Abstract
Human lymphoblastoid cells derived from different healthy individuals display considerable variation in their transcription profiles. Here we show that such variation in gene expression underlies interindividual susceptibility to DNA damaging agents. The results demonstrate the massive differences in sensitivity across a diverse cell line panel exposed to an alkylating agent. Computational models identified 48 genes with basal expression that predicts susceptibility with 94% accuracy. Modulating transcript levels for two member genes, MYH and C21ORF56, confirmed that their expression does indeed influence alkylation sensitivity. Many proteins encoded by these genes are interconnected in cellular networks related to human cancer and tumorigenesis.
Keywords: DNA damage susceptibility, alkylating agents, prediction
The interindividual differences in disease susceptibility, responsiveness to chemotherapeutics, and susceptibility to environmental exposures across human populations are influenced by a combination of gene–environment interactions. Over the past few years, studies aimed at dissecting the genetic basis underlying human phenotypic variation have built off of the dense genotyping established by the HapMap Consortium. A wealth of genome-wide association studies (GWAS) have described how human genetic variation, at the level of single nucleotide differences, is linked to such complex diseases as diabetes and breast cancer (Hunter et al. 2007; Zeggini and McCarthy 2007). In addition, GWAS have also linked DNA polymorphic variants to gene expression variation across populations (Cheung et al. 2005; Stranger et al. 2005, 2007; Dixon et al. 2007).
However, while it is known that human lymphoblastoid cells derived from different healthy individuals display considerable variation in their transcription profiles (Cheung et al. 2003, 2005; Stranger et al. 2005, 2007; Dixon et al. 2007), the influence this variation has on the response to environmental and chemotherapeutic agents is unknown. In this study, a panel of 24 cell lines previously derived from unrelated, healthy individuals with diverse ancestry (Collins et al. 1998) was tested for variation in sensitivity to the DNA damaging agent, N-methyl-N′-nitro-N-nitrosoguanidine (MNNG). MNNG induces a variety of alkylated DNA bases, among which O6-methylguanine (O6MeG) is known to be particularly toxic as well as mutagenic because it pairs with thymine during replication. O6MeG can be repaired by the MGMT DNA repair methyltransferase (Pegg 1990, 2000), but left unrepaired, the ensuing O6MeG:T base pair can be processed by the DNA mismatch repair (MMR) pathway, and such processing actually triggers apoptotic cell death and cytotoxicity (Kaina et al. 1997; Hickman and Samson 1999, 2004). Therefore, cells deficient in MGMT but proficient for MMR are extremely sensitive to MNNG-induced killing, whereas cells deficient in both MGMT and MMR are extremely resistant or tolerant to MNNG, but at the cost of increased mutation (Karran 2001). While MGMT and MMR status are thus known to be associated with alkylation sensitivity, we questioned whether the expression level of these two repair pathways is sufficient to explain interindividual variation in alkylation sensitivity.
We show that there is extensive interindividual variation in the response of cell lines derived from a healthy, genetically diverse population upon exposure to the DNA alkylating agent MNNG. The differences in susceptibility to MNNG were associated with variation in gene expression to identify genomic predictors of cellular sensitivity. A set of 48 genes was identified that can predict, with a remarkable 94% accuracy, differences in cellular sensitivity to MNNG in a test population. The basal gene expression, rather than MNNG-treated expression level, was found to be the better predictor of cellular sensitivity. To validate the computational models, the expression level of two members of the predictive gene set, C21ORF56 and MYH, were modulated and shown to alter cellular sensitivity to MNNG. These findings may have profound implications in the clinical setting, where the collective set of 48 genes may be used as predictors and modulators of cellular sensitivity to cancer chemotherapeutics.
