Abstract
A temperature-sensitive mutant, sec34-2, is defective in the late stages of endoplasmic reticulum (ER)-to-Golgi transport. A high-copy suppressor screen that uses the sec34-2 mutant has resulted in the identification of the SEC34 structural gene and a novel gene called GRP1. GRP1 encodes a previously unidentified hydrophilic yeast protein related to the mammalian Golgi protein golgin-160. Although GRP1 is not essential for growth, the grp1Δ mutation displays synthetic lethal interactions with several mutations that result in ER accumulation and a block in the late stages of ER-to-Golgi transport, but not with those that block the budding of vesicles from the ER. Our findings suggest that Grp1p may facilitate membrane traffic indirectly, possibly by maintaining Golgi function. In an effort to identify genes whose products physically interact with Sec34p, we also tested the ability of overexpressed SEC34 to suppress known secretory mutations that block vesicular traffic between the ER and the Golgi. This screen revealed that SEC34 specifically suppresses sec35-1. SEC34 encodes a hydrophilic protein of ∼100 kDa. Like Sec35p, which has been implicated in the tethering of ER-derived vesicles to the Golgi, Sec34p is predominantly soluble. Sec34p and Sec35p stably associate with each other to form a multiprotein complex of ∼480 kDa. These data indicate that Sec34p acts in conjunction with Sec35p to mediate a common step in vesicular traffic.
INTRODUCTION
The SNAREs, a family of cytoplasmically oriented membrane proteins, are key players in the fusion of vesicles with their acceptor membranes (reviewed by Ferro-Novick and Jahn, 1994). These membrane proteins interact with each other to form a stable complex that binds the soluble factors NSF and α-SNAP (yeast SEC18 and SEC17 gene products, respectively). Subsequent to membrane fusion, NSF disassembles the SNARE complex and releases α-SNAP to allow the SNAREs to participate in a new round of transport (Ungermann et al., 1998). Although it is clear that the SNAREs play a critical role in fusing membranes (Weber et al., 1998), the machinery that correctly targets a vesicle to its acceptor membrane has been more elusive.
In endoplasmic reticulum (ER)-to-Golgi transport, several factors (Ypt1p, Uso1p, and Sec35p) and a large novel complex, called TRAPP, are candidates for participating in the initial targeting or tethering of vesicles to the Golgi (Lian et al., 1994; Rossi et al., 1995; Sapperstein et al., 1996; Sacher et al., 1998; Vanrheenen et al., 1998). Uso1p, which is homologous to the mammalian vesicle-docking protein p115 (Nakamura et al., 1997), is a large cytosolic protein (206 kDa) with a large globular head and a long coiled-coil tail. In a cell-free assay, Sec35p acts in the Uso1p-dependent docking of ER-derived vesicles to the Golgi (Vanrheenen et al., 1998). TRAPP is a highly conserved novel complex that contains 10 subunits. It resides on the cis-Golgi, where it acts upstream of the SNAREs (Sacher et al., 1998). The activity of Uso1p, Sec35p, and TRAPP may be regulated by the small ras-like GTP-binding protein Ypt1p (Sapperstein et al., 1996; Sacher et al., 1998; Vanrheenen et al., 1998).
To identify additional players that regulate the targeting and fusion activity of ER-to-Golgi transport vesicles with the Golgi apparatus, we performed two different high-copy suppressor screens. A screen for suppressors of sec34-2, a mutant defective in these processes (Wuestehube et al., 1996), resulted in the identification of GRP1 (golgin-160–related protein), a nonessential yeast gene whose product is related to the putative mammalian Golgi matrix protein golgin-160 (Fritzler et al., 1993). We propose that Grp1p may act indirectly to facilitate the late stages of ER-to-Golgi transport. A second screen revealed that SEC34 specifically suppresses sec35-1 and not other mutations that block ER-to-Golgi transport. Coprecipitation studies demonstrate that Sec34p forms a complex with Sec35p. These findings imply that Sec34p acts in conjunction with Sec35p to mediate the targeting of ER-to-Golgi transport vesicles to the Golgi apparatus.
MATERIALS AND METHODS
Strains and Growth Conditions
Bacterial strains used in this study were DH5α and XL2-Blue. They were grown in Luria-Bertani medium or on Luria-Bertani plates with 2% agar. Transformants carrying plasmids were grown in the presence of 100 μg/ml ampicillin. Yeast strains used (see Table 1) were grown in either YPD or minimal medium containing the appropriate amino acids (20 μg/ml histidine, 100 μg/ml leucine, 30 μg/ml lysine, and 20 μg/ml uracil).
Table 1.
Yeast strains used in this study
| Strain | Genotype | Source |
|---|---|---|
| ANY112 | MATa bet1-1 ura3-52 | Ferro-Novick Lab Collection |
| NY3 | MATa sec1-1 ura3-52 | Novick Lab Collection |
| NY17 | MATa sec6-4 ura3-52 | Novick Lab Collection |
| NY57 | MATa sec9-4 ura3-52 | Novick Lab Collection |
| NY61 | MATa sec10-2 ura3-52 | Novick Lab Collection |
| NY64 | MATa sec15-1 ura3-52 | Novick Lab Collection |
| NY130 | MATa sec2-41 ura3-52 | Novick Lab Collection |
| NY402 | MATa sec5-24 ura3-52 | Novick Lab Collection |
| NY405 | MATa sec4-8 ura3-52 | Novick Lab Collection |
| NY410 | MATa sec8-9 ura3-52 | Novick Lab Collection |
| NY413 | MATα sec13-1 ura3-52 | Novick Lab Collection |
| NY416 | MATα sec16-2 ura3-52 | Novick Lab Collection |
| NY417 | MATα sec17-1 ura3-52 | Novick Lab Collection |
| NY420 | MATa sec19-1 ura3-52 | Novick Lab Collection |
| NY422 | MATa sec20-1 ura3-52 | Novick Lab Collection |
| NY424 | MATα sec21-1 ura3-52 | Novick Lab Collection |
| NY425 | MATα sec22-3 ura3-52 | Novick Lab Collection |
| NY431 | MATa sec18-1 ura3-52 | Novick Lab Collection |
| NY435 | MATa ypt1-1 ura3-52 | Novick Lab Collection |
| NY806 | MATa sec23-1 ura3-52 | Novick Lab Collection |
| NY1222 | MATa sec3-2 ura3-52 | Novick Lab Collection |
| NY1262 | MATa ypt1-3 ura3-52 | Novick Lab Collection |
| NY1282 | MATa bos1-1 ura3-52 | Novick Lab Collection |
| SFNY26-3A | MATa ura3-52 | Ferro-Novick Lab Collection |
| SFNY26-4C | MATa ura3-52 his4-619 | Ferro-Novick Lab Collection |
| SFNY26-12C | MATα ura3-52 | Ferro-Novick Lab Collection |
| SFNY48-4A | MATα sec7-1 ura3-52 | Ferro-Novick Lab Collection |
| SFNY77 | MATa sec22-3 ura3-52 | Ferro-Novick Lab Collection |
| SFNY293 | MATα uso1-1 ura3-52 trp1-Δ901 his7 | Ferro-Novick Lab Collection |
| SFNY413 | MATα bos1-1 ura3-52 leu2-3, 112 | Ferro-Novick Lab Collection |
| SFNY562 | MATa/α Gal+ leu2-3, 112/leu2-3, 112 ura3-52/ura3-52 | Ferro-Novick Lab Collection |
| SFNY571 | MATα sed5-1 ura3-52 leu2-3, 112 his3-Δ200 trp1-Δ901 lys2-801 suc2-Δ9 | Ferro-Novick Lab Collection |
| SFNY596 | MATa bet3-1 ura3-52 leu2-3, 112 | Ferro-Novick Lab Collection |
| SFNY688 | MATa sec34-1 lys2-801 | Ferro-Novick Lab Collection |
| SFNY691 | MATα sec34-2 ura3-52 leu2-3, 112 | Ferro-Novick Lab Collection |
| SFNY692 | MATa sec35-1 lys2-801 | Ferro-Novick Lab Collection |
| SFNY760 | MATα Gal+ leu2-3, 112 ura3-52 grp1Δ∷URA3 | This study |
| SFNY761 | MATa/α Gal+ leu2-3, 112/leu2-3, 112 ura3-52/ura3-52 GRP1/grp1Δ∷URA3 | This study |
| SFNY769 | MATa sec34-2 ura3-52 | This study |
| SFNY770 | MATα sec34-2 ura3-52 | This study |
| SFNY771 | MATa Gal+ leu2-3, 112 ura3-52 grp1Δ∷URA3 | This study |
| SFNY772 | MATa ura3-52 SEC34 (with three c-myc tags) | This study |
| SFNY794 | MATa sec34-1 ura3-52 | This study |
| SFNY795 | MATα sec34-1 ura3-52 | This study |
| SFNY816 | MATa sec35-1 ura3-52 | This study |
| SFNY919 | MATa/α Gal+ leu2-3, 112/leu2-3, 112 ura3-52/ura3-52 SEC34/sec34Δ∷URA3 | This study |
| SFNY947 | MATa ura3-52 SEC35 (with three c-myc tags) | This study |
Screen for Multicopy Suppressors of sec34-2
High-copy suppressors of sec34-2 were isolated by transforming the mutant strain with a yeast genomic high-copy library (Carlson and Botstein, 1982), followed by screening for transformants that grow at 38.5°C. This was done in several steps. First, plasmid DNA was transformed by the lithium acetate method (Ito et al., 1983) into sec34-2, and Ura+ transformants were selected on minimal medium lacking uracil at 24°C. After 5 d, the transformants were replica plated onto YPD plates and incubated overnight at 38.5°C. The large colonies that grew were purified on minimal medium lacking uracil, and the purified transformants were tested for suppression on YPD plates at 38.5°C. Of the 22,000 transformants screened, 8 were found to suppress sec34-2 at this temperature. Plasmids from the 8 transformants were retrieved and reintroduced into sec34-2 to confirm suppression. DNA sequence analysis revealed that the 8 transformants represented three different regions of genomic DNA. These plasmids were placed into three groups. The subcloning of the inserts for two of these groups is shown in Figures 1A and 2. One group contained the SEC34 structural gene (Figure 1A), and the other group included GRP1 (Figure 2). The third group was not studied further because members of this group were found to suppress secretory mutations that block membrane traffic at all stages of the exocytic pathway.
