Abstract
We present a mixture model-based analysis for identifying differences in the distribution of RNA polymerase II (Pol II) in transcribed regions, measured using ChIP-seq (chromatin immunoprecipitation following massively parallel sequencing technology). The statistical model assumes that the number of Pol II-targeted sequences contained within each genomic region follows a Poisson distribution. A Poisson mixture model was then developed to distinguish Pol II binding changes in transcribed region using an empirical approach and an expectation-maximization (EM) algorithm developed for estimation and inference. In order to achieve a global maximum in the M-step, a particle swarm optimization (PSO) was implemented. We applied this model to Pol II binding data generated from hormone-dependent MCF7 breast cancer cells and antiestrogen-resistant MCF7 breast cancer cells before and after treatment with 17β-estradiol (E2). We determined that in the hormone-dependent cells, ~9.9% (2527) genes showed significant changes in Pol II binding after E2 treatment. However, only ~0.7% (172) genes displayed significant Pol II binding changes in E2-treated antiestrogen-resistant cells. These results show that a Poisson mixture model can be used to analyze ChIP-seq data.
Introduction
Massively parallel sequencing is a high-throughput technology capable of sequencing hundreds of thousands of DNA fragments in a single experiment. Combined with antibody-based chromatin immunoprecipitation assay (or ChIP-seq assay), this technology has been demonstrated to be a comprehensive, quantitative and cost-effective approach for mapping protein-DNA interaction on a genome-wide scale [1]. One ChIP-seq run can generate more than 20 million sequence tags of up to 36 bps each, which can then be definitively mapped to the human genome.
Due to the large amount of data generated in high-throughput sequencing experiments, innovative computational and statistical approaches are required to identify biological signals from ChIP-seq data. To date, several approaches have been applied to identify genomic regions containing a high concentration of sequence hits, i.e., "ChIP-seq peaks" [1]. An underlying assumption of current approaches is that DNA-binding proteins, such as transcription factors, contain sequence-specific, DNA binding domains that target a cluster of cis-acting DNA elements sharing certain sequence features. While such algorithms can identify DNA binding sites for highly specific transcription factors, current approaches are not appropriate for identifying binding sites for the general transcriptional machinery, such as RNA polymerase II (Pol II), which typically does not display high sequence specificity. In addition, as Pol II activity likely extends beyond the promoter/transcription start site of active genes, algorithms for assessing long-range Pol II binding are needed. Therefore, in this study, we have proposed a mixture model-based analysis for identifying differences in Pol II distribution in combined 5'-end, open reading frame (ORF), and 3'-untranscribed regions of active genes. Our strategy is based on the underlying assumption that the number of Pol II-target sequences follows a Poisson distribution and can be used to identify differentially transcribed genes under different experimental conditions. Furthermore, our proposed methodology can be used for making statistical inferences in experiments for which replicates are not available.
To date statistical models for ChIP-seq data are very limited. Nevertheless, new algorithms for ChIP-seq can be developed using existing framework for identifying differentially expressed genes from microarray data. For example, Kerr et al. [2] and Wolfinger et al. [3] employed analysis of variance (ANOVA) models to conduct a hypothetical test for different expression levels of individual genes in multiple microarray experiments. Dudoit et al. [4] used a t-statistic to address the problem of multiple comparisons through permutation analysis. These approaches yielded only p-values representing the probability of having an observed expression difference of a given gene, if the status is assumed to be the same before and after a treatment. However, for Pol II binding data derived from ChIP-seq, these approaches cannot be used to estimate the status change. A more appropriate choice analyzing ChIP-seq Pol II binding data may be a Bayes or an empirical Bayes approach. In this regard, Efron et al. [5] proposed an empirical Bayes model for calculating the probability of a differentially expressed gene given the observed data. As the empirical distribution for their t-like statistic for each gene does not share variation information, it works well only in situations involving at least a few replicates. Newton et al. [6] proposed another empirical Bayes model for cDNA microarray experiments with only one replicate. According to their method, if a gene is differentially expressed under two conditions, its level of expression is independently generated from the same distribution [6]; otherwise, the level of expression is the same between experimental and control samples. In their work, the observational component follows a Gamma distribution with mean μg, and μg itself follows an inverse Gamma distribution (prior component). This hierarchical model is often referred to as the Gamma-Gamma model. Specifically, all genes share the same distribution for the within-gene sampling errors, a crucial feature in their method, as no replicates were available in their data example. Kendziorski et al. [7] further extended the Gamma-Gamma model to situations where replicates were available. In addition, they developed a log-normal model for the observational component and a normal model for the prior component. They demonstrated a comparable performance for a lognormal-normal model and a Gamma-Gamma model. A major advantage in the methods proposed by Newton et al. [6] and Kendziorski et al. [7] is that information sharing is a consequence of the empirical Bayes approach. The model pools the variation information across all genes, making it well suited for data sets containing only a few replicates (e.g., 2 replicates), and we have successfully utilized this model framework to test the correlation among genome wide gene expression, DNA methylation, and histone acetylation [8,9].
