Abstract
Fission yeast rad22+, a homologue of budding yeast RAD52, encodes a double-strand break repair component, which is dispensable for proliferation. We, however, have recently obtained a cell division cycle mutant with a temperature-sensitive allele of rad22+, designated rad22-H6, which resulted from a point mutation in the conserved coding sequence leading to one amino acid alteration. We have subsequently isolated rad22+ and its novel homologue rti1+ as multicopy suppressors of this mutant. rti1+ suppresses all the defects of cells lacking rad22+. Mating type switch-inactive heterothallic cells lacking either rad22+ or rti1+ are viable, but those lacking both genes are inviable and arrest proliferation with a cell division cycle phenotype. At the nonpermissive temperature, a synchronous culture of rad22-H6 cells performs DNA synthesis without delay and arrests with chromosomes seemingly intact and replication completed and with a high level of tyrosine-phosphorylated Cdc2. However, rad22-H6 cells show a typical S phase arrest phenotype if combined with the rad1-1 checkpoint mutation. rad22+ genetically interacts with rad11+, which encodes the large subunit of replication protein A. Deletion of rad22+/rti1+ or the presence of rad22-H6 mutation decreases the restriction temperature of rad11-A1 cells by 4–6°C and leads to cell cycle arrest with chromosomes incompletely replicated. Thus, in fission yeast a double-strand break repair component is required for a certain step of chromosome replication unlinked to repair, partly via interacting with replication protein A.
INTRODUCTION
In the budding yeast Saccharomyces cerevisiae, homologous recombination, double-strand break repair, and gene conversion are performed by a system involving Rad52, Rad51, and Rad54 proteins, whose molecular functions have lately begun to be understood (Resnick, 1975; Game, 1993). Rad51 is a RecA-like protein (Shinohara et al., 1992) that catalyzes strand exchanges between homologous sequences in cooperation with replication protein A (RPA) (Namsaraev and Berg, 1997). Rad52 and Rad54 assist the Rad51-catalyzed strand exchanges by direct interactions (Sung, 1997a; New et al., 1998; Shinohara and Ogawa, 1998). Rad52 binds to both Rad51 and RPA (Fimenich et al., 1995; Hays et al., 1998) as well as single-stranded DNA (Mortensen et al., 1996) and forms Rad51-nucleoprotein filaments (Gasior et al., 1998). In addition, Rad52 has an ability to promote reannealing of RPA-bound complementary single-strand DNAs (Sugiyama et al., 1998). At least two additional factors are known to cooperate for strand exchanges. Rad55 and Rad57 form a heterodimer and cooperate with RPA to promote Rad51-catalyzed strand exchanges (Sung, 1997b). Budding yeast contains the RAD52 homologue called RAD59, which is involved in RAD51-independent mitotic recombination (Bai and Symington, 1996).
The double-strand break repair system involving these factors is evolutionarily conserved throughout eukaryotes. Various organisms including the fission yeast Schizosaccharomyces pombe and mammals contain counterparts of these factors (Bezzubova et al., 1993; Muris et al., 1993, 1994; Ostermann et al., 1993; Shinohara et al., 1993; Kanaar et al., 1996; Albala et al., 1997). In budding and fission yeast, these factors are dispensable for viability although required for mating type switching and repair of chemically or physically induced double-strand breaks (McKee and Lawrence, 1980; Borts et al., 1986; Kezenman et al., 1992; Schlake et al., 1993). However, in higher eukaryotes, some of these factors are indispensable for viability because of requirement for repair of the double-strand breaks that are spontaneously produced during chromosomal replication at least in some cells (Lim and Hasty, 1996; Tsuzuki et al., 1996). RAD51-disrupted chicken DT40 cells die of chromosomal fragmentation, and RAD51-disrupted murine fertilized eggs fail to develop properly, resulting in embryonic lethality (Sonoda et al., 1998). On the other hand, gene knockout mice inactivated for a RAD54 homologue gene develop normally but exhibit an increased susceptibility to double-strand breaks, like yeast (Essers et al., 1997).
Recently we isolated a typical cell division cycle mutant of S. pombe that resulted from a point mutation of rad22+. This was totally unexpected because, as reported previously, cells lacking rad22+ are viable. We subsequently found that fission yeast contained a functional homologue of rad22+. In this communication, we report that in fission yeast this double-strand break repair component plays an essential role in a certain step of chromosome replication seemingly unrelated to repair during regular cell cycling.
MATERIALS AND METHODS
Strains and Media
The S. pombe strains used in this study are listed in Table 1. The pombe minimal (PM) medium was described previously (Nurse, 1975) and contains routinely 2% glucose unless otherwise indicated. PM + leu medium contains 50 μg of leucine/ml in PM medium. YE medium was described elsewhere (Alfa et al., 1993).
Table 1.
Strains used in this study
| Strain | Genotype |
|---|---|
| DP2 | h−/h+ ade6-M210/ade6-M216 leu1-32/leu1-32 ura4-D18/ura4-D18 |
| K135-B25 | h90 leu1-32 |
| K153-B4 | h90 leu1-32 ura4-D18 |
| ATCC38399 | h− leu1-32 |
| K150-A13 | h+ leu1-32 |
| NRC349 | h+ rad11-A1 |
| HM370 | h+ rad11-A1 leu1-32 |
| AN1 | h− rad22-H6 leu1-32 |
| HM366 | h+ rad22-H6 leu1-32 |
| HM367 | h+ rad22∷ura4+ leu1-32 ura4-D18 |
| HM368 | h+ rti1∷ura4+ leu1-32 ura4-D18 |
| HM369 | h+ rad22-H6 rti1∷ura4+ leu1-32 ura4-D18 |
| HM73 | h− rad1-1 leu1-32 |
| HM105 | h+ rad1-1 rad22-H6 leu1-32 |
| HM128 | h+ cdc21-M63 leu1-32 |
| HM132 | h− cdc25-22 leu1-32 |
| HM372 | h+ rad11-A1 rad22-H6 leu1-32 |
| HM373 | h+ rad11-A1 rti1∷ura4+ leu1-32 ura4-D18 |
| HM374 | h+ rad11-A1 rad22∷ura4+ leu1-32 ura4-D18 |
| HM365 | h− rad22∷ura4+ leu1-32 ura4-D18 |
| KS-1 | h−/h+ ade6-M210/ade6-M216 leu1-32/leu1-32 |
| KS-2 | h−/h+ rad22+/rad22-H6 ade6-M210/ade6-M216 leu1-32/leu1-32 |
| KS-3 | h−/h+ rad22-H6/rad22-H6 ade6-M210/ade6-M216 leu1-32/leu1-32 |
| pB9 | h+ mat1-p Δ17 leu1-32 ura4-D18 |
Isolation of Temperature-sensitive Cell Division Cycle Mutants
General culture media and routine genetic methods were used (Egel and Egel-Mitani, 1974; Moreno et al., 1991). Homothallic h90 leu1-32 (K153-B25) cells were mutagenized with 1 mg of nitrosoguanidine (NG)/ml for 15 min to obtain a 30% survival. Approximately 6 × 104 NG-treated viable cells were plated on molt extract medium and incubated at 25°C for 4–6 d to induce conjugation and sporulation, followed by treatment with acetone vapor to kill vegetative cells (Egel, 1977). Spores were germinated, and formed colonies were replica plated and tested for thermosensitive proliferation at 36°C. The colonies that arrested proliferation with cell elongation were isolated as candidates for cell division cycle mutants and back-crossed to h90 leu1-32 (K153-B25), h− leu1-32 (ATCC38399), or h+ leu1-32 (K150-A13) at least three times to eliminate irrelevant mutations.
Isolation of rti1+ and Sequence Determination
S. pombe transformation and gene cloning were carried out as described (Okazaki et al., 1990). The h+ rad22-H6 leu1-32 cells were transfected with an S. pombe HindIII genomic library that was constructed with the pALSK vector containing ars and the LEU2 marker gene (Okazaki et al., 1990). The cells were incubated at 23°C for 24 h on minimum agar plates and selected at 35°C for 4 d. Among 2 × 105 stable leu+ transfectants selected, dozens of colonies grew at 35°C, from which the rad22+ and rti1+ genes were recovered. An rti1+ cDNA spanning the entire coding region was obtained by reverse transcription of the mRNA prepared from the rti1+-transformed colony followed by PCR amplification with specific primers. DNA sequences were determined by the dideoxy method (Sanger et al., 1977).