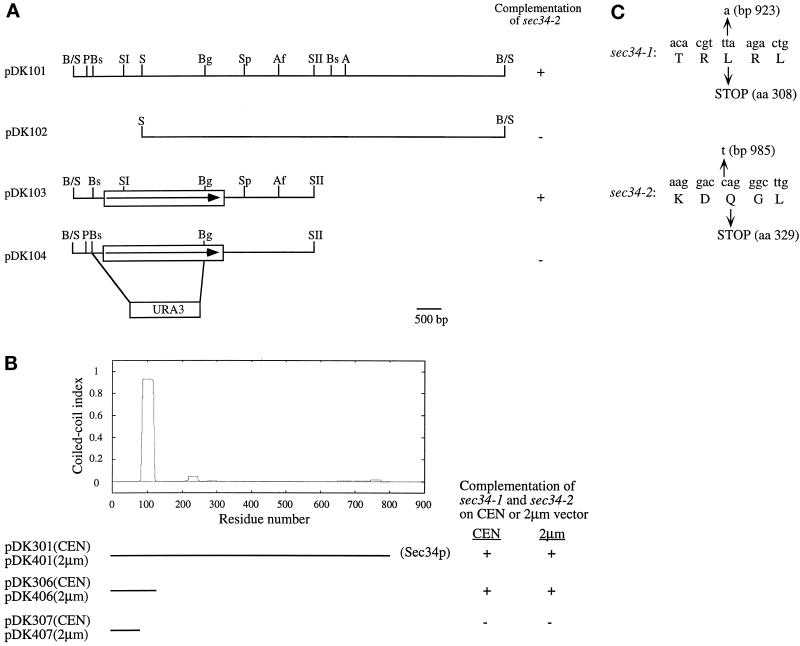
Figure 1.
(A) Complementing activity of clones containing the SEC34 gene. Only the cloned insert is shown. B/S, BamHI-Sau3A junction; P, PvuII; Bs, BstEII; SI, SacI; S, SalI; Bg, BglII; Sp, SphI; Af, AflII; SII, SacII; A, AgeI. (B) The N-terminal coiled-coil region of Sec34p is important for its function. Sec34p has a high probability of forming a coiled-coil region between amino acids 87 and 114. Plasmids pDK301 and pDK401 contain the entire SEC34 gene, whereas pDK307 and pDK407 harbor the extreme N terminus of Sec34p (amino acids 1–85) lacking the coiled-coil domain. This domain is present in pDK306 and pKD406 (which contain amino acids 1–128). The sec34-1 and sec34-2 mutants were transformed with these constructs and incubated for 2 d at 37°C to test for suppression. All constructs, except for pDK307 and pDK407, in which the coiled-coil region was disrupted, suppressed sec34-1 and sec34-2. Only the ORF of SEC34 is shown. (C) The sec34-1 and sec34-2 mutants encode truncated proteins of Sec34p. The base pairs and the corresponding amino acids of Sec34p that are changed in sec34-1 and sec34-2 are indicated with arrows.
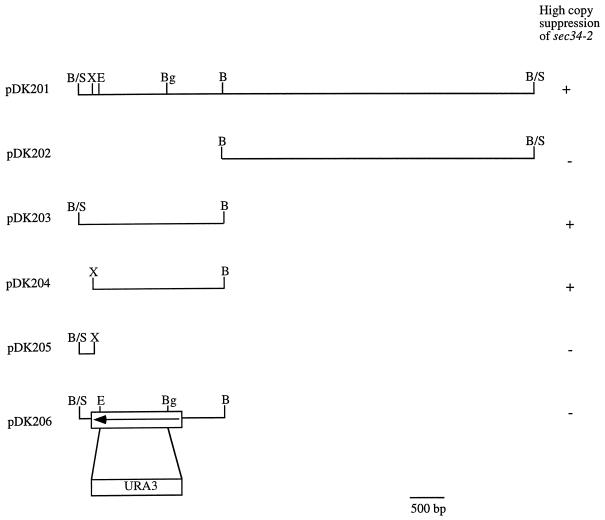
Figure 2.
Ability of clones containing YOR216C to suppress sec34-2. Only the cloned insert is shown. B/S, BamHI-Sau3A junction; X, XhoI; E, Eco47III; Bg, BglII; B, BamHI.
Disruption of GRP1 and SEC34
A chromosomal disruption of GRP1 was created by replacing base pairs (bp) 187-1383 of GRP1 with the URA3 gene. This was done as follows: a 2.3-kilobase (kb) BamHI-BamHI fragment containing the GRP1 gene (Figure 2, pDK203) was inserted into the BamHI site of Bluescript II KS−. The resulting plasmid was digested with Eco47III and BglII to delete most of the GRP1 coding region, which was then replaced with a PvuII-BamHI fragment harboring the URA3 gene to yield pBSΔ18. A diploid strain, with one disrupted copy of GRP1, was created by digesting pBSΔ18 with BamHI and transforming the 2.6-kb linear fragment into SFNY562 (Table 1). PCR was used to confirm that GRP1 was disrupted. A purified transformant containing the disruption was sporulated and subjected to tetrad analysis. To construct pDK206 (Figure 2), pBSΔ18 was digested with BamHI, and the resulting 2.6-kb linear fragment was inserted into the BamHI site of pRS426.
The SEC34 gene was disrupted by replacing the promotor and part of the coding region of SEC34 (bp 1–2100) with the URA3 gene. Briefly, the 2.2-kb BstEII-BglII fragment from pDK103 (Figure 1A) was replaced with a 1.2-kb HindIII-HindIII fragment harboring the URA3 gene to yield pDK104. The pDK104 plasmid was digested with PvuII and SacII, and the resulting 3.4-kb linear fragment was transformed into SFNY562 (Table 1) to yield a diploid with one disrupted copy of SEC34 (SFNY 919). PCR was used to confirm the disruption before the strain was subjected to tetrad analysis.
DNA Constructions
All restriction enzymes were obtained from New England Biolabs (Beverly, MA). The plasmids shown in Figures 1 and 2 were constructed as described below. Briefly, pDK101 (Figure 1A) was digested with SalI, and the resulting 14.8-kb fragment was ligated to produce pDK102. Plasmid pDK103 was constructed by inserting the 5.4-kb EagI-SacII fragment from pDK101 into the EagI-SacII sites of pRS426. Plasmid pDK203 (Figure 2) was constructed by digesting pDK201 with BamHI. The resulting 2.3-kb fragment was inserted into the BamHI site of pRS426 to yield pDK203. A larger 12.7-kb fragment was ligated to generate pDK202. To construct pDK204 and pDK205, plasmid pDK203 was digested with XhoI. The 2.2-kb fragment was inserted into the XhoI site of pRS426 to yield pDK204, and the larger 5.9-kb fragment was ligated to generate pDK205. Plasmid pDK401 (Figure 1B) was generated by digesting pDK101 with AgeI and XmaI and ligating the resulting 12.8-kb fragment. Plasmids pDK406 and pDK407 were amplified by PCR with pDK103 as a template and the following primers (5′–3′): sense primers, CTA CAT AAG CTT TAT CAA TGA ATA CAG TAA TCC AAA G; antisense primers, CTA CAT GGA TCC TTA GAA AGC TCC AGT ATC CTG AGA G (pDK406) and CTA CAT GGA TCC TTA GAA TTT ACT GTA CAA GAA GGC GTC (pDK407). The amplified fragments were inserted into the HindIII site of pRS426 to generate pDK406 and pDK407. Plasmid pDK301 was constructed by inserting the 5.4-kb EagI-SacII fragment from pDK101 into the same sites of pRS316. pDK306 and pDK307 were constructed by inserting the 0.7- and 0.5-kb HindIII-BamHI fragments, respectively, from pDK406 and pDK407 into the HindIII-BamHI sites of pRS316.