In the current paper, we propose a different model, Poisson mixture model, within the same empirical Bayes framework for identifying gene targets with differential Pol II binding activities in breast cancer MCF7 cell line under various conditions. ChIP-seq data processing, normalization, and statistical methods are proposed in the method section; analysis of ChIP-seq data from breast cancer cell lines (MCF7 and its tamoxifen-resistant subline OHT-MCF7) before and after treatment with 17β-estradial (E2), are presented in the result section, with conclusions at the end of the paper.
Methods
As described by Fan et al [10], MCF7 human breast cancer cells (American Type Culture Collection, Manassas, VA) and MCF7-OTH cells were cultured and treated with E2 (10-8 mol/L) for three hours. Then, cells were cross-linked with 1% formaldehyde and chromatin immunoprecipitation was done as previously described [11]. The antibodies against Pol II were purchased from Santa Cruz Biotechnology (Santa Cruz, sc-899 X and sc-8005 X). After immunoprecipitation and purification, ChIP DNA sample was run in 12% PAGE and the 100–300 bp DNA fraction was excised and eluted from the gel slice. Then, Illumina library was constructed and sequenced with Illumina/Solexa Genome Analyzer.
Most DNA binding proteins, such as transcription factors, bind to cis-acting DNA element with specific sequence features usually described by a position weight matrix (PWM). A hypothetical distribution of ChIP-seq-derived DNA fragments corresponding to transcription factors and RNA polymerase II (Pol II) is shown in Figs. 1A and 1B, respectively. For transcription factors, ChIP-seq detects a set of fragments that cluster and center around distinct biological binding sites, forming a "peak" around the binding locus (Fig. 1A). In contrast, Pol II binds throughout promoter regions, the 5'- and 3'- untranslated regions, the open reading frame (ORF), and downstream regions of the activated gene (Fig. 1B). Although Pol II can form a distinct peak around the transcription start site under certain circumstances, commonly-used peak-finding algorithms are not able to identify Pol II-enriched regions in gene transcript region derived from ChIP-seq experiments.
Figure 1.
Schematics of ChIP-seq-derived DNA fragments targeting transcription factors and RNA polymerase II. Blue and red ellipses indicate transcription factors binding on specific cis-acting DNA element, and RNA polymerase II not targeting certain binding sites, respectively. Blue and red lines under the ellipses illustrate sheared DNA fragments bound by the DNA-binding protein and pulled down by the immunoprecipitation assay. Green box indicates the fragment derived by Solexa sequencing (~25–36 bp).
