells were placed on slides and imaged on a Zeiss Axioskop by phase and epifluorescence microscopy using 365-nm excitation and blue emission filters, a 40× Fluar oil-immersion objective, and 16× eyepieces. The cell shapes and the positions and forms of nuclei in each budded cell were determined. Each cell was classified into groups by apparent bud size/mother cell size ratio and into one of five nuclear morphology classes of the budding cycle: 1) central nucleus, S phase; 2) nucleus at neck in mother cell, S phase/G2; 3) nucleus distributed across neck, G2/metaphase; 4) nucleus stretched along mitotic spindle, anaphase; or 5) separate nuclear masses but no septum formed, telophase. Both as a control for staining and to evaluate nuclear phenotypes, propidium iodide–stained cells used for flow cytometry were examined by epifluorescence microscopy as above but with 546-nm excitation and orange-red emission filters.
Fixed cells grown on rich media were stained with rhodamine or fluorescein phalloidin (Molecular Probes, Eugene, OR) and DAPI as previously described (Pringle et al., 1991) to evaluate nuclear position and reveal altered actin distribution patterns during the cell cycle. Cells from liquid cultures in rich media were prepared for immunofluorescence microscopy and stained with YOL 1/35 anti-tubulin antibody (Kilmartin et al., 1982) (Accurate Chemical and Scientific) and Cy3 goat anti-rat immunoglobulin G polyclonal secondary antibody (Jackson Immunoresearch) and counterstained with DAPI to stain nuclear and mitochondrial DNA essentially as described (Pringle et al., 1991), except that all steps were performed with cells in suspension and gentle centrifugation was used to exchange solutions. Cells were imaged with 546-nm excitation and orange-red emission filters to evaluate tubulin, 365-nm excitation and blue emission filters for DNA, and Nomarski differential interference contrast optics for cell morphology. Calcofluor staining (Pringle, 1991) was performed on exponentially growing cells. Samples were fixed in 3.7% formaldehyde at 25°C for 30 min. Aliquots of ∼107 cells were pelleted, resuspended in 1 ml of 0.1 μg/ml Calcofluor staining solution, and incubated for 5 min. The cells were washed three times with 1 ml of water and resuspended to a final volume of 50 μl. Cells were examined to determine bud scar patterns using 365-nm excitation and blue emission filters and Nomarski differential interference contrast optics. Images were recorded using epifluorescence illumination on a Zeiss Axioskop using a 63× PlanApochromat objective, a Sensys 1600 charge-coupled device camera, and IPLab image-acquisition software.
Percentage Elongation on Agar
Yeast strains (SKY757, wild type, and L5535, ste7/ste7) were grown overnight to saturation at 30°C in 5 ml of synthetic minimal medium. Cells were washed, diluted to 1000 cells/ml, and spread onto plates of SLAD with uracil. Analysis of cell elongation was performed by microscopic examination using the Zeiss Axiovert 25 and a 32× LD Achroplan objective to image cells through the agar and plastic Petri dish. Scoring was performed by visual estimation of the length-to-width ratios of at least 400 cells. For 6 h, cells were counted in three categories: unbudded, round buds (bud length ≤ twofold bud width), and elongated buds (bud length ≥ twofold bud width).
Flow Cytometry
Cells from log-phase cultures were fixed and stained with propidium iodide to prepare samples for flow cytometry as described (Hutter and Eipel, 1978). To disrupt cell aggregates, samples were sonicated until judged well dispersed by phase microscopy. Staining was evaluated by epifluorescence imaging using 546-nm excitation and orange emission filters. Samples were analyzed on a FacsScan (Becton-Dickinson, Franklin Lakes, NJ) flow cytometer, and 60,000 events were collected on each sample. The plots presented here represent the gated fluorescence area data. The gated data were analyzed to determine relative DNA content.
Whole Cell Extracts and Protein Analysis
Whole cell extracts were obtained by a method based on that of Amon (1992). Strains were grown overnight to saturation in YPD rich liquid media, diluted 1:50, and regrown to a density of ∼107 cells/ml in YPD at 30°C. Cells were chilled, collected by centrifugation, washed in 10 mM Tris-HCl (pH 8.0), and resuspended in 200 μl of buffer A (50 mM Tris-HCl, pH 7.5, 1 mM EDTA, 5 mM DTT, 1 mM PMSF, 0.2 mM l-1-chloro-3-[4-tosylamido]-4-phenyl-2-butanone, 0.025 mM l-1-chloro-3-[4-tosylamido]-7-amino-2-heptanone-HCl, 2 mg/ml pepstatin in DMSO). Approximately 0.2 ml of 0.2-mm-diameter acid-washed glass beads was added, cells were vortexed for 5 min, and the extract was cleared by a 5-min centrifugation at 16,000 × g at 4°C. The supernatant was collected and stored at −80°C. Protein concentrations were determined by the Bradford protein assay (Bio-Rad, Richmond, CA) using BSA as a standard.
For Western analysis, protein samples were solubilized with SDS/DTT sample buffer, boiled, and loaded onto 12% bis:acrylamide gels (1:19) with 50 μg of total yeast protein per lane. Samples were electrophoretically separated and transferred to nylon-supported nitrocellulose (Magna). Primary antibodies were, for Cdc28, a mouse anti-PSTAIRE mAb (Calbiochem, La Jolla, CA), and for Clb2, a rabbit polyclonal antibody raised against a TrpE-Clb2 fusion protein (Amon et al., 1994) and affinity purified using the antigen. Immunoreactivity was detected with 125I-protein A (ICN, Costa Mesa, CA), and blots were scanned using a phosphorimager (Molecular Dynamics, Sunnyvale, CA) and analyzed using Imagequant and IPLab (Scanalytics). For analysis of expression from the pIL30 (Laloux et al., 1994) Ty1::lacZ reporter, extracts were obtained, assayed for total β-galactosidase activity, and normalized to total protein as described (Alfa et al., 1993).
RESULTS
Activation of the STE MAPK Signal for Filamentous Growth Induces a Delay in G2/M
Wild-type, prototrophic strains constructed in the Σ1278b background exhibit filamentous development when subjected to low-nitrogen stress. When such cells are transferred to nitrogen starvation media (SLAD agar), many cells show a striking change in bud morphology within their first budding cycle, immediately adopting a filamentous growth pattern. In the first cell cycle under nitrogen starvation conditions, 91% of budded wild-type cells (n = 400) formed an elongated bud when examined 4 h after inoculation onto SLAD media. This phenotype is reminiscent of the arrest of cells carrying the cdc28-1N mutation (Surana et al., 1991) overexpressing the Swe1 tyrosine kinase (Booher et al., 1993) or overexpressing a G1 cyclin (Lew and Reed, 1993), each of which blocks cell cycle progression at the metaphase-to-anaphase transition by antagonizing Clb1- and Clb2-dependent Cdc28 complexes. When cells lacking elements of the RAS/STE filamentous development pathway are inoculated onto SLAD, the proportion of elongated buds formed is significantly decreased. Only 10% (n = 400) of cells lacking the STE7 MEK kinase, and thereby defective in RAS/STE signaling, formed elongated buds in their first cell cycles. Rather, most buds emerging from Ste7-deficient mother cells formed small ovoid shapes and soon detached from the mother cell (Figure 1A). This pattern is typical of yeast cell growth on YPD agar media (rich media containing yeast extract, peptone, and dextrose). Nonetheless, the rate of bud emergence in the RAS/STE-signaling mutant was indistinguishable from that in the wild type (wild type, 78% budded at 4 h; ste7/ste7, 76% budded at 4 h). A simple inference is that the RAS/STE pathway may antagonize processes that are limiting for completion of mitosis (e.g., accumulation of Clb1,2/Cdc28 complexes) but has little effect on factors limiting Start (e.g., accumulation of Cln1,2/Cdc28 complexes).