Construction of Epitope-tagged SEC34 and SEC35
Triple c-myc–tagged Sec34p and Sec35p were constructed according to the method of Schneider et al. (1995). Briefly, hybrid sequences containing the URA3 gene and epitope tags flanked by a part of the SEC34 or SEC35 gene were amplified by PCR. These products were then transformed into wild-type cells (SFNY26-3A) to direct integration at the SEC34 or SEC35 locus, and Ura+ transformants were selected. After confirming integration by PCR, the URA3 gene was popped out on plates containing 5-FOA, leaving the myc epitope tag at the C terminus of SEC34 or SEC35. Finally, those colonies containing triple myc-tagged Sec34p or Sec35p were confirmed by PCR and Western blot analysis. Cells containing epitope-tagged proteins showed the same growth properties as cells that lacked the tag.
Sequencing the sec34 Mutations
The technique of gap repair (Orr-Weaver et al., 1983) was used to sequence two different chromosomal mutant alleles of SEC34. Briefly, pDK103 (Figure 1A) was digested with AflII and BstEII to completely remove the ORF of SEC34. The resulting linear fragment, which contained the selectable marker URA3, was purified and transformed into the sec34-1 and sec34-2 mutants. The gaps in the plasmids were repaired with the use of the chromosomal mutant alleles as templates, and Ura+ transformants were selected at 30°C on minimal medium lacking uracil. Plasmids were recovered, amplified in bacteria, and sequenced.
Production of Antibodies Directed against Sec34p and Sec35p
Antibodies to Sec34p and Sec35p were prepared against purified His6-tagged recombinant forms of these proteins. To construct the plasmid encoding Sec34p-His6, the SEC34 ORF was amplified by PCR, placing a NdeI site at the sequence encoding the first amino acid of SEC34 and a XhoI site replacing the stop codon (sense primer [5′–3′], ATA CAT CAT ATG GCG AGA AGT AGA AAG AAT TCA; antisense primer [5′–3′], ATA CAT CTC GAG TTT CGT TAT GGT ATC AAT ATC ACC). The PCR product was digested with NdeI and XhoI and then ligated into the NdeI-XhoI sites of pET-29a (Novagen, Madison, WI). The plasmid encoding Sec35p-His6 was generated as follows: the SEC35 ORF was amplified by PCR, placing a NcoI site 5′ to the start codon and with a XhoI site replacing the stop codon (sense primer [5′–3′], CGT ACC ATG GTC AAC AGT CAT AGT CGC; antisense primer [5′–3′], GGT GCT CGA GTG CCG TTT TTA TAA TGG AGA C). The PCR product was digested with NcoI and XhoI and ligated into the NcoI-XhoI sites of pET-28b (Novagen).
The constructs were transformed into BL21 cells (Novagen) and induced at OD600 = 0.5 for 2.5 h at 37°C by the addition of isopropylthio-β-galactoside (1 mM final concentration). Sec35p-His6 recombinant protein was purified on a nickel-agarose column (Qiagen, Hilden, Germany), whereas Sec34p-His6 was purified by electroelution. Briefly, the gel containing Sec34p-His6 was stained with 0.3 M CuCl2 and the appropriate band was excised, cut into small pieces, and destained three times with 0.25 M EDTA in 0.25 M Tris-HCl, pH 9.0. After equilibration in SDS-PAGE running buffer, Sec34p-His6 was electroeluted in a Bio-Rad (Richmond, CA) electroelution apparatus according to the instructions provided by the manufacturer. The concentration of the purified proteins was determined by a gel assay with BSA as a standard. Antibodies to the recombinant proteins were raised in female New Zealand White rabbits with the use of the lymph node protocol.
Fractionation and Extraction Studies
Cells were grown in YPD medium to an OD599 of 2.0. A total of 150 OD units were washed with cold 10 mM NaN3 and converted to spheroplasts during a 1-h incubation at 37°C, as described previously (Shim et al., 1991). The spheroplasts were layered onto a cold sorbitol cushion (1.7 M sorbitol, 50 mM potassium phosphate, pH 7.5) and centrifuged at 3000 × g for 10 min at 4°C. The pellet was lysed in 6 ml of cold lysis buffer (20 mM HEPES/KOH, pH 7.4) containing 1× protease inhibitor cocktail (Ruohola et al.,1988) and homogenized six times with a Dounce tissue grinder (Wheaton Science Products, Millville, NJ). The crude lysate was centrifuged at 200 × g for 3 min at 4°C to generate lysed (S1) and unlysed (P1) cell fractions. The S1 fraction was centrifuged at 200,000 × g for 60 min to yield supernatant (S2) and pellet (P2) fractions. All pellet fractions were resuspended in the same volume of lysis buffer as the supernatant. For protein extractions, 0.5 ml of the S1 fraction was mixed with an equal volume of one of the following: lysis buffer, 2% Triton X-100 in lysis buffer, 2 M NaCl in lysis buffer, and 200 mM Na2CO3 in water. After a 45-min incubation on ice, the samples were centrifuged at 200,000 × g for 60 min and then separated into supernatant and pellet fractions. Equal volumes of the supernatant and pellet were boiled in sample buffer and resolved on a 10% SDS-polyacrylamide gel.
Immunoprecipitation Studies
Lysates were prepared from two different wild-type strains, SFNY26-3A and SFNY772 (Sec34p-myc). A total of 300 OD units of cells were converted to spheroplasts during a 1-h incubation at 37°C, as described previously (Shim et al., 1991). The spheroplasts were resuspended in 200 ml of recovery medium (1% Bacto-yeast extract, 2% Bacto-peptone, 0.1% glucose, 1.4 M sorbitol) and incubated for 1 h at 37°C. After a 5-min spin at 1600 × g, the pellet was lysed in 840 μl of cold lysis buffer (20 mM HEPES, pH 7.4, 500 mM KCl, 1 mM DTT, 2 mM EDTA, 2% Triton X-100, and protease inhibitor cocktail) and centrifuged at 120,000 × g for 1 h at 4°C. The protein concentration of the supernatant was determined by the Bradford method with bovine immunoglobulin G as the standard. The lysate (3 mg) was diluted with buffer A (20 mM HEPES, pH 7.2, 100 mM KCl, 1 mM DTT, 2 mM EDTA, 0.5% Triton X-100, and protease inhibitor cocktail) to a final volume of 1 ml. One sample was treated with 1% SDS and heated to 100°C for 5 min before it was diluted with buffer A. Then, 42 μl of anti-myc antibody was added to each sample and incubated for 2.5 h at 4°C. The immune complexes were collected onto protein A–Sepharose beads during a 1.5-h incubation at 4°C. The beads were washed two times with buffer B (20 mM HEPES, pH 7.2, 500 mM KCl, 1 mM DTT, 2 mM EDTA, 0.5% Triton X-100, and protease inhibitor cocktail), three times with buffer A, and one time with buffer C (20 mM Tris-HCl, pH 7.5). The samples were heated to 100°C in SDS-PAGE sample buffer (75 μl), and the eluted protein was resolved on a 10% SDS-polyacrylamide gel that was subjected to Western blot analysis with the use of anti-Sec34p (1:5000 dilution) and anti-Sec35p (1:5000 dilution) antibodies.
For immunoprecipitation experiments from radiolabeled extracts, 15 OD units of cells were radiolabeled in 7.5 ml of synthetic medium with 100 μCi ProMix/ml for 2 h at 25°C. Cells were converted to spheroplasts at 37°C during a 30-min incubation in spheroplasting buffer (1.4 M sorbitol, 50 mM potassium phosphate, pH 7.5, 50 mM β-mercaptoethanol, 30 μg/ml zymolyase) and lysed in 600 μl of buffer D (20 mM HEPES, pH 7.2, 1% Triton X-100, 150 mM KCl, 0.5 mM DTT, 2 mM EDTA, and protease inhibitor cocktail). The cell lysate was centrifuged at 100,000 × g for 1 h in an SW50.1 rotor (Beckman, Fullerton, CA), and the radiolabeled supernatant was diluted with buffer D to 50 × 106 cpm/ml. To identify Sec35p-myc, some samples were boiled in 1% SDS before dilution. The supernatant was precleared during a 1-h incubation with 30 μl of a 50% slurry of protein A–Sepharose beads, and then the sample was transferred to a new tube containing 2 μl of 9E10 (anti-c-myc epitope) ascites fluid. The antigen/antibody complexes that formed during a 1-h incubation at 4°C were precipitated onto protein A–Sepharose beads at 4°C for 1 h. The beads were washed three times with buffer E (same as buffer A but with 500 mM KCl) and two times with buffer D. After the final wash, the samples were heated in sample buffer and the entire sample was analyzed on a 13% SDS-polyacrylamide gel. To quantitate the ratio between Sec34p and Sec35p, the autoradiogram in Figure 7B was scanned and Sec34p and Sec35p were quantitated with the use of Intelligent software version 2.1.2 (BioImage, Ann Arbor, MI).
