Abstract
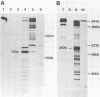
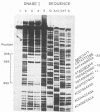
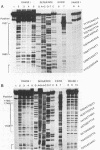
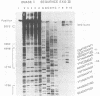
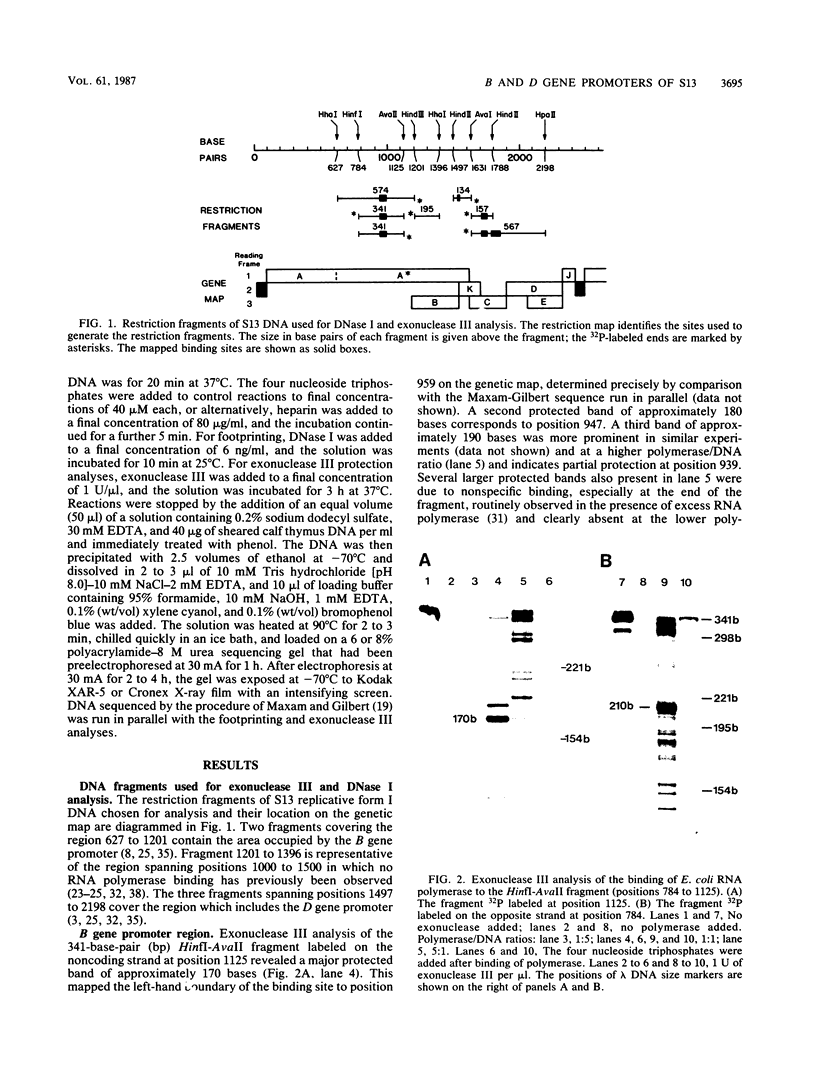
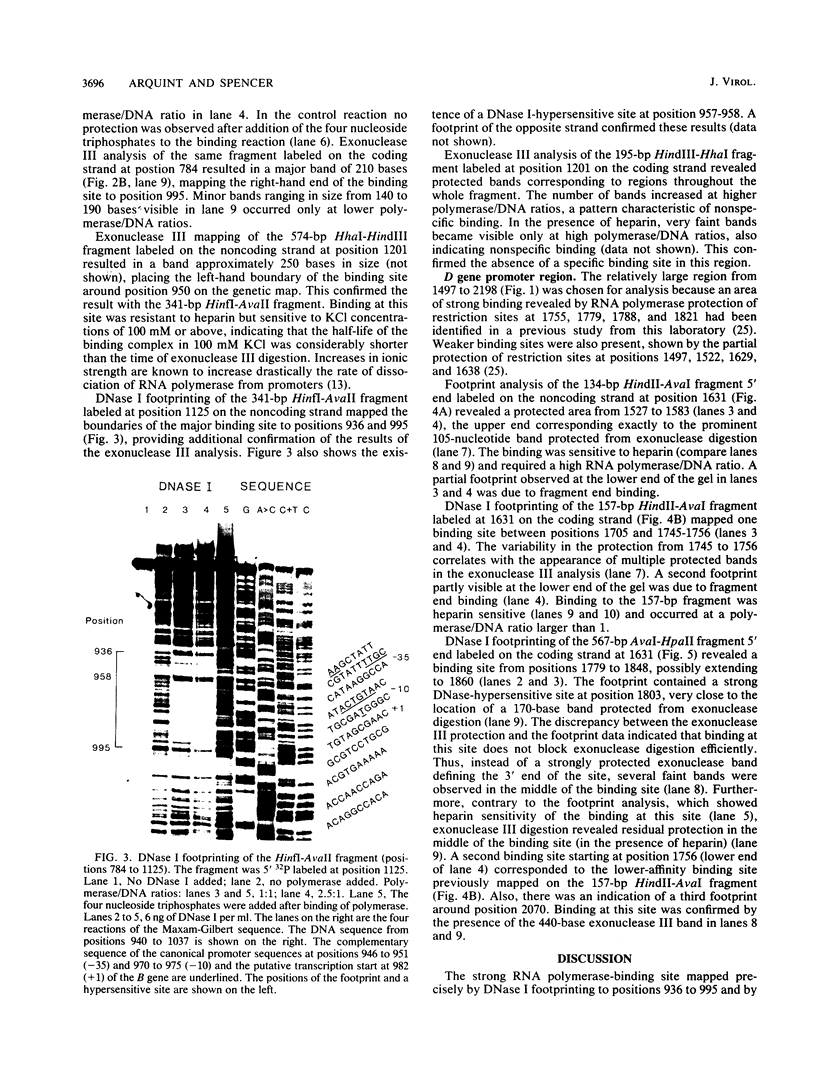
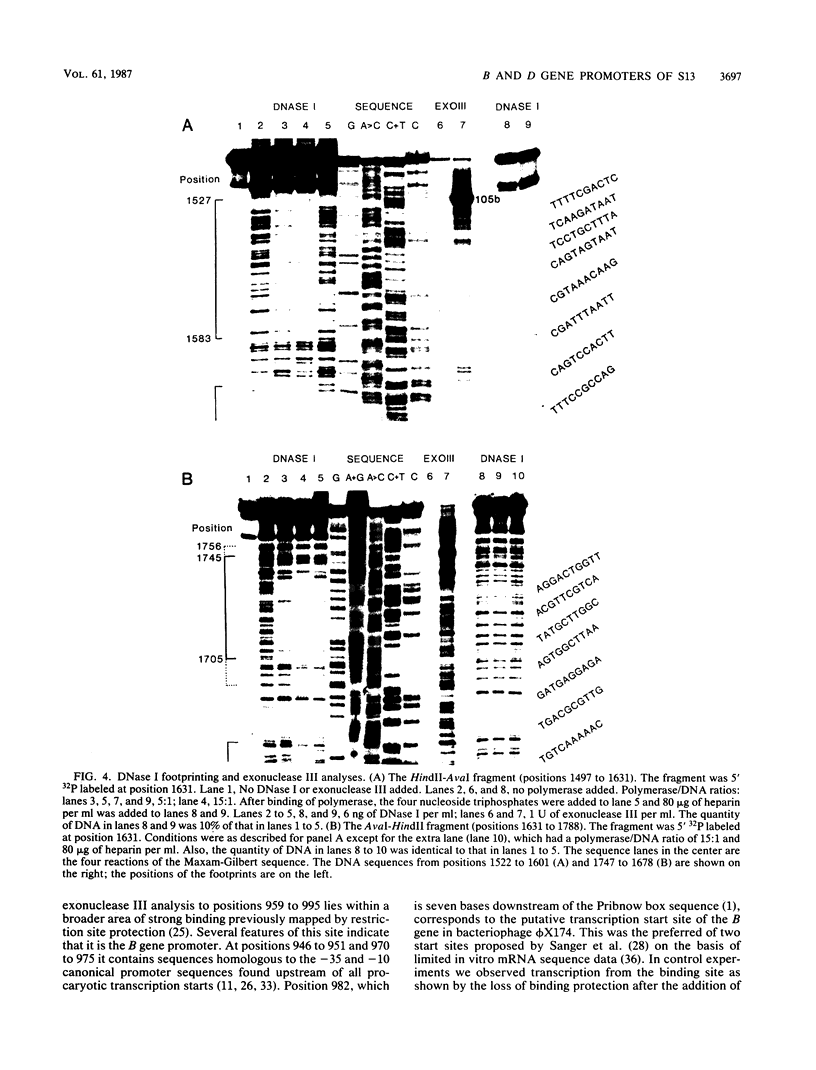
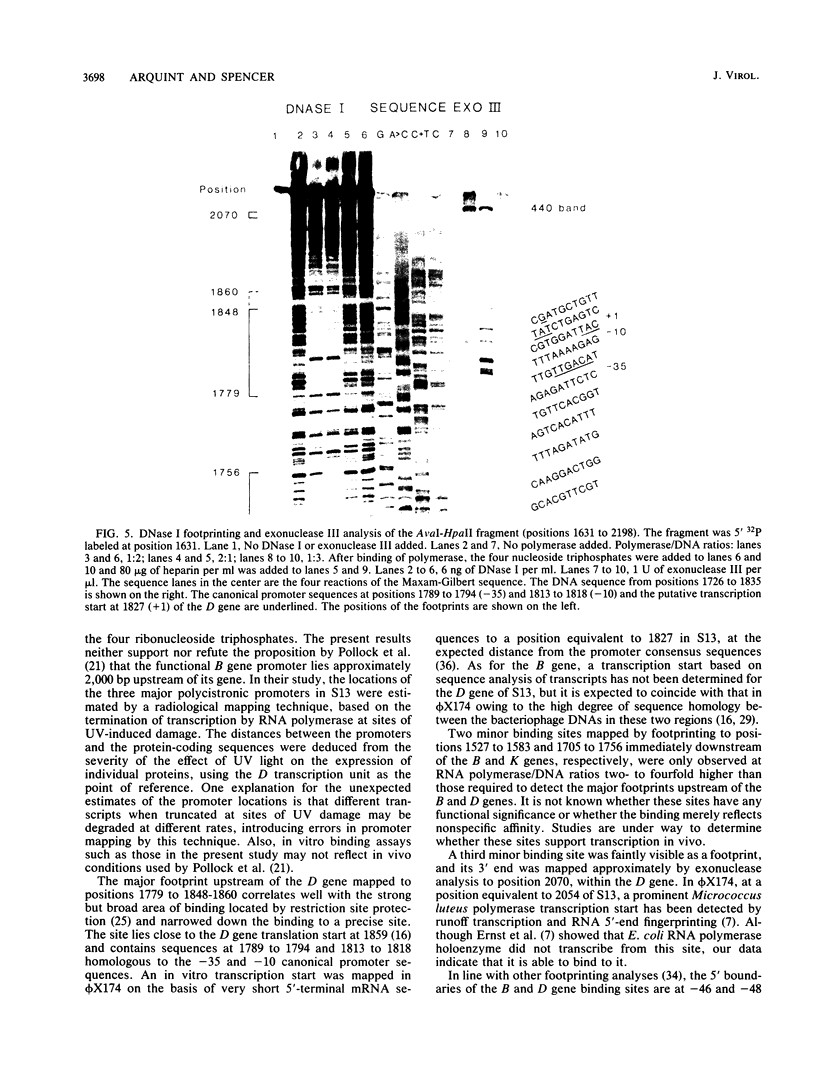
The region of the bacteriophage S13 genome which contains the B, K, and C genes and most of the A, D, and E genes (map positions 627 to 2198) was analyzed for Escherichia coli RNA polymerase-binding sites by combined DNase I footprinting, exonuclease III analysis, and DNA sequencing. Two high-affinity binding sites that correspond to the B and D gene promoters were mapped at positions 936 to 995 and 1779 to 1848, respectively. Positions 936 to 995 are in gene A, preceding the B gene, and positions 1779 to 1848 are in gene C, just preceding the D gene. In addition, two lower-affinity binding sites were identified at positions 1527 to 1538 and 1705 to 1756 preceding the C and D genes, respectively. The footprinting studies described here, in combination with previous studies on transcription, provide definitive evidence on the position of the B and D gene promoters in S13 and the closely related phage phi X174.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Aoyama T., Takanami M. Essential structure of E. coli promoter II. Effect of the sequences around the RNA start point on promoter function. Nucleic Acids Res. 1985 Jun 11;13(11):4085–4096. doi: 10.1093/nar/13.11.4085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Axelrod N. Transcription of bacteriophage phi-X174 in vitro: selective initiation with oligonucleotides. J Mol Biol. 1976 Dec 25;108(4):753–770. doi: 10.1016/s0022-2836(76)80115-5. [DOI] [PubMed] [Google Scholar]
- Axelrod N. Transcription of bacteriophage phiX174 in vitro: analysis with restriction enzymes. J Mol Biol. 1976 Dec 25;108(4):771–779. doi: 10.1016/s0022-2836(76)80116-7. [DOI] [PubMed] [Google Scholar]