Gene Replacement and Integration
Cells with rad22+ or rti1+ deleted were constructed as follows. The 1.4-kb KpnI–NruI fragment containing 95% of the rad22+ coding region and the 1.4-kb HindIII–EcoRI fragment containing the entire rti1+ coding sequence were replaced with the 1.8-kb ura4+ gene. The SpeI fragment containing the disrupted rad22 and the HindIII fragment containing the disrupted rti1 were transfected into the diploid strain h−/h+ ade6-M210/ade6-M216 leu1-32/leu1-32 ura4-D18/ura4-D18 (DP2), and stable ura+ transformants were selected as described (Tanaka et al., 1992). Successful gene disruptions were confirmed by Southern blot analysis.
Flow Cytometry
Flow cytometry was performed as described previously (Tanaka et al., 1992), using the FACScan system and the CellFIT cell cycle analysis program with the software LYSIS (Becton Dickinson, Mountain, View, CA).
Pulsed Field Gel Electrophoresis
Cells were prepared as described (Kelly et al., 1993). Pulsed field gel electrophoresis was carried out in a 0.8% chromosomal grade agarose gel at 45 V for 100 h in 20 mM Tris-acetate, pH 8.0, containing 0.5 mM EDTA, with alternating currency at 60-min intervals.
Preparation of the rad11-A1 Mutant
The original S. pombe rad11-404 strain (NRC2349) harbors an extragenic suppressor mutation of the thermosensitive growth (Phipps et al., 1985; Parker et al., 1997). Therefore, the parental rad11-404 strain was backcrossed three times to a wild-type strain. A resulting cell clone showing both UV sensitivity and thermosensitive proliferation was isolated and used for this study as rad11-A1 (Parker et al., 1997).
RESULTS
Isolation of a Novel rad22+ Homologue
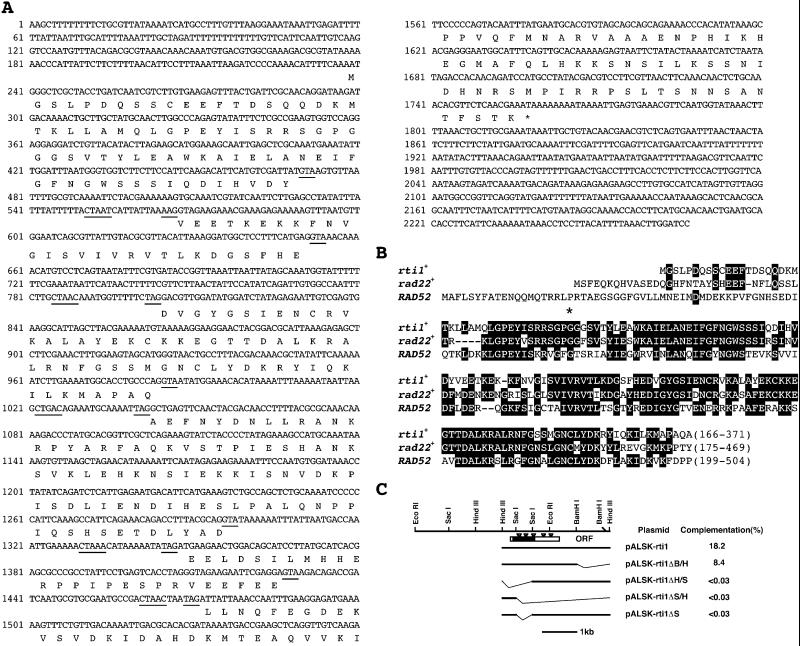
In a search for new elements controlling the cell cycle in S. pombe, we obtained a temperature-sensitive cell division cycle mutant that arrested with a 2C DNA content upon a shift to 35°C. Crossing with the existing mutants indicated that this mutant was allelic to rad22-67, and it was named rad22-H6. Such a mutant was totally unexpected because cells lacking rad22+ are viable (Ostermann et al., 1993). Screening an S. pombe genomic library for multicopy suppressors of this mutant led to isolation of two active genes. Nucleotide sequencing revealed that one was rad22+ itself, and the other was a novel gene homologous to rad22+. The latter gene, named rti1+ (rad twenty-two isogene 1), contains six exons and five short introns with typical splicing consensus sequences (Figure 1A), which were identified by sequencing reverse-transcribed, PCR-amplified rti1+ mRNA. rti1+ is capable of encoding a 371-amino-acid protein with a calculated molecular mass of 42 kDa. The N-terminal half of Rti1 was highly homologous (65% amino acid identity) to the corresponding region of Rad22 protein (Figure 1B) and was required for the suppression of the thermosensitivity of rad22-H6 cells at 36°C. Deletion of this region (Figure 1C) completely abrogated such suppression.
Figure 1.
Structure of rti1+. (A) Nucleotide sequence of rti1+ and the deduced amino acid sequence of the putative gene product. Underlines within introns indicate consensus splicing sequences. Splice junctions were determined by comparing the genomic and cDNA sequences of rti1+. The predicted 371-amino-acid sequence is indicated in single-letter code. (B) Amino acid alignment of the predicted Rti1 protein with Rad22 and Rad52. The aa residues identical among them are boxed. The asterisk indicates the position of mutation in the rad22-H6 gene. In this mutant allele, there is a G to A conversion at nucleotide 441 resulting in an amino acid change from glycine to aspartic acid at amino acid 110. (C) Organization and deletion analysis of rti1+. The activity of variously deleted rti1+ gene was expressed as percent complementation of the thermosensitivity of rad22-H6 cells. The filled box indicates the highly homologous region, and inverted triangles indicate the positions of introns. Thin lines are deleted regions.
Point Mutation in rad22-H6 Allele
We identified the mutation in the temperature-sensitive rad22-H6 allele, which was of interest because cells lacking rad22+ are viable. Cloning and sequencing revealed that the rad22-H6 gene contained a G to A mutation at nucleotide 441 that changes a glycine at position 110 to aspartic acid in the highly conserved region (Figure 1, star). This glycine is conserved among the three cognates, rad22+, rti1+, and budding yeast RAD52.
Functional Similarity of rti1+ to rad22+
Not only in structure but also in function rti1+ resembled rad22+. When expressed from a plasmid vector, rti1+ suppressed the short UV and bleomycin sensitivities of mating type switch-inactive heterothallic rad22 disruptant (Δrad22) cells as well as the lethality of the mating type switch-active homothallic Δrad22 cells, albeit it was less potent than rad22+ (Table 2). Cells lacking rad22+ were highly sterile presumably because of facilitated G2 arrest upon nitrogen starvation. This sterility was also suppressed by overexpression of rti1+.
Table 2.
Rescue of rad22-H6 and Δrad22 cells by rti1+
| Plasmid | Colony formation at 36°C of rad22-H6 | Suppression of Δrad22 sensitivity to
|
Suppression of Δrad22 sterility | Suppression of mating type switch lethality of Δrad22 | |
|---|---|---|---|---|---|
| UV | bleomycin (%) | ||||
| pALSK | <0.03 | <0.04 | 0.05 | <0.01 | <0.02 |
| pALSK-rad22+ | 37.0 | 50.3 | 19.9 | 8.5 | 14.0 |
| pALSK-rti1+ | 18.2 | 15.5 | 4.8 | 2.7 | 5.1 |
The rad22-H6 (HM366) cells were transfected with the indicated plasmids and selected on MMA plates at 36°C. The ratio of the number of leu+ colonies formed at 36°C to that formed at 23°C was expressed as percent colony formation. The h+ Δrad22 (HM367) cells stably transfected with the indicated plasmids were plated on MMA plates at a density of 500 cells per plate, exposed to 120 J/m2 of UV, and incubated at 23°C for 7 d. Sensitivity to 0.1 mg/ml bleomycin was assayed as described in Figure 2A. The h+ Δrad22 (HM367) and h− Δrad22 (HM375) cells each harboring the indicated plasmid were mixed together and incubated in nitrogen-free PM medium at 23°C for 15 h. Suppression of sterility is expressed as percentage of mated cells in the total population. The h+ Δrad22 (HM367) cells harboring the indicated plasmid were mated with the h90 rad22+ (K153-B4) cells in nitrogen-free PM medium at 28°C for 24 h. Formed asci were treated with 30% ethanol, and ura+ leu+ spores were allowed to grow on MMA plates. Each colony was determined for mating type. A total 500 colonies for each plasmid were examined. The ratio of the number of h90 colonies to the number of h+ colonies was expressed as percent suppression of mating type switch lethality.