A Poisson mixture model to identify transcripts with different Pol II binding quantity
Based on the assumption that the number of Pol II-binding fragments detected within gene transcript region, including 5'- and 3'-untranslated regions and open reading frames, follows a Poisson distribution, we developed a mixture model to identify differences in Pol II binding under two conditions, control vs. treatment. Denote yij as the Pol II quantity for gene i (i = 1,..., n) under condition j (j = 1, 2). In this application, j = 1, 2 indicates two biological conditions, MCF7 control (vehicle-treated) and MCF7 treated (3 hour E2 treatment), respectively. Marginally, yij follows a Poisson distribution,
| (1) |
where λij denotes the expected quantity of Pol II binding for gene i under condition j (j = 1, 2). For the i-th gene, if Pol II binding quantities within the gene transcript region are statistically different between the two biological conditions, yij follows marginal distributions with different parameters λi1 and λi2; conversely, if the number of Pol II binding does not demonstrate a significant difference, yij follows the same distribution with a unified λ. A mixture model is proposed to estimate the posterior probability of differential Pol II binding quantity.
For unified Pol II binding quantity between two conditions, λi1 and λi2 follow the same Gamma distribution (2). The selection of Gamma distribution is based on two considerations. Firstly, the two parameter Gamma distribution is a very flexible function which can be used to describe a wide range of distribution shapes. Secondly, the Gamma distribution is a conjugate distribution of Poisson for the λ parameters. Hence, the follow-up expectation step in an E-M algorithm has a close form solution.
| (2) |
Otherwise, (λi1, λi2) are distributed independently.
| (3) |
Denote Zi as the Bernoulli random variable with probability p, i.e., it is equal to 1 if (λi1, λi2) are independently distributed (and equal to 0 if not). Therefore, the joint distribution of Pol II binding quantity is modeled by a mixture of uniform binding events (p = 0) and differential binding events (p = 1).
| (4) |
Based on equation (4), we implement E-M algorithm by treating Zi as missing data.
The E-step of the algorithm is specified as:
| (5) |
In the M-step, the parameters in the Gamma distribution (α1, β1, α2, β2) are estimated by maximizing the two likelihood functions in Equation 6.
| (6) |
In the M-step, the optimization procedure is challenging, because searching for the optimal solutions for Gamma parameters can be trapped into local optimum, causing either slow convergence or failure to converge on the global optimal solution. In order to overcome these difficulties, we utilize particle swarm optimization (PSO), an artificial intelligence approach that mimics a behavior of swarm-forming agents, providing a good balance between global optimum searching and computation efficiency [12].
Because the likelihood functions of (α1, β1) and (α2, β2) are factorized in equation 6, the PSO optimization procedures are conducted independently using the following four steps.
Step 1: 100 particles (potential solution) were initially randomly distributed in 2-dimensional parameter spaces (α1, β1) or (α2, β2).
Step 2: the likelihood of each of the 100 particles are calculated by following Equation 6.
Step 3: the velocity vector of the particle, serving as the guide to search for the optimal solution, was calculated using Equation 7.
| (7) |
where Pglobal is the global optimal solution achieved so far; Pk-local is the local optimal solution achieved by particle k; and C0, C1, C2 are adjustable weight factors used to control searching speed.
Step 4: in the solution space, all the particles are re-positioned based on their current positions and movement velocities calculated in Equation 8.
| (8) |
Steps 1–4 will be iterated until further particle movement cannot result in higher likelihood (defined in Equation 6).
At the convergence, Zi can be interpreted as the probability of differential Pol II binding between two conditions. Although the model derivation is based on ChIP-seq data from MCF7 cells before and after treatment with vehicle or E2 for 3 hours, it can also be equally applied to OHT-MCF7 (+/- E2 treatment). In practice, the solution of the E-M algorithm converges in only 5 to 6 cycles.
Results and discussion
Genome-wide identification of Pol II binding in breast cancer cell lines
We tested our model on the Pol II binding quantity in MCF7 and OHT-MCF7 breast cancer cell lines (+/- E2 treatment) derived from the use of ChIP-seq technology. Among all the DNA fragments detected in each sample, we selected only those with high sequencing and matching quality that could be mapped to unique genomic locus. This pre-filtering step sufficiently removed background detection noise, and 2.59, 2.52, 3.00, and 1.33 million reads passed the above filter in MCF7 control, MCF7 E2-treated, OHT-MCF7 control, and OHT-MCF7 E2-treated samples, respectively. In order to compare Pol II binding quantity within a specific genomic region across multiple samples, the number of detected Pol II fragments was normalized using the total number of matched fragments in each sample, based on the assumption that the total number of DNA-binding Pol II would be similar in different cell types under different biological conditions.