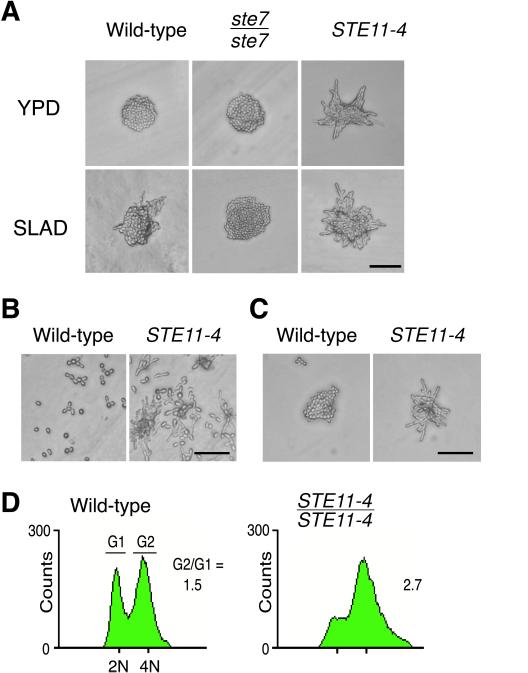
Figure 1.
Ectopic activation of the RAS/STE pathway via the STE11-4 mutation. (A) The dominant active STE11-4 mutation promotes enhanced filamentous differentiation. Microcolonies formed by the indicated strains (STE11-4 is the STE11:: STE11-4:: LEU2/STE11 strain SKY2183) on rich YPD media and low-nitrogen SLAD media reveal that the requirement for nutrient starvation is bypassed by activation of RAS/STE signaling. A cell deficient for STE7 (ste7/ste7, L5535), an element of the RAS/STE pathway, does not exhibit a filamentous phenotype on YPD or SLAD media. Photographs show representative colonies of each strain after 24 h at 22°C. (B) Diploid cell agar invasion assay. Strains were inoculated on YPD agar and grown for 72 h at 30°C. Photographs were taken after the nonadherent cells were washed away with water. The STE11-4 mutant is hyperinvasive even in the rich media. (C) Cell morphology of mutant cultured in liquid YPD nitrogen-rich media. The STE11-4 mutant exhibits highly elongated cells that grow as adherent cell aggregates reminiscent of filamentous colonies. This phenotype suggests that the agar signal requirement is also bypassed by ectopic RAS/STE signals. Bars, 50 μm. (D) Cell cycle kinetics of the STE11-4/STE11-4 homozygous diploid (SKY2226). Flow cytometry was performed on asynchronous cultures growing on YPD rich media to determine the distribution of nuclear DNA content. The STE11-4 mutant reveals a predominance of cells with G2/M DNA content, which correlates to a large G2/G1 ratio, reflecting the relative percentages of cells falling in the indicated gating intervals of fluorescence intensity.
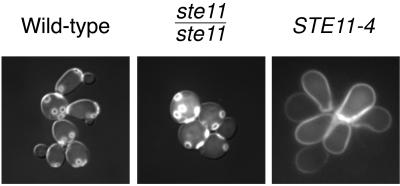
To investigate this phenomenon further, we examined the behavior of cells in which the RAS/STE pathway was ectopically induced. We investigated the effect of the dominant activated STE11-4 mutant allele, T596I (Stevenson et al., 1992), on cell growth in rich media. When introduced as a single copy into diploid cells via integration at the STE11 locus, STE11-4 confers enhanced filamentous growth on SLAD media, as previously reported by Liu et al. (1993) for the plasmid-borne allele (Figure 1A). We examined several markers for filamentous development in these stable STE11-4 cells. When inoculated on YPD agar media, this strain (SKY2183) displayed phenotypes characteristic of nitrogen starvation and activation of the RAS/STE pathway, including striking filamentous colony morphology (Figure 1A) and elevated expression of the pIL30 Ty1-lacZ FRE reporter. After 2 d of growth on YPD agar, the STE11-4 cells invaded the agar surface to a greater degree than a wild-type diploid control (Figure 1B). Even in suspension culture in rich media, the STE11-4 mutant grew as highly elongated cells that remained attached in clusters, forming florets reminiscent of filamentous colonies (Figure 1C). As in bona fide filamentous cells (Kron et al., 1994), STE11-4 cells exhibited apical polar budding. In the cell clusters and in colonies on agar, buds arose preferentially from the apical end of the mother cell. However, when these cells were examined with the chitin stain Calcofluor to confirm bud-site selection pattern, rather than the usual chitin rings at bud scars, a delocalized distribution of stain was apparent (Figure 2), perhaps consistent with a loss of temporal and/or spatial restriction of chitin deposition. However, staining of STE11-4 cells with fluorescein phalloidin revealed persistence of an apically polarized distribution of actin cortical patches in large budded cells, as observed in wild-type cells responding to nitrogen starvation (Kron et al., 1994; Cali et al., 1998). Thus, via ectopic activation of the RAS/STE pathway, many features of the filamentous cell phenotype can be dissociated from nitrogen starvation.
Figure 2.
Fluorescence imaging of Calcofluor staining of chitin distribution. The STE11-4 mutant shows a delocalized distribution of chitin rather than the expected chitin rings that mark bud scars, as is seen in the wild type and the ste11/ste11 mutant.
Flow cytometry of a STE11-4/STE11-4 homozygous diploid (SKY2226) growing in rich media demonstrated a predominance of cells with G2/M DNA content (Figure 1D). DAPI staining to determine the distribution of nuclear DNA suggested that most cells with large buds do not have separated nuclear masses or an elongated spindle (Figure 3). These data were confirmed by examination of spindles by anti-tubulin immunofluorescence microscopy. These assays indicated that the STE11-4 cells do not have any increase in content of anaphase cells, despite their long budded period and G2/M shift. From these data, we infer that the predominant cell type in STE11-4 cells grown on rich media is a cell carrying an elongated bud that has completed DNA synthesis but has yet to leave G2/metaphase, much like filamentous cells cultured on nitrogen starvation media.
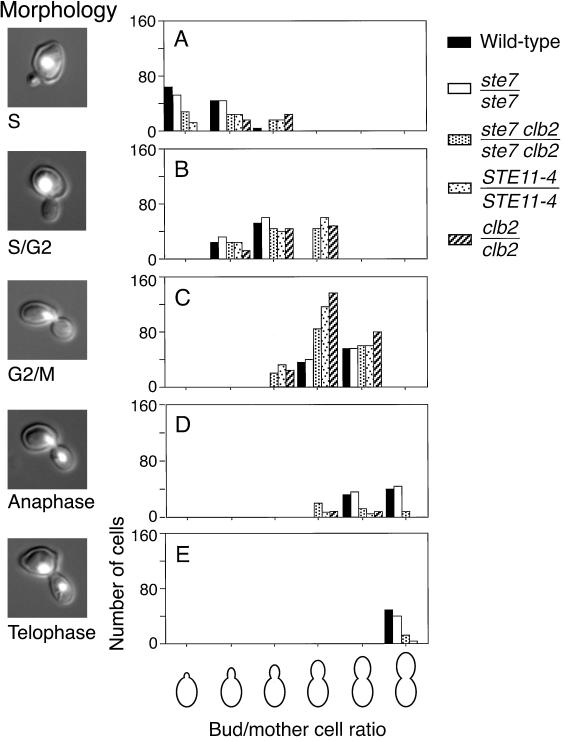
Figure 3.
Distribution of nuclear DNA through the budding cycle. Cells were grown on YPD rich media and fixed, and nuclear DNA was stained with DAPI as described in MATERIALS AND METHODS. The cell shapes and the positions and forms of nuclei in 400 budded cells were determined. Each cell was scored individually and classified into groups by apparent bud size/mother cell size ratio and into nuclear morphology classes corresponding to stages in the budding cycle. The histograms plot the number of cells that fall in one morphology class, divided into six groups by their bud size/mother cell size ratio. The five morphology classes plotted are: central nucleus, S phase (A); nucleus at neck in mother cell, S phase/G2 (B); nucleus distributed across neck, G2/metaphase (C); nucleus stretched along mitotic spindle, anaphase (D); and separate nuclear masses but no septum formed, telophase (E). Representative cells are shown to the left of each histogram. These data show that mitotic progression correlates well with bud growth in wild-type cells. By contrast, STE11-4 and clb2/clb2 mutants display an overabundance of G2/metaphase nuclei throughout the budded period and a decreased proportion of anaphase and telophase morphologies. This phenotype is suggestive of a postreplication delay analogous to that of filamentous cells cultured on nitrogen starvation media. The ste7/ste7 clb2/clb2 mutant reveals a slight increase in the proportion of cells in anaphase compared with clb2/clb2 but still significantly fewer compared with the wild-type control or the ste7/ste7 mutant.