Results and Discussion
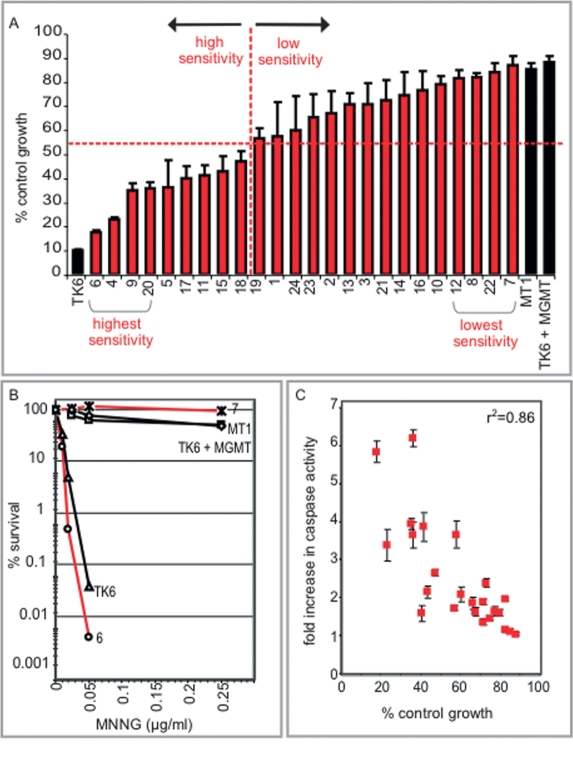
To assess the range of interindividual differences in sensitivity to a DNA alkylating agent, we used a high-throughput growth inhibition assay (percentage of control growth) across the panel of cell lines. Control cell lines with very high MNNG sensitivity (TK6) or very low sensitivity (TK6 + MGMT, MT1) were included (Kat et al. 1993). The panel of cell lines displayed MNNG sensitivities spanning the entire range between the control cell lines (Fig. 1A). These large differences were even more apparent when measured by the lower throughput killing curve assay that has a greater dynamic range (Fig. 1B). Importantly, the MNNG sensitivities of the cell lines are not associated with individual differences in growth characteristics (e.g., cell doubling time) (Supplemental Fig. S1). We also monitored MNNG-induced apoptosis in the cell lines (by caspase-3 activation) and found a positive correlation with MNNG sensitivity (Fig. 1C). Thus, the growth inhibition, survival, and apoptosis assays each underscore the extensive range of interindividual responses to MNNG among genetically diverse cells.
Figure 1.
A considerable range of interindividual sensitivity to a DNA alkylating agent. (A) The percentage of control growth of the cell lines at 72 h after treatment with MNNG (0.5 μg/mL) using a growth inhibition assay. The division between high and low sensitivity among the cell lines is demarcated at 53% with a red dotted line. (B) The percentage of survival of cell lines 6 and 7 was determined 10 d after treatment with MNNG and compared with three control cell lines (TK6, TK6 + MGMT, and MT1) using a killing curve assay. (C) Fold increase in caspase-3 activity was determined 72 h post-treatment with MNNG across the cell line panel.
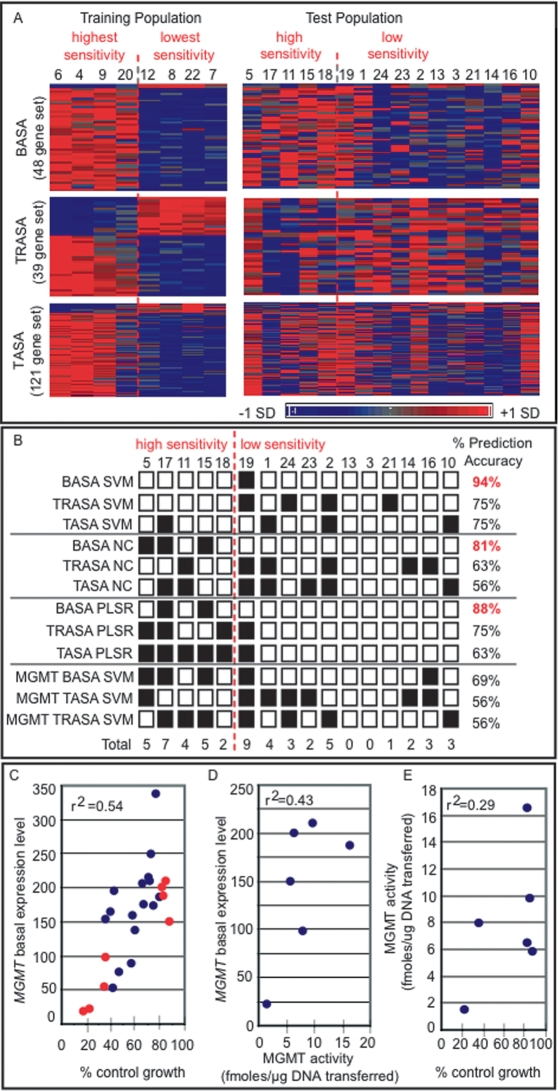
To determine whether transcriptional profiles could predict cellular response to MNNG, a two-class prediction algorithm was applied to the gene expression profiles of the 24 cell lines under both untreated (basal) and MNNG-treated conditions. The cell lines were divided into two classes with either high sensitivity or low sensitivity, with a cut point of 53% control growth based on the midpoint between the most sensitive and least sensitive cell line (Fig. 1A, lines 6,7). For the two-class prediction model, a training population composed of the four most sensitive and the four least sensitive cell lines was selected and analyzed to identify genes that were not only differentially expressed but also showed significant positive or negative correlation with increasing MNNG sensitivity. Three alkylation sensitivity-associated (ASA) gene sets were identified as follows: (1) a set of 48 genes derived from basal gene expression (the BASA set), (2) a set of 39 genes derived from treatment-to-basal expression ratio (the TRASA set), and (3) a set of 121 genes derived from treatment-induced expression (the TASA set) (Supplemental Tables 1–3). The expression patterns of the ASA gene sets across the training population and the test population (i.e., the 16 cell lines not included in the training population) are visualized in Figure 2A.
