Abstract
Cleft lip with or without cleft palate (CLP) is the most common craniofacial birth defect in humans. Recently, mutations in the WNT3 and Wnt9b genes, encoding two members of the Wnt family of signaling molecules, were found associated with CLP in human and mice, respectively. To investigate whether Wnt3 and Wnt9b directly regulate facial development, we analyzed their developmental expression patterns and found that both Wnt3 and Wnt9b are expressed in the facial ectoderm at critical stages of midfacial morphogenesis during mouse embryogenesis. Whereas Wnt3 mRNA is mainly expressed in the maxillary and medial nasal ectoderm, Wnt9b mRNA is expressed in maxillary, medial nasal and lateral nasal ectoderm. During lip fusion, Wnt9b, but not Wnt3, is expressed in the epithelial seam between the fusing medial and lateral nasal processes. Furthermore, we found that expression of TOPGAL, a transgenic reporter of activation of canonical Wnt signaling pathway, is specifically activated in the distal regions of the medial nasal, lateral nasal and maxillary processes prior to lip fusion. During lip fusion, the epithelial seam between the medial and lateral nasal processes as well as the facial mesenchyme directly beneath the fusing epithelia strongly expresses TOPGAL. These data, together with the CLP lip phenotype in WNT3−/− humans and Wnt9b−/− mutant mice, indicate that Wnt3 and Wnt9b signal through the canonical Wnt signaling pathway to regulate midfacial development and lip fusion.
Keywords: cleft lip, cleft palate, craniofacial development, facial morphogenesis, midface, lip fusion, TOPGAL, Wnt3, Wnt9b, Wnt signaling
Introduction
Cleft lip with or without cleft palate (CLP) has an occurrence of 1 in 500 to 2500 live births worldwide, which represents the most common craniofacial birth defect in humans (Vanderas, 1987; Schutte and Murray, 1999; Gorlin et al., 2001). The majority of CLP cases are nonsyndromic and the etiology is complex. Epidemiological studies consistently showed strong familial clustering and increased concordance in monozygotic twins (reviewed in Mitchell, 2002), indicating genetic predisposition to CLP. Genetic studies indicated that CLP is a multifactorial trait and recurrence patterns analysis suggested that there are likely three to fourteen CLP susceptibility loci in humans (Schliekelman and Slatkin 2002). The identity of these major CLP genes has remained elusive.
To facilitate identification of CLP susceptibility genes, several laboratories have carried out genetic analysis of CLP formation in mouse models because the morphogenetic and molecular processes of craniofacial development are strikingly similar in the mouse and human (e.g., Diehl and Erickson, 1997; Juriloff et al., 2001; Bush et al., 2004). One family of inbred mouse strains, the “A” strains, provides a particularly good model for understanding the genetic basis of nonsyndromic clefting because 5% to 30% of the newborns exhibit spontaneous CLP in different substrains and the genetic cause of this phenotype is determined by at least two interacting loci (Juriloff and Mah, 1995; Juriloff et al., 2001; 2004). A recessive gene, clf1, essential for CLP occurrence in the A strains was genetically mapped to distal chromosome 11 (Juriloff and Mah, 1995; Juriloff et al., 1996; 2001; 2004). Interestingly, the clf1 region of mouse chromosome 11 is syntenic to human Chromosome 17q21, a chromosome region strongly associated with nonsyndromic CLP in some human populations (Chenevix-Trench et al., 1992; Shaw et al. 1993; Mitchell 1994; Peanchitlertkajorn et al., 2003; Marazita et al. 2004; Moreno et al., 2004). The clf1 region contains two closely linked genes, Wnt3 and Wnt9b, encoding members of the Wnt family of secreted cysteine-rich glycoproteins (Juriloff et al., 2004; 2005). Interestingly, Niemann et al. (2004) recently showed the correlation of a nonsense mutation (Q83X) in the WNT3 gene with tetra-amelia and CLP in a large consanguineous family, indicating that Wnt3 plays important roles in craniofacial development.
