Abstract
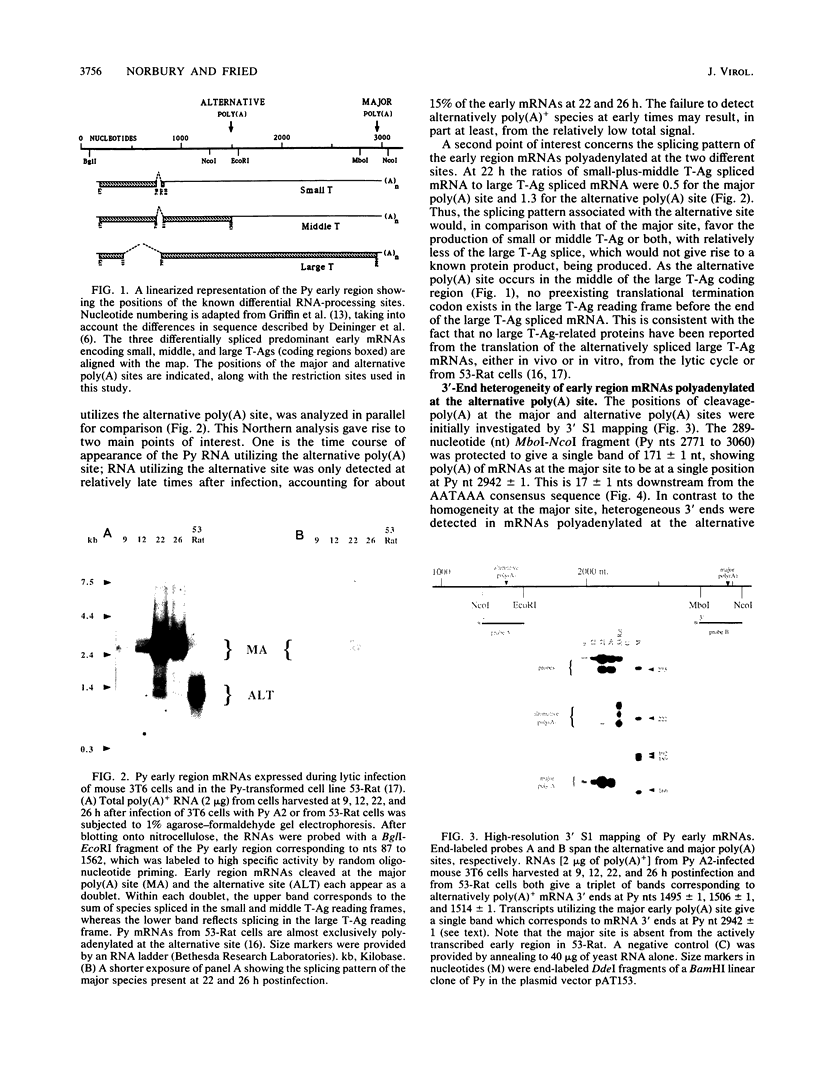
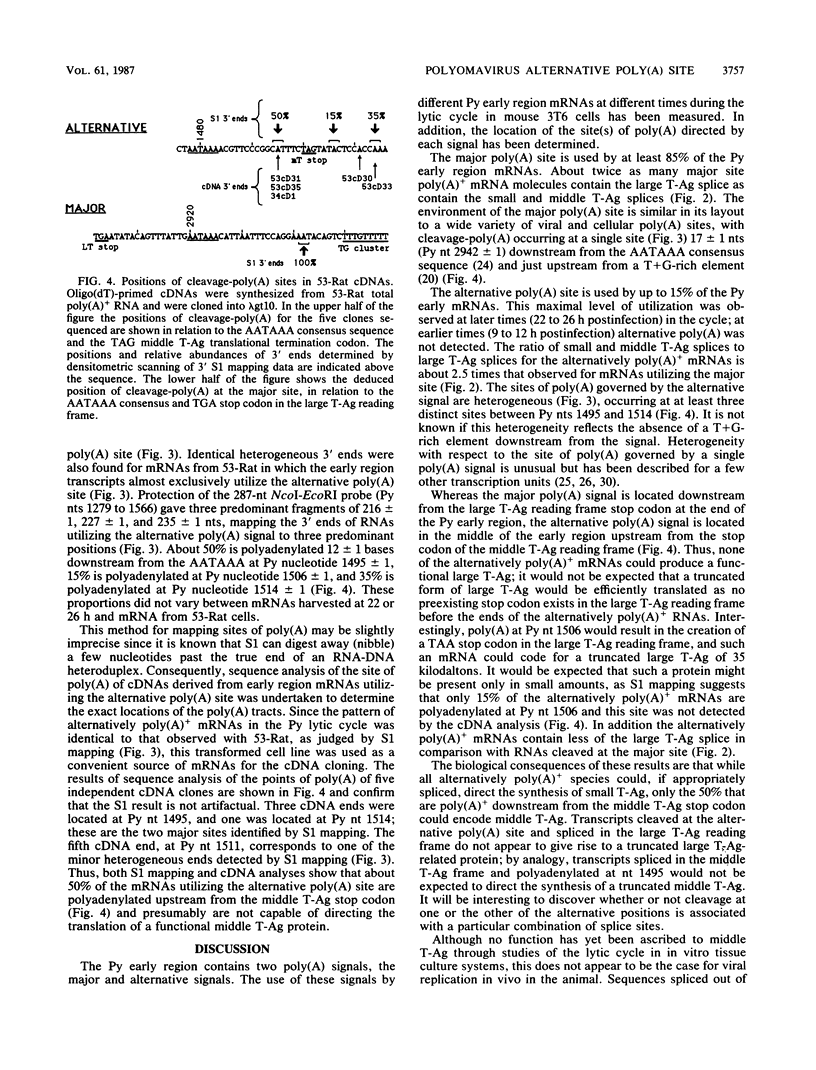
The position of an alternative polyadenylation [poly(A)] site at the 3' end of the polyomavirus middle T antigen (T-Ag) coding sequences suggests the possibility of a functional role for this site in early gene regulation. The fine structure of this alternative poly(A) site was determined by cDNA sequence and 3' S1 analyses. Cleavage-poly(A) was found to be heterogeneous, occurring at multiple CA dinucleotides downstream from the AATAAA signal sequence. About 50% of the alternative poly(A) takes place upstream from the middle T-Ag stop codon. In addition, the pattern of splicing of transcripts with the alternative poly(A) site differed from that with the major poly(A) site at the end of the early region. The ratio of the small and middle T-Ag splices to the large T-Ag splice for the alternative poly(A)+ mRNAs is about 2.5 times that found for mRNAs with the major poly(A) site. The altered splicing pattern and 3'-end heterogeneity of the alternative poly(A)+ mRNAs would result in preferential translation of small T-Ag (to a greater degree) and middle T-Ag over large T-Ag at later times in the polyomavirus lytic cycle.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Berger H., Wintersberger E. Polyomavirus small T antigen enhances replication of viral genomes in 3T6 mouse fibroblasts. J Virol. 1986 Nov;60(2):768–770. doi: 10.1128/jvi.60.2.768-770.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cogen B. Virus-specific early RNA in 3T6 cells infected by a tsA mutant of polyoma virus. Virology. 1978 Mar;85(1):222–230. doi: 10.1016/0042-6822(78)90426-9. [DOI] [PubMed] [Google Scholar]
- Courtneidge S. A., Smith A. E. Polyoma virus transforming protein associates with the product of the c-src cellular gene. Nature. 1983 Jun 2;303(5916):435–439. doi: 10.1038/303435a0. [DOI] [PubMed] [Google Scholar]
- Delmas V., Bastien C., Scherneck S., Feunteun J. A new member of the polyomavirus family: the hamster papovavirus. Complete nucleotide sequence and transformation properties. EMBO J. 1985 May;4(5):1279–1286. doi: 10.1002/j.1460-2075.1985.tb03773.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deninger P. L., Esty A., LaPorte P., Hsu H., Friedmann T. The nucleotide sequence and restriction enzyme sites of the polyoma genome. Nucleic Acids Res. 1980 Feb 25;8(4):855–860. [PMC free article] [PubMed] [Google Scholar]