These data are consistent with a mechanism by which the RAS/STE pathway negatively regulates the onset of mitotic anaphase, independent of any direct effects of nitrogen starvation. A reasonable target is the abundance or activity of mitosis-promoting factor (MPF), the mitotic form of CDK. We asked whether yeast mitotic cyclins Clb1 and Clb2 and/or the CDK Cdc28 might be regulated by RAS/STE signals.
STE MAPK Signaling Can Promote Filamentous Growth in the Absence of Cdc28 Tyrosine Phosphorylation
Mitotic onset in most eukaryotes is controlled by the reversible inhibitory phosphorylation of a conserved tyrosine and threonine adjacent to the active site in CDKs (Feilotter et al., 1992). Phosphorylation sequesters preformed CDK/cyclin complexes as preMPF and delays anaphase. Our previous studies showed that cells carrying the constitutively active CDC28-T18A, Y19F (AF) mutant allele in which the Thr and Tyr residues are substituted with Ala and Phe still form filamentous colonies when inoculated onto low-nitrogen agar, although they are attenuated in their development. This result does not eliminate a pathway whereby the RAS/STE signal for filamentous differentiation promotes phosphorylation of this tyrosine.
We reexamined the behavior of mutants with altered Cdc28 tyrosine phosphorylation and/or altered RAS/STE function exposed to low-nitrogen agar media. We found that cells in which tyrosine phosphorylation is decreased (CDC28-T18A, Y19F/CDC28-T18A, Y19F, swe1/swe1, [2 μm MIH1]) are impaired in filamentous development and yield colonies on SLAD media in which cells at the colony periphery lack the degree of cell elongation observed in wild-type strains (data not shown). On the other hand, mutants with constitutively increased tyrosine phosphorylation (mih1/mih1, [2 μm SWE1]) are moderately enhanced in filamentous growth and produce markedly elongated cells at the colony periphery (data not shown). We confirmed published results that cells lacking certain elements of the RAS/STE pathway (ste7/ste7, ste12/ste12 [see Figure 8A], or ste11/ste11 [data not shown]) display markedly reduced filamentous development and cells carrying mutations that increase RAS/STE signaling ([CEN RAS2-Val19], [CEN STE11-4], [2 μm STE12] [data not shown]) display dramatically enhanced filamentous growth.
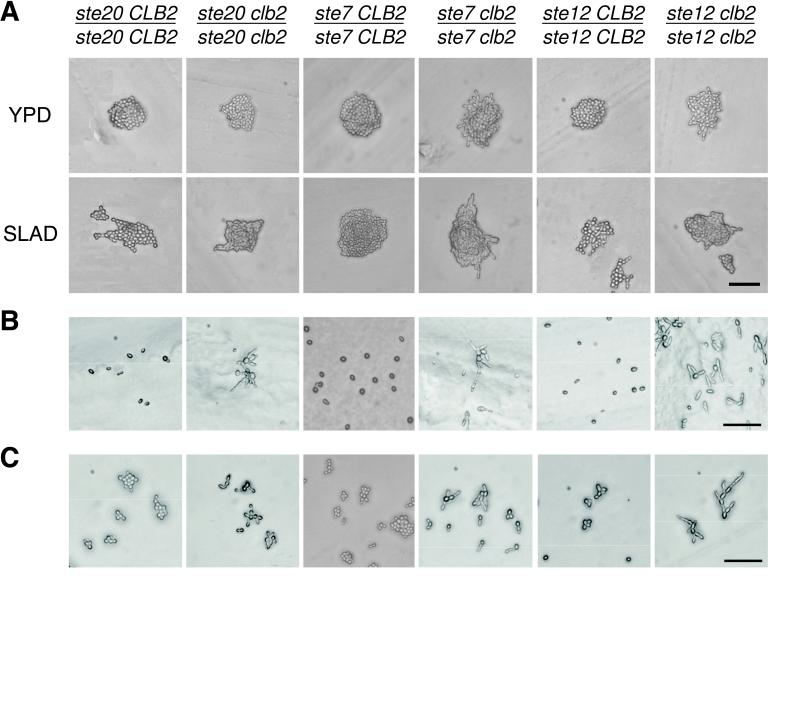
Figure 8.
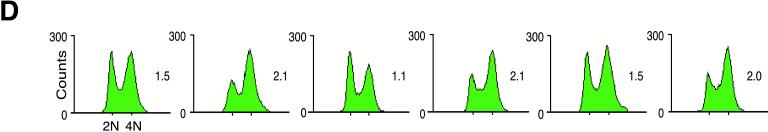
Bypass of mutations in the RAS/STE pathway by Clb2 deficiency. (A) STE20-, STE7-, and STE12-deficient mutants have a decreased filamentous development phenotype. Cells lacking both the Ste20 PAK kinase and the Clb2 mitotic cyclin show only a very subtle suppression, whereas double mutants lacking Ste20 and Clb2, Ste12 and Clb2, or Ste7 and Clb2 exhibit limited filamentous growth on both rich YPD media and low-nitrogen SLAD media. These phenotypes suggest partial bypass of the RAS/STE pathway. Photographs show representative colonies of each strain after 24 h at 22°C. (B) Agar invasion assay. Double mutants lacking both Clb2 and components of the RAS/STE pathway exhibit partial invasion, but ste7/ste7, ste12/ste12, and ste20/ste20 mutants confer decreased agar invasion. (C) Cell morphology in liquid YPD media. ste7/ste7 clb2/clb2, ste12/ste12 clb2/clb2, and ste20/ste20 clb2/clb2 double mutants display limited cell elongation, but to a greater degree than ste7/ste7, ste12/ste12, and ste20/ste20 mutants. Bars, 50 μm. (D) Cell cycle kinetics. A shift toward G2/M DNA content is apparent in cells deficient in both Clb2 mitotic cyclin and elements of the RAS/STE pathway.
One model consistent with these data, as suggested by Edgington et al. (1999), is that inhibition of anaphase by the RAS/STE pathway is mediated by hyperphosphorylation of Thr-18 and/or Tyr-19 of the Cdc28 CDK, affected by activation of the Swe1 kinase or inhibition of the Mih1 phosphatase. Alternatively, the RAS/STE pathway and Cdc28 tyrosine phosphorylation may contribute independently to differentiation. In support of the latter possibility, we observed that ectopic activation of the RAS/STE pathway via plasmids containing the STE11-4 or RAS2-Val19 mutations or overexpressing STE12 suppressed both the impaired filamentation and the decreased cell elongation of swe1/swe1 and CDC28-T18A, Y19F mutants (Figure 4A). Finally, the double-mutant diploid carrying CDC28-T18A, Y19F and STE11-4 showed enhanced filamentous growth (Figure 4B). Like the STE11-4 strain, it showed enhanced agar invasion, highly elongated cells that remain attached in clusters, and a G2/M cell cycle shift (Figure 4, C–E). Furthermore, we confirmed our previous observations (Kron et al., 1994) that CDC28-T18A, Y19F is dominant with respect to SWE1. Overexpression of SWE1 via an integrated copy of a fusion between the inducible GAL1,10 promoter and SWE1 (Booher et al., 1993) did not significantly delay cell division or alter filamentous growth in the CDC28-T18A, Y19F mutant. These data suggest that the RAS/STE pathway and Cdc28 tyrosine phosphorylation are independent pathways modulating filamentous growth.
Figure 4.