The Wnt family contains 19 members in humans and mice. Wnts can bind cell surface receptors of the Frizzled (Fzd) family and signal through several different intracellular signal transduction pathways to regulate diverse developmental processes, including cell proliferation, cell polarity, cell fate determination, and patterning of the neural tube and limbs (reviewed in Cadigan and Nusse, 1997; Wodarz and Nusse, 1998). The best characterized Wnt signaling pathway, termed the canonical Wnt pathway, involves stabilization and nuclear transportation of β-catenin. In cells that do not receive a Wnt signal, cytoplasmic β-catenin is rapidly degraded through the ubiquitin-proteasome pathway. In cells responding to canonical Wnt signaling, β-catenin is stabilized and enters the nucleus to activate the Tcf/Lef family transcription factors and regulate transcription of downstream genes. Although several Wnt and Fzd genes as well as Tcf1 and Lef1 have been shown to be expressed in the developing facial primordia (Gavin et al., 1990; Oosterwegel et al. 1993; Parr et al., 1993; Christiansen et al., 1995; Wang and Shackleford, 1996; Borello et al., 1999), a role for Wnt signaling in facial morphogenesis was not known until the identification of the WNT3 mutation in humans (Niemann et al., 2004) and a recent report of the Wnt9b−/− mutant mouse phenotype that included incomplete penetrance of CLP (Carroll et al., 2005). Whereas no coding mutation in either the Wnt3 or Wnt9b genes has been found in the A strains, a retrotransposon insertion has been identified approximately 6.6 kb 3′ to the Wnt9b gene specifically in the clf1 allele (Juriloff et al., 2005). This retrotransposon is present in all the cleft mouse strains containing the clf1 allele and is not present in CBA/J, the ancestral strain to the A strains, indicating that the clf1 mutation most likely causes CLP by interfering with Wnt9b gene expression (Juriloff et al., 2005). However, a report of the spatiotemporal expression patterns of either Wnt3 or Wnt9b during craniofacial development is lacking and it is not known whether these genes directly regulate facial morphogenesis or whether the CLP phenotype results from secondary effects of the mutations. Moreover, although Tcf1 and Lef1 are expressed in the developing facial primordia (Oosterwegel et al. 1993), it is not known whether the canonical Wnt signaling pathway is actively involved in facial morphogenesis. We report here that both Wnt3 and Wnt9b are expressed in the developing facial ectoderm and that the canonical Wnt signaling pathway is activated during facial outgrowth and lip fusion.
Results and Discussion
Expression of Wnt3 and Wnt9b during craniofacial development
Since the clf1 mutation mostly likely affects the Wnt9b gene and since targeted disruption of Wnt9b causes CLP in mice (Carroll et al., 2005; Juriloff et al., 2005), we first examined Wnt9b mRNA expression during craniofacial development by using whole mount in situ hybridization of mouse embryos. By whole mount in situ hybridization analysis, Wnt9b mRNA expression is detected in the surface ectoderm covering the head and the branchial arches at E9.5 (Fig. 1A). Whereas the whole mount hybridization signal appears diffuse at this stage, specific Wnt9b mRNA expression in the surface ectoderm of the head and branchial arches is confirmed by in situ hybridization of paraffin sections of E9.5 embryos (Fig. 2A). By E10.5, Wnt9b mRNA is highly expressed in the facial primordia (Fig. 1B), in addition to strong expression in the Wolffian duct and the genital tubercle of the urogenital system as previously reported (Carroll et al., 2005). Facial views of the embryonic head region at this stage shows that Wnt9b mRNA is most strongly expressed in the distal regions of the medial and lateral nasal processes and of the adjacent maxillary processes (Fig. 1C). Wnt9b mRNA is also strongly expressed in the surface ectoderm of the mandibular processes (Fig. 1C).
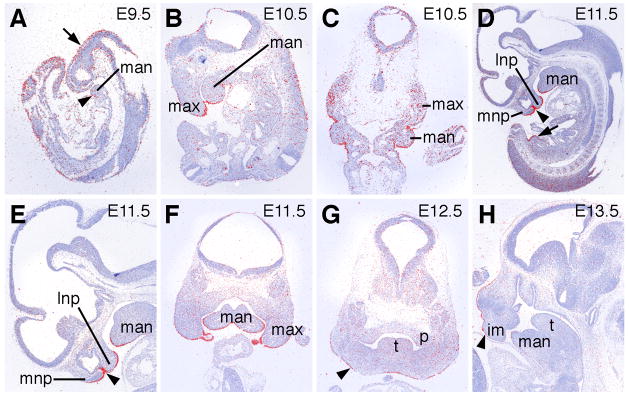
Fig. 1.
Expression of Wnt9b and Wnt3 mRNAs during craniofacial development in mice. (A) Lateral view of an E9.5 embryo showing Wnt9b mRNA expression in the head ectoderm. (B) Lateral view of an E10.5 embryo showing Wnt9b mRNA expression in the nephric duct (arrow), the genital tubercle (arrowhead), and in the facial ectoderm. (C) Facial view of the embryo in B showing strong Wnt9b mRNA expression in the distal epithelium of the medial nasal, lateral nasal, and maxillary processes as well as in the rostral surface of the mandibular processes. Arrowhead points to site of future lip fusion. (D) Lateral view of an E9.5 embryo hybridized with the Wnt3 cRNA probe. Wnt3 mRNA is expressed in the tailbud but not in the craniofacial region at this stage. (E) Lateral view of an E10.5 embryo hybridized with the Wnt3 cRNA probe. Wnt3 mRNA expression at this stage is weak except in the distal facial ectoderm. (F) Facial view of the embryo in E showing relatively abundant Wnt3 mRNA expression in the maxillary and rostral mandibular processes. Weak expression is seen on the oral surface of the medial nasal processes but not at the site of contact between the medial and lateral nasal processes (arrowhead). (G) Frontal section through the pre-fusion medial and lateral nasal processes of an E10.5 embryo shows that Wnt9b mRNA expression (blue staining) is restricted to the distal epithelia (arrowheads) in both processes. (H) At E11.0, when the medial and lateral nasal processes begin to fuse with each other, Wnt9b mRNA is expressed in the epithelial seam (arrowhead). ba2, second brancial arch; fl, forelimb bud; fnp, frontonasal process; hl, hindlimb bud; lnp, lateral nasal process; man, mandibular process; max, maxillary process; mnp, medial nasal process.