Figure 2.
Identification of ASA genes that predict interindividual differences in alkylation sensitivity. (A) Three ASA gene sets were identified from a training population comprising the four most sensitive and the four least sensitive cell lines including (1) 48 genes derived from basal gene expression (the BASA set), (2) 39 genes derived from treatment-to-basal expression ratio (the TRASA set), and (3) 121 genes derived from treatment-induced expression (the TASA set). Expression patterns for the gene sets are shown for both the training and the test populations of cell lines. Expression values are mean centered with high relative expression indicated in red and low relative expression indicated in blue. (B) The sensitivity of the test population of cell lines to MNNG was predicted using three algorithms: SVM, NC, and PLSR. The two-class prediction algorithms were used with each of the three ASA gene sets as well as the MGMT transcript alone. Correct prediction is indicated with a white box; incorrect prediction, with a black box. (C) MGMT expression level is plotted versus the percentage of control growth of the cell lines treated with MNNG. Red circles indicate cell lines of the training population, and blue circles indicate cell lines of the test population. (D) O6-MeG DNA methyltransferase activity was determined in protein extracts derived from cell lines 4, 7, 20, 12, 22, and 8. Methyltransferase activity is plotted versus the baseline expression level of MGMT in each cell line. (E) Methyltransferase activity is plotted versus the percentage of control growth for the same cell lines as in (D).
The ability of the three ASA gene sets to predict MNNG sensitivity was assessed using the support vector machine (SVM) algorithm. The TRASA and TASA gene sets predicted the MNNG sensitivity of the test population with 75% accuracy (Fig. 2B). Remarkably, the BASA gene set accurately predicted sensitivity in 15 of 16 cell lines (94% accuracy), with only cell line 19 misclassified (Fig. 2B); note that cell line 19 falls on the boundary of the cut point between high and low sensitivity (Fig. 1A). To validate the SVM results, we applied two other prediction algorithms, namely, a nearest centroid (NC) and a partial least squares regression (PLSR) model (Fig. 2B). For all algorithms, the BASA gene set provided maximal prediction of MNNG sensitivity, with SVM providing the highest accuracy (Fig. 2B). The lack of prediction of the TRASA and TASA gene sets may be a result of time point selection, a feature that a more global assessment of temporal responses would potentially capture. That basal gene expression is the most accurate predictor of alkylation sensitivity bodes well for translating these findings to a clinical setting; for example, to predict whether a tumor will respond to alkylation chemotherapy.
The BASA gene set contained two genes that showed positive association of expression with lower MNNG sensitivity; namely, MGMT and the C21ORF56 (Supplemental Table S1). MGMT efficiently repairs O6MeG (Pegg 1990, 2000), and its activity is known to vary among individuals (Vahakangas et al. 1991; Margison et al. 2003). Likewise, we identified considerable variation in the expression level of MGMT across the 24 cell lines, and we demonstrate a positive association of MGMT expression with lower MNNG sensitivity (Fig. 2C). Although MGMT expression level was positively associated with MGMT activity (Fig. 2D), and activity was positively associated with MNNG resistance (Fig. 2E), the correlations were relatively weak. This likely explains why MGMT expression alone is not as strong a predictor of alkylation sensitivity as the set of 48 transcripts together (Fig. 2B).
MGMT silencing is currently being used as a prognostic indicator of successful alkylation chemotherapy for gliobastoma (Hegi et al. 2005); our results suggest that expression levels for the 48 genes described here may prove a more accurate indicator. It should be mentioned that MGMT is the only member of the BASA gene set previously known to influence alkylation sensitivity. Interestingly, upon testing each member of the BASA gene set independently for predictive capacity, genes with equal or greater accuracy than MGMT were identified (Supplemental Table S1). However, while some genes showed higher prediction accuracy than MGMT, it was the collective set of 48 transcripts that provided the maximal prediction of 94% accuracy. The probability of 48 transcripts chosen at random from the pool of expressed genes predicting MNNG sensitivity with such accuracy is <0.002.
In addition to MGMT, the only other member of the BASA gene set with positive association of expression with lower MNNG sensitivity was C21ORF56 (Supplemental Table S1). The C21ORF56 protein exists in two isoforms that are highly conserved across mammals and show homology with SPATC1 (spermatogenesis and centriole-associated protein 1) (Supplemental Fig. S2A,B). Supporting our finding of the variation in the expression level of C21ORF56 in these cell lines, its expression has been documented as highly variable across four separate populations of cell lines. Specifically, C21ORF56 showed variation in expression level across three of the four HapMap populations, namely, the populations of European (CEU), Chinese (CHB), and Japanese (JPT) origin (Stranger et al. 2005, 2007) and in a separate population of British descent (Dixon et al. 2007).