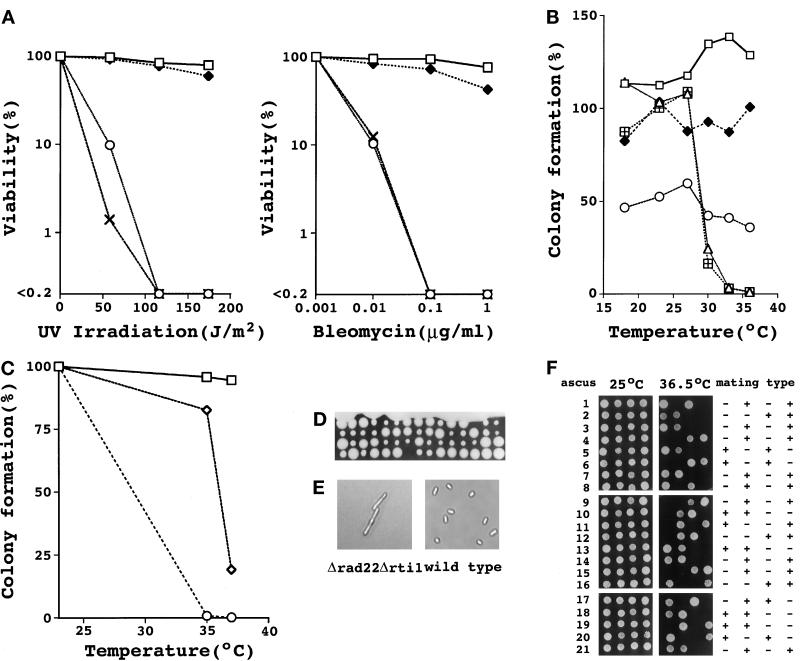
To further investigate rti1+ function, we disrupted the rti1+ gene by replacing the entire coding region with the ura4+ gene cassette. The complete inactivation of rti1+ function in the disrupted gene was confirmed by its inability to suppress the thermosensitivity of rad22-H6 cells. Cells inactivated for rti1+ were then generated by homologously integrating the disrupted gene into one rti1+ locus in diploid cells. After Southern blot confirmation, disruptants were induced for meiosis and sporulation followed by germination. Germinating haploid cells that lacked rti1+ (Δrti1) were viable. As already known, mating type switch-inactive heterothallic Δrad22 cells were viable yet highly sensitive to short UV and bleomycin, whereas mating type switch-active homothallic counterparts were inviable (Ostermann et al., 1993). By contrast, the Δrti1 cells were almost as insensitive to these agents as wild-type cells (Figure 2A) and viable at all the temperatures tested irrespective of their mating type. These results show that rti1+ is a rad22+ homologue playing an auxiliary role. Given this fact, the inability of rad22-H6 but not Δrad22 cells to proliferate at 36°C indicates that rad22-H6 is likely to be a dominant negative type of mutation. This was demonstrated by analysis of a rad22+/rad22-H6 diploid strain and cells lacking both rad22+ and rti1+.
Figure 2.
UV and bleomycin sensitivities and thermosensitive colony formation of rad22 and rti1 mutants. (A) UV and bleomycin sensitivities of Δrti1 and Δrad22 cells. Wild-type (ATCC38399; square), rad1-1 (HM73; cross), Δrad22 (HM367; circle), and Δrti1 (HM368; closed diamond) cells were plated on MMA + leu plates at 500 cells per plate, irradiated with various doses of short UV, and incubated at 23°C for 7 d. Sensitivity to bleomycin was determined by plating on MMA + leu plates containing the indicated concentrations of bleomycin. Percent cell viability was calculated by dividing the number of formed colonies by the number of plated cells. (B) Temperature-sensitive colony formation of rad22 and rti1 mutants. Wild-type (K150-A13; square), Δrad22 (HM367; circle), Δrti1 (HM368; closed diamond), rad22-H6 (HM366; triangle), and rad22-H6 Δrti1 (HM369; square with plus) cells were grown in YEA at 23°C for 1 d, transferred in YE medium at 23°C, and incubated for 14 h. One thousand cells were plated onto YEA plates and incubated at the temperatures specified for 7 d. Percent colony formation was calculated by dividing the number of formed colonies by the number of plated cells. (C) rad22+/rad22+ diploid (KS-1; square), rad22+/rad22-H6 diploid (KS-2; diamond), and rad22-H6/rad22-H6 diploid (KS-3; circle) cells were grown in PM + leu at 23°C for 16 h. Five hundred cells of each strain were plated on to MMA + leu plates and incubated at the indicated temperatures for 5 d. Percent colony formation was calculated by dividing the number of formed colonies by the number of plated cells. (D) Tetrad analysis of Δrad22 Δrti1 double disruptants. The h− rad22::ura4+ leu1-32 ura4-D18 cells (HM375) and the h+ rti1::ura4+ leu1-32 ura4-D18 cells (HM3368) were mixed and plated on a MEA plate and incubated at 25°C for 3 d for conjugation, meiosis, and sporulation. Spores were then isolated from each ascus and placed on YEA followed by incubation at 25°C for 5 d. (E) Morphology of geminated Δrad22 Δrti1 double disruptant cells. Double disruptant spores were germinated at 25°C for 5 d and photographed. Germinated wild-type cells in the same experiment were suspended in YE and photographed for comparison of cell size. (F) Tetrad analysis of rad22-H6 mat1-p Δ17 double mutants. The h− rad22-H6 leu1-32 (AN1) cells were crossed to h+ mat1-p Δ17 leu1-32 ura4-D18 cells. Formed spore asci were tetrad dissected, grown on YEA plates at 25°C, and examined for the ability to grow at 36.5°C and mating type. The mating type was determined by crossing to h− leu1-32 or h+ leu1-32 cells.
Unlike the wild-type diploid strain, the rad22+/rad22-H6 strain showed a marked reduction in colony-forming ability at 37°C, whereas rad22-H6/rad22-H6 cells were unable to grow already at 35°C (Figure 2C). Additionally, as anticipated, cells lacking both rad22+ and rti1+ were nonviable. To prepare Δrad22 Δrti1 double disruptants, diploid cells in which one allele each of rad22+ and rti1+ was deleted were constructed and induced to sporulate, and spore asci were analyzed by tetrad dissection. Among 60 asci analyzed, 56 contained four viable spores that formed two small Δrad22 and two large Δrti1 colonies at 25°C (Figure 2D). The remaining four asci contained three viable and one inviable spores. Each inviable spore was assigned to a Δrad22 Δrti1 double disruptant by determining the genetic markers of the remaining three viable spores. Microscopic examination revealed that the Δrad22 Δrti1 double disruptant spores germinated but ceased proliferation with cell elongation after one division (Figure 2E). Thus, rad22+/rti1+ that encodes a critical component of the major double-strand break repair system in fission yeast was essential for mitotic cell cycling.
Requirement for rad22+/rti1+ in S Phase Completion
To understand the reason for the requirement of rad22+/rti1+ for cell cycling, we first examined the viability of a rad22-H6 mutant also null in double-strand breaks in the mat1 locus, because even in the regular heterothallic strains a double-strand break occurs in the mat1 locus at a low frequency (Beach, 1983), and we thought that this might partly cause rad22+/rti1 to be indispensable. The strain used was h+ mat1-p Δ17, in which no double-strand breaks occur at the mat1-p locus because of a 122-bp deletion near the HO endonuclease cut site (Arcangioli and Klar, 1991). The h− rad22-H6 cells were crossed to the h+ mat1-p Δ17 strain, and the ability to grow at 36.5°C and the mating type of the spores in 21 asci were examined after tetrad dissection. As shown in Figure 2F, two spores from any ascus examined were unable to grow at 36.5°C no matter whether they were null in mat1-p double-strand breaks, which is indicated by the mating type. We thus concluded that the inability of rad22-H6 cells to grow at the nonpermissive temperature was not due to a failure to repair a double-strand break at the mat1 locus that infrequently occurs in heterothallic cells.