The RAS/STE pathway and Cdc28 tyrosine phosphorylation independently modulate filamentous growth. (A) Filament formation of Cdc28 tyrosine phosphorylation mutants with altered RAS/STE function on synthetic nitrogen (SC) or nitrogen starvation (SLAD) agar. Mutants with decreased tyrosine phosphorylation lacking the inhibitory Swe1 kinase or its phosphorylation site on Cdc28 (swe1/swe1 and CDC28-T18A, Y19F [AF]) show impaired filamentous development on SC and SLAD media compared with the wild-type strain. On the other hand, lack of the reactivating phosphatase Mih1 (mih1/mih1) confers constitutively increased Cdc28 tyrosine phosphorylation and a moderately enhanced filamentous phenotype. Plasmids expressing the STE11-4 or RAS2-Val19 allele suppress the impaired filamentation of both the swe1/swe1 and CDC28-T18A, Y19F strains and further enhance the filamentous phenotype of mih1/mih1 strains. (B) STE11-4/STE11-4 CDC28-AF/CDC28-AF double mutants on YPD or SLAD agar. The nonphosphorylatable CDK mutant CDC28-T18A, Y19F does not block an ectopic RAS/STE signal. (C) Agar invasion assay. Cells were treated as described in Figure 1B. Like the STE11-4 mutant, the double mutant is hyperinvasive in YPD agar. (D) Cell morphology in liquid YPD media. Highly elongated cells that grow as adherent cell aggregates reminiscent of the STE11-4 florets are apparent in STE11-4/STE11-4 CDC28-AF/CDC28-AF double mutant cultures. Bars, 50 μm. (E) Flow cytometry. Cells were prepared as described in MATERIALS AND METHODS. The STE11--4/STE11-4 CDC28-AF/CDC28-AF double mutant reveals a marked shift to 4N DNA content relative to the CDC28-AF/CDC28-AF control and comparable to that of the STE11-4/STE11-4 mutant alone.
Altered Cyclin Expression Modulates Filamentous Growth
Although control of mitotic anaphase by the RAS/STE pathway may not involve regulation at the level of tyrosine phosphorylation, other modes of regulation of MPF cannot be eliminated. One limiting factor for completion of mitosis is the accumulation of the functionally redundant mitotic cyclins Clb1 and Clb2. We examined whether altering the activity of mitotic kinase by changing the expression of mitotic cyclins might have a significant effect on filamentous development.
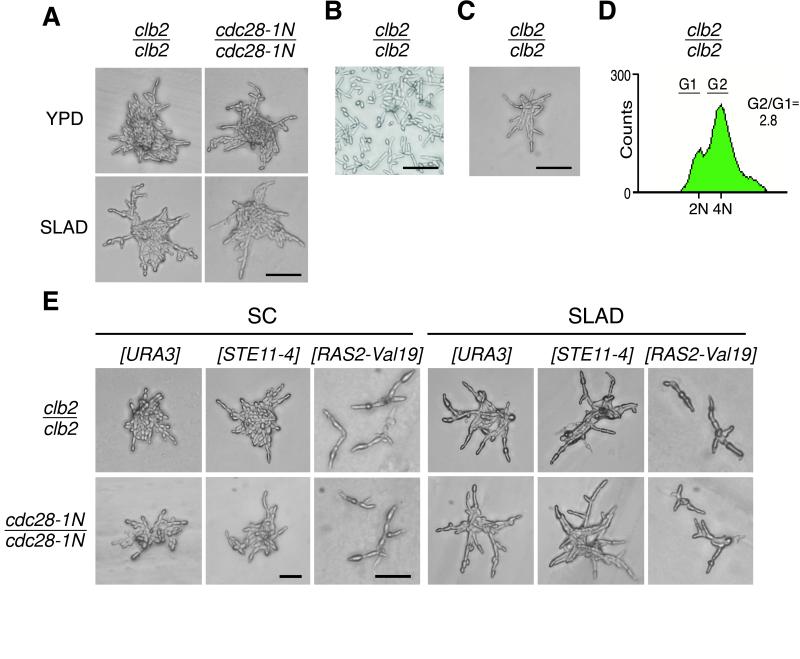
Previous studies have shown that deletion or moderate overexpression of either CLB1 or CLB2 has only subtle effects on cell growth on rich media (but see Stueland et al., 1993). To test whether altered mitotic cyclin expression might differentially affect filamentous development, we constructed diploid cells deficient for either the CLB1 or CLB2 gene. In contrast to other genetic backgrounds, diploid Σ1278b cells carrying a homozygous deletion of CLB2 exhibited several characteristics of filamentous development even when grown on rich complete media. When inoculated on YPD agar, clb2/clb2 cells initially formed microcolonies consisting of mostly filamentous cells (Figure 5A). When patches of Clb2-deficient cells were inoculated on YPD agar and allowed to grow for several days, they displayed increased cell adhesion (rough surface and granular consistency) and enhanced agar invasion (Figure 5B) comparable to that displayed by the STE11-4 strain.
Figure 5.
Mutations in mitotic control genes modulate filamentous differentiation. The clb2/clb2 mutant exhibits exaggerated filamentous development and a growth pattern similar to that of the STE11-4 mutant. (A) Both the strain carrying the ts mitotic arrest allele cdc28-1N and the strain deficient in CLB2 exhibit filamentous growth on YPD media and are further induced by nitrogen starvation. Photographs show representative colonies of each strain after 24 h at 22°C. (B) Agar invasion assay. Cells lacking CLB2 have enhanced YPD agar invasion comparable to that displayed by the STE11-4 mutant. (C) Cell morphology in liquid YPD media. The clb2/clb2 strain grows as highly elongated cells that remain attached in clusters, reminiscent of the STE11-4 mutant. Bars, 50 μm. (D) Cell cycle kinetics. Like STE11-4 cells, CLB2-deficient cells demonstrate a predominance of cells with G2/M DNA content. (E) Deficiency in mitotic Cdc28 activity conferred by deficiency in the Clb2 mitotic cyclin dramatically enhances filamentous growth on both synthetic nitrogen (SC) and nitrogen starvation (SLAD) media. Activation of the RAS/STE pathway via plasmids containing STE11-4 or RAS2-Val19 augments the striking cell elongation phenotype of these strains. Bars, 50 μm.
In liquid YPD media, the clb2/clb2 mutant cells were heterogeneous, with an unusual number of elongated cells that grew as adherent cell aggregates reminiscent of the STE11-4 florets (Figure 5C). Flow cytometry revealed a marked shift to cells with 4N DNA content (Figure 5D). When inoculated onto SLAD, the clb2/clb2 mutant exhibited remarkable filamentous growth characterized by highly elongated cells and very few rounded cells in the colonies (Figure 5A), a growth pattern similar to that of the STE11-4 mutant.
Cells lacking Clb2 are specifically sensitized to the RAS/STE signal. When [CEN RAS2-Val19] or [CEN STE11-4] was introduced into the clb2/clb2 mutant cells, a compound effect of dramatic cell elongation and striking filamentation was observed on SLAD media (Figure 5E). Further supporting the hypothesis that diploid cells lacking Clb2 are sensitized to the filamentous growth-inducing signal, we observed that when this mutant is grown in the presence of high-ammonia-containing synthetic media, the cells adopt a rounded shape like that of a wild-type control strain. This suggests that 1) the elongated phenotype on YPD media, a nitrogen source relatively low in ammonia but rich in short peptides and other complex forms of nitrogen, may be due to low but significant basal activity of the signaling pathway, and 2) the presence of ammonia nitrogen might actively repress the signaling pathway, rather than starvation for ammonia nitrogen inducing its function.
Using the same logic that prompted examination of the Clb2 mutant, we tested whether a strain lacking the Clb1 cyclin might also demonstrate induced filamentous growth. A diploid Clb1 mutant constructed by deletion of both CLB1 alleles showed few or no rich media phenotypes and less cell cycle shift but a moderate induction of filamentous growth on SLAD. This induction was further increased by transformation with the [CEN RAS2-Val19] or [CEN STE11-4] inducer.
To confirm that the effects of the CLB1 and CLB2 deletions were mediated via reduced activation of the Cdc28 CDK, we examined the effect of substituting the temperature-sensitive G2 arrest allele, cdc28-1N, for wild-type CDC28 in an otherwise wild-type strain. Like the Clb2-deficient strain, when grown at a permissive temperature the cdc28-1N/cdc28-1N strain exhibited significant cell elongation on YPD media and dramatic filamentous growth on SLAD low-nitrogen media (Figure 5A). The [CEN STE11-4]– and [CEN RAS2-Val19]–inducing plasmids further enhanced this filamentous phenotype (Figure 5E).