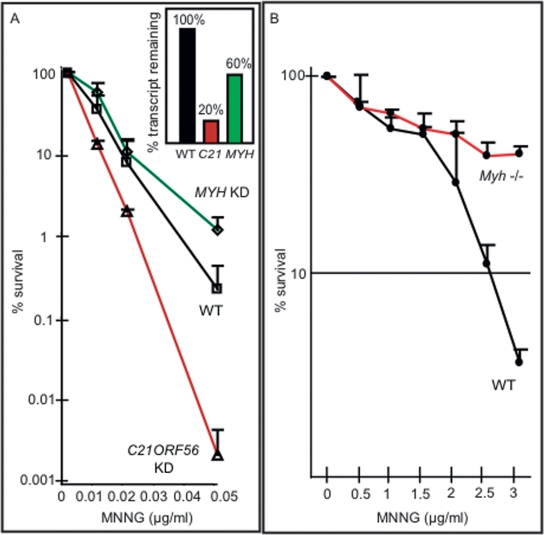
Given the significant association of C21ORF56 gene expression with MNNG sensitivity across the cell line panel, our analysis suggested that C21ORF56, like MGMT, might play a role in protecting cells against MNNG-induced killing. To confirm the role of C21ORF56 in modulating alkylation sensitivity, we show that TK6 cells with 80% knockdown of the C21ORF56 transcript show greatly increased MNNG sensitivity relative to control cells expressing a nontargeting control shRNA (Fig. 3A). In addition, using an alternate C21ORF56-targeting shRNA, an empty vector control, and individually established clones with differential expression of C21ORF56, we show that decreased expression of C21ORF56 is associated with increased sensitivity to MNNG (Supplemental Fig. S3). These results validate the computational prediction that C21ORF56 expression modulates the response of human cells to alkylating agents.
Figure 3.
Modulation of C21ORF56, MYH, and Myh influences MNNG sensitivity. (A) TK6 cells expressing a control shRNA (WT) or shRNA specifically targeting the C21ORF56 transcript (v1), or the MYH transcript (v1), were assessed for the percentage of survival after exposure to MNNG. The inset shows the percentage of transcript remaining C21ORF56 and MYH in knockdown cells. (B) Percentage of survival of Myh−/− or wild-type MEFs determined after treatment with MNNG.
The gene with the most significant positive association of expression with alkylation sensitivity in the BASA gene set was MYH (Supplemental Table S1), a DNA glycosylase that initiates base excision repair by removing adenines mispaired opposite 8-oxoguanine lesions (Slupska et al. 1996; Parker and Eshleman 2003) and not previously known to modulate alkylation sensitivity. In support of our finding of high variance of MYH expression across a genetically diverse population, its expression levels were found to vary among individuals of a separate population (Dixon et al. 2007). Importantly, however, to date no link between the variation in expression level of MYH and interindividual differences in sensitivity upon exposure to a DNA alkylating agent has been made.
As MYH expression was higher in the more sensitive cells, we hypothesized that MYH deficiency might confer resistance, in much the same way that MMR deficiency confers MNNG resistance (Kaina et al. 1997; Karran 2001). This was tested by decreasing the expression of MYH in TK6 cells. The MYH knockdown cells were indeed significantly less sensitive than the parent TK6 cells expressing a control shRNA to cell killing (Fig. 3A). In addition, using an alternate MYH-targeting shRNA, an empty vector control, and individually established clones with differential expression of MYH, we show that increased expression of MYH is associated with MNNG sensitivity (Supplemental Fig. S3). Importantly, the influence of Myh in modulating MNNG sensitivity was further established in mouse cells by showing that Myh−/− mouse embryonic fibroblasts (MEFs) were much less sensitive than wild type to MNNG-induced cell killing (Fig. 3B). These results indicate that MYH expression indeed correlates with increased alkylation sensitivity. Likewise, MYH expression was also significantly higher in the alkylation-sensitive TK6 cell line relative to the much less sensitive MT1 cell line (Fig. 1A,B; data not shown) compatible with the trend seen across the panel of genetically diverse cell line (Fig. 1A). Interestingly, although not known to repair damage induced by MNNG, MYH is known to interact with the MutSα heterodimer (Gu et al. 2002) that binds O6MeG mispairs in DNA to initiate the triggering of apoptotic cell death (Ceccotti et al. 1996; Hickman and Samson 2004). It remains to be determined whether MYH influences alkylation sensitivity via its interaction with the MMR machinery. Finally, it was surprising that no MMR transcripts were represented in the BASA gene set. It turned out that the differences in expression for MSH2, MSH6, and MLH1 did not exceed 1.5-fold, eliminating them from the gene set; however, all three transcripts were in fact higher in the most sensitive versus the least sensitive cell line (data not shown).