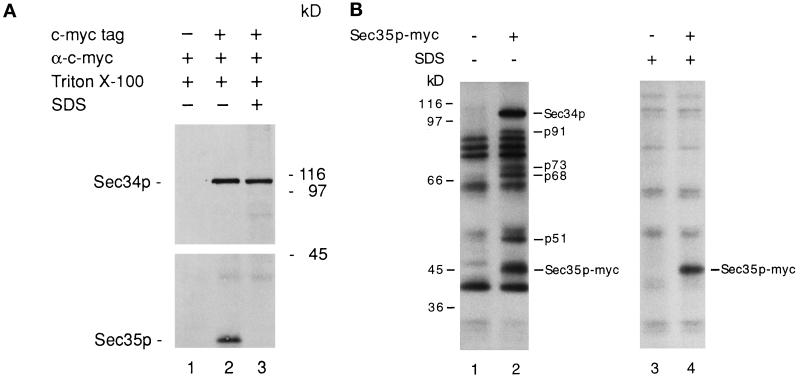
Figure 7.
(A) Sec35p coprecipitates with Sec34p. Lysates from strains containing tagged and untagged Sec34p were diluted with buffer A. Anti-c-myc antibody was added to each sample and incubated at 4°C to form immune complexes. One sample prepared from the tagged strain (lane 3) was pretreated with 1% SDS before the addition of antibody. The immune complexes were collected on protein A–Sepharose beads and then washed. The washed beads were boiled in SDS-PAGE sample buffer, and the eluted proteins were resolved on a 10% SDS-polyacrylamide gel that was subjected to Western blot analysis with the use of anti-Sec34p and anti-Sec35p antibodies. Lane 1, untagged strain; lane 2, Sec34p-myc–tagged strain; and lane 3, Sec34p-myc–tagged strain pretreated with 1% SDS. (B) Sec34p and Sec35p are members of a multiprotein complex. Radiolabeled lysates were prepared as described in MATERIALS AND METHODS from untagged (lanes 1 and 3) and Sec35p-myc–tagged (lanes 2 and 4) strains. Aliquots were left untreated (lanes 1 and 2) or boiled in 1% SDS (lanes 3 and 4) and then immunoprecipitated with anti-c-myc ascites fluid and fractionated by SDS-PAGE. Molecular mass makers are shown to the left of lane 1; p51, p68, p73, p91, and Sec34p refer to the polypeptides specifically immunoprecipitated with native Sec35p-myc.
Gel Filtration Analysis
Lysates were prepared from a wild-type strain, SFNY26-3A. A total of 300 OD units of cells were converted to spheroplasts during a 30-min incubation at 37°C. After a 5-min spin at 1600 × g, the pellet was lysed in 840 μl of cold lysis buffer (20 mM HEPES, pH 7.4, 150 mM KCl, 1 mM DTT, 2 mM EDTA, and protease inhibitor cocktail) and centrifuged at 120,000 × g for 1 h at 4°C. The protein concentration of the supernatant was determined by the Bradford method with bovine immunoglobulin G as the standard. To analyze the total cellular pool of Sec34p, the 1600 × g pellet was lysed in cold lysis buffer that contained 2% Triton X-100. A total of 5 mg of protein was applied to a Superose 6 HR 10/30 column (Pharmacia, Piscataway, NJ), and fractionation was performed at a flow rate of 0.3 ml/min. Fractions of 0.5 ml were collected, and 60 μl of each fraction was analyzed on a 10% polyacrylamide gel with the use of Western blot analysis. Molecular size standards (Pharmacia) used to calibrate the column were thyroglobulin (669 kDa), catalase (232 kDa), BSA (67 kDa), ovalbumin (43 kDa), and chymotrypsinogen A (25 kDa).
Database Search and Sequence Analysis
Plasmids that suppress the growth defect of sec34-2 were amplified in Escherichia coli and sequenced at the Keck Foundation at Yale University. The following primer sets (5′–3′) were used for sequencing: sense primer, GCT CGC TTC GCT ACT TGG AGC; antisense primer, TAT AGG CGC CAG CAA CCG CAC. DNA sequences of the insert clones were analyzed by a BLAST search of the Saccharomyces Genome Database from Stanford Genomic Resources (Stanford, CA). Homology searches and protein sequence analyses were performed with the use of the database of the National Center for Biotechnology Information and software from the Swiss Institute for Experimental Cancer Research Bioinformatics Group (Epalinges, Switzerland) or the Wisconsin Genetics Computer Group (version 8.1) (Madison, WI).
RESULTS
Identification of High-Copy Suppressors of the sec34-2 Mutation
The temperature-sensitive sec34 mutant accumulates ER-modified precursor forms of secretory proteins and small vesicles at its restrictive temperature, implying that Sec34p is required for the targeting and/or fusion of ER-to-Golgi transport vesicles (Wuestehube et al., 1996). To identify the SEC34 structural gene as well as new genes that may interact with SEC34, we transformed the sec34-2 mutant with a 2-μm yeast genomic library and screened for genes that suppress the growth defect of sec34-2 at 38.5°C. Of the 22,000 transformants examined, 8 grew at 38.5°C. Plasmids retrieved from these transformants were found to suppress sec34-2 when reintroduced into the mutant, indicating that suppression was plasmid dependent. DNA sequence analysis revealed that these plasmids contained three different regions of genomic DNA.
The plasmids were placed into three groups based on their sequence, and a member of each group was analyzed further. The insert in the first group (2 plasmids) was subcloned to 4.8 kb (Figure 1A, pDK103). It contained a single hypothetical ORF that conferred suppression. A search of the database revealed that this ORF resides on yeast chromosome V (YER157W). The insert in the second group (4 plasmids) was subcloned to a 2.3-kb fragment that retained full suppression activity (Figure 2, pDK203). DNA sequence analysis indicated that it contained an ORF on yeast chromosome XV (YOR216C). The insert in the third group (2 plasmids) contained four ORFs that were subcloned into two fragments. One fragment contained SBPI and RPL8A. SBPI encodes a single-stranded nucleic acid–binding protein, whereas RPL8A encodes the ribosomal protein L8A. The other fragment included the GOS1 gene, which encodes a putative 28-kDa SNARE (McNew et al., 1997). GOS1 did not significantly suppress sec34-2 at 38.5°C. However, transformants containing this gene grew somewhat faster at 24°C, but not at higher temperatures. Members of the third group were found to suppress secretory mutations that block membrane traffic at all stages of the exocytic pathway, implying that suppression was indirect. As a consequence, this group was not studied further.
Overproduction of YOR216C, a Nonessential Gene Whose Product Is Related to Mammalian Golgin-160, Specifically Suppresses Mutations That Block the Late Stages of ER-to-Golgi Transport
To gain insight into the role of YOR216C, we tested its ability to suppress known secretory mutations that block membrane traffic at different stages of the secretory pathway. In addition to sec34-2 (Figure 3A), the overexpression of YOR216C was found to suppress sec34-1 as well as several other mutations that result in a block in the targeting or fusion of ER-to-Golgi transport vesicles with their acceptor compartment (sec22-3 > sec35-1 > bos1-1 > uso1-1), but not sec17-1, sec18-1, bet1-1, bet3-1, bet5-1, or ypt1-1. Bos1p and Sec22p are vesicle SNAREs (Lian and Ferro-Novick, 1993; Lian et al., 1994), whereas Uso1p and Sec35p are peripheral membrane proteins that mediate an earlier step in transport. Secretory mutations that block the budding of vesicles from the ER (sec12-4, sec13-1, sec16-2, and sec23-1) or other stages of the pathway (sec1-1, sec2-41, sec3-2, sec4-8, sec5-24, sec6-4, sec7-1, sec8-9, sec9-4, sec10-2, sec15-1, sec19-1, sec20-1, and sec21-1) were not suppressed by the overproduction of YOR216C, indicating that YOR216C specifically suppresses mutations that block ER-to-Golgi transport at late stages.
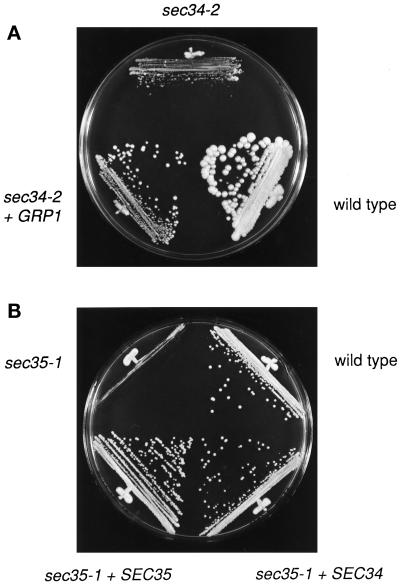
Figure 3.