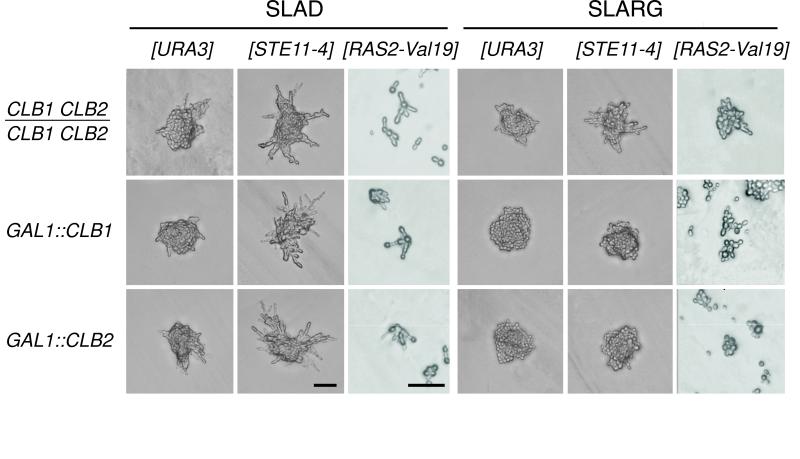
To study the effect of overexpression of mitotic cyclins, we constructed strains carrying integrated copies of fusions between the inducible GAL1,10 promoter and the CLB1 and CLB2 cyclin genes. Although diploid cells carrying an integrated GAL1::CLB2 construct exhibited normal filamentous growth on repressing SLAD agar media, induction of Clb2 overexpression by inoculating cells onto synthetic low-ammonia galactose agar media abrogated filamentous growth. Rather than forming the chains of elongated cells characteristic of filamentous growth, these cells formed piles of rounded cells (Figure 6). Diminution of filamentous growth was also observed in cells overexpressing the Clb1 cyclin, whereas overexpression of the Clb3, Clb4, or Clb5 cyclin had only modest effects on PH growth. Finally, overexpression of Clb1 and Clb2 suppressed the filamentous phenotype of cdc28-1N cells (data not shown).
Figure 6.
Overexpression of the Clb1 and Clb2 mitotic cyclins abrogates filamentous development and antagonizes RAS/STE signaling. Strains containing integrated copies of the CLB1 and CLB2 genes under an inducible GAL1,10 promoter and a control plasmid or the STE11-4 or RAS2-Val19 allele were grown on nitrogen starvation (SLAD) or synthetic low-ammonia galactose (SLARG) media for 24 h. Both basal and enhanced filamentous growth are abolished by Clb1 or Clb2 overexpression on SLARG media, indicating that the RAS/STE pathway may function upstream of Clb1 and Clb2. Representative colonies of each strain were photographed after 24 h at 22°C. Bars, 50 μm.
These data suggest that the mitotic cyclin Clb2 performs a function in direct antagonism to the RAS/STE pathway. However, although mitotic cyclins may be negatively regulated at some level by the RAS/STE pathway, these results do not indicate that either Clb1 or Clb2 is the sole target of the RAS/STE pathway in the control of anaphase.
One biochemical mechanism consistent with the genetic results described above is that the cell cycle shift observed upon ectopic activation of the RAS/STE pathway, as in the STE11-4 mutant, might be due to down-regulation of the Clb2 cyclin. A lower cellular content of Clb2 would result in a proportional decrease in Clb2-dependent Cdc28 kinase and thereby slow mitotic entry. Furthermore, low cellular Clb2 content would predict the observed similarity in phenotypes between STE11-4–expressing cells and Clb2-deficient cells. Protein extracts were prepared from asynchronous cultures of cells grown on rich YPD media. When we compared levels of Clb2 protein in extracts derived from the wild-type, Ste11-deficient, Clb2-deficient, and STE11-4–expressing strains, we observed the expected absence of Clb2 protein from the Clb2-deficient strain. However, the other strains showed equal levels of expression of Clb2 irrespective of genotype (Figure 7), suggesting that the RAS/STE pathway might have little or no effect on Clb2 cyclin expression or stability.
Figure 7.
Modulating RAS/STE signaling regulates filamentous growth without altering Clb2 abundance. The anti-PSTAIRE mAb recognizing Cdc28 was used as a loading control. Western analysis of Clb2 and Cdc28 reveals that Clb2 levels are relatively unaffected by RAS/STE signals. WT, wild type.
Epistasis Experiments Place Clb2 Downstream of the RAS/STE Pathway
Were the RAS/STE pathway genetically upstream of Cdc28 and Clb2, one inference would be that loss of function in Clb2, which confers enhanced filamentous growth, might suppress the decreased filamentous growth observed in mutants lacking components of the RAS/STE pathway. We examined mutants lacking the STE20 MEK kinase kinase, the STE11 MEK kinase, and the STE7 MEK. As previously reported (Liu et al., 1993), ste7/ste7 and ste12/ste12 mutants each confer decreased filamentous development, and ste20/ste20 mutants display complete loss of filamentous growth (Figure 8A). Furthermore, all of these mutants were readily washed off the agar with flowing water (Figure 8B) and failed to grow in suspension culture as adherent cell aggregates (Figure 8C). We constructed mutant strains that combined each of the STE kinase mutations with deletion of the CLB2 cyclin gene and examined their filamentous phenotypes. We found that the ste20/ste20 clb2/clb2, ste7/ste7 clb2/clb2, and ste12/ste12 clb2/clb2 double mutants all exhibited a limited form of filamentous differentiation when placed under inducing conditions of low-nitrogen agar (Figure 8A). All of these mutants showed partial agar invasion (Figure 8B) and an elongated cell phenotype (Figure 8C), even in liquid media. By comparison, the ste20/ste20 clb2/clb2 double mutant showed only a very subtle suppression. The flow cytometry profiles for each double mutant showed a significant shift toward the 4N DNA content (Figure 8D), and we observed a decrease in the population of anaphase cells (Figure 3). Thus, the bypass of the STE pathway blockade effected by a deletion of CLB2 was partial but nonetheless significant.
A complementary test to reveal order of function was to examine whether the induction of filamentous growth by a dominant active allele of a component of the RAS/STE pathway could override the inhibitory effect of Clb1 or Clb2 overexpression. We found that cells carrying the integrated GAL1::CLB1 and GAL1::CLB2 constructs failed to form filaments on low-nitrogen galactose-containing media, even if they also carried a [CEN RAS2-Val19] or [CEN STE11-4] plasmid (Figure 6). These data suggest that the RAS/STE pathway indeed acts upstream of mitotic cyclins as part of the filamentous differentiation response.
DISCUSSION
Physiological study of filamentous growth has suggested that a change in cell cycle control is critical to the differentiation of rounded yeast cells into elongated filamentous cells. We set out to examine whether modulation of the cell cycle machinery might be both necessary and sufficient for proper morphogenesis in filamentous growth. In particular, we asked whether other cell-autonomous features of filamentous differentiation, such as elongated cell morphology, polar budding, persistent cell connections, and agar invasion, might be downstream of the cell cycle switch.
The STE MAPK Pathway Regulates the G2/M Transition in Diploid Yeast Cells
Previous studies have shown that the low-nitrogen signal for filamentous growth is transduced via a dual signaling pathway involving elements of the STE MAPK– and RAS GTPase/cAMP–dependent kinase cascades. A critical question is whether the cell cycle shift to G2/M in filamentous cells is downstream of the RAS/STE-signaling pathway or is an independent response to nitrogen starvation. The results described here establish a pathway linking the MAPK-signaling cascade directly to the cell cycle machinery.
Our logic is based on the following argument. We have used a dominant activated mutant, STE11-4, to produce an ectopic, constitutive signal that stimulates the filamentous development pathway, even in the absence of nitrogen starvation or an agar substrate. Previous studies (Liu et al., 1993) found that the STE11-4 mutation confers enhanced filamentous growth and promotes expression of the filamentous signaling reporter, Ty1-lacZ (Mosch et al., 1996), via its FRE element in a low-nitrogen-independent manner.
We extended these observations by creating diploid yeast strains in which the STE11-4 mutation is stably expressed from its genomic locus. As expected, these strains exhibit enhanced filamentous growth on low-nitrogen media and display high constitutive expression of the Ty1-lacZ reporter. However, we found that such STE11-4 strains express an ectopic filamentous growth phenotype, even in rich liquid media. Because of persistent attachment to the mother cell after septation, they grow as florets that may contain dozens of cells. STE11-4 florets formed in rich liquid media are indistinguishable from microcolonies of wild-type cells growing on the surface of low-nitrogen agar. Thus, STE11-4 is sufficient to replace at least the nitrogen starvation and agar components of the conditions conducive to filamentous growth and to provide a model system that reveals the RAS/STE pathway–specific aspects of filamentous growth.