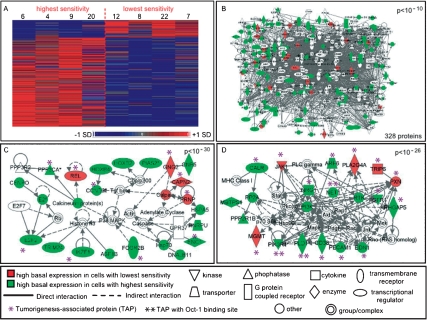
To gain a more comprehensive view of the various pathways that influence alkylation sensitivity, all genes that were differentially expressed under basal conditions between cell lines with the highest and lowest MNNG sensitivity (this time not imposing a requirement for trend significance) were analyzed for network properties (Fig. 4A; Supplemental Table S4). For the 240 genes identified as differentially expressed, 148 gene products are found in the Ingenuity database, and a remarkable ∼85% of these are contained in a single significant interacting network (P < 10−10) (Fig. 4B; Supplemental Tables S5, S6). It thus seems that proteins likely to play a role in determining interindividual differences in alkylation sensitivity are highly connected, a phenomenon previously observed in Saccharomyces cerevisiae (Begley et al. 2004; Said et al. 2004). Contained within the large interacting network are subnetworks that integrate 18 BASA transcripts (including MGMT and MYH), and these subnetworks are enriched for proteins that are associated with tumorigenesis and cancer predisposition (Fig. 4C,D; Supplemental Tables S1, S6). In general, the expression level of the tumorigenesis-associated transcripts showed elevated basal expression in the cell lines with high MNNG sensitivity. Finally, by analyzing the promoter regions of the ASA genes for enriched transcription factor-binding sites, we find evidence for a common regulatory factor, namely, the octamer-binding transcription factor, Oct-1 (Supplemental Table S7). Oct-1 is known to respond to DNA alkylation damage (Zhao et al. 2000) and is a known regulator of stress responses (Tantin et al. 2005). Here we find Oct-1-binding sites significantly enriched in transcripts with elevated basal expression in cell lines with high MNNG sensitivity as well as in tumorigenesis-associated proteins that show higher basal expression in cells with high sensitivity to MNNG (P = 2.05 × 10−5) (Fig. 4D).
Figure 4.
Basal expression networks associated with interindividual differences in sensitivity to MNNG. (A) A heat map of the 240 basally differentially expressed genes identified between two classes of the training population, those with highest and lowest MNNG sensitivity. Expression values are mean centered with high relative expression indicated in red and low relative expression indicated in blue. (B) Of the 240 genes from A, 148 were present in the Ingenuity database. These 148 proteins were analyzed for significant enrichment of molecular interactions. A significant (P < 10−10) interactome of 328 total proteins containing 125 of the 148 proteins was identified. (C) The most significant subnetwork (P < 10−30) of ASA proteins. (D) The second most significant subnetwork (P < 10−26) of ASA proteins. Proteins in red are encoded by transcripts with high basal expression in cells with low MNNG sensitivity, and proteins in green are encoded by transcripts with high basal expression in cells with high MNNG sensitivity; proteins in white are associated with these ASA proteins. Tumorigenesis-associated proteins (TAPs) are indicated with an asterisk and TAP proteins containing Oct-1-binding sites are indicated with two asterisks.
To conclude, our findings may have profound implications in the clinical setting, where the expression of the 48 transcripts encompassing MGMT, C21ORF56, MYH, and many others may not only predict interindividual responses to alkylating agents but could be modulated to affect cancer treatment response. Furthermore, as cell lines derived from different individuals indeed preserve genetic diversity at the level of gene expression (Cheung et al. 2003; Correa and Cheung 2004; Morley et al. 2004), they serve as an ideal tool for establishing interindividual differences in DNA damage responses. We propose that upon exposure of these cell lines to other environmental toxicants and cancer chemotherapeutics we will discover more genes of hitherto unknown function responsible for interindividual differences in sensitivity to DNA damaging agents.
Materials and methods
Cell line panel, drug treatment, and RNA extraction
The 24 lymphoblastoid cell lines, established using EBV transformation, were obtained from the Coriell Institute (New Jersey) and were designated 1–24 as follows: #1 (GM15029), #2 (GM15036), #3 (GM15215), #4 (GM15223), #5 (GM15245), #6 (15,224), #7 (GM15236), #8 (GM15510), #9 (GM15213), #10 (GM15221), #11 (GM15227), #12 (GM15385), #13 (GM15590), #14 (GM15038), #15 (GM15056), #16 (GM15072), #17 (GM15144), #18 (GM15216), #19 (GM15226), #20 (GM15242), #21 (GM15268), #22 (GM15324), #23 (GM15386), and #24 (GM15061). Percentage of control growth was measured using logarithmically growing cells. Cells were treated with 0.5 μg/μL MNNG or untreated, and viable cells were counted 72 h after treatment using a coulter counter coupled with trypan blue staining (total number of viable treated cells/total number of viable untreated cells). Percentage of survival was determined using a killing curve assay counted 10 d after treatment with MNNG as described (Furth et al. 1981). Total RNA was isolated from log phase cells according to the mammalian cell protocol (Qiagen) and labeled according to the Affymetrix protocol. RNA was hybridized to HGU133 Plus 2.0 full genome human arrays in technical duplicate totaling 96 arrays.