Estrogen-induced changes in Pol II binding quantity in two cell types
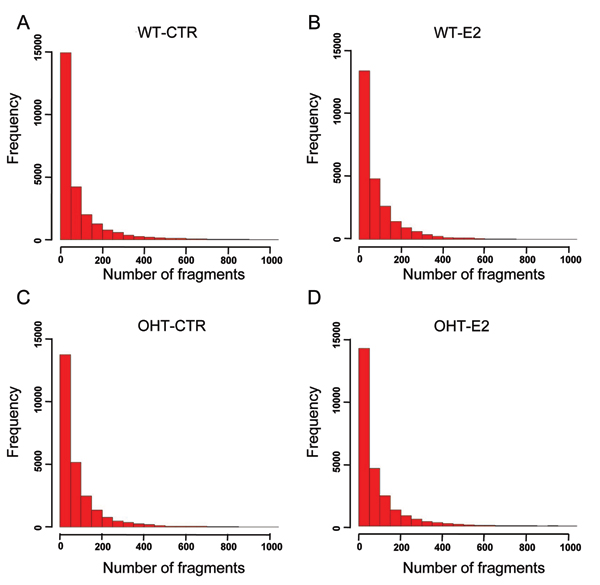
As the Pol II binding quantity in each gene transcript region reflects the expression level of the corresponding gene, we analyzed whole genome Pol II binding distributions for the four breast cancer cell samples. For MCF7, E2 treatment resulted in a slightly higher Pol II quantity distribution (Fig. 2, upper panels). Global gene profiles tended to be higher after E2 treatment, as reflected by a decrease in the number of genes showing a lower level of expression and an increase in the more highly expressed genes (Fig. 2). In control MCF7 samples, ~15,000 genes contained less than 50 ChIP-seq-derived DNA fragments in the gene transcript region (Fig. 2A), decreasing to ~13,000 after E2 treatment (Fig. 2B), a trend not observed in OHT MCF7 cells.
Figure 2.
Histogram of the number of Pol II binding fragments found within over 20,000 open reading frames in four samples including (A) wild type MCF7 control, (B) wild type MCF7 treated with E2, (C) OHT-resistant MCF7 control, and (D) OHT-resistant MCF7 treated with E2 samples.
This observation is consistent with the nature of the MCF7 cell line and the OHT-MCF7 subline, representing hormone-dependent and -independent breast cancer, respectively, and was also seen in the mean values and standard deviations for these samples (Table 1). After normalization, all four MCF7 cell lines had the same mean value for Pol II quantity; however, the slight decrease in standard deviation for Pol II quantity after E2 treatment of wild type cells indicates that a greater number of genes were expressed at a higher level. Furthermore, in the OHT cells, which are less sensitive to E2 stimulation compared to the parental MCF7 cell line [13], the standard deviation of Pol II quantity distribution remained essentially unchanged (Table 1).
Table 1.
The mean value and standard deviation of Pol II quantity in gene transcript regions in MCF7 and OHT-resistant MCF7 cells before and after E2 treatment.