Fig. 2.
Section in situ hybridization analysis of Wnt9b mRNA expression during craniofacial development. mRNA signal is shown in red and cell nuclei are counterstained blue. (A) Sagittal section of an E9.5 embryo showing Wnt9b mRNA expression in the head ectoderm (arrow) as well as in the surface and neural ectoderm in the tail region. Weak expression is also detected in the ectoderm of the mandibular processes (arrowhead). (B) Sagittal section of an E10.5 embryo showing strong Wnt9b mRNA expression in the ectoderm covering the maxillary and mandibular processes. (C) Frontal section of an E10.5 embryo showing Wnt9b mRNA expression in the maxillary ectoderm and mesenchyme as well as in the mandibular ectoderm. (D) At E11.5, Wnt9b mRNA is strongly expressed and highly restricted in the facial ectoderm, including the epithelial seam between the fusing medial and lateral processes (arrowhead). Arrow points to moderate levels of Wnt9b mRNA in the ectoderm covering the genital tubercle. (E) Higher magnification view of the head region of the embryo section shown in D. Arrowhead points to the epithelial seam between the fusing medial and lateral nasal processes. (F) Frontal section through the distal region of the facial primordia of an E11.5 embryo showing highly restricted Wnt9b expression domain in the facial ectoderm of the maxillary and mandibular processes. (G) Frontal section of an E12.5 embryo showing decreased Wnt9b mRNA expression in the facial ectoderm (arrowhead) and the absence of Wnt9b expression in the secondary palate. (H) Sagittal section of an E13.5 embryo shows that Wnt9b mRNA is expressed at low but detectable levels in the facial ectoderm (arrowhead) at this stage. im, intermaxillary segment of the medial nasal processes; lnp, lateral nasal process; man, mandibular process; max, maxillary process; mnp, medial nasal process; p, palatal shelf; t, tongue.
Since CLP is part of the phenotype in WNT3−/− human fetuses with tetra-amelia and since the Wnt3 gene is closely linked to and shares the upstream region with the Wnt9b gene in both mice and humans, we also investigated the expression pattern of the Wnt3 gene. At E9.5, Wnt3 mRNA expression is detected in the tailbud but not in the developing head region (Fig. 1D). At E10.5, Wnt3 mRNA is expressed in the surface ectoderm of the distal medial nasal processes and the maxillary and mandibular processes, partially overlapping with Wnt9b mRNA expression in the facial primordia (Fig. 1E, F). In contrast to Wnt9b expression, Wnt3 mRNA is absent from the ectodermal contact sites between the medial and lateral nasal processes (arrowheads in Fig. 1C, F). This is further demonstrated by in situ hybridization of frozen sections of E10.5 and E11.5 mouse embryos. At E10.5, Wnt9b mRNA is expressed in the pre-fusion medial and lateral nasal ectoderm (Fig. 1G). This ectodermal expression of Wnt9b mRNA persists during fusion between the medial and lateral nasal processes at E11.0 (Fig. 1H). Together with the craniofacial defects observed in WNT3−/− human and Wnt9b−/− mice (Niemann et al., 2004; Carroll et al., 2005), these data indicate that Wnt3 and Wnt9b play distinct roles in midfacial development.
To gain further insight into the role of Wnt9b in craniofacial morphogenesis, we carried out in situ hybridization of paraffin sections of mouse embryos at different stages using radioactively labeled cRNA probes. Consistent with the whole mount in situ hybridization data shown in Fig. 1, Wnt9b mRNA is detected in the surface ectoderm of the head and the branchial arches at E9.5 (Fig. 2A). By E10.5, Wnt9b mRNA expression is significantly increased in the ectoderm of the facial primordia, including the maxillary and mandibular processes (Fig. 2B, C). Expression of Wnt9b mRNA is also detected in the maxillary mesenchyme at this stage (Fig. 2C). By E11.5, Wnt9b mRNA is strongly expressed in a highly restricted pattern in the facial ectoderm of the nasal, maxillary, and mandibular processes, including the epithelial seam between the fusing medial and lateral nasal processes (Fig. 2, D–F). Wnt9b mRNA expression is not detectable from the facial mesenchyme by E11.5 (Fig. 2E, F). By E12.5, Wnt9b mRNA expression in the facial ectoderm is substantially down-regulated but remained detectable above background levels throughout late embryogenesis (Fig. 2G, H). The spatiotemporal pattern of Wnt9b mRNA expression, together with the CLP phenotype in the Wnt9b−/− mutant mice, indicate that Wnt9b plays critical roles in midfacial morphogenesis through regulating outgrowth of the facial processes and/or fusion between the medial and lateral nasal processes. Wnt9b mRNA expression is not detected in the secondary palate throughout its development (Fig. 2G, and data not shown), suggesting that the cleft palate phenotype observed in the Wnt9b−/− mutant mice is a secondary effect of disruption of earlier facial developmental processes in these mutants.