Microarray data analysis
Data were normalized using a PLIER algorithm and filtered for nonexpressed transcripts across all arrays as described in Fry et al. (2007), resulting in a reduction of the probesets from the original 54,675 to 19,290. ASA gene sets were determined as follows. The four cell lines with highest MNNG sensitivity and four cell lines with lowest MNNG sensitivity were used as a training population. For the ASA sets, genes with differential expression between the two groups were identified with (1) significant fold change (≥1.5 or ≤−1.5, P < 0.05 t-test), and (2) a significant positive or negative trend for association of gene expression with increasing growth inhibition (percentage of control growth) using a correlation measurement (r ≥ 0.7 or ≤−0.7, P < 0.01 Trend) calculated using a linear regression model in S-PLUS 7.0 (http://www.insightful.com). For the fourth gene set (Fig. 4), a P for trend was not imposed. Three methods for two-class prediction were used, including SVM algorithm carried out using Gene Pattern Software (version 2.0.1) (http://www.broad.mit.edu), the NC algorithm based in R, and PLSR analysis programmed in MATLAB (Mathworks, Inc.) and adapted from Geladi et al. (1996). Network and gene ontology analysis was performed using Ingenuity software (http://www.ingenuity.com). Cancer and tumorigenesis-associated proteins were identified using the Ingenuity database as well as the Genomica module analysis (http://genie.weizmann.ac.il). Transcription factor-binding site analysis was performed as described (Fry et al. 2007) using the EXPANDER program. Microarray data have been deposited in the Gene Expression Omnibus repository (http://www.ncbi.nlm.nih.gov/geo) under accession number GSE10313.
Sequence analysis
Orthologs of human C21ORF56 (NM_032261) were identified by comparing Homo sapiens protein sequence to those existing sequences currently available in the University of California at Santa Cruz Genome Browser (http://genome.ucsc.edu). Sequences were aligned using ClustalX.
Caspase 3 activation
After treatment with MNNG, caspase-3 activity was measured using a caspase activity assay kit (Promega). Cells were resuspended in PBS and incubated for 45 min with the proluminescent caspase-3 substrate containing the DEVD sequence. Luminesence was measured with a luminometer.
MGMT activity assay
Lymphoblastoid cells in log phase growth were resuspended in MTase buffer (50 mM HEPES at pH 7.8, 10 mM DTT, 1 mM EDTA, 5% glycerol), sonicated, and lysate cleared by centrifugation. Protein concentration was determined using the Pierce Better Bradford assay. MTase activity for each cell line was determined using calf thymus DNA methylated in vitro with [3H] MNU as described (Glassner et al. 1999).
MEF colony forming assay
MEFs were cultured in DMEM media (10% fetal bovine serum, 1% penicillin, and streptomycin). Cells were seeded at 200 cells per 10 mL of culture in 100-mm dishes. Media was replaced 24 h after seeding, and cells were preincubated with O6-benzylguanine (10 mM) for 2 h. Cells were then treated with MNNG (0–3 μg/mL) and colonies counted 5 d after treatment.
shRNA knockdown cell line generation and treatment
shRNAs expressed in a lentiviral plasmid (pGIPZ) were purchased from Open Biosystems to target the C21ORF56 transcript (v1:#RHS4430-98844079 and v2:#RHS4430-98477469) or the MYH (v1:#RHS4430-98904053 and v2:#RHS4430-99140608) transcript. Knockdown cells were compared with TK6 cells expressing a nontargeting shRNA (#RHS4346) or an empty vector control (#RHS4349). Virus was generated in 293T cells using packaging plasmids (psPAX2, pMD2.G Addgene). The TK6 cell line was infected with virus and stable clones selected using Puromycin. The percentage of survival was measured using a killing curve assay 14 d after treatment with MNNG as described (Furth et al. 1981).
Acknowledgments
We are grateful to Margherita Bignami for the generous gift of the Myh−/− MEFs. This work was supported by the National Institute of Environmental Health Sciences (ES11399 and ES002109) and the National Cancer Institute (CA055042 and U54-CA112967). J.P.S. was supported by a Swedish Research Council Fellowship. C.V. received support from NIH training grant 1-R90-DK071503-03. L.D.S. is an American Cancer Society Research Professor.
Footnotes
Supplemental material is available at http://www.genesdev.org.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.1688508.
