Abstract
HBO1 histone acetylase is important for DNA replication licensing. In human cells, HBO1 associates with replication origins specifically during the G1 phase of the cell cycle in a manner that depends on the replication licensing factor Cdt1, but is independent of the Cdt1 repressor Geminin. HBO1 directly interacts with Cdt1, and it enhances Cdt1-dependent rereplication. Thus, HBO1 plays a direct role at replication origins as a coactivator of the Cdt1 licensing factor. As HBO1 is also a transcriptional coactivator, it has the potential to integrate internal and external stimuli to coordinate transcriptional responses with initiation of DNA replication.
Keywords: HBO1, DNA replication, Cdt1, origin licensing, histone acetylation, Geminin
The initial step of DNA replication involves the formation of the prereplication complex (pre-RC) on origins of replication distributed throughout the genome. Sequential assembly of two multiprotein complexes, ORC (origin recognition complex) and MCM (minichromosome maintenance), result in a pre-RC that is “licensed” for replication that will occur in the subsequent S phase (Bell and Dutta 2002). ORC associates with replication origins throughout the entire cell cycle, whereas the MCM complex is specifically loaded during late mitosis through G1 phase under the control of ORC and ORC-associated licensing factors Cdt1 and Cdc6 (Thommes and Blow 1997; Bell and Dutta 2002). As cells enter S phase, origins are activated to initiate DNA replication, whereupon the pre-RC disassembles. Relicensing of replication origins does not occur in S phase, thereby restricting DNA synthesis to once per cell cycle.
Regulation of Cdt1 plays a critical role in the licensing process. Inhibition of Cdt1 activity occurs by multiple mechanisms, including ubiquitin-dependent proteolysis and binding to its potent inhibitor Geminin (Wohlschlegel et al. 2000; Tada et al. 2001; Arias and Walter 2007). Misregulation of the licensing process and defects in the ordered assembly and disassembly of the pre-RC alter the integrity of the genome (Vaziri et al. 2003; Saxena and Dutta 2005; Niida and Nakanishi 2006; Tatsumi et al. 2006; Hook et al. 2007). Thus, cell cycle regulation of Cdt1 expression and function is responsible for licensing replication origins in G1 and preventing relicensing and hence rereplication in S phase.
As expected for any molecular process involving DNA in eukaryotic cells, DNA replication initiation is influenced by chromatin structure. In particular, histone acetylation is linked to pre-RC assembly and the control of initiation of DNA replication. In yeast and mammalian cells, early-firing origins are typically localized in genomic regions that are transcribed and contain hyperacetylated chromatin, whereas late-firing origins lie in silenced heterochromatic domains (Kemp et al. 2005; Zhou et al. 2005; Karnani et al. 2007; Lucas et al. 2007; Goren et al. 2008). In addition, histone acetylation is involved in origin activation during early development in Xenopus (Danis et al. 2004) and at the chorion gene loci in Drosophila follicle cells (Aggarwal and Calvi 2004; Hartl et al. 2007). However, it is unknown whether and how histone acetylases or deacetylases activities are targeted at origins to promote pre-RC assembly and/or origin activity.
HBO1 (human acetylase binding to ORC1) is a MYST family histone acetylase that interacts both with transcriptional activator proteins (Georgiakaki et al. 2006; Miotto and Struhl 2006; Miotto et al. 2006) and with MCM2 and ORC1 (Iizuka and Stillman 1999; Burke et al. 2001; Doyon et al. 2006). HBO1 depletion reduces the rate of DNA synthesis, the amount of MCM complex bound to chromatin, and progression through S phase (Doyon et al. 2006; Iizuka et al. 2006). In addition, the Drosophila HBO1 homolog (Chameau) increases origin activity when artificially recruited to a synthetic replication origin (Aggarwal and Calvi 2004). However, it is unknown whether HBO1 associates with origins in vivo, and the observation that HBO1 copurifies with MCM subunits in the absence of ORC suggests that HBO1 may control DNA replication during S phase rather than in G1 (Doyon et al. 2006).
Here, we show that HBO1 is targeted to origins through a direct interaction with the licensing regulator Cdt1, and that HBO1 enhances Cdt1-dependent rereplication. As HBO1 is not required for Cdt1 association with replication origins, these observations indicate that HBO1 functions as a coactivator of Cdt1 during the licensing process.