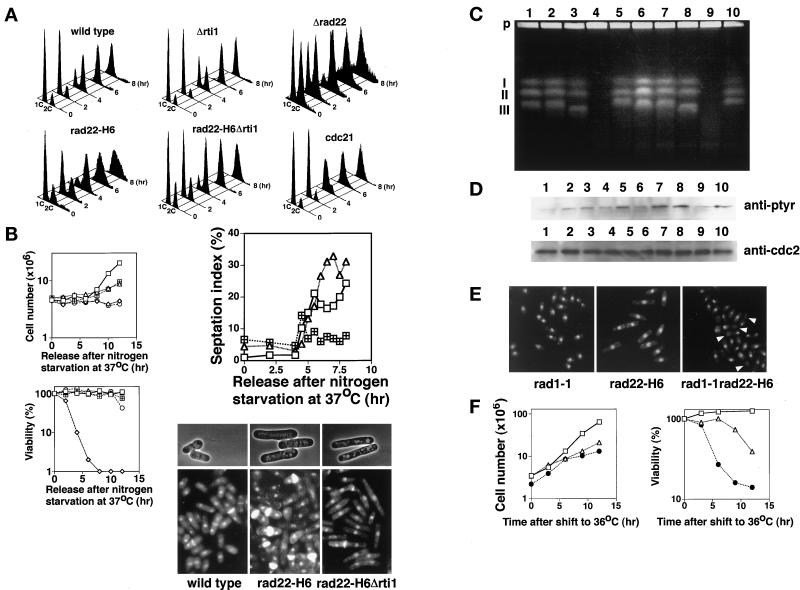
Obviously, this experimental result does not exclude another possibility: that rad22+/rti1+ is required for the repair of double-strand breaks that might be generated spontaneously during cell cycling, as in chicken DT cells (Sonoda et al., 1998). This possibility, however, seems to be remote because as exemplified by mating type switching, introduction of even one double-strand break in a chromosome during cell cycling is lethal to the cells without rad22+, yet heterothallic cells are viable without rad22+. To understand the function of rad22+/rti1+ in cell cycling, we determined the cell cycle phase in which rad22+/rti1+ is required. Rapidly growing heterothallic cells of wild-type, rad22-H6, Δrti1, Δrad22, and rad22-H6 Δrti1 strains were arrested in G1 by nitrogen starvation (Figure 3A). Similarly arrested heterothallic cdc21-M63 cells were used as the positive control for a defect in DNA synthesis (Coxon et al., 1992). Upon nitrogen starvation, all but the Δrad22 cells predominantly arrested in G1, as expected. The cells were then transferred to 37°C, incubated for 12 h, and released at 37°C to resume cell cycling. The Δrti1 cells progressed through S phase as rapidly as wild-type cells and continued to proliferate. Similarly, the Δrad22 cells, which tended to arrest with a 2C DNA content upon nitrogen starvation, showed no apparent S phase retardation and proliferated, although slowly and with a broad 2C DNA content. Even the rad22-H6 and rad22-H6 Δrti1 cells, both of which eventually arrested cell cycling at this temperature, performed bulk DNA synthesis as rapidly as wild-type cells and slowly increased in cell number with broad 2C–4C and 2C peaks, respectively (Figure 3, A and B). The concurrent analysis of septation index showed that rad22-H6 cells were septated with a significant delay (1.5 h) followed by an accumulation of septated cells (Figure 3B), which perhaps reflected the broad 2C–4C peak seen at 8 h for this mutant in flow cytometry (Figure 3A). Delay in the onset of mitosis was more profound for rad22-H6 Δrti1 cells. In the same analysis, the rad22-H6 Δrti1 cells showed no significant increase in septation index (Figure 3B). The small spike at 4.5 h may reflect septation of the G2-arrested cells that occupied ∼30% of the total population (Figure 3A). The majority of the mutant cells at 8 h were elongated, particularly with few septations and seeming interphase nuclei for the double mutant (Figure 3B). These results suggest that cells inactivated for rad22+/rti1+ were arrested or significantly slowed in progression before the onset of M phase. The delayed septation and the accumulation of septated cells with 2C–4C DNA contents seen for rad22-H6 but not rad22-H6 Δrti1 cells was perhaps a consequence of entry into M phase caused by the presence of active Rti1 followed by arrest without efficient cell separation in the next cycle.
Figure 3.
Cells defective in rad22+/rti1+ are not delayed in bulk DNA synthesis but arrest before G2 phase. (A) Flow cytometry of rad22 and rti1 mutants released from G1. Wild-type, Δrad22 (HM367), Δrti1(HM368), rad22-H6 (HM366), rad22-H6 Δrti1 (HM369), and cdc21-M63 (HM128) cells were grown in PM + leu medium to middle log phase at 23°C. Each strain was incubated in nitrogen-free PM medium containing leucine (50 μg/ml) at 23°C for 24 h and then at 37°C for 12 h. The cells were transferred to PM + leu medium at 37°C for starting cell cycling. Incubation was continued at 37°C for the indicated times. (B) Proliferation, viability, and septation of rad22/rti1 mutant cells released from G1 at 37°C. Wild-type (square), rad22-H6 (triangle), rad22-H6 Δrti1 (square with plus), cdc21-M63 (diamond), and cdc25–22 (circle) cells were incubated as described in A, and their cell number and viability were determined. Septation indexes were determined after double staining with DAPI and calcofluor (Alfa et al., 1993). For photographs, cells at 8 h were fixed with 70% ethanol, and some were stained with DAPI and calcofluor. (C) Pulsed field gel electrophoresis. Wild-type (lanes 1 and 6), rad22-H6 (lanes 2 and 7), rad22-H6 Δrti1(lanes 3 and 8), cdc21-M63 (lanes 4 and 9), and cdc25–22 (lane 5 and 10) cells were collected at 6 h (lanes 1–5) and 8 h (lanes 6–10), and their chromosomal DNA was prepared in agarose plugs and separated by pulsed field gel electrophoresis as described in MATERIALS AND METHODS. P, agarose plug at the origin of electrophoresis; I–III, positions of S. pombe chromosomes 1–3, respectively. (D) Levels of tyrosine-phosphorylated Cdc2 in rad22/rti1 mutant cells at the stage of post-DNA synthesis. Wild-type (lanes 1 and 6), rad22-H6 (lanes 2 and 7), rad22-H6 Δrti1 (lanes 3 and 8), cdc21-M63 (lanes 4 and 9) and cdc25–22 (lanes 5 and 10) cells were collected at 6 h (lanes 1–5) and 8 h (lanes 6–10). Cell extracts were prepared and electrophoresed on 12% SDS-polyacrylamide gels with loading of 60 μg of protein per lane for the detection of tyrosine-phosphorylated Cdc2 (upper lanes) and 15 μg of protein per lane for the detection of Cdc2 (lower lanes), transferred to nitrocellulose membranes, and probed with anti-phosphotyrosine antibody (purchased from NBL) and anti-Cdc2 antibody, respectively. The anti-Cdc2 rabbit antibody was raised against the C-terminal seven amino acids of S. pombe Cdc2 protein. (E) When combined with the checkpoint rad1-1 mutation, rad22-H6 cells enter premature mitosis. Rapidly growing rad1-1(HM73), rad22-H6, and rad22-H6 rad1-1 (HM105) cells were incubated in YE at 36°C for 12 h. Arrows show cut cells, those in typical premature mitosis. (F) Proliferation and viability of rad22-H6 rad1-1 cells. Rapidly growing rad1-1 (MH73; square), rad22-H6 (HM366; triangle), and rad22-H6 rad1-1 (MH105; filled circle) cells were incubated in YE at 36°C for various times, and the cell number and viability were determined.