- FRIED M. CELL-TRANSFORMING ABILITY OF A TEMPERATURE-SENSITIVE MUTANT OF POLYOMA VIRUS. Proc Natl Acad Sci U S A. 1965 Mar;53:486–491. doi: 10.1073/pnas.53.3.486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinberg A. P., Vogelstein B. A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal Biochem. 1983 Jul 1;132(1):6–13. doi: 10.1016/0003-2697(83)90418-9. [DOI] [PubMed] [Google Scholar]
- Francke B., Eckhart W. Polyoma gene function required for viral DNA synthesis. Virology. 1973 Sep;55(1):127–135. doi: 10.1016/s0042-6822(73)81014-1. [DOI] [PubMed] [Google Scholar]
- Fried M. Characterization of a temperature-sensitive mutant of polyoma virus. Virology. 1970 Mar;40(3):605–617. doi: 10.1016/0042-6822(70)90205-9. [DOI] [PubMed] [Google Scholar]
- Garcea R. L., Ballmer-Hofer K., Benjamin T. L. Virion assembly defect of polyomavirus hr-t mutants: underphosphorylation of major capsid protein VP1 before viral DNA encapsidation. J Virol. 1985 May;54(2):311–316. doi: 10.1128/jvi.54.2.311-316.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gubler U., Hoffman B. J. A simple and very efficient method for generating cDNA libraries. Gene. 1983 Nov;25(2-3):263–269. doi: 10.1016/0378-1119(83)90230-5. [DOI] [PubMed] [Google Scholar]
- Kamen R., Favaloro J., Parker J., Treisman R., Lania L., Fried M., Mellor A. Comparison of polyoma virus transcription in productively infected mouse cells and transformed rodent cell lines. Cold Spring Harb Symp Quant Biol. 1980;44(Pt 1):63–75. doi: 10.1101/sqb.1980.044.01.009. [DOI] [PubMed] [Google Scholar]
- Lania L., Gandini-Attardi D., Griffiths M., Cooke B., De Cicco D., Fried M. The polyoma virus 100K large T-antigen is not required for the maintenance of transformation. Virology. 1980 Feb;101(1):217–232. doi: 10.1016/0042-6822(80)90497-3. [DOI] [PubMed] [Google Scholar]
- McCance D. J. Growth and persistence of polyoma early region deletion mutants in mice. J Virol. 1981 Sep;39(3):958–962. doi: 10.1128/jvi.39.3.958-962.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLauchlan J., Gaffney D., Whitton J. L., Clements J. B. The consensus sequence YGTGTTYY located downstream from the AATAAA signal is required for efficient formation of mRNA 3' termini. Nucleic Acids Res. 1985 Feb 25;13(4):1347–1368. doi: 10.1093/nar/13.4.1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proudfoot N. J., Brownlee G. G. 3' non-coding region sequences in eukaryotic messenger RNA. Nature. 1976 Sep 16;263(5574):211–214. doi: 10.1038/263211a0. [DOI] [PubMed] [Google Scholar]