Results and Discussion
HBO1 associates with replication origins in mammalian cells
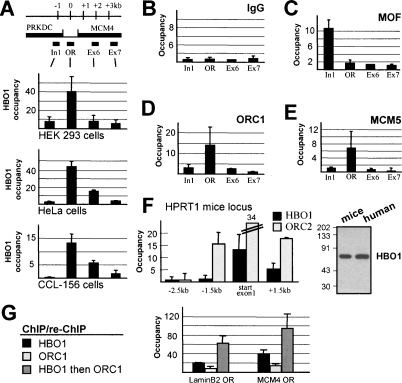
Although HBO1 interacts with the ORC and MCM complexes and affects DNA replication (Supplemental Fig. S1), it is still unknown whether HBO1 directly associates with replication origins in vivo. As assayed by chromatin immunoprecipitation (ChIP), HBO1, but not the related MYST acetylase hMOF, associates with origins in several human cell lines (Fig. 1A–E; Supplemental Fig. S2). In addition, HBO1 associates with the origin located within the HPRT1 coding sequence (Cohen et al. 2004) in mouse lymphocytes (Fig. 1F). Sequential ChIP assays demonstrate that HBO1 and ORC1 coexist on origins in human cells (Fig. 1G). As ORC1 (unlike other ORC subunits) is degraded after initiation of DNA replication (Mendez et al. 2002), this observation suggests that HBO1 is bound at the time of DNA licensing.
Figure 1.
HBO1 associates with mammalian origins of replication. (A) Binding profile of HBO1 at the MCM4 origin (OR) locus in HEK293 kidney, HeLa adenocarcinoma, and CCL-156 lymphoblastoid cells. Control IgG (B), MOF (C), ORC1 (D), and MCM5 (E) binding profile at the MCM4 origin in the same chromatin sample. (F) ChIP analysis of HBO1 and ORC2 binding profile at the mice HPRT1 origin. (Insert) Western blot analysis of HBO1 antibody specificity against human and mice whole cellular extracts. (G) Re-ChIP analysis of HBO1 and ORC1 co-occupancy on MCM4 and LaminB2 origins in HeLa cells.
HBO1 can be copurified with ING4 and ING5 proteins, and it has been suggested that such complexes regulate replication (Doyon et al. 2006). However, despite successful recovery of ING4 and ING5 in immunoprecipitates from HeLa and HEK 293 cells using different commercially available antibodies, we were unable to observe selective association with replication origins (data not shown). Although not conclusive, this observation suggests that HBO1 may associate with origins independently of ING proteins, and that ING/HBO1 complexes may regulate later stages of replication (Doyon et al. 2006).
HBO1 association with origins is restricted to G1
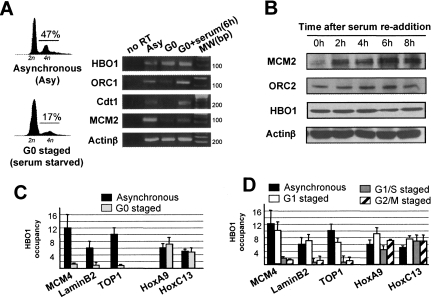
Expression and/or chromatin association of several replication factors is maximal in G1, when DNA licensing occurs, and reduced upon cell cycle exit or progression in S phase (Bell and Dutta 2002). Expression of HBO1 mRNA and protein are not differentially regulated in response to serum deprivation and cell cycle exit, nor are they affected when cells re-enter the proliferative cycle following serum readdition (Fig. 2A,B). The persistence of HBO1 expression in nonproliferating cells is consistent with the important biological roles of HBO1 that are unrelated to DNA replication (Doyon et al. 2006; Georgiakaki et al. 2006; Miotto and Struhl 2006). Further, as previously reported (Iizuka et al. 2006), HBO1 mRNA expression and protein stability are also not cell cycle-regulated (Supplemental Fig. S3).
Figure 2.
HBO1 association with origins is restricted to G1. (A) Expression level of HBO1, ORC1, Cdt1, and MCM2 mRNAs in CCL-156 cells in culture (asynchronously), following serum deprivation and cell cycle exit (G0), and following readdition of 20% serum in the media for 6 h after serum deprivation (G0 + serum). The left panel illustrates the cell cycle profile of samples used in the experiment. “no RT” referrs to the control condition where cDNA synthesis was omitted prior to PCR analysis. (B) Expression level of HBO1, MCM2, and ORC2 proteins upon readdition of serum for indicated number of hours. (C) ChIP analysis of HBO1 binding at origins and control sites in growing and serum-starved cells. (D) ChIP analysis of HBO1 binding at origins in cells staged at different phase of the cycle.
Although HBO1 protein levels are unregulated by growth conditions or cell cycle status, HBO1 does not associate with replication origins in nonproliferating cells (Fig. 2C). Furthermore, HBO1 strongly associates with origins in cells staged in G1 by mimosine treatment, but not in cells staged in early S or in M phase by hydroxyurea (HU) and nocodazole treatments, respectively (Fig. 2D). In contrast, levels of HBO1 bound to bulk chromatin (Supplemental Fig. S3) and HBO1 association with target sites unrelated to DNA replication (HoxA9 and HoxC13) are unaffected by cell growth and not restricted to G1 (Fig. 2D), indicating that cell cycle regulation of HBO1 association is specific to origins. Therefore, HBO1 is cyclically assembled onto origins in G1 and disassembled following origin firing, strongly suggesting that HBO1 function at origins is restricted to G1. As ORC, but not the MCM complex, is loaded onto chromatin in the absence of HBO1 (Iizuka et al. 2006), HBO1 may mediate and/or stabilize the interaction between ORC and MCM complexes at the origin.