References
- Begley T.J., Rosenbach A.S., Ideker T., Samson L.D. Hot spots for modulating toxicity identified by genomic phenotyping and localization mapping. Mol. Cell. 2004;16:117–125. doi: 10.1016/j.molcel.2004.09.005. [DOI] [PubMed] [Google Scholar]
- Ceccotti S., Aquilina G., Macpherson P., Yamada M., Karran P., Bignami M. Processing of O6-methylguanine by mismatch correction in human cell extracts. Curr. Biol. 1996;6:1528–1531. doi: 10.1016/s0960-9822(96)00758-0. [DOI] [PubMed] [Google Scholar]
- Cheung V.G., Conlin L.K., Weber T.M., Arcaro M., Jen K.Y., Morley M., Spielman R.S. Natural variation in human gene expression assessed in lymphoblastoid cells. Nat. Genet. 2003;33:422–425. doi: 10.1038/ng1094. [DOI] [PubMed] [Google Scholar]
- Cheung V.G., Spielman R.S., Ewens K.G., Weber T.M., Morley M., Burdick J.T. Mapping determinants of human gene expression by regional and genome-wide association. Nature. 2005;437:1365–1369. doi: 10.1038/nature04244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins F.S., Brooks L.D., Chakravarti A. A DNA polymorphism discovery resource for research on human genetic variation. Genome Res. 1998;8:1229–1231. doi: 10.1101/gr.8.12.1229. [DOI] [PubMed] [Google Scholar]
- Correa C.R., Cheung V.G. Genetic variation in radiation-induced expression phenotypes. Am. J. Hum. Genet. 2004;75:885–890. doi: 10.1086/425221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon A.L., Liang L., Moffatt M.F., Chen W., Heath S., Wong K.C., Taylor J., Burnett E., Gut I., Farrall M., et al. A genome-wide association study of global gene expression. Nat. Genet. 2007;39:1202–1207. doi: 10.1038/ng2109. [DOI] [PubMed] [Google Scholar]
- Fry R.C., Navasumrit P., Valiathan C., Svensson J.P., Hogan B.J., Luo M., Bhattacharya S., Kandjanapa K., Soontararuks S., Nookabkaew S., et al. Activation of inflammation/NF-κB signaling in infants born to arsenic-exposed mothers. PLoS Genet. 2007;3:e207. doi: 10.1371/journal.pgen.0030207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furth E.E., Thilly W.G., Penman B.W., Liber H.L., Rand W.M. Quantitative assay for mutation in diploid human lymphoblasts using microtiter plates. Anal. Biochem. 1981;110:1–8. doi: 10.1016/0003-2697(81)90103-2. [DOI] [PubMed] [Google Scholar]
- Geladi P., Martens H., Hadjiski L., Hopke P.A. A calibration tutorial for spectral data. Part 2. Partial least squares regression using Matlab and some neural network results. J. Near Infrared Spectrosc. 1996;4:243–255. [Google Scholar]
- Glassner B.J., Weeda G., Allan J.M., Broekhof J.L., Carls N.H., Donker I., Engelward B.P., Hampson R.J., Hersmus R., Hickman M.J., et al. DNA repair methyltransferase (Mgmt) knockout mice are sensitive to the lethal effects of chemotherapeutic alkylating agents. Mutagenesis. 1999;14:339–347. doi: 10.1093/mutage/14.3.339. [DOI] [PubMed] [Google Scholar]
- Gu Y., Parker A., Wilson T.M., Bai H., Chang D.Y., Lu A.L. Human MutY homolog, a DNA glycosylase involved in base excision repair, physically and functionally interacts with mismatch repair proteins human MutS homolog 2/human MutS homolog 6. J. Biol. Chem. 2002;277:11135–11142. doi: 10.1074/jbc.M108618200. [DOI] [PubMed] [Google Scholar]
- Hegi M.E., Diserens A.C., Gorlia T., Hamou M.F., de Tribolet N., Weller M., Kros J.M., Hainfellner J.A., Mason W., Mariani L., et al. MGMT gene silencing and benefit from temozolomide in glioblastoma. N. Engl. J. Med. 2005;352:997–1003. doi: 10.1056/NEJMoa043331. [DOI] [PubMed] [Google Scholar]
- Hickman M.J., Samson L.D. Role of DNA mismatch repair and p53 in signaling induction of apoptosis by alkylating agents. Proc. Natl. Acad. Sci. 1999;96:10764–10769. doi: 10.1073/pnas.96.19.10764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hickman M.J., Samson L.D. Apoptotic signaling in response to a single type of DNA lesion, O6-methylguanine. Mol. Cell. 2004;14:105–116. doi: 10.1016/s1097-2765(04)00162-5. [DOI] [PubMed] [Google Scholar]