The canonical Wnt signaling pathway is activated during lip fusion
Previous reports suggested that both Wnt3 and Wnt9b act through the β-catenin-mediated canonical signaling pathway during limb and urogenital development, respectively (Barrow et al., 2003; Carroll et al., 2005). To investigate whether the canonical Wnt signaling pathway is activated during facial primordial outgrowth and lip fusion, we examined the developmental expression patterns of TOPGAL, a transgenic lacZ reporter gene specifically activated in response to Tcf/Lef-mediated transcriptional activation by nuclear β-catenin (DasGupta and Fuchs, 1999; Merrill et al., 2004). At E9.5, expression of β-galactosidase activity from the TOPGAL transgene was detected in the midbrain and the migrating neural crest cells, including the trigeminal and facioacoustic ganglia and the neural crest streams migrating toward the branchial arches (Fig. 3A). Patchy expression of β-galactosidase activity was also detected in the forelimb ectoderm at this stage (Fig. 3A). By E10.5, strong β-galactosidase activity was detected in the dorsal telencephalon and mesencephalon, the cranial ganglia, and the apical ectodermal ridge of the limb buds (Fig. 3B). Strong β-galactosidase activity was also detected in the distal regions of the nasal, maxillary and mandibular processes at this stage (Fig. 3B, D). X-gal staining of cryostat sections of E10.5 embryos showed that TOPGAL is expressed in the ectoderm of the medial and lateral nasal processes as well as in the facial mesenchyme immediately adjacent to the lacZ-positive ectoderm (Fig. 3G). Between E11.0 and E11.5, the medial and lateral nasal processes fuse to each other in a proximal to distal zippering sequence (Trasler, 1968). Strong lacZ staining was detected in the pre-fusion medial and lateral nasal ectoderm as well as in the epithelial seam between the fusing nasal processes (Fig. 3H–J). Strong lacZ expression was also detected in the nasal mesenchyme adjacent to the expressing ectoderm in E11.5 and E12.5 embryos (Fig. 3E, F, I, J). However, no expression of the TOPGAL reporter transgene was detected in the developing palatal shelves of the secondary palate at these stages (data not shown). Thus, the expression pattern of the TOPGAL transgene in the developing facial primordia correlates with the Wnt9b mRNA expression pattern and indicates that the canonical Wnt signaling pathway is activated in both the facial ectoderm and adjacent facial mesenchyme during midfacial morphogenesis.
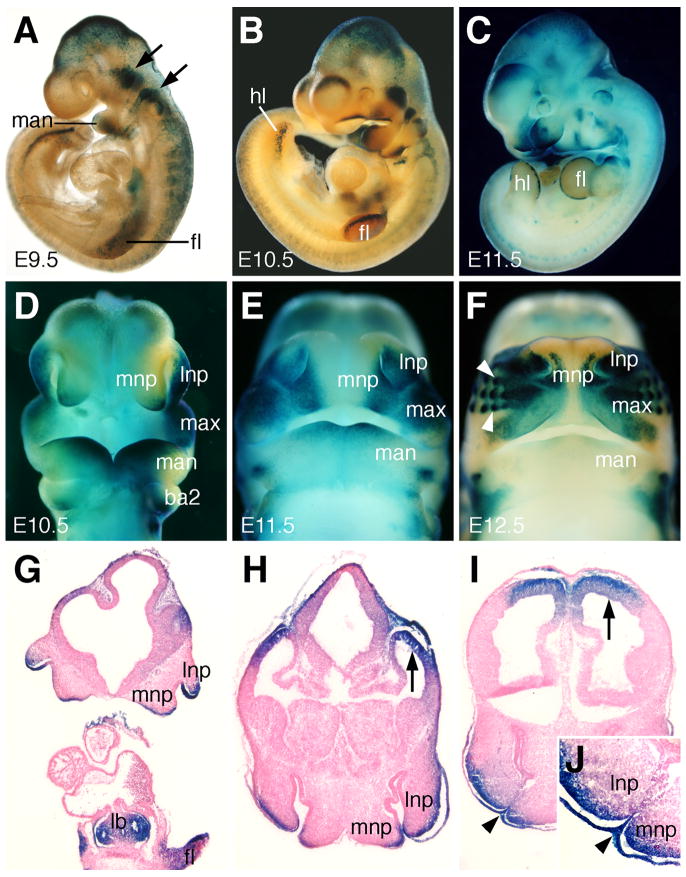
Fig. 3.