Licensing factor Cdt1 stabilizes HBO1 at origins
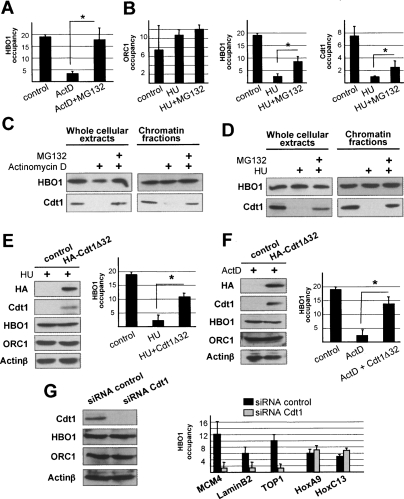
The results above strongly suggest that HBO1 association with replication origins is mediated by a G1-specific factor(s). Aside from the ORC and MCM complexes, Cdc6 and Cdt1 are essential for DNA licensing in G1 (Bell and Dutta 2002). In addition, HBO1 dissociates from origins shortly (2 h) after cells are treated with the DNA damaging agent actinomycin D (Fig. 3A; data not shown), while the overall association of HBO1 with chromatin is unaffected (Fig. 3C). Therefore, HBO1 dissociation from origins is either triggered by proteolytic degradation of a key licensing cofactor or by post-translational event(s) induced by HU and/or actinomycin D treatment(s).
Figure 3.
Licensing factor Cdt1 stabilizes HBO1 at origins. (A) Impact of proteasome inhibition on HBO1 association with the laminB2 origin in cells previously treated with DNA damaging agent actinomycin D. (B) Impact of proteasome inhibition on ORC1, HBO1, and Cdt1 association with the LaminB2 origin in cells previously G1/S staged with HU. (C) Impact of proteasome inhibition on Cdt1 and HBO1 expression level and chromatin association in HeLa cells treated with ActD. (D) Impact of proteasome inhibition on Cdt1 and HBO1 expression level and chromatin association in HeLa cells treated with HU. (E) Impact of Cdt1Δ32 overexpression on HBO1 association with LaminB2 origin in HeLa cells treated with HU. (F) Impact of Cdt1Δ32 overexpression on HBO1 association with LaminB2 origin in HeLa cells treated with ActD. (G) Impact of Cdt1 depletion on HBO1 association with replication origins in HeLa cells.
To address these issues, we analyzed HBO1 binding at origins under conditions where the proteasome machinery is inactivated (by MG132 treatment) prior to actinomycin D treatment or following HU treatment. Treatment with MG132 partially restores HBO1 binding on origins in both cases (Fig. 3A,B), strongly suggesting that HBO1 association with origins involves a factor that is destroyed by the proteasome. Treatment with HU or actinomycin D did not affect the level of MCM2–7 and ORC1 bound to chromatin (Supplemental Fig. S3). In addition, treatment with actinomycin D did not affect the level of chromatin-bound Cdc6, and a significant proportion of the protein is still bound during S phase. Thus, association of HBO1 with origins in the presence of MG132 is not correlated with increased association of ORC1, MCM2–7, or Cdc6.
In contrast, Cdt1, the key regulator of DNA licensing in metazoans, is proteolytically degraded at the G1/S transition (i.e., HU staged) and in response to DNA damage (Supplemental Fig. S3; Arias and Walter 2007). When proteasome activity is inhibited, Cdt1 degradation is blocked and HBO1 associates with origins (Fig. 3B–D). Furthermore, overexpression of Cdt1Δ32, a constitutive nondegradable Cdt1 protein that is otherwise fully functional (Takeda et al. 2005), rescues HBO1 binding at origins following HU and actinomycin D treatment (Fig. 3E,F). Thus, selective and rapid (2 h) degradation of Cdt1 upon actinomycin D treatment impairs HBO1 loading, and this effect is prevented by overexpression of Cdt1Δ32. Lastly, selective depletion of Cdt1 with siRNA duplexes impairs HBO1 binding at origins (Fig. 3G). Therefore, HBO1 loading correlates with Cdt1 presence at origins, and Cdt1 recruits HBO1 to replication origins.