To pinpoint the cell cycle position of retardation, we analyzed cells at 6 and 8 h after release for the status of chromosome replication by pulsed field gel electrophoresis. At both time points the chromosomes of the rad22-H6 and rad22-H6 Δrti1 cells migrated to the same positions as those of wild-type cells, with no sign of fragmentation (Figure 3C). Chromosome III of the rad22-H6 Δrti1 cells as well as of rad11-A1 and rad11-A1 rad22-H6 cells (see Figure 5) migrated slightly faster for unknown reasons. However, there was no link between this migration anomaly and the phenotype of rad22-H6 or rad11-A1 cells. In this experiment, the chromosomes of cdc21-M63 cells used as positive control failed to enter the gel. Thus, the chromosomes of the cells lacking rad22+ function replicated completely or nearly completely without any noticeable fragmentation, further confirming that spontaneous chromosome breaks during S phase progression are unlikely to be the cause of cell cycle arrest.
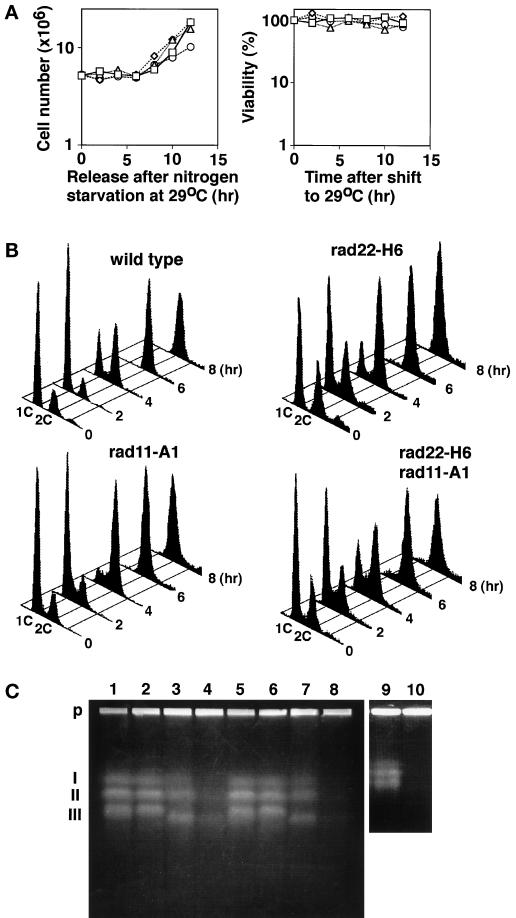
Figure 5.
rad22-H6 rad11-A1 double mutant cells arrest without completion of DNA synthesis. (A) Proliferation and viability of wild-type (square), rad22-H6 (triangle), rad11-A1 (diamond), and rad22-H6 rad11-A1 (circle) cells. Each strain was grown in PM + leu medium to middle log phase at 23°C and arrested in G1 by incubating in nitrogen-free PM + leu for 24 h. The cells were then released to start cell cycling by incubating in PM + leu at 29°C and sampled at the indicated times for the determination of the cell number and viability. (B) Cell cycle progression of wild-type, rad22-H6, rad11-A1 (HM370), and rad22-H6 rad11-A1 (HM372) cells released at 29°C after G1 arrest. Each strain was arrested and released from G1 at 29°C as in A. Cells were then harvested and analyzed by flow cytometry for cell cycle progression. (C) Pulsed field gel electrophoresis. Wild-type (lanes 1, 5, and 9), rad22-H6 (lanes 2 and 6), rad11-A1 (lanes 3, 7, and 10), and rad22-H6 rad11-A1 (lanes 4 and 8) cells were arrested at G1 as in A, released at 27°C (lanes 1–4), 29°C (lanes 5–8), or 35°C (lanes 9 and 10), and incubated for 6 h. P, agarose plugs at the origin of electrophoresis; I–III, positions of S. pombe chromosomes 1–3, respectively.
As already shown, the delayed septation particularly for rad22-H6 Δrti1 cells suggested that they were arrested or slowed in progression before M phase. This was supported by the next experiment. During progression through S and G2 phases, Cdc2 kinase is held inactive by phosphorylation on tyrosine 15 and activated by dephosphorylation at the onset of mitosis (Nurse, 1994). We examined the levels of tyrosine-phosphorylated Cdc2 in the mutant cells at 6 and 8 h after release by direct Western blot of cell lysates with an anti-phosphotyrosine antibody (Figure 3D). Unlike in wild-type cells, but just like in Cdc25 phosphatase-deficient G2-arrested cells, Cdc2 kinase in these cells remained phosphorylated on tyrosine. These results led us to conclude that cells lacking rad22+ function were arrested or slowed in progression at late S or G2 phase.
Distinction between late S or G2 arrest is difficult, but it is generally known that if combined with a checkpoint rad mutation, S phase mutants enter abnormal mitosis, typically with the production of anucleate cells and cells septated on nuclei (“cut” phenotype) (Al-Khodairy and Carr, 1992; Rowley et al., 1992). We used this assay. When combined with the checkpoint-defective rad1-1 mutation, rad22-H6 cells showed a cut phenotype with a concomitantly accelerated viability loss upon a shift to the nonpermissive temperature (Figure 3, E and F). These results suggest that cells lacking rad22+ function were arrested or markedly slowed in progression at late S phase, during which replication of the bulk of the chromosomes was completed.
Replication Factor A Is a Critical Target for S Phase-promoting Action of rad22+/rti1+
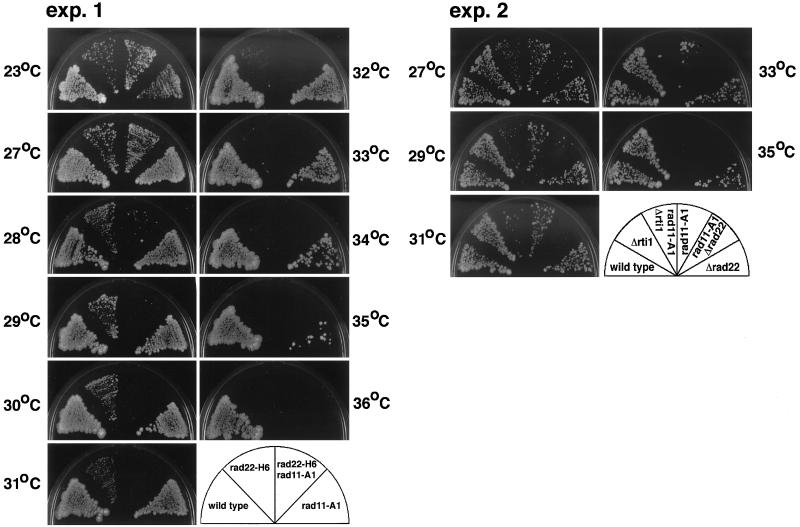
During the process of homologous recombination performed by the Rad51 system, Rad52 directly interacts with the large and middle subunits of RPA as well as Rad51, which catalyzes strand exchanges (Fimenich et al., 1995; Park et al., 1996; Sung, 1997; Hays et al., 1998). Because RPA is also essential for DNA replication, we suspected that RPA might be a target for the S phase-promoting action of rad22+/rti1+ and examined a possible genetic link between rad22-H6 and rad11-A1, the latter of which encodes the large subunit of RPA (Ishiai et al., 1996; Parker et al., 1997). rad22-H6 rad11-A1 double mutant cells were generated by crossing, streaked on plates along with rad22-H6 and rad11-A1 single mutants having otherwise identical genetic backgrounds, and compared for their ability to form colonies at various temperatures. Under the conditions used, rad11-A1 cells were unable to proliferate at 35°C, and rad22-H6 cells were unable to proliferate at 32–33°C. As shown in Figure 4, unlike rad22-H6 and rad11-A1 cells, the double mutant was found unable to grow at 28–29°C. This synthetic effect was not specific to a certain mutation allele of rad22+ and was observed with the entire loss of rad22+. As shown in Figure 4, exp.2, rad11-A1 Δrad22 cells were unable to proliferate at 30°C, 5°C below the restriction temperature for rad11-A1 cells. Such a synthetic effect was also observed between rad11-A1 and deletion of rt1+, albeit it was less. The restriction temperature of rad11-A1 cells dropped by 2°C upon deletion of rti1+. Thus, there was a remarkable functional link between Rad22/Rti1 and the large subunit of RPA. This functional link suggests that either Rad22/Rti1 promotes S phase progression via activation of RPA, or RPA activates the S phase-promoting function of Rad22/Rti1.