(A) GRP1 is a high-copy suppressor of sec34-2. Yeast cells were grown on a YPD plate and incubated for 4 d at 38.5°C. GRP1 is on a 2-μm plasmid. (B) SEC34 is a high-copy suppressor of the sec35-1 mutation. Yeast cells were grown on a YPD plate and incubated for 2 d at 34°C. SEC34 and SEC35 are on 2-μm plasmids.
YOR216C encodes a highly hydrophilic protein of 484 amino acids with a predicted molecular mass of 56 kDa (apparent size is ∼80 kDa on SDS-PAGE [our unpublished results]). It has no predicted signal peptide and no significant hydrophobic stretch of amino acids that may serve as a transmembrane domain. Interestingly, ∼57% of the protein is predicted to form a coiled-coil structure. YOR216C is related to the mammalian Golgi protein golgin-160 (160 kDa) (Figure 4), which is one of two autoantigens that cross-react with the sera of patients with autoimmune diseases (Fritzler et al., 1993). Like YOR216C, a large portion of the protein (amino acids 708-1124) is predicted to form a coiled-coil structure. Golgin-160 resides on the Golgi and is part of a matrix that cannot be extracted from membranes with the detergent Triton X-100 (Fritzler et al., 1993). We have named this ORF GRP1 (golgin-160–related protein) because of its similarity to golgin-160.
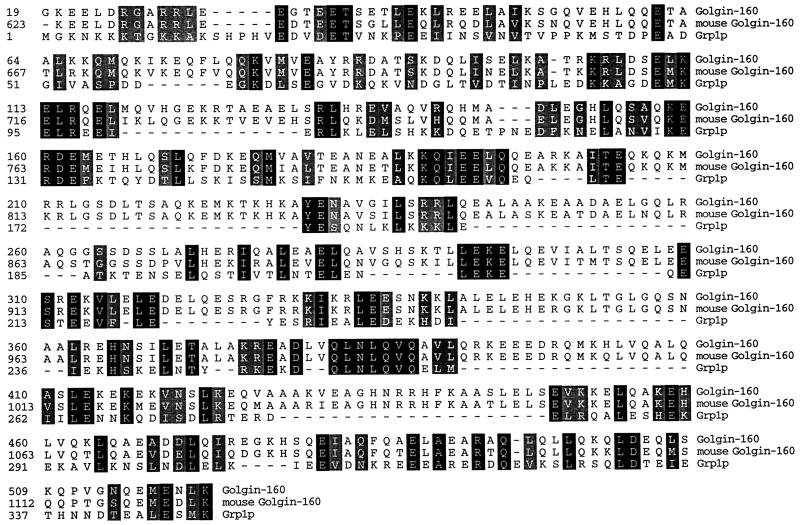
Figure 4.
Grp1p is related to golgin-160. Alignment of human and mouse golgin-160 with Grp1p. To determine if the similarity between Grp1p and golgin-160 is due to the composition of the coils, golgin-160 was randomized and aligned repeatedly to the predicted coiled-coil region in Grp1p, and a score was computed for each successive alignment. The unrandomized alignment score was 119, and the average randomized alignment score was 21.5 ± 12.9. Thus, the similarity observed between these two proteins is greater than expected for two unrelated proteins containing coiled-coil domains. In the unrandomized alignment, the percent identity between the sequences over this region was 34%, indicating that the two sequences are distantly related. Identical residues shared among the three proteins are shaded in black, and conserved residues are shaded in gray.
To determine if GRP1 encodes a protein essential for the vegetative growth of yeast, a diploid strain (SFNY761; see Table 1) with one disrupted copy of GRP1 was created by transforming SFNY562 with a linear fragment of DNA that harbored the disrupted gene. This construct was made by replacing most of GRP1 with the URA3 gene (Figure 2, pDK206), as described in MATERIALS AND METHODS, and confirming the replacement by PCR. The diploid, with one disrupted copy of GRP1, was sporulated and subjected to tetrad analysis to yield SFNY760, a haploid strain that lacks GRP1. Of the 21 tetrads examined, all contained four viable spores, and the Ura+ phenotype segregated 2:2 in each tetrad. Furthermore, the Ura+ colonies did not display an appreciable growth defect on plates at 25, 30, or 37°C, indicating that GRP1 is not essential for the growth of yeast cells.
The grp1Δ Mutation Does Not Block the Transit of Carboxypeptidase Y but Interacts with Mutations That Block ER-to-Golgi Trafficking at Late Stages
To determine the relationship of GRP1 to other genes whose products function in ER-to-Golgi transport, we crossed SFNY760 (grp1Δ::URA3) to all known mutants that accumulate ER and looked for synthetic lethal interactions (Table 2). Synthetic lethality or inviability of double mutants results when the effect of combining two mutations in the same haploid cell causes lethality under normally permissive conditions (Salminen and Novick, 1987; Botstein, 1988; Newman et al., 1990; Rossi et al., 1991; Sacher et al., 1997). Such interactions may indicate that the proteins encoded by the mutated genes are functionally related. When SFNY760 was crossed to mutants blocked in ER-to-Golgi traffic, such as sec34-2, sec34-1, and bos1-1, the majority of the tetrads yielded three viable colonies, with some having two or four viable colonies. None of the viable colonies was Ura+ and Ts−, indicating that the double mutants were inviable. In crosses with the sec17-1, bet1-1, sec22-3, sec35-1, and uso1-1 mutants, the majority of the tetrads yielded four viable colonies, although the double mutants were sick at 25 or 34°C. In contrast, no synthetic growth defects were observed with any ER-accumulating mutants defective in vesicle budding or with two different post-Golgi mutants (sec1-1 and sec10-2) (Table 2).
Table 2.
Synthetic growth effects between grp1Δ∷URA3 and the other mutations
| grp1Δ∷URA3 in combination with | 25°C | 30°C | 34°C |
|---|---|---|---|
| sec34-2 | Inviable | ||
| sec34-1 | Inviable | ||
| bos1-1 | Inviable | ||
| sec22-3 | Sick | ||
| sec35-1 | Sick | ||
| sec17-1 | Sick | ||
| uso1-1 | No effect | Sick | |
| bet1-1 | No effect | Sick | |
| ypt1-1 | No effect | No effect | |
| ypt1-3 | No effect | No effect | |
| sec7-1 | No effect | No effect | |
| sec18-1 | No effect | No effect | |
| sec12-4 | No effect | No effect | |
| sec13-1 | No effect | No effect | |
| sec16-2 | No effect | No effect | |
| sec23-1 | No effect | No effect | |
| sec1-1 | No effect | No effect | |
| sec10-2 | No effect | No effect |
Although the grp1Δ mutation displayed synthetic lethal interactions with mutations that result in a block in the late stages of ER-to-Golgi traffic, pulse-chase analysis of Grp1p-depleted cells did not reveal a significant delay in the transit of the vacuolar protease carboxypeptidase Y. This finding suggests that Grp1p may not play a direct role in membrane traffic. Additionally, coprecipitation studies with myc-tagged Sec34p did not reveal an interaction between Grp1p and Sec34p. Thus, although SEC34 interacts genetically with GRP1, the products of these genes do not appear to interact stably with each other.
The Loss of Sec34p Results in a Severe Growth Defect
Members of the first suppressor group were the strongest suppressors of sec34-2, suggesting that they contained the SEC34 structural gene. This was confirmed by an integration experiment. A 3.6-kb SphI-SphI fragment from pDK103 (Figure 1A) was ligated into the SphI site of YIp5, a vector that must integrate into the genome. The resulting plasmid was then cut at the SacI site within the ORF and transformed into SFNY26-4C to direct integration in the genome. This event placed the URA3 gene adjacent to the cloned sequence. One transformant was crossed to SFNY691, and the diploid was sporulated and dissected. The Ura+ and Ts+ phenotypes were found to cosegregate in all 15 tetrads analyzed, indicating that the identified ORF (YER157W) was tightly linked to the SEC34 gene.
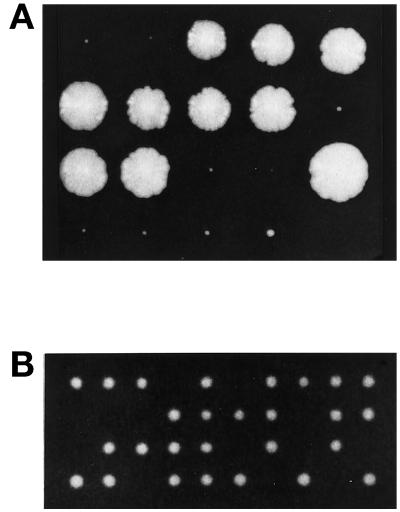
To determine if SEC34 encodes a protein that is essential for the growth of yeast cells, a diploid (SFNY919; see Table 1) with one disrupted copy of SEC34 was created by transforming SFNY562 with a linear fragment of DNA that harbored the disruption. This construct was made by replacing the promotor and coding regions of SEC34 with URA (Figure 1A, pDK104). The diploid, with one disrupted copy of SEC34, was sporulated and dissected on YPD plates that were incubated at 25 or 30°C. After 3 d, the 33 tetrads examined showed 2+:2− segregation for viability. After 5 d at 25 or 30°C, each of the tetrads contained two large and one or two tiny colonies (Figure 5A). The small colonies were Ura+, indicating that they contained a disrupted copy of SEC34. These findings demonstrate that SEC34 is not essential for growth, but in its absence yeast cells display a severe growth defect at 25 or 30°C. The disrupted colonies failed to grow at 14 and 37°C.