We examined these STE11-4 cells for markers of cell cycle progression known to be altered during filamentous differentiation. STE11-4 filamentous cells grown on rich liquid media are shifted into G2/M. Most cells were budded, many with a single nucleus lodged in the bud neck and a short mitotic spindle spanning the nucleus. Even though many cells had buds nearly equal in size to the mother cell, very few STE11-4 cells were observed with the elongated mitotic spindle or separated nuclear masses suggestive of anaphase. This finding suggests that a RAS/STE signal alone is sufficient to produce the characteristic cell cycle of filamentous growth.
The RAS/STE-dependent Mitotic Delay Is Not Mediated by Cdc28 Tyr-19 Phosphorylation
A simple model, as suggested by Edgington et al. (1999), might explain the link between signal and cell cycle: the physiological pathway from nitrogen starvation to cell cycle may be via a MAPK cascade that promotes increased Tyr-19 phosphorylation of Cdc28. In fact, many observations are consistent with this model. Ectopic activation of the Swe1 kinase by mutation of an inhibitory kinase (Edgington et al., 1999) or overexpression of the Swe1 kinase or lack of the Mih1 phosphatase, as shown here, promote hyperphosphorylation of Cdc28 Tyr-19, delay mitotic onset, and enhance filamentous growth. Mutation of the Cdc28 Tyr-19 residue to Phe or deficiency of the Swe1 kinase each promote mitosis and, as shown here, attenuate filamentous growth. Nonetheless, we observed that the RAS/STE signal can still induce ectopic filamentous development, even in cells lacking the Swe1 kinase or expressing the dominant active CDC28-T18A, Y19F mutant lacking the site for inhibitory tyrosine phosphorylation. Although our data and those of Edgington et al. (1999) demonstrate that regulation of Cdc28 tyrosine phosphorylation modulates filamentous development, we find that this regulation is neither necessary nor sufficient to confer the mitotic delay or other effects induced by RAS/STE pathway activation. Furthermore, although nitrogen starvation may promote Cdc28 tyrosine phosphorylation and thereby contribute to differentiation, this regulation is not essential for most aspects of filamentous differentiation, including the G2/M cell cycle shift.
Altered Cyclin Expression, Mitotic Cdc28 Activity, and Filamentous Development
Perhaps the most remarkable findings reported here and by Edgington et al. (1999) are that simple genetic manipulations to create strains deficient in or overexpressing a single mitotic cyclin are so altered in their developmental responses. Cells lacking the mitotic cyclin Clb2 are highly induced for filamentous growth. When inoculated onto low-nitrogen agar, Clb2-deficient cells form colonies of almost exclusively filamentous cells. In turn, when inoculated onto rich complex agar media (YPD) or even when grown on rich complex liquid media, Clb2 mutants remain highly elongated, bud in a polar pattern, remain attached to their mother cells, and show a dramatic cell cycle shift to G2/M when assayed by flow cytometry. We find that these phenotypes derive in part from supersensitivity to the low-nitrogen signal. Activation of the RAS/STE pathway, as by the STE11-4 mutation, or expression of RAS2-Val19 exacerbates the Clb2 mutant phenotype on low-nitrogen media.
That these effects are due to decreased activation of the mitotic forms of Cdc28 is indicated by the phenotypes conferred by the cdc28-1N ts mutation at a permissive temperature. At the nonpermissive temperature (37°C), cells carrying only cdc28-1N arrest before mitotic anaphase but with completed DNA synthesis and an assembled mitotic spindle (Surana et al., 1991). Overexpression of mitotic cyclins suppresses this ts arrest. In turn, cells carrying both cdc28-1N and a deficiency for CLB1 or CLB2 are nonviable even at 22°C. This synthetic lethality demonstrates a critical defect in mitotic kinase activity in cdc28-1N mutants even at the permissive temperature. When yeast cells carrying the cdc28-1N mutation were inoculated onto low-nitrogen media and cultured at a permissive temperature, like the Clb2-deficient cells, they were hyperfilamentous. Furthermore, when superstimulated by expression of the STE11-4– or RAS2-Val19–inducing mutations, like the Clb2-deficient cells, the cdc28-1N mutants displayed a dramatically enhanced filamentous phenotype.
As noted above, Myers and colleagues (Edgington et al., 1999) have shown that filamentous growth is modulated by Cdc28 function, but they focus on mitotic functions. They isolated a Cdc28 mutant, cdc28-127 C127Y, that causes constitutive expression of most filamentous growth characteristics, such as enhanced cell polarity, elongated shape, and G2/M cell cycle delay. Like cdc28-1N, cdc28-127 is defective in mitotic progression. Their data confirm our observations that altered Clb1,2/Cdc28 activity can modulate filamentous growth independent of signal transduction. In contrast to the results reported here, their data indicate a significant role for Cdc28 Tyr-19 phosphorylation by Swe1 kinase in filamentous differentiation. However, as cdc28-127 remains subject to regulation by Tyr-19 phosphorylation, their data also suggest that there may be multiple means of regulating CDK activity to promote filamentous differentiation.
In reciprocal experiments, we determined that hyperactivity of mitotic cyclins and, by inference, high activity of mitotic kinase can abrogate filamentous development directed by RAS/STE signaling. Cells overexpressing Clb1 or Clb2 maintain a round cell shape and a nonfilamentous growth pattern even in the face of maximal stimulation provided by the nutritional signal of low-nitrogen media combined with dominant active mutations in the RAS/STE signaling pathway. In turn, overexpression of the Clb1 or Clb2 cyclin does not repress the low-nitrogen-induced expression of a FRE reporter (our unpublished results). These data suggest that 1) mitotic functions of the Cdc28 CDK are a critical target of the RAS/STE signal, and 2) repression of mitotic kinase is both necessary and sufficient to induce a coordinated developmental switch characterized by elongated cell shape, polar budding, persistent cell attachment, and G2/M cell cycle delay. This pattern recapitulates key features of filamentous differentiation as stimulated by the physiological signal.
Rather than proving a direct pathway that links RAS/STE to mitotic cyclins, we cannot eliminate the possibility that the effects of altered cyclin activity on filamentous growth, like the effects of modulating Cdc28 Tyr-19 phosphorylation, are simply evidence for a parallel pathway regulating morphogenesis. Other data linking the Cln1 and Cln2 G1 cyclins to filamentous differentiation suggest that their effects may be quite direct, serving as modulators of the RAS/STE response in the signaling pathway (Oehlen and Cross, 1998) and/or as pathway-independent activators of gene expression (Loeb et al., 1999). Nonetheless, neither mechanism adequately addresses the origin of the mitotic delay or its regulation by RAS/STE signals. We propose an alternative model whereby the RAS/STE pathway directly effects repression of mitotic kinase activity in diploid cells. Our hypothesis is based on the paradigm of pheromone response, in which distinct gene products induced by the STE MAPK pathway mediate cell surface (Fus1) and cell cycle (Far1) aspects of differentiation. Far1 serves as an inhibitor of Cln1,Cln2-dependent Cdc28 activity and thereby enforces the G1 arrest of conjugating haploid cells. We imagine a functional equivalent for Far1 acting downstream of the RAS/STE pathway to enforce its cell cycle effects. Coordinately with expression of the cell surface flocculin Flo11 that is required for the increased cell adhesion in filamentous growth, this hypothesized gene product would be under FRE control so that it would be induced in diploid cells upon activation of the RAS/STE pathway to mediate the mitotic delay. As we did not observe any decrease in the steady-state abundance of Clb2 protein in our STE11-4 cells, we suppose that activation of the RAS/STE pathway neither decreases expression nor increases destruction of mitotic cyclins. An alternative biochemical activity of this factor might be direct, stoichiometric inhibition of Clb1- and Clb2-dependent Cdc28 mitotic kinase activity. As a preliminary experiment to detect regulation at the level of Cdc28 activity, we assayed precipitates of extracts derived from asynchronous cultures of wild-type and RAS/STE pathway mutant yeast strains for histone H1 phosphorylation to quantitate CDK activity. Initial results suggest that although total, p13suc1-associated CDK activity may be unaltered, Clb2-associated Cdc28 activity in STE11-4 cells appears relatively depressed (our unpublished results). Confirmation and characterization of this apparent mitotic inhibition remain to be pursued. In turn, genetic studies may reveal mutations that do not block Flo11 expression but prevent down-regulation of the mitotic activity of Cdc28. Such studies would provide the critical evidence for the hypothesized factor coupling the gene expression program induced by the RAS/STE filamentous signaling pathway to its effects on cell cycle progression.