| Wild Type | OHT-resistant | |||
| Control | Treatment | Control | Treatment | |
| Mean Value | 90.2 | 90.2 | 90.2 | 90.2 |
| Standard Deviation | 202.8 | 162.8 | 172.7 | 171.9 |
Genome Pol II quantity changing level analysis
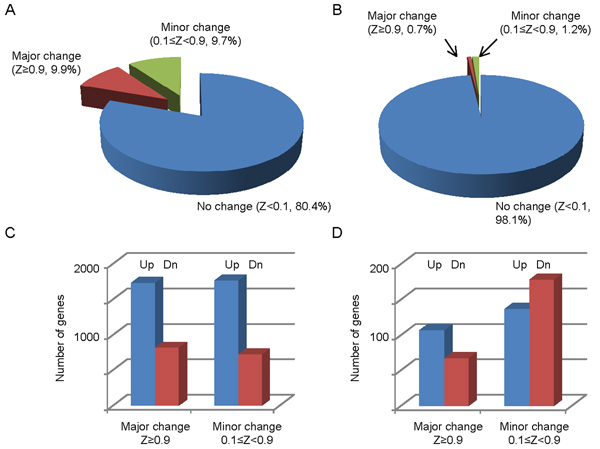
Because no replicates were available for the test data, a Poisson mixture model was used to identify estrogen-induced differences in Pol II binding quantity in the two cell lines. The results are shown as a scatter plot of the (log2) number of fragments in control and E2-treated samples (Fig. 3; each dot in the figure denotes a gene). This figure demonstrates a clear trend that with the increase of the number of Pol II binding quantity in the gene transcript region, a less relative change is required for a gene to be considered as major change (red dots, Zi ≥ 0.9), or minor change (green dots, 0.1 ≤ Zi < 0.9), where Zi is a posterior probability that the Pol II binding quantity changed after E2 treatment. This result demonstrates a critical feature of the Poisson mixture model: more weight is given to high abundant signals, while additional penalties are imposed on genes with low abundant quantities. The motivation is that additional relative changes are required to separate low abundant signals from background noise, because high signals are less sensitive to background noise. Consistent with previous observations [13], wild-type MCF7 cells have more gene targets with an altered quantity of Pol II than OHT MCF7 cells.
Figure 3.
Scatter plot of Pol II binding quantity in control and E2 treated samples: (A) wild type MCF7 cells and (B) OHT-resistant MCF7 cells. Blue, green, and red dots indicate genes with no change (Z < 0.1), minor change (0.1 ≤ Z < 0.9), and major change (Z ≥ 0.9), respectively.
Fragment distribution in the gene transcript region of two genes falling in the "major change" catergories, PgR (progesterone receptor) and MYC, two well known ERα targets in hormone-dependent breast cancer [14,15] are shown in Fig. 4. Pol II binding quantity in the gene transcript region of PgR was significantly increased by E2 in wild-type MCF7, but no change was detected in OHT-MCF7 (Fig. 4B). Pol II binding in MYC, however, was significantly increased in both MCF7 and OHT-MCF7 cells. Overall, in wild type MCF7 cells, Pol II binding quantities in the gene transcript region of 9.9% and 9.7% of the genes were classified as major (Z ≥ 0.9) and minor (0.1 ≤ Z < 0.9) changes, respectively (Fig. 5A). These percentages, however, dropped almost 10 fold to 0.7% and 1.2% in OHT-resistant MCF7 cells, while 98.1% of genes had the posterior probability of less than 0.1 (Fig. 5B). Furthermore, in wild type MCF7 cells, among the 2,527 and 2,464 genes showing major and minor changes, 68.1% (1,721) and 71.2% (1,754) of the genes demonstrated increased Pol II binding, respectively. In contrast, an increase in Pol II binding was observed in only 61.6% (106) of the major changed genes in the OHT-MCF7 cells. Strikingly, this number decreased to 43.5% (136) in the minor change group, while 56.5% (177) genes contain decreased Pol II quantity in OHT-MCF7 (Table 2).
Figure 4.
Examples of Pol II binding quantity in the open reading frame of (A) PGR, progesterone receptor and (B) MYC in all four samples.
Figure 5.
Changes of Pol II binding quantity in wild type MCF7 cells and OHT-resistant MCF7 cells before and after E2 treatment. Percentage of genes with no change (Z < 0.1), minor change (0.1 ≤ Z < 0.9), and major change (Z ≥ 0.9) in (A) wild type MCF7 cells and (B) OHT-resistant MCF7 cells. Number of genes with increased and decreased Pol II binding quantity in (C) wild type MCF7 cells and (D) OHT-resistant MCF7 cells. Dn = Down.