TOPGAL reporter gene expression during facial morphogenesis. (A–C) Lateral view of whole mount E9.5 (A), E10.5 (B) and E11.5 (C) hemizygous transgenic mouse embryos stained with X-gal. The TOPGAL expressing tissues are stained green/blue. (A) At E9.5, TOPGAL expression is detected in the migrating neural crest cells and cranial ganglia (arrows). A few cells in the distal mandibular arch are also TOPGAL-positive. (B, D) Lateral and facial views of E10.5 embryos show TOPGAL expression in the distal regions of maxillary, mandibular, medial nasal and lateral nasal processes. (C, E) At E11.5, TOPGAL is still expressed in the distal maxillary, medial nasal and lateral nasal processes as these processes fuse to form the upper lip. Expression in the mandibular process is restricted to the rostromedial region adjacent to the median groove. (F) At E12.5, as lip fusion is completed, TOPGAL expression is activated in the newly formed vibrissae (arrowheads). Expression in the mandible is significantly down regulated. (G) Frontal section of an E10.5 embryo showing TOPGAL expression in the lateral and medial nasal ectoderm, as well as in the limb bud and lung bud. (H) Frontal section of the ventral region of the medial and lateral nasal processes showing TOPGAL expression in the pre-fusion ectoderm and underlying mesenchyme of both nasal processes. Arrow points to expression in the dorsal telencephalon. (I, J) Frontal section of an E11.5 embryo face during the fusion between the medial and lateral nasal processes. J is a higher magnification view of the fusing medial and lateral nasal processes in I. TOPGAL is expressed strongly in the epithelial seam (arrowheads) between the nasal processes. Expression is also detected in the mesenchyme directly beneath the expressing ectoderm. Arrow points to TOPGAL expression domain in the dorsal telencephalon. ba2, second branchial arch; fl, forelimb bud; hl, hindlimb bud; lb, lung bud; lnp, lateral nasal process; man, mandibular process; max, maxillary process; mnp, medial nasal process.
Development of the midface and formation of the intact upper lip depend on reciprocal signaling interactions between the ectoderm and the neural crest-derived mesenchyme in the facial primordia (Wedden, 1987; Richman and Tickle, 1989; Ashique et al., 2002; reviewed in Francis-West et al., 1998; 2003). Several major signaling pathways, including Bmp, Fgf, and Shh pathways have been shown to play essential roles in midfacial outgowth and upper lip morphogenesis (Hu and Helms, 1999; Trumpp et al., 1999; Ashique et al., 2002; Trokovic et al., 2003; Jeong et al., 2004; Liu et al., 2005). In particular, tissue specific inactivation of either Bmp4 or Bmpr1a results in failure of lip fusion in mice (Liu et al., 2005). Our data showing expression of both Wnt9b mRNA and TOPGAL activity in the pre-fusion and fusing lip ectoderm, together with the CLP phenotype in Wnt9b−/− mutant mice (Carroll et al., 2005), suggest that Wnt9b activates the canonical Wnt signaling pathway to regulate lip fusion. Interestingly, both Wnt9b mRNA and TOPGAL expression overlap with the previously reported domains of Bmp4 expression in the distal ectoderm of the facial primordia prior to and during lip fusion (Gong and Guo, 2003; Liu et al., 2005). In several developmental processes, including during development of the lung and limbs, Wnt/β-catenin signaling has been shown to act upstream of Bmp4 (Barrow et al., 2003; Soshnikova et al., 2003; Shu et al., 2005). Moreover, the Bmp4 gene promoter contains several Tcf/Lef binding sites and cell transfection studies have shown direct activation of the Bmp4 promoter by activated canonical Wnt signaling (Shu et al., 2005). Thus, it is tempting to suggest that Wnt9b may act upstream of Bmp4 signaling to regulate lip fusion.
Methods
In situ hybridization and X-gal staining
Mouse embryos were collected at E9.5, E10.5 and E11.5, fixed in 4% paraformaldehyde overnight at 4 °C. Whole mount in situ hybridization was carried out as previously described (Jiang et al., 1998). For non-radioactive section in situ hybridization, embryos were washed in cold PBS, embedded in Tissue-Tek O.C.T. compound (Sakura Finetechnology Inc.), and cryosectioned at 10 μm thickness. Sections were refixed with 4% paraformaldehyde for 20 minutes, digested with proteinase K, refixed, washed and hybridized with digoxigenin-labeled Wnt3 or Wnt9b cRNA probes. Unbound probes were removed with stringent washes and the hybridization signals detected as previously described (Li and Joyner, 2001). The slides were further counterstained with nuclear fast red to visualize morphology. Section in situ hybridization using radioactive cRNA probes was carried out as previously described (Lan et al., 2001).
Expression of the TOPGAL transgene was detected by using X-gal staining. Timed matings of wildtype ICR female mice and TOPGAL transgenic male mice (purchased from the Jackson Laboratory, Bar Harbor, Maine) were set up and embryos dissected at E8.5 to E13.5. Fixation and X-gal staining procedures were performed as described previously (Jiang et al., 1998; Lan et al., 2004).