ACKNOWLEDGMENTS
We thank Ankit Desai for technical assistance. Preliminary studies leading to the results reported here were completed by S.J.K. while a postdoctoral fellow with Gerald R. Fink at the Whitehead Institute (Cambridge, MA). S.J.K. gratefully acknowledges Dr. Fink for his generous support, guidance, and scientific insights and Cora Styles for help with strain constructions and valuable suggestions. We thank A. Amon, R. Booher, B. Errede, D. Lew, H. Liu, H.-U. Mosch, S. Reed, and P. Russel for reagents and helpful comments. This work was supported by grants to S.J.K. from the Arnold and Mabel Beckman Foundation, the Horace W. Goldsmith Foundation, the Cancer Research Foundation, and funds from a Howard Hughes Medical Institute Research Resources for Medical Schools grant to the University of Chicago.
REFERENCES
- Alfa C, Fantes P, Hyams J, McLeod M, Warbrick E. Experiments with Fission Yeast. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1993. [Google Scholar]
- Amon A. Characterization of the Mitotic Kinase in the Budding Yeast S. cerevisiae. Ph.D. Thesis. Vienna, Austria: University of Vienna; 1992. [Google Scholar]
- Amon A, Irniger S, Nasmyth K. Closing the cell cycle circle in yeast: G2 cyclin proteolysis initiated at mitosis persists until the activation of G1 cyclins in the next cycle. Cell. 1994;77:1037–1050. doi: 10.1016/0092-8674(94)90443-x. [DOI] [PubMed] [Google Scholar]
- Amon A, Surana U, Muroff I, Nasmyth K. Regulation of p34CDC28 tyrosine phosphorylation is not required for entry into mitosis in S. cerevisiae. Nature. 1992;355:368–371. doi: 10.1038/355368a0. [DOI] [PubMed] [Google Scholar]
- Banuett F. Signaling in the yeasts: an informational cascade with links to the filamentous fungi. Microbiol Mol Biol Rev. 1998;62:249–274. doi: 10.1128/mmbr.62.2.249-274.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Booher RN, Deshaies RJ, Kirschner MW. Properties of Saccharomyces cerevisiae wee1 and its differential regulation of p34CDC28 in response to G1 and G2 cyclins. EMBO J. 1993;12:3417–3426. doi: 10.1002/j.1460-2075.1993.tb06016.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cali BM, Doyle TC, Botstein D, Fink GR. Multiple functions for actin during filamentous growth of Saccharomyces cerevisiae. Mol Biol Cell. 1998;9:1873–1889. doi: 10.1091/mbc.9.7.1873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang F, Herskowitz I. Identification of a gene necessary for cell cycle arrest by a negative growth factor of yeast: FAR1 is an inhibitor of a G1 cyclin, CLN2. Cell. 1990;63:999–1011. doi: 10.1016/0092-8674(90)90503-7. [DOI] [PubMed] [Google Scholar]
- Cook JG, Bardwell L, Thorner J. Inhibitory and activating functions for MAPK Kss1 in the S. cerevisiae filamentous-growth signaling pathway. Nature. 1997;390:85–88. doi: 10.1038/36355. [DOI] [PubMed] [Google Scholar]
- Edgington NP, Blacketer MJ, Bierwagen TA, Myers AM. Control of Saccharomyces cerevisiae filamentous growth by cyclin-dependent kinase Cdc28. Mol Cell Biol. 1999;19:1369–1380. doi: 10.1128/mcb.19.2.1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards MC, Liegeois N, Horecka J, DePinho RA, Sprague GF, Jr, Tyers M, Elledge SJ. Human CPR (cell cycle progression restoration) genes impart a Far− phenotype on yeast cells. Genetics. 1997;147:1063–1076. doi: 10.1093/genetics/147.3.1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feilotter H, Lingner C, Rowley R, Young PG. Regulation of the G2-mitosis transition. Biochem Cell Biol. 1992;70:954–971. doi: 10.1139/o92-140. [DOI] [PubMed] [Google Scholar]
- Gavrias V, Andrianopoulos A, Gimeno CJ, Timberlake WE. Saccharomyces cerevisiae TEC1 is required for pseudohyphal growth. Mol Microbiol. 1996;19:1255–1263. doi: 10.1111/j.1365-2958.1996.tb02470.x. [DOI] [PubMed] [Google Scholar]
- Gimeno CJ, Ljungdahl PO, Styles CA, Fink GR. Unipolar cell divisions in the yeast S. cerevisiae lead to filamentous growth: regulation by starvation and RAS. Cell. 1992;68:1077–1090. doi: 10.1016/0092-8674(92)90079-r. [DOI] [PubMed] [Google Scholar]
- Grenson M, Mousset M, Wiame JM, Bechet J. Multiplicity of the amino acid permeases in Saccharomyces cerevisiae. I. Evidence for a specific arginine-transporting system. Biochim Biophys Acta. 1966;127:325–338. doi: 10.1016/0304-4165(66)90387-4. [DOI] [PubMed] [Google Scholar]
- Hagen DC, McCaffrey G, Sprague GF., Jr Pheromone response elements are necessary and sufficient for basal and pheromone-induced transcription of the FUS1 gene of Saccharomyces cerevisiae. Mol Cell Biol. 1991;11:2952–2961. doi: 10.1128/mcb.11.6.2952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutter KJ, Eipel HE. Flow cytometric determinations of cellular substances in algae, bacteria, molds and yeasts. Antonie Leeuwenhoek. 1978;44:269–282. doi: 10.1007/BF00394305. [DOI] [PubMed] [Google Scholar]
- Kilmartin JV, Wright B, Milstein C. Rat monoclonal antitubulin antibodies derived by using a new nonsecreting rat cell line. J Cell Biol. 1982;93:576–582. doi: 10.1083/jcb.93.3.576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kron SJ, Gow NA. Budding yeast morphogenesis: signaling, cytoskeleton and cell cycle. Curr Opin Cell Biol. 1995;7:845–855. doi: 10.1016/0955-0674(95)80069-7. [DOI] [PubMed] [Google Scholar]
- Kron SJ, Styles CA, Fink GR. Symmetric cell division in pseudohyphae of the yeast Saccharomyces cerevisiae. Mol Biol Cell. 1994;5:1003–1022. doi: 10.1091/mbc.5.9.1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laloux I, Jacobs E, Dubois E. Involvement of SRE element of Ty1 transposon in TEC1-dependent transcriptional activation. Nucleic Acids Res. 1994;22:999–1005. doi: 10.1093/nar/22.6.999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambrechts MG, Bauer FF, Marmur J, Pretorius IS. Muc1, a mucin-like protein that is regulated by Mss10, is critical for pseudohyphal differentiation in yeast. Proc Natl Acad Sci USA. 1996;93:8419–8424. doi: 10.1073/pnas.93.16.8419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leberer E, Thomas DY, Whiteway M. Pheromone signaling and polarized morphogenesis in yeast. Curr Opin Genet Dev. 1997;7:59–66. doi: 10.1016/s0959-437x(97)80110-4. [DOI] [PubMed] [Google Scholar]
- Lew DJ, Reed SI. Morphogenesis in the yeast cell cycle: regulation by Cdc28 and cyclins. J Cell Biol. 1993;120:1305–1320. doi: 10.1083/jcb.120.6.1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lew DJ, Weinert T, Pringle JR. Cell cycle control in Saccharomyces cerevisiae. In: Pringle JR, E.W.J. JRB, editors. The Molecular and Cellular Biology of the Yeast Saccharomyces: Cell Cycle and Cell Biology. III. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1997. , 1131. [Google Scholar]