Figure 4.
rad22+/rti1+ genetically interacts with rad11+ in cell proliferation. Wild-type, rad22-H6, rad11-A1, and rad22-H6 rad11-A1 cells (exp. 1) and wild-type, Δrt1, Δrad22, Δrti1 rad11-A1 (HM373), and Δrad22 rad11-A1 (HM374) cells (exp. 2) were streaked on PM + leu plates and incubated for 7 d at the indicated temperatures.
If the former possibility held true, the double mutant would have been arrested with the phenotype of defective RPA. If the latter held true, the double mutant would have been arrested with the phenotype of defective Rad22. Because cells with defective RPA were severely deficient in DNA synthesis, these two phenotypes could be distinguished by pulsed field gel electrophoresis. The rad22-H6 rad11-A1 double mutant and each single mutant were arrested in G1 by nitrogen starvation and released to start cell cycling at 29 or 27°C. As shown in Figure 5A, at 8 h after release, both the single mutants started to increase in cell number with the same rate as wild-type cells and retained viability at least until 12 h. The double mutant behaved similarly but displayed a slightly reduced growth rate at 29°C as only a noticeable difference. Flow cytometric analysis of the progression of DNA synthesis showed that none of these mutants had any apparent retardation in bulk DNA synthesis at 29°C (Figure 5B). Next, to examined whether their chromosomes were replicated completely, cells were collected at 6 h after release, time enough for wild-type cells to complete chromosome replication, and their chromosomes were analyzed by pulsed field gel electrophoresis. Unlike those of wild-type cells and the single mutants, the chromosomes of the double mutant failed to enter the gel (see Figure 5C, lane 8), showing that their replication was incomplete. Incomplete chromosome replication in some cell fraction was also observed even at 27°C, a temperature permissive to the double mutant (Figure 5C, lane 4). Because incomplete chromosome replication is characteristic of the rad11-A1 mutant and was seen originally at 35°C for this single mutation (Figure 5C, lane 10), these results show that the presence of rad22-H6 mutation drastically enhanced the phenotype of rad11-A1. We therefore concluded that RPA was a critical target for the S phase-promoting action of Rad22/Rti1, and that at least the mutated RPA absolutely required Rad22/Rti1 for DNA replication in S phase at the regular growth temperature.
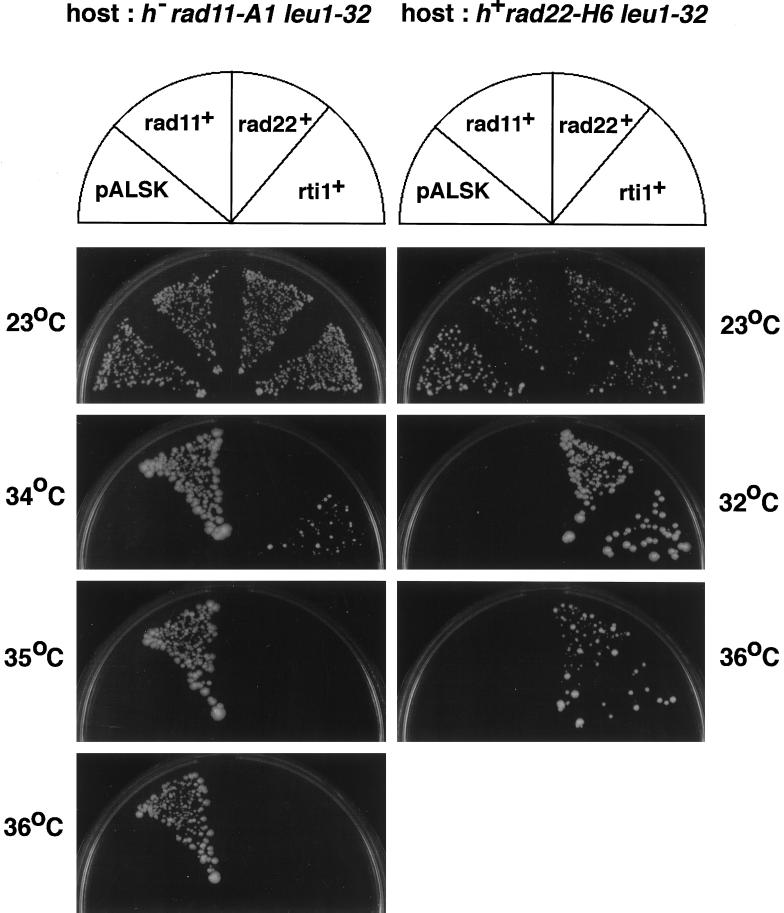
The functional interaction between rad22+/rti1+ and RPA was also detected in a different assay. Overexpression of rti1+ but not of rad22+ suppressed the thermosensitivity of rad11-A1 cells, albeit marginally (Figure 6). However, overexpression of rad11+ did not apparently suppress the thermosensitivity of rad22-H6 cells. This result supports the conclusion that RPA is a critical target for the S phase-promoting action of Rad22/Rti1.
Figure 6.
Overexpression of rti1+ suppresses the thermosensitivity of rad11-A1 cells. The rad22-H6 and rad11-A1 cells stably transfected with the empty pALSK vector, pALSK-rad11+, pALSK-rad22+, or pALSK-rti1+ were streaked on PM agar plates and incubated at the indicated temperatures.
DISCUSSION
All the data presented here show that Rad22/Rti1, a key component of the major double-strand break repair system in fission yeast, is required intrinsically for S phase completion in cycling cells rather than solely for repair of the double-strand breaks that might spontaneously or forcedly be generated during the replication of chromosomes. At the rad22-H6 arrest point, the chromosomes were replicated and lacked detectable fragmentation, as indicated by pulsed field gel electrophoresis patterns. However, S phase was not completed, as indicated by the occurrence of mitotic catastrophe when the rad22-H6 mutation was combined with the rad1 checkpoint mutation. The mechanism by which Rad22/Rti1 promotes S phase completion is unclear. However, the strong genetic interaction between this mutation and rad11-A1 and the ability of overexpressed rti1+ to rescue rad11-A1 cells indicate that at least RPA is a critical target for the S phase-promoting action of Rad22/Rti1. Consequently, given that RPA is essential for DNA synthesis, the lack of a defect in bulk chromosome replication in Rad22/Rti1-deficient cells may in turn suggest two possibilities concerning the regulation of RPA. A Rad22/Rti1-like factor might be present in the cell and specifically used to activate RPA for replication of the bulk of the chromosomes, whereas Rad22/Rti1 might be used for replication of some specific parts of the chromosomes or for DNA synthesis at a certain stage of late S phase during which the Rad22/Rti1-like factor might be inactive. Alternatively, RPA might require Rad22/Rti1 specifically for replication of certain DNA sequences after completion of bulk chromosome replication. Regardless of which possibility is correct, the strong synthetic effect of the rad22-H6 mutation on rad11-A1 leading to defective chromosome replication indicates that Rad22/Rti1 could function as a universal activator of RPA throughout S phase in regular mitotic cell cycling. Although it is clear that at least RPA is a critical target for Rad22/Rti1 to promote S phase completion, our data are also consistent with the possibility that there may be other targets. First, at the rad22-H6 arrest point, chromosome replication was seemingly completed. Second, we failed to detect any ability of overexpressed rad11+ to rescue rad22-H6 cells. Although consistent with the existence of other targets, these results could also be explained if RPA is the sole target, because the active RPA is a heterotrimer (Sibenaller et al., 1998). Therefore, supplying only the large subunit may not be sufficient to compensate for partial inactivation of Rad22.
The mechanism by which Rad22/Rti1 activates RPA function in regular mitotic cell cycling is unclear at present but may be similar to that for homologous recombination. Budding yeast Rad52 has the ability to bind DNA and the large and middle subunits of RPA (Mortensen et al., 1996; Hays et al., 1998). Given this biochemical property of the molecule, three mechanisms may be conceivable. First, fission yeast Rad22/Rti1 might promote assembly of the RPA subunits into an active complex. Second, it might facilitate or stabilize binding of RPA to single-strand DNA. Third, it might promote removal of RPA from single-strand DNAs, as shown for Rad52 in Rad51-catalyzed strand exchanges (Benson et al., 1998; New et al., 1998; Shinohara and Ogawa, 1998). Further studies are needed to resolve this question.
rti1+ seems to slightly differ from rad22+ in biological role. rti1+ is less potent than rad22+ in rescue of rad22-H6 cells, yet it is more potent in rescue of rad11-A1 cells. Moreover, deletion of rti1+ resulted in little impairment of double-strand break repair but significantly influenced the thermosensitivity of rad11-A1 cells. These results suggest that rti1+ might be more specialized for activating RPA during regular cell cycling.