Acknowledgments
Grant Sponsor: NIH; Grant number: DE015207; Grant number: DE016215; grant number DE015291.
RCR is co-first author for this paper. We thank Tom Carroll and Andy McMahon for the Wnt3 and Wnt9b cDNA probes. We thank Hiroyuki Mishima and Brandon Newell for assistance. This work was supported by NIH/NIDCR grant DE015207 to RJ and by NIH/NIDCR grants P50DE016215 and KO2DE015291 along with a University of Iowa College of Dentistry seed grant to ACL.
References
- Ashique AM, Fu K, Richman JM. Endogenous bone morphogenetic proteins regulate outgrowth and epithelial survival during avian lip fusion. Development. 2002;129:4647–4660. doi: 10.1242/dev.129.19.4647. [DOI] [PubMed] [Google Scholar]
- Barrow JR, Thomas KR, Boussadia-Zahui O, Moore R, Kemler R, Capecchi MR, McMahon AP. Ectodermal Wnt3/beta -catenin signaling is required for the establishment and maintenance of the apical ectodermal ridge. Genes Dev. 2003;17:394–409. doi: 10.1101/gad.1044903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borello U, Buffa V, Sonnino C, Melchionna R, Vivarelli E, Cossu G. Differential expression of the Wnt putative receptors Frizzled during mouse somitogenesis. Mech Dev. 1999;89:173–177. doi: 10.1016/s0925-4773(99)00205-1. [DOI] [PubMed] [Google Scholar]
- Bush JO, Lan Y, Jiang R. The cleft lip and palate defects in the Dancer mutant mice result from gain of function of the Tbx10 gene. Proc Natl Acad Sci USA. 2004;101:7022–7027. doi: 10.1073/pnas.0401025101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cadigan KM, Nusse R. Wnt signaling: a common theme in animal development. Genes Dev. 1997;11:3286–3305. doi: 10.1101/gad.11.24.3286. [DOI] [PubMed] [Google Scholar]
- Carroll TJ, Park JS, Hayashi S, Majumdar A, McMahon AP. Wnt9b plays a central role in the regulation of mesenchymal to epithelial transitions underlying organogenesis of the mammalian urogenital system. Dev Cell. 2005;9:283–292. doi: 10.1016/j.devcel.2005.05.016. [DOI] [PubMed] [Google Scholar]
- Chenevix-Trench G, Jones K, Green AC, Duffy DL, Martin NG. Cleft lip with or without cleft palate: associations with transforming growth factor alpha and retinoic acid receptor loci. Am J Hum Genet. 1992;51:1377–1385. [PMC free article] [PubMed] [Google Scholar]
- Christiansen JH, Dennis CL, Wicking CA, Monkley SJ, Wilkinson DG, Wainwright BJ. Murine Wnt-11 and Wnt-12 have temporally and spatially restricted expression patterns during embryonic development. Mech Dev. 1995;51:341–350. doi: 10.1016/0925-4773(95)00383-5. [DOI] [PubMed] [Google Scholar]
- DasGupta R, Fuchs E. Multiple roles for activated LEF/TCF transcription complexes during hair follicle development and differentiation. Development. 1999;126:4557–4568. doi: 10.1242/dev.126.20.4557. [DOI] [PubMed] [Google Scholar]
- Diehl SR, Erickson RP. Genome scan for teratogen-induced clefting susceptibility loci in the mouse: evidence of both allelic and locus heterogeneity distinguishing cleft lip and cleft palate. Proc Natl Acad Sci USA. 1997;94:5231–5236. doi: 10.1073/pnas.94.10.5231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francis-West P, Ladher R, Barlow A, Graveson A. Signalling interactions during facial development. Mech Dev. 1998;75:3–28. doi: 10.1016/s0925-4773(98)00082-3. [DOI] [PubMed] [Google Scholar]
- Francis-West PH, Robson L, Evans DJ. Craniofacial development: the tissue and molecular interactions that control development of the head. Adv Anat Embryol Cell Biol. 2003;169(III–VI):1–138. doi: 10.1007/978-3-642-55570-1. [DOI] [PubMed] [Google Scholar]
- Gavin BJ, McMahon JA, McMahon AP. Expression of multiple novel Wnt-1/int-1-related genes during fetal and adult mouse development. Genes Dev. 1990;4:2319–2332. doi: 10.1101/gad.4.12b.2319. [DOI] [PubMed] [Google Scholar]
- Gong SG, Guo C. Bmp4 gene is expressed at the putative site of fusion in the midfacial region. Differentiation. 2003;71:228–236. doi: 10.1046/j.1432-0436.2003.710304.x. [DOI] [PubMed] [Google Scholar]
- Gorlin RJ, Cohen MM, Jr, Hennekam RCM. Syndromes of the Head and Neck. Oxford University Press; New York: 2001. [Google Scholar]