Table 2.
The gene numbers and percentage of whole genome in wild type and OHT-resistant MCF7 cells with minor change (0.1 ≤ Z < 0.9) or major change (Z ≥ 0.9)Pol II quantity.
| Wilde type MCF7 Cell Genes | OHT-resistant MCF7 cell Genes | |||||||
| Up-regulate | Dn-regulate | Up-regulate | Dn-regulate | |||||
| Number | Percent | Number | Percent | Number | Percent | Number | Percent | |
| Minor change (0.1 ≤ Z < 0.9) | 1754 | 6.9% | 710 | 2.8% | 136 | 0.5% | 177 | 0.7% |
| Major change (Z ≥ 0.9) | 1721 | 6.7% | 806 | 3.2% | 106 | 0.4% | 66 | 0.3% |
| Sum | 3475 | 13.6% | 1516 | 6.0% | 172 | 0.9% | 243 | 1.0% |
Conclusion
We report a Poisson mixture model to identify estrogen-induced changes in Pol II binding quantity in wild type MCF7 cells and OHT-resistant MCF7 cells. Despite having only one replicate available, our model successfully identified genes with different Pol II binding quantities from data derived using ChIP-seq technology. This model can distinguish differentially expressed Pol II activities from unchanged Pol II activities using a posterior probability calculated through an empirical Bayes approach. The empirical Bayes approach utilizes a combination of E-M and PSO algorithms for estimation and optimization in detection of the differential Pol II binding in two biological conditions. In this model, small signals require large changes in binding quantity to reach the same level of significance. The proposed model is unique in its ability to handle the ChIP-seq data without replicates and thus is an excellent tool for laboratories to evaluate preliminary ChIP-seq results. However, it is important to point out that despite the fact that no replicates are required to calculate changing probability, the use of biological replicates to capture persistent measurements in response to certain treatments are strongly encouraged.
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
YL and LL designed the study. WF, YL and LL designed and performed the computational modeling and drafted the manuscript. JW, THH, and KPN performed ChIP-seq experiment. All the authors read and approved the final manuscript.
Acknowledgments
Acknowledgements
This work is supported by National Cancer Institute grants CA085289 (KPN) and CA113001 (TH-MH), CSC (China Scholarship Council) Scholarship Programs (WF), and the Indiana Genomics Initiative of Indiana University (supported in part by the Lilly Endowment, Inc., YL).
This article has been published as part of BMC Genomics Volume 9 Supplement 2, 2008: IEEE 7th International Conference on Bioinformatics and Bioengineering at Harvard Medical School. The full contents of the supplement are available online at http://www.biomedcentral.com/1471-2164/9?issue=S2
Contributor Information
Weixing Feng, Email: wfeng@compbio.iupui.edu.
Yunlong Liu, Email: yunliu@iupui.edu.
Jiejun Wu, Email: jiejun.wu@osumc.edu.
Kenneth P Nephew, Email: knephew@indiana.edu.
Tim HM Huang, Email: tim.huang@osumc.edu.
Lang Li, Email: lali@iupui.edu.