Like fission yeast, budding yeast contains a Rad52 homologue, Rad59, which is involved in Rad51-independent mitotic recombination and double-strand break repair (Bai and Symington, 1996). The Rad52/Rad59 pair, however, seems to functionally differ from the Rad22/Rti1 pair. Unlike Δrad22 Δrti1 cells, budding yeast ΔRAD52 ΔRAD59 cells are still viable despite severe defects in double-strand break repair. Moreover, unlike Rad59, Rti1 is likely to be involved in Rad51-dependent double-strand break repair, because cells lacking rhp51+ (fission yeast homologue of RAD51) are profoundly more sensitive to x-rays than Δrad22 cells (Muris et al., 1997).
It is unknown at present whether Rad22/Rad52 homologues play a role promoting S phase progression in mitotic cell cycling in other organisms. In the organisms studied to date, Rad52 or its counterpart in other organisms is dispensable for cell proliferation (Bai and Symington, 1996; Lim et al., 1996; Tsuzuki et al., 1996). This might be due to the presence of a functional homologue, like in the fission yeast, or to the presence of a hypothetical Rad22-like factor that has been evolved to be specialized for the activation of replication protein A during S phase. In this regard, it is noteworthy that in S. cerevisiae cells, Rad52 protein molecules localize at chromosomes even during mitotic cell cycling (Gasior et al., 1998) and that human Rad52 is specifically expressed in S phase (Chen et al., 1997), suggesting that at least budding yeast and human Rad52 may share S phase-promoting function with fission yeast Rad22/Rti1 to a certain extent.
ACKNOWLEDGMENTS
We thank P. Nurse for various cdc mutants, B. Arcangioli for pB9, and C. Shimoda for advice on isolation of cell mutants. H.M is a recipient of JSPS Postdoctoral Fellowships for Research Abroad. This work was supported by grants from the Ministry of Education, Science and Culture of Japan.
REFERENCES
- Albala JS, Thelen MP, Prange C, Fan W, Christensen M, Thompson LH, Lennon GG. Identification of a novel human RAD51 homolog, RAD52B. Genomics. 1997;46:476–479. doi: 10.1006/geno.1997.5062. [DOI] [PubMed] [Google Scholar]
- Alfa C, Fantes P, Hyams J, McLeod M, Warbrick E. Experiment with Fission Yeast: A Laboratory Course Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1993. [Google Scholar]
- Al-Khodairy F, Carr AM. DNA repair mutants defining G2 checkpoint pathways in Schizosaccharomyces pombe. EMBO J. 1992;11:1343–1350. doi: 10.1002/j.1460-2075.1992.tb05179.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arcangioli B, Klar AJS. A novel switch-activating site (SAS1) and its cognate binding factor (SAP1) required for efficient mat1 switching in Schizosaccharomyces pombe. EMBO J. 1991;10:3025–3032. doi: 10.1002/j.1460-2075.1991.tb07853.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai Y, Symington LS. A Rad52 homolog is required for RAD51-independent mitotic recombination in Saccharomyces cerevisiae. Genes & Dev. 1996;10:2025–2037. doi: 10.1101/gad.10.16.2025. [DOI] [PubMed] [Google Scholar]
- Beach DH. Cell type switching by DNA transposition in fission yeast. Nature. 1983;305:682–688. [Google Scholar]
- Benson FE, Baumann P, West SC. Synergistic actions of Rad51 and Rad52 in recombination and DNA repair. Nature. 1998;391:401–404. doi: 10.1038/34937. [DOI] [PubMed] [Google Scholar]
- Bezzubova OY, Schmidt H, Ostermann K, Heyer W-D, Buerstedde J-M. Identification of a chicken RAD52 homologue suggests conservation of the RAD52 recombination pathway throughout the evolution of higher eukaryotes. Nucleic Acids Res. 1993;21:5945–5949. doi: 10.1093/nar/21.25.5945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borts RH, Lichten M, Haber JE. Analysis of meiosis- defective mutations in yeast by physical monitoring of recombination. Genetics. 1986;113:551–567. doi: 10.1093/genetics/113.3.551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen F, Nastasi A, Shen Z, Brenneman M, Crissman H, Chen DJ. Cell cycle-dependent protein expression of mammalian homologs of yeast DNA double-strand break repair genes Rad51 andRad52. Mutat Res, 1997;384:205–211. doi: 10.1016/s0921-8777(97)00020-7. [DOI] [PubMed] [Google Scholar]
- Coxon A, Maundrell K, Kearsey SE. Fission yeast cdc21+ belongs to a family of proteins involved in an early step of chromosome replication. Nucleic Acids Res. 1992;20:5571–5577. doi: 10.1093/nar/20.21.5571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egel R, Egel-Mitani M. Premeiotic DNA synthesis in fission yeast. Exp Cell Res. 1974;88:127–134. doi: 10.1016/0014-4827(74)90626-0. [DOI] [PubMed] [Google Scholar]
- Egel R. Selective spore survival during replica-plating of fission yeast. Arch Microbiol. 1977;112:109–110. doi: 10.1007/BF00446662. [DOI] [PubMed] [Google Scholar]
- Essers J, Hendriks RW, Swagemakers AM, Troelstra C, de Wit J, Bootsma D, Hoeijmakers JH, Kanaar R. Disruption of mouse RAD54 reduces ionizing radiation resistance and homologous recombination. Cell. 1997;89:195–204. doi: 10.1016/s0092-8674(00)80199-3. [DOI] [PubMed] [Google Scholar]
- Fimenich AA, Elias-Arnanz M, Berg P. A novel allele of Saccharomyces cerevisiae RFA1 that is deficient in recombination and repair and suppressible by. Rad52. Mol Cell Biol. 1995;15:1620–1631. doi: 10.1128/mcb.15.3.1620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Game JC. DNA double-strand breaks and the RAD50-RAD57 genes in Saccharomyces. Semin Cancer Biol. 1993;4:73–83. [PubMed] [Google Scholar]
- Gasior SL, Wong AK, Kora Y, Shinohara A, Bishop DK. Rad52 associates with RPA and functions with Rad55 and Rad57 to assemble meiotic recombination complexes. Genes & Dev. 1998;12:2208–2221. doi: 10.1101/gad.12.14.2208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hays SL, Firmenich SL, Massey SL, Banerjee R, Burg P. Studies of the interaction between Rad52 protein and the yeast single-stranded DNA binding protein RPA. Mol Cell Biol. 1998;18:4400–4406. doi: 10.1128/mcb.18.7.4400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishiai M, Sanchez JP, Amin AA, Murakami Y, Hurwitz J. Purification, gene cloning, and reconstitution of the heterotrimeric single-stranded DNA-binding protein from Schizosaccharomyces pombe. J Biol Chem. 1996;271:20868–20878. doi: 10.1074/jbc.271.34.20868. [DOI] [PubMed] [Google Scholar]
- Kanaar R, et al. Human and mouse homologs of the Saccharomyces cerevisiae RAD54 DNA repair gene: evidence for functional conservation. Curr Biol. 1996;6:828–838. doi: 10.1016/s0960-9822(02)00606-1. [DOI] [PubMed] [Google Scholar]
- Kelly TJ, Martin GS, Forsburg SL, Stephen RJ, Russo A, Nurse P. The fission yeast cdc18+ gene product couples S phase to START and mitosis. Cell. 1993;74:371–382. doi: 10.1016/0092-8674(93)90427-r. [DOI] [PubMed] [Google Scholar]