Figure 5.
(A) The loss of Sec34p results in a severe growth defect. The sec34Δ/sec34Δ strain was sporulated and dissected on a YPD plate that was incubated at 30°C for 7 d. (B) sec34-1 sec35-1 double mutants display a synthetic growth defect. The sec34-1 mutant was crossed to sec35-1, and the resulting diploid was sporulated, dissected on a YPD plate, and incubated at 25°C for 3 d. The colonies that failed to grow were inferred to be double mutants.
The C Terminus of Sec34p Is Dispensable at 25°C
The sequence of SEC34 encodes a hydrophilic protein of 801 amino acids with a predicted molecular mass of 92.5 kDa (actual size is ∼100 kDa on SDS-PAGE). The size of Sec34p was confirmed by Western blot analysis with the use of an antibody directed against this protein (Figures 6 and 7). A BLAST search of the database did not reveal any significant homologies, although with a window of 28 amino acids the N terminus is predicted to contain a coiled-coil domain between amino acids 87 and 114. This domain appears to be important for function because its deletion leads to the inability of SEC34 to complement the sec34 mutations (Figure 1B).
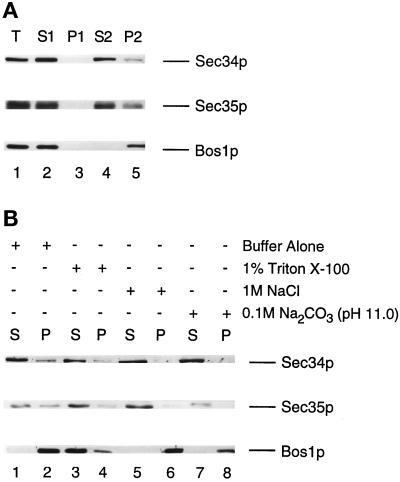
Figure 6.
(A) Sec34p and Sec35p are largely cytosolic. Wild-type cells were grown in YPD medium, converted to spheroplasts, and lysed as described in MATERIALS AND METHODS. Proteins were resolved on a 10% SDS-polyacrylamide gel and then subjected to Western blot analysis with the use of anti-Sec34p, anti-Sec35p, or anti-Bos1p antibodies. A total lysate (T) (lane 1) was centrifuged at 200 × g for 3 min to generate the S1 supernatant (lane 2) and P1 pellet (lane 3) fractions. The S1 fraction was centrifuged at 200,000 × g to obtain the S2 (lane 4) and P2 (lane 5) fractions. Equivalent amounts of the supernatant and pellet fractions were loaded onto the gel. (B) A fraction of Sec34p and Sec35p is peripherally associated with membranes. The S1 fraction was incubated for 45 min at 4°C with lysis buffer alone (lanes 1 and 2), 1% Triton X-100 in buffer (lanes 3 and 4), 1 M NaCl in buffer (lanes 5 and 6), or 0.1 M Na2CO3, pH 11.0 (lanes 7 and 8), and centrifuged at 200,000 × g to generate soluble (S) and insoluble (P) fractions. Equal amounts of each sample were electrophoresed on a 10% SDS-polyacrylamide gel and immunoblotted with anti-Sec34p, anti-Sec35p, and anti-Bos1p antibodies.
When we sequenced the two different mutations of SEC34, sec34-1 and sec34-2, by the method of gap repair (Orr-Weaver et al., 1983), we found that the sec34-1 mutation changed a single base pair at position 923 (T to A), altering amino acid 308 from a leucine to an ochre stop codon (Figure 1C). In the case of sec34-2, a base pair change at position 985 (C to T) altered amino acid 329 (glutamine) to an amber codon (Figure 1C). Thus, the sec34-1 and sec34-2 mutants encode truncated proteins of Sec34p with predicted molecular masses of ∼35 and ∼38 kDa, respectively. Strains containing these truncations grew somewhat slower than wild-type strains at the permissive temperature. These findings, together with the observation that the N-terminal coiled-coil domain of Sec34p is important for function, support the hypothesis that the C terminus of Sec34p is dispensable.
SEC34 Specifically Suppresses the Growth Defect of the sec35-1 Mutation
In an effort to identify genes whose products may interact with Sec34p, we used a second genetic screen. This screen relied on the ability of SEC34 (URA3, 2 μm) to suppress the growth defect of known mutations whose defective products may stably or transiently interact with Sec34p. When the ability of SEC34 to suppress all secretory mutations that block membrane traffic between the ER and the Golgi complex was tested (Table 3), we observed that SEC34 suppressed only sec35-1 and not mutations blocked at other stages of the pathway. This suppression was strong and specific (Figure 3B), suggesting a possible physical interaction between Sec34p and Sec35p. Surprisingly, the overproduction of SEC34 was also found to inhibit the growth of the sec9-4 mutant at 34°C (Table 3). SEC9 encodes the yeast exocytic t-SNARE (SNARE on the target membrane homologous to the mammalian SNAP-25 protein) that binds to the post-Golgi vesicle SNAREs (Brennwald et al., 1994).
Table 3.
Overproduction of SEC34 suppresses the sec35-1 mutation
| Mutant | Vector
|
SEC34
|
||||
|---|---|---|---|---|---|---|
| 30°C | 34°C | 37°C | 30°C | 34°C | 37°C | |
| ER accumulating | ||||||
| Vesicle budding | ||||||
| sec12-4 | + | − | − | + | − | − |
| sec13-1 | + | − | − | + | − | − |
| sec16-2 | − | − | − | − | − | − |
| sec23-1 | + | − | − | + | − | − |
| Vesicle targeting/fusion | ||||||
| sec17-1 | + | + | − | + | + | − |
| sec18-1 | + | − | − | + | − | − |
| sec22-3 | − | − | − | − | − | − |
| bos1-1 | − | − | − | − | − | − |
| sed5-1 | + | − | − | + | − | − |
| uso1-1 | + | − | − | + | − | − |
| sec34-1 | + | + | − | + | + | + |
| sec34-2 | + | + | − | + | + | + |
| sec35-1 | + | − | − | + | + | − |
| bet1-1 | + | + | − | + | + | − |
| bet3-1 | − | − | − | − | − | − |
| bet5-1 | + | − | − | + | − | − |
| ypt1-1 | + | + | − | + | + | − |
| Golgi complex accumulating | ||||||
| sec7-1 | + | + | − | + | + | − |
| Vesicle accumulating (post-Golgi complex) | ||||||
| sec1-1 | + | − | − | + | − | − |
| sec2-41 | + | − | − | + | − | − |
| sec3-2 | + | + | − | + | + | − |
| sec4-8 | − | − | − | − | − | − |
| sec5-24 | + | − | − | + | − | − |
| sec6-4 | + | − | − | + | − | − |
| sec8-9 | + | + | − | + | + | − |
| sec9-4 | + | + | − | + | − | − |
| sec10-2 | + | + | − | + | + | − |
| sec15-1 | + | − | − | + | − | − |
| Multiple steps | ||||||
| sec19-1 | + | − | − | + | − | − |
| Recycling (Golgi to ER) | ||||||
| sec20-1 | + | − | − | + | − | − |
| sec21-1 | + | − | − | + | − | − |
To further define the genetic interactions between SEC34 and SEC35, we crossed sec34-1 to sec35-1 and performed tetrad analysis. As shown in Figure 5B, the resulting tetrads displayed a pattern indicative of synthetic lethality. That is, of the 12 tetrads analyzed, 6 had three viable spores (2 of which were temperature sensitive) and 5 had two viable spores (none of which was temperature sensitive). One tetrad had four viable spores, all of which were temperature sensitive. This result, together with the suppression data, implies that SEC34 acts in conjunction with SEC35.
Both Sec34p and Sec35p Are Predominantly Soluble
Previous studies (Vanrheenen et al., 1998) have shown that Sec35p partitions with the cytosol and membranes. To determine if Sec34p behaves the same, differential centrifugation studies were performed (Figure 6A). Cells were converted to spheroplasts and lysed (lane 1), and the unlysed cells (lane 3) were removed during a low-speed spin. The supernatant (lane 2) was then centrifuged at 200,000 × g for 60 min to separate the soluble (lane 4) from the insoluble (lane 5) material. Sec34p was largely found in the S2 fraction (lane 4) along with Sec35p, whereas the integral membrane protein Bos1p was found in the P2 fraction (lane 5).