- Hu D, Helms JA. The role of sonic hedgehog in normal and abnormal craniofacial morphogenesis. Development. 1999;126:4873–4884. doi: 10.1242/dev.126.21.4873. [DOI] [PubMed] [Google Scholar]
- Jeong J, Mao J, Tenzen T, Kottmann AH, McMahon AP. Hedgehog signaling in the neural crest cells regulates the patterning and growth of facial primordia. Genes Dev. 2004;18:937–951. doi: 10.1101/gad.1190304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang R, Lan Y, Norton CR, Sundberg JP, Gridley T. The Slug gene is not essential for mesoderm or neural crest development in mice. Dev Biol. 1998;198:277–285. [PubMed] [Google Scholar]
- Juriloff DM, Harris MJ, Brown CJ. Unravelling the complex genetics of cleft lip in the mouse model. Mamm Genome. 2001;12:426–435. doi: 10.1007/s003350010284. [DOI] [PubMed] [Google Scholar]
- Juriloff DM, Harris MJ, Dewell SL. A digenic cause of cleft lip in A-strain mice and definition of candidate genes for the two loci. Birth Defects Res A Clin Mol Teratol. 2004;70:509–518. doi: 10.1002/bdra.20041. [DOI] [PubMed] [Google Scholar]
- Juriloff DM, Harris MJ, Dewell SL, Brown CJ, Mager DL, Gagnier L, Mah DG. Investigations of the genomic region that contains the clf1 mutation, a causal gene in multifactorial cleft lip and palate in mice. Birth Defects Res A Clin Mol Teratol. 2005;73:103–113. doi: 10.1002/bdra.20106. [DOI] [PubMed] [Google Scholar]
- Juriloff DM, Harris MJ, Mah DG. The clf1 gene maps to a 2- to 3-cM region of distal mouse chromosome 11. Mamm Genome. 1996;7:789. doi: 10.1007/s003359900298. [DOI] [PubMed] [Google Scholar]
- Juriloff DM, Mah DG. The major locus for multifactorial nonsyndromic cleft lip maps to mouse chromosome 11. Mamm Genome. 1995;6:63–69. doi: 10.1007/BF00303246. [DOI] [PubMed] [Google Scholar]
- Lan Y, Kingsley PD, Cho ES, Jiang R. Osr2, a new mouse gene related to Drosophila odd-skipped exhibits dynamic expression patterns during craniofacial, limb, and kidney development. Mech Dev. 2001;107:175–179. doi: 10.1016/s0925-4773(01)00457-9. [DOI] [PubMed] [Google Scholar]
- Lan Y, Ovitt CE, Cho E-S, Maltby KM, Wang Q, Jiang R. Odd-skipped related 2 (Osr2) encodes a key intrinsic regulator of secondary palate growth and morphogenesis. Development. 2004;131:3207–3216. doi: 10.1242/dev.01175. [DOI] [PubMed] [Google Scholar]
- Li JY, Joyner AL. Otx2 and Gbx2 are required for refinement and not induction of mid-hindbrain gene expression. Development. 2001;128:4979–4991. doi: 10.1242/dev.128.24.4979. [DOI] [PubMed] [Google Scholar]
- Liu P, Wakamiya M, Shea MJ, Albrecht U, Behringer RR, Bradley A. Requirement for Wnt3 in vertebrate axis formation. Nat Genet. 1999;22:361–365. doi: 10.1038/11932. [DOI] [PubMed] [Google Scholar]
- Liu W, Sun X, Braut A, Mishina Y, Behringer RR, Mina M, Martin JF. Distinct functions for Bmp signaling in lip and palate fusion in mice. Development. 2005;132:1453–1461. doi: 10.1242/dev.01676. [DOI] [PubMed] [Google Scholar]
- Marazita ML, Murray JC, Lidral AC, Arcos-Burgos M, Cooper ME, Goldstein T, Maher BS, et al. Meta-Analysis of 13 Genome Scans Reveals Multiple Cleft Lip/Palate Genes with Novel Loci on 9q21 and 2q32–35. Am J Hum Genet. 2004;75:161–173. doi: 10.1086/422475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merrill BJ, Pasolli HA, Polak L, Rendl M, Garcia-Garcia MJ, Anderson KV, Fuchs E. Tcf3: a transcriptional regulator of axis induction in the early embryo. Development. 2004;131:263–274. doi: 10.1242/dev.00935. [DOI] [PubMed] [Google Scholar]
- Mitchell LE. Interpreting the evidence for an association between the retinoic acid receptor locus and non-syndromic cleft lip with or without cleft palate. J Med Genet. 1994;31:425. doi: 10.1136/jmg.31.5.425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell LE. Twin studies in oral cleft research. In: Wyszynski DF, editor. Cleft Lip & Palate, From Origin to Treatment. Oxford University Press; New York: 2002. pp. 234–239. [Google Scholar]
- Moreno LM, Arcos-Burgos M, Marazita ML, Krahn K, Maher BS, Cooper ME, Valencia-Ramirez CR, Lidral AC. Genetic analysis of candidate loci in non-syndromic cleft lip families from Antioquia-Colombia and Ohio. Am J Med Genet A. 2004;125:135–144. doi: 10.1002/ajmg.a.20425. [DOI] [PubMed] [Google Scholar]