References
- Johnson DS, Mortazavi A, Myers RM, Wold B. Genome-Wide Mapping of in Vivo Protein-DNA Interactions. Science. 2007;316:1497. doi: 10.1126/science.1141319. [DOI] [PubMed] [Google Scholar]
- Kerr MK, Martin M, Churchill GA. Analysis of Variance for Gene Expression Microarray Data. Journal of Computational Biology. 2000;7:819–837. doi: 10.1089/10665270050514954. [DOI] [PubMed] [Google Scholar]
- Wolfinger RD, Gibson G, Wolfinger ED, Bennett L, Hamadeh H, Bushel P, Afshari C, Paules RS. Assessing Gene Significance from cDNA Microarray Expression Data via Mixed Models. Journal of Computational Biology. 2001;8:625–637. doi: 10.1089/106652701753307520. [DOI] [PubMed] [Google Scholar]
- Dudoit S, Yang YH, Callow MJ, Speed TP. Statistical methods for identifying differentially expressed genes in replicated cDNA microarray experiments. Statistica Sinica. 2002;12:111–139. [Google Scholar]
- Efron B, Tibshirani R, Storey JD, Tusher V. Empirical Bayes Analysis of a Microarray Experiment. Journal of the American Statistical Association. 2001;96:1151–1161. doi: 10.1198/016214501753382129. [DOI] [Google Scholar]
- Newton MA, Kendziorski CM, Richmond CS, Blattner FR, Tsui KW. On Differential Variability of Expression Ratios: Improving Statistical Inference about Gene Expression Changes from Microarray Data. Journal of Computational Biology. 2001;8:37–52. doi: 10.1089/106652701300099074. [DOI] [PubMed] [Google Scholar]
- Kendziorski C, Newton M, Lan H, Gould M. On parametric empirical bayes methods for comparing multiple groups using replicated gene expression profiles. Presented at the Twenty-second Annual Conference of the International Society for Clinical Biostatistics. p. 23.
- Khalili A, Potter D, Yan P, Li L, Gray J, Huang T, Lin S. Gamma-Normal-Gamma Mixture Model for Detecting Differentially Methylated Loci in Three Breast Cancer Cell Lines. Cancer Informatics. 2007;2:43–54. [PMC free article] [PubMed] [Google Scholar]
- Li L, Shi H, Yiannoutsos C, Huang THM, Nephew KP. Epigenetic Hypothesis Tests for Methylation and Acetylation in a Triple Microarray System. Journal of Computational Biology. 2005;12:370–390. doi: 10.1089/cmb.2005.12.370. [DOI] [PubMed] [Google Scholar]
- Fan M, Yan PS, Hartman-Frey C, Chen L, Paik H, Oyer SL, Salisbury JD, Cheng AS, Li L, Abbosh PH, et al. Diverse gene expression and DNA methylation profiles correlate with differential adaptation of breast cancer cells to the antiestrogens tamoxifen and fulvestrant. Cancer Res. 2006;66:11954–11966. doi: 10.1158/0008-5472.CAN-06-1666. [DOI] [PubMed] [Google Scholar]
- Lee TI, Johnstone SE, Young RA. Chromatin immunoprecipitation and microarray-based analysis of protein location. Nat Protoc. 2006;1:729–748. doi: 10.1038/nprot.2006.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsopoulos KE, Vrahatis MN. Recent approaches to global optimization problems through Particle Swarm Optimization. Natural Computing. 2002;1:235–306. doi: 10.1023/A:1016568309421. [DOI] [Google Scholar]
- Fan MYPS, Hartman-Frey C, Chen L, Paik H, Abbosh P, Cheng ASL, Li L, Huang HMT, Nephew K. Breast cancer cells with acquired resistance to 4-hydroxytamoxifen and fulvestrant display divergent gene expression and DNA methylation signatures. Cancer Research. 2006;66:11964–11966. doi: 10.1158/0008-5472.CAN-06-1666. [DOI] [PubMed] [Google Scholar]
- Carroll JS, Meyer CA, Song J, Li W, Geistlinger TR, Eeckhoute J, Brodsky AS, Keeton EK, Fertuck KC, Hall GF. Genome-wide analysis of estrogen receptor binding sites. Nat Genet. 2006;38:1289–1297. doi: 10.1038/ng1901. [DOI] [PubMed] [Google Scholar]
- Li L, Cheng ASL, Jin VX, Paik HH, Fan M, Li X, Zhang W, Robarge J, Balch C, Davuluri RV. A mixture model-based discriminate analysis for identifying ordered 18 transcription factor binding site pairs in gene promoters directly regulated by estrogen receptor-{alpha} Bioinformatics. 2006;22:2210. doi: 10.1093/bioinformatics/btl329. [DOI] [PubMed] [Google Scholar]