- Leza MA, Elion EA. POG1, a novel yeast gene, promotes recovery from pheromone arrest via the G1 cyclin CLN2. Genetics. 1999;151:531–543. doi: 10.1093/genetics/151.2.531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Styles CA, Fink GR. Elements of the yeast pheromone response pathway required for filamentous growth of diploids. Science. 1993;262:1741–1744. doi: 10.1126/science.8259520. [DOI] [PubMed] [Google Scholar]
- Lo HJ, Kohler JR, DiDomenico B, Loebenberg D, Cacciapuoti A, Fink GR. Nonfilamentous C. albicans mutants are avirulent. Cell. 1997;90:939–949. doi: 10.1016/s0092-8674(00)80358-x. [DOI] [PubMed] [Google Scholar]
- Lo WS, Dranginis AM. The cell surface flocculin Flo11 is required for pseudohyphae formation and invasion by Saccharomyces cerevisiae. Mol Biol Cell. 1998;9:161–171. doi: 10.1091/mbc.9.1.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loeb, J.D.J., Kerentseva, T.A., Pan, T., Sepulveda-Becerra, M., and Liu, H. (1999). Saccharomyces cerevisiae G1 cyclins are differentially involved in invasive and pseudohyphal growth independent of the filamentation MAP kinase pathway. Genetics (in press). [DOI] [PMC free article] [PubMed]
- Madden K, Snyder M. Cell polarity and morphogenesis in budding yeast. Annu Rev Microbiol. 1998;52:687–744. doi: 10.1146/annurev.micro.52.1.687. [DOI] [PubMed] [Google Scholar]
- Madhani HD, Fink GR. Combinatorial control required for the specificity of yeast MAPK signaling. Science. 1997;275:1314–1317. doi: 10.1126/science.275.5304.1314. [DOI] [PubMed] [Google Scholar]
- Madhani HD, Fink GR. The control of filamentous differentiation and virulence in fungi. Trends Cell Biol. 1998a;8:348–353. doi: 10.1016/s0962-8924(98)01298-7. [DOI] [PubMed] [Google Scholar]
- Madhani HD, Fink GR. The riddle of MAP kinase signaling specificity. Trends Genet. 1998b;14:151–155. doi: 10.1016/s0168-9525(98)01425-5. [DOI] [PubMed] [Google Scholar]
- Madhani HD, Styles CA, Fink GR. MAP kinases with distinct inhibitory functions impart signaling specificity during yeast differentiation. Cell. 1997;91:673–684. doi: 10.1016/s0092-8674(00)80454-7. [DOI] [PubMed] [Google Scholar]
- Mendenhall MD, Hodge AE. Regulation of Cdc28 cyclin-dependent protein kinase activity during the cell cycle of the yeast Saccharomyces cerevisiae. Microbiol Mol Biol Rev. 1998;62:1191–1243. doi: 10.1128/mmbr.62.4.1191-1243.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosch H-U, Fink GR. Dissection of filamentous growth by transposon mutagenesis in Saccharomyces cerevisiae. Genetics. 1997;145:671–684. doi: 10.1093/genetics/145.3.671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosch HU, Kubler E, Krappmann S, Fink GR, Braus GH. Cross-talk between the Ras2p-controlled mitogen-activated protein kinase and cAMP pathways during invasive growth of Saccharomyces cerevisiae. Mol Biol Cell. 1999;10:1325–1335. doi: 10.1091/mbc.10.5.1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosch HU, Roberts RL, Fink GR. Ras2 signals via the Cdc42/Ste20/mitogen-activated protein kinase module to induce filamentous growth in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1996;93:5352–5356. doi: 10.1073/pnas.93.11.5352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oehlen LJ, Cross FR. G1 cyclins CLN1 and CLN2 repress the mating factor response pathway at Start in the yeast cell cycle. Genes Dev. 1994;8:1058–1070. doi: 10.1101/gad.8.9.1058. [DOI] [PubMed] [Google Scholar]
- Oehlen LJ, Cross FR. Potential regulation of Ste20 function by the Cln1-Cdc28 and Cln2-Cdc28 cyclin-dependent protein kinases. J Biol Chem. 1998;273:25089–25097. doi: 10.1074/jbc.273.39.25089. [DOI] [PubMed] [Google Scholar]
- Pan X, Heitman J. Cyclic AMP-dependent protein kinase regulates pseudohyphal differentiation in Saccharomyces cerevisiae. Mol Cell Biol. 1999;19:4874–4887. doi: 10.1128/mcb.19.7.4874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peter M, Herskowitz I. Direct inhibition of the yeast cyclin-dependent kinase Cdc28-Cln by Far1. Science. 1994;265:1228–1231. doi: 10.1126/science.8066461. [DOI] [PubMed] [Google Scholar]
- Pringle JR. Staining of bud scars and other cell wall chitin with calcofluor. Methods Enzymol. 1991;194:732–735. doi: 10.1016/0076-6879(91)94055-h. [DOI] [PubMed] [Google Scholar]
- Pringle JR, Adams AE, Drubin DG, Haarer BK. Immunofluorescence methods for yeast. Methods Enzymol. 1991;194:565–602. doi: 10.1016/0076-6879(91)94043-c. [DOI] [PubMed] [Google Scholar]
- Roberts RL, Fink GR. Elements of a single MAP kinase cascade in Saccharomyces cerevisiae mediate two developmental programs in the same cell type: mating and invasive growth. Genes Dev. 1994;8:2974–2985. doi: 10.1101/gad.8.24.2974. [DOI] [PubMed] [Google Scholar]
- Robertson LS, Fink GR. The three yeast A kinases have specific signaling functions in pseudohyphal growth. Proc Natl Acad Sci USA. 1998;95:13783–13787. doi: 10.1073/pnas.95.23.13783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rupp S, Summers E, Lo HJ, Madhani H, Fink G. MAP kinase and cAMP filamentation signaling pathways converge on the unusually large promoter of the yeast FLO11 gene. EMBO J. 1999;18:1257–1269. doi: 10.1093/emboj/18.5.1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiestl RH, Gietz RD. High efficiency transformation of intact yeast cells using single stranded nucleic acids as a carrier. Curr Genet. 1989;16:339–346. doi: 10.1007/BF00340712. [DOI] [PubMed] [Google Scholar]
- Sikorski RS, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevenson BJ, Rhodes N, Errede B, Sprague GF., Jr Constitutive mutants of the protein kinase STE11 activate the yeast pheromone response pathway in the absence of the G protein. Genes Dev. 1992;6:1293–1304. doi: 10.1101/gad.6.7.1293. [DOI] [PubMed] [Google Scholar]
- Stueland CS, Lew DJ, Cismowski MJ, Reed SI. Full activation of p34CDC28 histone H1 kinase activity is unable to promote entry into mitosis in checkpoint-arrested cells of the yeast Saccharomyces cerevisiae. Mol Cell Biol. 1993;13:3744–3755. doi: 10.1128/mcb.13.6.3744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surana U, Robitsch H, Price C, Schuster T, Fitch I, Futcher AB, Nasmyth K. The role of CDC28 and cyclins during mitosis in the budding yeast S. cerevisiae. Cell. 1991;65:145–161. doi: 10.1016/0092-8674(91)90416-v. [DOI] [PubMed] [Google Scholar]
- Tjandra H, Compton J, Kellogg D. Control of mitotic events by the Cdc42 GTPase, the Clb2 cyclin and a member of the PAK kinase family. Curr Biol. 1998;8:991–1000. doi: 10.1016/s0960-9822(07)00419-8. [DOI] [PubMed] [Google Scholar]
- Wassmann K, Ammerer G. Overexpression of the G1-cyclin gene CLN2 represses the mating pathway in Saccharomyces cerevisiae at the level of the MEKK Ste11. J Biol Chem. 1997;272:13180–13188. doi: 10.1074/jbc.272.20.13180. [DOI] [PubMed] [Google Scholar]
- Wu C, Leeuw T, Leberer E, Thomas DY, Whiteway M. Cell cycle- and Cln2p-Cdc28p-dependent phosphorylation of the yeast Ste20p protein kinase. J Biol Chem. 1998;273:28107–28115. doi: 10.1074/jbc.273.43.28107. [DOI] [PubMed] [Google Scholar]