- Bertrand-Burggraf E., Lefèvre J. F., Daune M. A new experimental approach for studying the association between RNA polymerase and the tet promoter of pBR322. Nucleic Acids Res. 1984 Feb 10;12(3):1697–1706. doi: 10.1093/nar/12.3.1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan P. T., Lebowitz J. The coupled use of 'footprinting' and exonuclease III methodology for RNA polymerase binding and initiation. Application for the analysis of three tandem promoters at the control region of colicin El. Nucleic Acids Res. 1983 Feb 25;11(4):1099–1116. doi: 10.1093/nar/11.4.1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLorbe W. J., Surzycki S., Gussin G. Inactivation of E. coli RNA polymerase by polyriboinosinic acid: heterogeneity of RS complexes. Mol Gen Genet. 1979 May 23;173(1):51–59. doi: 10.1007/BF00267690. [DOI] [PubMed] [Google Scholar]
- Ernst H., Hartmann G. R., Domdey H. Species specificity of promoter recognition by RNA polymerase and its transfer by the sigma factor. Eur J Biochem. 1982 Jun;124(3):427–433. doi: 10.1111/j.1432-1033.1982.tb06610.x. [DOI] [PubMed] [Google Scholar]
- Fedchenko V. I., Chestukhin A. V., Shemiakin M. F. Zavisimost' aktivnosti B-promotora bakteriofaga phiX174 v ékspressii gal-operona Escherichia coli ot chisla ego kopii i ikh orientatsii. Mol Biol (Mosk) 1983 Mar-Apr;17(2):418–429. [PubMed] [Google Scholar]
- Galas D. J., Schmitz A. DNAse footprinting: a simple method for the detection of protein-DNA binding specificity. Nucleic Acids Res. 1978 Sep;5(9):3157–3170. doi: 10.1093/nar/5.9.3157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawley D. K., McClure W. R. Compilation and analysis of Escherichia coli promoter DNA sequences. Nucleic Acids Res. 1983 Apr 25;11(8):2237–2255. doi: 10.1093/nar/11.8.2237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi M., Fujimura F. K., Hayashi M. Mapping of in vivo messenger RNAs for bacteriophage phiX-174. Proc Natl Acad Sci U S A. 1976 Oct;73(10):3519–3523. doi: 10.1073/pnas.73.10.3519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadesch T. R., Rosenberg S., Chamberlin M. J. Binding of Escherichia coli RNA polymerase holoenzyme to bacteriophage T7 DNA. Measurements of binding at bacteriophage T7 promoter A1 using a template competition assay. J Mol Biol. 1982 Feb 15;155(1):1–29. doi: 10.1016/0022-2836(82)90489-2. [DOI] [PubMed] [Google Scholar]
- Lau P. C., Spencer J. H. Nucleotide sequence and genome organization of bacteriophage S13 DNA. Gene. 1985;40(2-3):273–284. doi: 10.1016/0378-1119(85)90050-2. [DOI] [PubMed] [Google Scholar]
- Lomonossoff G. P., Butler P. J., Klug A. Sequence-dependent variation in the conformation of DNA. J Mol Biol. 1981 Jul 15;149(4):745–760. doi: 10.1016/0022-2836(81)90356-9. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. A new method for sequencing DNA. Proc Natl Acad Sci U S A. 1977 Feb;74(2):560–564. doi: 10.1073/pnas.74.2.560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeffer S. R., Stahl S. J., Chamberlin M. J. Binding of Escherichia coli RNA polymerase to T7 DNA. Displacement of holoenzyme from promoter complexes by heparin. J Biol Chem. 1977 Aug 10;252(15):5403–5407. [PubMed] [Google Scholar]
- Pollock T. J., Tessman I., Tessman E. S. Radiological mapping of functional transcription units of bacteriophages phiX174 and S13. The function of proximal and distal promoters. J Mol Biol. 1978 Sep 5;124(1):147–160. doi: 10.1016/0022-2836(78)90153-5. [DOI] [PubMed] [Google Scholar]