- Hunter D.J., Kraft P., Jacobs K.B., Cox D.G., Yeager M., Hankinson S.E., Wacholder S., Wang Z., Welch R., Hutchinson A., et al. A genome-wide association study identifies alleles in FGFR2 associated with risk of sporadic postmenopausal breast cancer. Nat. Genet. 2007;39:870–874. doi: 10.1038/ng2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaina B., Ziouta A., Ochs K., Coquerelle T. Chromosomal instability, reproductive cell death and apoptosis induced by O6-methylguanine in Mex-, Mex+ and methylation-tolerant mismatch repair compromised cells: Facts and models. Mutat. Res. 1997;381:227–241. doi: 10.1016/s0027-5107(97)00187-5. [DOI] [PubMed] [Google Scholar]
- Karran P. Mechanisms of tolerance to DNA damaging therapeutic drugs. Carcinogenesis. 2001;22:1931–1937. doi: 10.1093/carcin/22.12.1931. [DOI] [PubMed] [Google Scholar]
- Kat A., Thilly W.G., Fang W.H., Longley M.J., Li G.M., Modrich P. An alkylation-tolerant, mutator human cell line is deficient in strand-specific mismatch repair. Proc. Natl. Acad. Sci. 1993;90:6424–6428. doi: 10.1073/pnas.90.14.6424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margison G.P., Povey A.C., Kaina B., Santibanez Koref M.F. Variability and regulation of O6-alkylguanine-DNA alkyltransferase. Carcinogenesis. 2003;24:625–635. doi: 10.1093/carcin/bgg005. [DOI] [PubMed] [Google Scholar]
- Morley M., Molony C.M., Weber T.M., Devlin J.L., Ewens K.G., Spielman R.S., Cheung V.G. Genetic analysis of genome-wide variation in human gene expression. Nature. 2004;430:743–747. doi: 10.1038/nature02797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker A.R., Eshleman J.R. Human MutY: Gene structure, protein functions and interactions, and role in carcinogenesis. Cell. Mol. Life Sci. 2003;60:2064–2083. doi: 10.1007/s00018-003-3053-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pegg A.E. Mammalian O6-alkylguanine-DNA alkyltransferase: Regulation and importance in response to alkylating carcinogenic and therapeutic agents. Cancer Res. 1990;50:6119–6129. [PubMed] [Google Scholar]
- Pegg A.E. Repair of O6-alkylguanine by alkyltransferases. Mutat. Res. 2000;462:83–100. doi: 10.1016/s1383-5742(00)00017-x. [DOI] [PubMed] [Google Scholar]
- Said M.R., Begley T.J., Oppenheim A.V., Lauffenburger D.A., Samson L.D. Global network analysis of phenotypic effects: Protein networks and toxicity modulation in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. 2004;101:18006–18011. doi: 10.1073/pnas.0405996101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slupska M.M., Baikalov C., Luther W.M., Chiang J.H., Wei Y.F., Miller J.H. Cloning and sequencing a human homolog (hMYH) of the Escherichia coli mutY gene whose function is required for the repair of oxidative DNA damage. J. Bacteriol. 1996;178:3885–3892. doi: 10.1128/jb.178.13.3885-3892.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stranger B.E., Forrest M.S., Clark A.G., Minichiello M.J., Deutsch S., Lyle R., Hunt S., Kahl B., Antonarakis S.E., Tavare S., et al. Genome-wide associations of gene expression variation in humans. PLoS Genet. 2005;1:e78. doi: 10.1371/journal.pgen.0010078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stranger B.E., Nica A.C., Forrest M.S., Dimas A., Bird C.P., Beazley C., Ingle C.E., Dunning M., Flicek P., Koller D., et al. Population genomics of human gene expression. Nat. Genet. 2007;39:1217–1224. doi: 10.1038/ng2142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tantin D., Schild-Poulter C., Wang V., Hache R.J., Sharp P.A. The octamer binding transcription factor Oct-1 is a stress sensor. Cancer Res. 2005;65:10750–10758. doi: 10.1158/0008-5472.CAN-05-2399. [DOI] [PubMed] [Google Scholar]
- Vahakangas K., Trivers G.E., Plummer S., Hayes R.B., Krokan H., Rowe M., Swartz R.P., Yeager H., Jr., Harris C.C. O(6)-methylguanine-DNA methyltransferase and uracil DNA glycosylase in human broncho-alveolar lavage cells and peripheral blood mononuclear cells from tobacco smokers and non-smokers. Carcinogenesis. 1991;12:1389–1394. doi: 10.1093/carcin/12.8.1389. [DOI] [PubMed] [Google Scholar]
- Zeggini E., McCarthy M.I. Identifying susceptibility variants for type 2 diabetes. Methods Mol. Biol. 2007;376:235–250. doi: 10.1007/978-1-59745-389-9_16. [DOI] [PubMed] [Google Scholar]
- Zhao H., Jin S., Fan F., Fan W., Tong T., Zhan Q. Activation of the transcription factor Oct-1 in response to DNA damage. Cancer Res. 2000;60:6276–6280. [PubMed] [Google Scholar]