The fraction of Sec35p associated with membranes is extractable by Triton X-100, NaCl, and Na2CO3 (Vanrheenen et al., 1998). To determine if Sec34p behaves similarly, the ability of these reagents to extract this protein from the pellet fraction was examined (Figure 6B). Peripherally associated membrane proteins are extracted with high concentrations of salt or high pH, whereas integral membrane proteins require detergents such as Triton X-100 to be released from membranes (Howell and Palade, 1982). Extraction studies were performed by incubating samples in one of four reagents (buffer alone [control], 1% Triton X-100, 1 M NaCl, and 0.1 M Na2CO3, pH 11) for 45 min on ice. At the end of the incubation, the samples were centrifuged at 200,000 × g for 60 min, and the distribution of Sec34p, Sec35p, and the integral membrane protein Bos1p was monitored in the supernatant and pellet fractions by Western blot analysis. Most of the Bos1p was solubilized by 1% Triton X-100, as was some of the Sec34p and Sec35p. Other reagents such as NaCl and Na2CO3 efficiently released Sec34p and Sec35p from the pellet. Neither NaCl nor Na2CO3 released Bos1p from membranes. Thus, like Sec35p, Sec34p behaves like a peripheral membrane protein.
Sec34p Forms a Complex with Sec35p
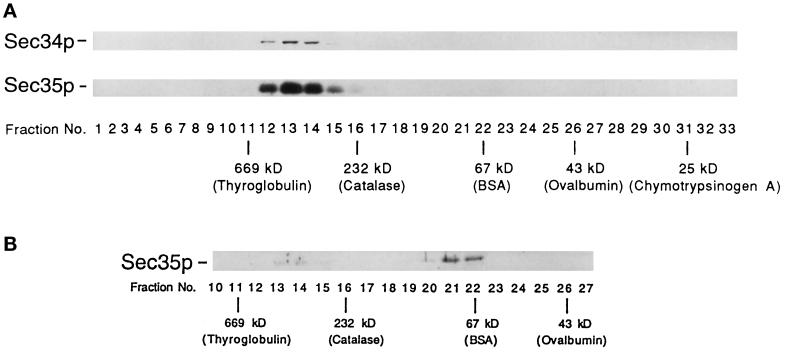
Several findings suggest that Sec34p and Sec35p may be members of the same complex. First, the depletion of Sec34p or Sec35p (Figure 5A; Vanrheenen et al., 1998) results in a severe growth defect. Second, the overproduction of SEC34 specifically suppresses sec35-1 but not other mutations that block membrane traffic between the ER and the Golgi complex (Table 3). Finally, sec35-1 displays synthetic lethal interactions when combined with sec34-1 (Figure 5B). To test the hypothesis that Sec34p physically interacts with Sec35p, we prepared yeast extracts from strains containing tagged (Sec34p-myc) and untagged Sec34p, as described in MATERIALS AND METHODS, and treated lysates with anti-c-myc antibody. Immunoprecipitates were subjected to electrophoresis, transferred to nitrocellulose, and blotted for the presence of Sec34p and Sec35p. As shown in Figure 7A, Sec35p was detected only in the immunoprecipitates from the strain that contained tagged protein (compare lane 2 and lane 1). When the lysate containing tagged Sec34p was pretreated with 1% SDS, Sec34p, but not Sec35p, was detected (lane 3), indicating that Sec35p coprecipitates with Sec34p only under nondenaturing conditions. Similar results were obtained when the same experiment was performed with a strain containing tagged Sec35p-myc. The size of the Sec34p/Sec35p complex was estimated by gel filtration chromatography of detergent-solubilized extracts on a Superose 6 column (Figure 8A). This analysis revealed that none of the cellular pool of Sec34p was monomeric, but instead it cofractionated with Sec35p at an estimated molecular mass of ∼480 kDa.
Figure 8.
Gel filtration analysis of the Sec34p/Sec35p complex. Lysates prepared from wild type (A) and the sec34-1 mutant (B) were chromatographed on a Superose 6 column. Wild-type fractions containing Sec34p and Sec35p (A) and mutant fractions containing Sec35p (B) were detected by Western blot analysis. The positions of the markers used to calibrate the column are shown. In B, Sec35p was not detected in any fractions other than those shown.
The native size of Sec34p suggests that it is a component of a multiprotein complex that contains Sec35p. To begin to identify other putative members of this complex, we prepared a radiolabeled lysate from a strain containing Sec35p-myc and precipitated the Sec35p-associated proteins with anti-c-myc antibody. In addition to Sec35p-myc, five other polypeptides (p91, p73, p68, p51, and Sec34p) were specifically precipitated (Figure 7B). These bands were precipitated only from a lysate that contained tagged Sec35p. Based on their cysteine and methionine content and quantitation of the radiolabeled Sec34p and Sec35p bands in Figure 7B, we estimated that these proteins are present in approximately equimolar amounts. Thus, Sec34p interacts stably with Sec35p and possibly several other proteins. The interaction between Sec34p and Sec35p appears to be important for function, because the majority of the Sec35p was not present in the 480-kDa complex in the sec34-1 mutant at 25°C (Figure 8B). The small amount of fully assembled complex (Figure 8B, lanes 13 and 14) may be sufficient to support growth at 25°C, although sec34-1 does grow somewhat slower than the wild type at this temperature.
DISCUSSION
Here we report the use of two different high-copy suppressor screens to identify genes whose products may physically interact with Sec34p. One screen was with sec34-2, a mutant defective in the docking or fusion of ER-derived vesicles with the Golgi apparatus. This screen resulted in the identification of the SEC34 structural gene as well as GRP1. The overexpression of GRP1 specifically suppresses a number of mutations that result in a defect in the targeting or fusion of ER-to-Golgi transport vesicles with the Golgi apparatus. Although GRP1 is not essential for the growth of yeast, the grp1Δ mutation displays synthetic lethal interactions with several mutations that result in ER accumulation and a block in the late stages of ER-to-Golgi transport, but not with those that block the budding of vesicles from the ER.
Grp1p contains a region that is homologous to a domain in golgin-160, a mammalian Golgi protein that is part of a Triton-insoluble matrix. Other putative Golgi matrix proteins include golgin-95 (Fritzler et al., 1993) and GM130. GM130 is the receptor for p115 (the mammalian homologue of Uso1p) (Nakamura et al., 1995, 1997), and golgin-95 is homologous to the C-terminal 616 residues of GM130 (76.3% identity). The vesicular docking protein p115 binds to GM130 on the Golgi and giantin on COPI-containing vesicles. This tethering event permits the regrowth of Golgi cisternae during interphase (Nakamura et al., 1997; Sönnichsen et al., 1998). Grp1p also resembles golgin-160 in that both proteins contain large domains that are predicted to form a coiled-coil structure. The homology shared between these proteins resides in these domains. Because the loss of Grp1p does not result in a significant delay in the transit of carboxypeptidase Y from the ER to the Golgi complex, our findings imply that Grp1p may facilitate membrane traffic indirectly, possibly by maintaining Golgi function. In support of this hypothesis, we have observed that a GFP (green fluorescent protein) fusion to Grp1p resides on punctate structures that are reminiscent of the Golgi (our unpublished results). Further experiments will be needed to precisely define the role of Grp1p.
In an effort to identify genes whose products may physically interact with Sec34p, we used a second genetic screen. This screen tested the ability of SEC34 to suppress known secretory mutations. SEC34 encodes a hydrophilic protein whose overexpression suppresses only sec35-1 and not other mutations that block ER-to-Golgi transport. This finding implies that SEC34 and SEC35 may function in conjunction with each other. Several lines of evidence support this hypothesis. First, both Sec35p and Sec34p are predominantly soluble. The fraction associated with membranes can be solubilized under the same conditions (Vanrheenen et al., 1998; Figure 6). Second, cells depleted of either Sec35p or Sec34p display similar growth phenotypes. The loss of either gene product severely hinders growth at 25 and 30°C and leads to the absence of growth at lower temperatures (Vanrheenen et al., 1998; Figure 5A). Finally, coprecipitation studies demonstrate that Sec34p and Sec35p are members of a complex that includes other components. The identity of these putative members (p91, p73, p68, and p51) awaits purification of the Sec34p/Sec35p complex.
Sec35p is one of several proteins that mediate the docking of vesicles to the Golgi apparatus. ER-derived vesicles formed in vitro have been shown to readily diffuse from permeabilized yeast cells that contain Golgi and other membranes. In the presence of functional Uso1p and Sec35p, vesicles no longer diffuse from these cells, presumably because they are tethered to Golgi retained within them (Barlowe, 1997; Vanrheenen et al., 1998). Based on the findings reported here, we conclude that Sec35p mediates this tethering event in conjunction with Sec34p. Although Sec34p and Sec35p physically interact with each other, SEC35 was not identified as a high-copy-number suppressor of the sec34-2 mutation. This is because the overexpression of SEC35 cannot suppress either sec34-2 or sec34-1 (our unpublished results). Thus, the suppression observed between SEC34 and SEC35 is not reciprocal
Genetic studies suggest that the Sec34p/Sec35p complex may act at other stages of the secretory pathway. For example, the overexpression of SEC34 was found to inhibit the growth of the sec9-4 mutant, which is defective in post-Golgi secretion (Table 3). Additionally, the overexpression of SNC2, a post-Golgi vesicle SNARE, has been reported to suppress sec35-1 (Vanrheenen et al., 1998). Together, these data suggest that the Sec34p/Sec35p complex may tether different classes of vesicles to their target membranes. Further studies are needed before any final conclusion can be drawn.