- Niemann S, Zhao C, Pascu F, Stahl U, Aulepp U, Niswander L, Weber JL, Muller U. Homozygous WNT3 mutation causes tetra-amelia in a large consanguineous family. Am J Hum Genet. 2004;74:558–563. doi: 10.1086/382196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oosterwegel M, van de Wetering M, Timmerman J, Kruisbeek A, Destree O, Meijlink F, Clevers H. Differential expression of the HMG box factors TCF-1 and LEF-1 during murine embryogenesis. Development. 1993;118:439–448. doi: 10.1242/dev.118.2.439. [DOI] [PubMed] [Google Scholar]
- Parr BA, Shea MJ, Vassileva G, McMahon AP. Mouse Wnt genes exhibit discrete domains of expression in the early embryonic CNS and limb buds. Development. 1993;119:247–261. doi: 10.1242/dev.119.1.247. [DOI] [PubMed] [Google Scholar]
- Peanchitlertkajorn S, Cooper ME, Liu YE, Field LL, Marazita ML. Chromosome 17: gene mapping studies of cleft lip with or without cleft palate in Chinese families. Cleft Palate Craniofac J. 2003;40:71–79. doi: 10.1597/1545-1569_2003_040_0071_cgmsoc_2.0.co_2. [DOI] [PubMed] [Google Scholar]
- Richman JM, Tickle C. Epithelia are interchangeable between facial primordia of chick embryos and morphogenesis is controlled by the mesenchyme. Dev Biol. 1989;136:201–210. doi: 10.1016/0012-1606(89)90142-5. [DOI] [PubMed] [Google Scholar]
- Schliekelman P, Slatkin M. Multiplex relative risk and estimation of the number of loci underlying an inherited disease. Am J Hum Genet. 2002;71:1369–1385. doi: 10.1086/344779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schutte BC, Murray JC. The many faces and factors of orofacial clefts. Hum Mol Genet. 1999;8:1853–1859. doi: 10.1093/hmg/8.10.1853. [DOI] [PubMed] [Google Scholar]
- Shaw D, Ray A, Marazita M, Field L. Further evidence of a relationship between the retinoic acid receptor alpha locus and nonsyndromic cleft lip with or without cleft palate. American Journal of Human Genetics. 1993;53:1156–1157. [PMC free article] [PubMed] [Google Scholar]
- Shu W, Guttentag S, Wang Z, Andl T, Ballard P, Lu MM, Piccolo S, Birchmeier W, Whitsett JA, Millar SE, Morrisey EE. Wnt/b-catenin signaling acts upstream of N-myc, BMP4, and FGF signaling to regulate proximal-distal patterning in the lung. Dev Biol. 2005;283:226–239. doi: 10.1016/j.ydbio.2005.04.014. [DOI] [PubMed] [Google Scholar]
- Soshnikova N, Zechner D, Huelsken J, Mishina Y, Behringer RR, Taketo MM, Crenshaw EB, 3rd, Birchmeier W. Genetic interaction between Wnt/beta-catenin and BMP receptor signaling during formation of the AER and the dorsal-ventral axis in the limb. Genes Dev. 2003;17:1963–1968. doi: 10.1101/gad.263003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trasler DG. Pathogenesis of cleft lip and its relation to embryonic face shape in A-J and C57BL mice. Teratology. 1968;1:33–49. doi: 10.1002/tera.1420010106. [DOI] [PubMed] [Google Scholar]
- Trokovic N, Trokovic R, Mai P, Partanen J. Fgfr1 regulates patterning of the pharyngeal region. Genes Dev. 2003;17:141–153. doi: 10.1101/gad.250703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trumpp A, Depew MJ, Rubenstein JL, Bishop JM, Martin GR. Cre-mediated gene inactivation demonstrates that FGF8 is required for cell survival and patterning of the first branchial arch. Genes Dev. 1999;13:3136–3148. doi: 10.1101/gad.13.23.3136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanderas AP. Incidence of cleft lip, cleft palate, and cleft lip and palate among races: a review. Cleft Palate J. 1987;24:216–225. [PubMed] [Google Scholar]
- Wang J, Shackleford GM. Murine Wnt10a and Wnt10b: cloning and expression in developing limbs, face and skin of embryos and in adults. Oncogene. 1996;13:1537–1544. [PubMed] [Google Scholar]
- Wedden SE. Epithelial-mesenchymal interactions in the development of chick facial primordia and the target of retinoid action. Development. 1987;99:341–351. doi: 10.1242/dev.99.3.341. [DOI] [PubMed] [Google Scholar]
- Wodarz A, Nusse R. Mechanisms of Wnt signaling in development. Annu Rev Cell Dev Biol. 1998;14:59–88. doi: 10.1146/annurev.cellbio.14.1.59. [DOI] [PubMed] [Google Scholar]