- Rassoulzadegan M., Cowie A., Carr A., Glaichenhaus N., Kamen R., Cuzin F. The roles of individual polyoma virus early proteins in oncogenic transformation. Nature. 1982 Dec 23;300(5894):713–718. doi: 10.1038/300713a0. [DOI] [PubMed] [Google Scholar]
- Ruley H. E., Lania L., Chaudry F., Fried M. Use of a cellular polyadenylation signal by viral transcripts in polyoma virus transformed cells. Nucleic Acids Res. 1982 Aug 11;10(15):4515–4524. doi: 10.1093/nar/10.15.4515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasavage N. L., Smith M., Gillam S., Woychik R. P., Rottman F. M. Variation in the polyadenylylation site of bovine prolactin mRNA. Proc Natl Acad Sci U S A. 1982 Jan;79(2):223–227. doi: 10.1073/pnas.79.2.223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonsen C. C., Levinson A. D. Analysis of processing and polyadenylation signals of the hepatitis B virus surface antigen gene by using simian virus 40-hepatitis B virus chimeric plasmids. Mol Cell Biol. 1983 Dec;3(12):2250–2258. doi: 10.1128/mcb.3.12.2250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Templeton D., Simon S., Eckhart W. Truncated forms of the polyomavirus middle T antigen can substitute for the small T antigen in lytic infection. J Virol. 1986 Jan;57(1):367–370. doi: 10.1128/jvi.57.1.367-370.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treisman R., Novak U., Favaloro J., Kamen R. Transformation of rat cells by an altered polyoma virus genome expressing only the middle-T protein. Nature. 1981 Aug 13;292(5824):595–600. doi: 10.1038/292595a0. [DOI] [PubMed] [Google Scholar]
- Wiedemann L. M., Perry R. P. Characterization of the expressed gene and several processed pseudogenes for the mouse ribosomal protein L30 gene family. Mol Cell Biol. 1984 Nov;4(11):2518–2528. doi: 10.1128/mcb.4.11.2518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Hoyningen-Huene V., Norbury C., Griffiths M., Fried M. Gene activation properties of a mouse DNA sequence isolated by expression selection. Nucleic Acids Res. 1986 Jul 25;14(14):5615–5627. doi: 10.1093/nar/14.14.5615. [DOI] [PMC free article] [PubMed] [Google Scholar]