ACKNOWLEDGMENTS
We thank Monica Andreoli, Elaine Downie, and Judy Burston for technical assistance, Jemima Barrowman and Chavela Carr for helpful discussions and comments on the manuscript, R. Schekman for the sec34 and sec35 mutants, and Joyce Anquillare for help in the preparation of the manuscript. D.-W. K. and M.S. are supported as Associates of the Howard Hughes Medical Institute.
REFERENCES
- Barlowe C. Coupled ER to Golgi transport reconstituted with purified cytosolic proteins. J Cell Biol. 1997;139:1097–1108. doi: 10.1083/jcb.139.5.1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botstein D. Why study the cytoskeleton in yeast? Harvey Lect. 1988;82:157–167. [PubMed] [Google Scholar]
- Brennwald P, Kearns B, Champion K, Keranen S, Bankaitis V, Novick P. Sec9 is a SNAP-25-like component of a yeast SNARE complex that may be the effector of Sec4 function in exocytosis. Cell. 1994;79:245–258. doi: 10.1016/0092-8674(94)90194-5. [DOI] [PubMed] [Google Scholar]
- Carlson M, Botstein D. Two differentially regulated mRNAs with different 5′ ends encode secreted with intracellular forms of yeast invertase. Cell. 1982;28:145–154. doi: 10.1016/0092-8674(82)90384-1. [DOI] [PubMed] [Google Scholar]
- Ferro-Novick S, Jahn R. Vesicle fusion from yeast to man. Nature. 1994;370:191–193. doi: 10.1038/370191a0. [DOI] [PubMed] [Google Scholar]
- Fritzler MJ, Hamel JC, Ochs RL, Chan EKL. Molecular characterization of two human autoantigens: unique cDNAs encoding 95- and 160-kDa proteins of a putative family in the Golgi complex. J Exp Med. 1993;178:49–62. doi: 10.1084/jem.178.1.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howell KE, Palade G. Hepatic Golgi fractions resolved into membrane and content subfractions. J Cell Biol. 1982;92:822–832. doi: 10.1083/jcb.92.3.822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito H, Fukuda Y, Murata K, Kimura A. Transformation of intact yeast cells treated with alkali cations. J Bacteriol. 1983;153:163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lian JP, Ferro-Novick S. Bos1p, an integral membrane protein of the endoplasmic reticulum to Golgi transport vesicles, is required for their fusion competence. Cell. 1993;73:735–745. doi: 10.1016/0092-8674(93)90253-m. [DOI] [PubMed] [Google Scholar]
- Lian JP, Stone S, Jiang Y, Lyons P, Ferro-Novick S. Ypt1p implicated in v-SNARE activation. Nature. 1994;372:698–701. doi: 10.1038/372698a0. [DOI] [PubMed] [Google Scholar]
- McNew JA, Søgaard M, Lampen NM, Machida S, Ye RR, Lacomis L, Tempst P, Rothman JE, Söllner TH. Ykt6p, a prenylated SNARE essential for endoplasmic reticulum-Golgi transport. J Biol Chem. 1997;272:17776–17783. doi: 10.1074/jbc.272.28.17776. [DOI] [PubMed] [Google Scholar]
- Nakamura N, Lowe M, Levine TP, Rabouille C, Warren G. The vesicle docking protein p115 binds GM130, a cis-Golgi matrix protein, in a mitotically regulated manner. Cell. 1997;89:445–455. doi: 10.1016/s0092-8674(00)80225-1. [DOI] [PubMed] [Google Scholar]
- Nakamura N, Rabouille C, Watson R, Nilsson T, Hui N, Slusarewicz P, Kreis TE, Warren G. Characterization of a cis-Golgi matrix protein, GM130. J Cell Biol. 1995;131:1715–1726. doi: 10.1083/jcb.131.6.1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman AP, Shim J, Ferro-Novick S. BET1, BOS1, and SEC22 are members of a group of interacting yeast genes required for transport from the endoplasmic reticulum to the Golgi complex. Mol Cell Biol. 1990;10:3405–3414. doi: 10.1128/mcb.10.7.3405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr-Weaver TL, Szostak JW, Rothstein RJ. Genetic applications of yeast transformation with linear and gapped plasmids. Methods Enzymol. 1983;101:228–245. doi: 10.1016/0076-6879(83)01017-4. [DOI] [PubMed] [Google Scholar]
- Rossi G, Jiang Y, Newman AP, Ferro-Novick S. Dependence of Ypt1 and Sec4 membrane attachment on Bet2. Nature. 1991;351:158–161. doi: 10.1038/351158a0. [DOI] [PubMed] [Google Scholar]
- Rossi G, Kolstad K, Stone S, Palluault F, Ferro-Novick S. BET3 encodes a novel hydrophilic protein that acts in conjunction with yeast SNAREs. Mol Biol Cell. 1995;6:1769–1780. doi: 10.1091/mbc.6.12.1769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruohola H, Kabcenell AK, Ferro-Novick S. Reconstitution of protein transport from the endoplasmic reticulum to the Golgi complex in yeast: the acceptor compartment is defective in the sec23 mutant. J Cell Biol. 1988;107:1465–1476. doi: 10.1083/jcb.107.4.1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sacher M, Jiang Y, Barrowman J, Scarpa A, Burston J, Zhang L, Schieltz D, Yates JR, III, Abeliovich H, Ferro-Novick S. TRAPP, a highly conserved novel complex on the cis-Golgi that mediates vesicle docking and fusion. EMBO J. 1998;17:2494–2503. doi: 10.1093/emboj/17.9.2494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sacher M, Stone S, Ferro-Novick S. The snaptobrevin-related domains of Bos1p and Sec22p bind to the syntaxin-like region of Sed5p. J Biol Chem. 1997;272:17134–17138. doi: 10.1074/jbc.272.27.17134. [DOI] [PubMed] [Google Scholar]
- Salminen A, Novick PJ. A ras-like protein is required for a post-Golgi event in yeast secretion. Cell. 1987;49:527–538. doi: 10.1016/0092-8674(87)90455-7. [DOI] [PubMed] [Google Scholar]
- Sapperstein SK, Lupashin VV, Schmitt HD, Waters MG. Assembly of the ER to Golgi SNARE complex requires Uso1p. J Cell Biol. 1996;132:755–767. doi: 10.1083/jcb.132.5.755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider BL, Seufert W, Steiner B, Yang QH, Futcher AB. Use of PCR epitope tagging for protein tagging in Saccharomyces cerevisiae. Yeast. 1995;11:1265–1274. doi: 10.1002/yea.320111306. [DOI] [PubMed] [Google Scholar]
- Shim J, Newman AP, Ferro-Novick S. The BOS1 gene encodes an essential 27-kDa putative membrane protein that is required for vesicular transport from the ER to the Golgi complex in yeast. J Cell Biol. 1991;113:55–64. doi: 10.1083/jcb.113.1.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sönnichsen B, Lowe M, Levine T, Jämsä E, Dirac-Svejstrup B, Warren G. A role for giantin in docking COPI vesicles to Golgi membranes. J Cell Biol. 1998;140:1013–1021. doi: 10.1083/jcb.140.5.1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ungermann C, Nichols BJ, Pelham HRB, Wickner W. A vacuolar v-t-SNARE complex, the predominant form in vivo and on isolated vacuoles, is disassembled and activated for docking and fusion. J Cell Biol. 1998;140:61–69. doi: 10.1083/jcb.140.1.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanrheenen SM, Cao X, Lupashin VV, Barlowe C, Waters MG. Sec35p, a novel peripheral membrane protein, is required for ER to Golgi vesicle docking. J Cell Biol. 1998;141:1107–1119. doi: 10.1083/jcb.141.5.1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber T, Zemelman BV, McNew JA, Westermann B, Gmachl M, Parlati F, Söllner TH, Rothman JH. SNAREpins: minimal machinery for membrane fusion. Cell. 1998;92:759–772. doi: 10.1016/s0092-8674(00)81404-x. [DOI] [PubMed] [Google Scholar]
- Wuestehube LJ, Duden R, Eun A, Hamamoto S, Korn P, Ram R, Schekman R. New mutants of Saccharomyces cerevisiae affected in the transport of proteins from the endoplasmic reticulum to the Golgi complex. Genetics. 1996;142:393–406. doi: 10.1093/genetics/142.2.393. [DOI] [PMC free article] [PubMed] [Google Scholar]