- Kezenman DJ, Salvo VA, Nunes E. Effect of bleomycin on growth kinetics and survival of Schizosaccharomyces pombe: a model of repair pathways. J Bacteriol. 1992;174:3125–3132. doi: 10.1128/jb.174.10.3125-3132.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim DS, Hasty P. A mutation in mouse RAD51 results in an early embryonic lethal that is suppressed by a mutation in p53. Mol Cell Biol. 1996;16:7133–7143. doi: 10.1128/mcb.16.12.7133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKee RH, Lawrence CW. Genetic analysis of gamma-ray mutagenesis in yeast. III. Double-mutant strains. Mutat Res. 1980;70:37–48. doi: 10.1016/0027-5107(80)90056-1. [DOI] [PubMed] [Google Scholar]
- Moreno S, Klar A, Nurse P. Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol. 1991;194:795–823. doi: 10.1016/0076-6879(91)94059-l. [DOI] [PubMed] [Google Scholar]
- Mortensen UH, Bendixen I, Sunjevaric I, Rothstein R. DNA strand annealing is promoted by the yeast Rad52 protein. Proc Natl Acad Sci USA. 1996;93:10729–10734. doi: 10.1073/pnas.93.20.10729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muris DF, et al. Cloning of human and mouse genes homologous to RAD52, a yeast gene involved in DNA repair and recombination. Mutat Res. 1994;315:295–305. doi: 10.1016/0921-8777(94)90040-x. [DOI] [PubMed] [Google Scholar]
- Muris DF, Vreeken K, Carr AM, Broughton BC, Lehmann AR, Lohman PH, Pastink A. Cloning the RAD51 homologue of Schizosaccharomyces pombe. Nucleic Acids Res. 1993;21:4586–4591. doi: 10.1093/nar/21.19.4586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muris DFR, Vreeken K, Schmidt H, Ostermann K, Clever B, Lohman PHM, Pastink A. Homologous recombination in the fission yeast Schizosaccharomyces pombe: different requirements for the rhp51+, rhp54+ and rad22+ genes. Curr Genet. 1997;31:248–254. doi: 10.1007/s002940050202. [DOI] [PubMed] [Google Scholar]
- Namsaraev E, Berg P. Characterization of strand exchange activity of yeast Rad51 protein. Mol Cell Biol. 1997;17:5359–5368. doi: 10.1128/mcb.17.9.5359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- New JA, Suglyama T, Zaitseva E, Kowalczykowski SC. Rad52 protein stimulates DNA strand exchange by Rad51 and replication protein A. Nature. 1998;391:407–410. doi: 10.1038/34950. [DOI] [PubMed] [Google Scholar]
- Nurse P. Genetic control of cell size at cell division in yeast. Nature. 1975;256:547–551. doi: 10.1038/256547a0. [DOI] [PubMed] [Google Scholar]
- Nurse P. Ordering S phase and M phase in the cell cycle. Cell. 1994;79:547–550. doi: 10.1016/0092-8674(94)90539-8. [DOI] [PubMed] [Google Scholar]
- Okazaki K, Okazaki N, Kume K, Jinno S, Tanaka K, Okayama H. High frequency transformation methods and library transducing vectors for cloning of mammalian cDNAs by trans-complementation of Schizosaccharomyces pombe. Nucleic Acids Res. 1990;18:6485–6489. doi: 10.1093/nar/18.22.6485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostermann K, Lorentz A, Schmidt H. The fission yeast rad22+ gene, having a function in mating-type switching and repair of DNA damages, encodes a protein homologous to Rad52 of Saccharomyces cerevisiae. Nucleic Acids Res. 1993;21:5940–5944. doi: 10.1093/nar/21.25.5940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker AE, Clyne RK, Carr AM, Kelly TJ. The Schizosaccharomyces pombe rad11+ gene encodes the large subunit of replication protein A. Mol Cell Biol. 1997;17:2381–2390. doi: 10.1128/mcb.17.5.2381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park MS, Ludwig DL, Stigger E, Lee S-H. Physical interaction between human RAD52 and RPA is required for homologous recombination in mammalian cells. J Biol Chem. 1996;271:18996–19000. doi: 10.1074/jbc.271.31.18996. [DOI] [PubMed] [Google Scholar]
- Phipps J, Nasim A, Miller DR. Recovery, repair, and mutagenesis in Schizosaccharomyces pombe. Adv Genet. 1985;23:1–72. doi: 10.1016/s0065-2660(08)60511-8. [DOI] [PubMed] [Google Scholar]
- Resnick MA. The repair of double-strand breaks in chromosomal DNA of yeast. Basic Life Sci. 1975;5B:549–556. doi: 10.1007/978-1-4684-2898-8_20. [DOI] [PubMed] [Google Scholar]
- Rowley R, Subramani S, Young PG. Checkpoint controls in Schizosaccharomyces pombe: rad1. EMBO J. 1992;11:1335–1342. doi: 10.1002/j.1460-2075.1992.tb05178.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F, Nicklen S, Coulson AR. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlake C, Ostermann K, Schmidt H, Gutz H. Analysis of DNA repair pathways of Schizosaccharomyces pombe by means of swi- rad double mutants. Mutat Res. 1993;294:59–67. doi: 10.1016/0921-8777(93)90058-o. [DOI] [PubMed] [Google Scholar]
- Shinohara A, Ogawa T. Stimulation by Rad52 of yeast Rad51-mediated recombination. Nature. 1998;391:404–407. doi: 10.1038/34943. [DOI] [PubMed] [Google Scholar]
- Shinohara A, Ogawa H, Matsuda Y, Ushio N, Ikeo K, Ogawa T. Cloning of human, mouse and fission yeast recombination genes homologous to RAD51 and RecA. Nat Genet. 1993;4:239–243. doi: 10.1038/ng0793-239. (erratum 5, 312). [DOI] [PubMed] [Google Scholar]
- Shinohara A, Ogawa H, Ogawa T. Rad51 protein involved in repair and recombination in S. cerevisiae is a RecA-like protein. Cell. 1992;69:457–470. doi: 10.1016/0092-8674(92)90447-k. (erratum 71, following 180). [DOI] [PubMed] [Google Scholar]
- Sibenaller ZA, Sorensen BR, Wold MS. The 32- and 14-kilodalton subunits of replication protein A are responsible for species-specific interactions with single-stranded DNA. Biochemistry. 1998;37:12496–12506. doi: 10.1021/bi981110+. [DOI] [PubMed] [Google Scholar]
- Sonoda E, Sasaki MS, Buerstedde J-M, Bezzubovo O, Shinohara A, Ogawa H, Takata M, Yamaguchi-Iwai Y, Takeda S. Rad51-deficient vertebrate cells accumulate chromosomal breaks before cell death. EMBO J. 1998;17:598–608. doi: 10.1093/emboj/17.2.598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugiyama T, New JH, Kowalczykowski SC. DNA annealing by Rad52 protein is stimulated by specific interaction with the complex of replication protein A and single-stranded DNA. Proc Natl Acad Sci USA. 1998;95:6049–6054. doi: 10.1073/pnas.95.11.6049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung P. Function of yeast Rad52 protein as a mediator between replication protein A and the Rad51 recombinase. J Biol Chem. 1997a;272:28194–28197. doi: 10.1074/jbc.272.45.28194. [DOI] [PubMed] [Google Scholar]
- Sung P. Yeast Rad55 and Rad57 proteins from a heterodimer that functions with replication protein A to promote DNA strand exchange by Rad51 recombinase. Genes & Dev. 1997b;11:1111–1121. doi: 10.1101/gad.11.9.1111. [DOI] [PubMed] [Google Scholar]
- Tanaka K, Okazaki K, Okazaki N, Ueda T, Sugiyama A, Nojima H, Okayama H. A new cdc gene required for S phase entry of Schizosaccharomyces pombe encodes a protein similar to the cdc10+ and SWI4 gene product. EMBO J. 1992;11:4923–4932. doi: 10.1002/j.1460-2075.1992.tb05599.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuzuki T, Fujii Y, Sakumi K, Tominaga Y, Nakao K, Sekiguchi M, Matsushiro A, Yoshimura Y, Morita T. Targeted disruption of the RAD51 gene leads to lethality in embryonic mice. Proc Natl Acad Sci USA. 1996;93:6236–6240. doi: 10.1073/pnas.93.13.6236. [DOI] [PMC free article] [PubMed] [Google Scholar]