- Queen C., Rosenberg M. A promoter of pBR322 activated by cAMP receptor protein. Nucleic Acids Res. 1981 Jul 24;9(14):3365–3377. doi: 10.1093/nar/9.14.3365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rassart E., Spencer J. H. Localization of Escherichia coli RNA polymerase binding sites on bacteriophage S13 and phiX174 DNA: alignment with restriction enzyme maps. J Virol. 1978 Sep;27(3):677–687. doi: 10.1128/jvi.27.3.677-687.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rassart E., Spencer J. H., Zollinger M. Localization of Escherichia coli RNA polymerase binding sites on bacteriophage S13 and 0X174 DNAs by electron microscopy. J Virol. 1979 Jan;29(1):179–184. doi: 10.1128/jvi.29.1.179-184.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ringuette M., Spencer J. H. Localization of Escherichia coli RNA polymerase-binding sites on bacteriophage S13 replicative form I DNA by protection of restriction enzyme cleavage sites. J Virol. 1987 Jul;61(7):2297–2303. doi: 10.1128/jvi.61.7.2297-2303.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg M., Court D. Regulatory sequences involved in the promotion and termination of RNA transcription. Annu Rev Genet. 1979;13:319–353. doi: 10.1146/annurev.ge.13.120179.001535. [DOI] [PubMed] [Google Scholar]
- Russell D. R., Bennett G. N. Characterization of the beta-lactamase promoter of pBR322. Nucleic Acids Res. 1981 Jun 11;9(11):2517–2533. doi: 10.1093/nar/9.11.2517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Air G. M., Barrell B. G., Brown N. L., Coulson A. R., Fiddes C. A., Hutchison C. A., Slocombe P. M., Smith M. Nucleotide sequence of bacteriophage phi X174 DNA. Nature. 1977 Feb 24;265(5596):687–695. doi: 10.1038/265687a0. [DOI] [PubMed] [Google Scholar]
- Sanger F., Coulson A. R., Friedmann T., Air G. M., Barrell B. G., Brown N. L., Fiddes J. C., Hutchison C. A., 3rd, Slocombe P. M., Smith M. The nucleotide sequence of bacteriophage phiX174. J Mol Biol. 1978 Oct 25;125(2):225–246. doi: 10.1016/0022-2836(78)90346-7. [DOI] [PubMed] [Google Scholar]
- Schmitz A., Galas D. J. The interaction of RNA polymerase and lac repressor with the lac control region. Nucleic Acids Res. 1979 Jan;6(1):111–137. doi: 10.1093/nar/6.1.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shanblatt S. H., Revzin A. Kinetics of RNA polymerase-promoter complex formation: effects of nonspecific DNA-protein interactions. Nucleic Acids Res. 1984 Jul 11;12(13):5287–5306. doi: 10.1093/nar/12.13.5287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shemiakin M. F., Shumilov V. Iu. Zashchita RNK-polimerazoi Escherichia coli uchastkov replikativnoi formy I DNK faga phiX174, uznavaemykh restriktazami HindII, BspRI i AluI. Mol Biol (Mosk) 1981 Sep-Oct;15(5):1144–1157. [PubMed] [Google Scholar]
- Siebenlist U., Simpson R. B., Gilbert W. E. coli RNA polymerase interacts homologously with two different promoters. Cell. 1980 Jun;20(2):269–281. doi: 10.1016/0092-8674(80)90613-3. [DOI] [PubMed] [Google Scholar]
- Smith L. H., Sinsheimer R. L. The in vitro transcription units of bacteriophage phiX174. II. In vitro initiation sites of phiX174 transcription. J Mol Biol. 1976 Jun 5;103(4):699–710. doi: 10.1016/0022-2836(76)90204-7. [DOI] [PubMed] [Google Scholar]
- Smith L. H., Sinsheimer R. L. The in vitro transcription units of bacteriophage phiX174. III. Initiation with specific 5' end oligonucleotides of in vitro phiX174 RNA. J Mol Biol. 1976 Jun 5;103(4):711–735. doi: 10.1016/0022-2836(76)90205-9. [DOI] [PubMed] [Google Scholar]
- Vanderbilt A. S., Borrás M. T., Germeraad S., Tessman I., Tessman E. S. A promoter site and polarity gradients in phage S13. Virology. 1972 Oct;50(1):171–179. doi: 10.1016/0042-6822(72)90357-1. [DOI] [PubMed] [Google Scholar]
- Williams R. C., Fisher H. W. Electron microscopic determination of the preferential binding sites of Escherichia coli RNA polymerase to phi X174 replicative form DNA. J Mol Biol. 1980 Jul 5;140(3):435–439. doi: 10.1016/0022-2836(80)90393-9. [DOI] [PubMed] [Google Scholar]