In addition to being regulated by proteolysis, Cdt1 licensing activity is restricted in G1 by association with a potent repressor, Geminin (Wohlschlegel et al. 2000; Tada et al. 2001). Overexpression of GemininL26A, a variant of Geminin that can accumulate in G1, prevents entry in S phase (Supplemental Fig. S4; Wohlschlegel et al. 2002). In cells overexpressing GemininL26A, and hence efficiently blocked in G0/G1, HBO1 still associates with origins, whereas MCM loading is impaired (Supplemental Fig. S4).
These observations strongly suggest that HBO1 association with origins requires Cdt1, and occurs prior to loading of the MCM complex. HBO1 loading does not appear to depend on the function of Cdt1 that is inhibited by Geminin, but rather on Cdt1 association. Lastly, as HBO1 and Cdt1 are both required for loading of the MCM complex onto origins during G1, our results suggest that HBO1 is directly involved in pre-RC formation as a recruited cofactor of Cdt1.
Cdt1 directly interacts with HBO1 in a manner that is independent of Geminin
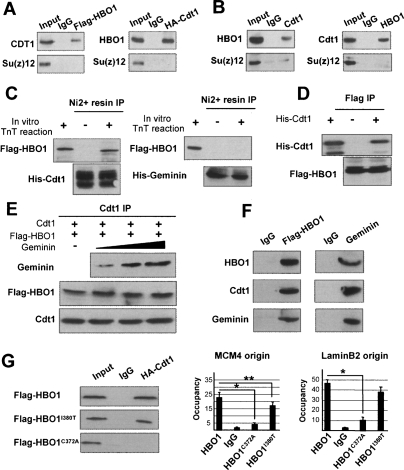
To address whether HBO1 directly interacts with Cdt1, we performed reciprocal coimmunoprecipitation assays in cell-free extracts and GST pulldown experiments in vitro. As shown in Figure 4A, endogenous HBO1 coprecipitates with HA-tagged Cdt1, and endogenous Cdt1 co precipitates with Flag-tagged HBO1. Other factors [Su(z)12, Sp1, HoxA10] were not coimmunoprecipitated, demonstrating the specificity of the interaction (Fig. 4A; data not shown). Furthermore, significant amounts of HBO1 are recovered in endogenous Cdt1 immunoprecipitates, and immunoprecipitation of HBO1 also pulls down Cdt1 (Fig. 4B). Addition of ethidium bromide to denature DNA did not affect the coimmunoprecipitation (data not shown), indicating that the interaction is mediated through protein/protein interaction(s). The interaction between HBO1 and Cdt1 is direct, because HBO1 translated in vitro interacts with a His-Cdt1(full-length) fusion, but not with His-Geminin (Fig. 4C). In a reverse experiment, in vitro translated HBO1 purified and immobilized on beads interacts with recombinant His-Cdt1 (Fig. 4D).
Figure 4.
Cdt1 directly interacts with HBO1. (A) Detection of HBO1 association with Cdt1 by reverse coimmunoprecipitation in HeLa cells overexpressing either HA-Cdt1 or Flag-HBO1. (B) Same experiment using antibodies directed against endogenous HBO1 and Cdt1. (C) Interaction of in vitro translated Flag-HBO1 with full-length His-Cdt1 and His-Geminin. (D) Interaction of purified in vitro translated Flag-HBO1 with His-Cdt1. (E) In vitro preformed Cdt1/HBO1 complex were challenged by increasing amount of bacterially produced Geminin. (F) Mix of Cdt1, Geminin, and HBO1 proteins were incubated with the indicated antibodies, and the purified complexes was determined by Western blot analysis. (G) Role of the MYST zinc finger. (Left panel) The indicated HBO1 derivatives were analyzed for interaction with Cdt1 or occupancy at the MCM4 and LaminB2 origins. (*) P < 0.05 ; (**) P < 0.15.
The interaction of Cdt1 with HBO1 is not affected by Geminin (Fig. 4E). In addition, HBO1 and Geminin coimmunoprecipitate in the presence of Cdt1 (Fig. 4F), indicating that HBO1, Cdt1, and Geminin can form a tripartite complex in vitro. Consistent with these observations, forcing Geminin expression in G1 does not abrogate HBO1 interaction with origins (Supplemental Fig. S4). Therefore, Cdt1 directly binds to HBO1 and its repressor Geminin does not regulate this interaction.
In addition, the Drosophila counterpart of HBO1, Chameau, interacts with human Cdt1 when overexpressed in human cells, and this interaction is observed with its MYST acetylase domain (74.4% identical; 92% similar to HBO1) in GST pulldown experiments (data not shown). The C372A derivative of HBO1, which disrupts the zinc finger in the MYST domain, abolishes Cdt1 interaction and impairs HBO1 loading onto origins (Fig. 4G). In contrast, the I380T derivative of HBO1, which selectively abolishes MCM2 interaction but does not disrupt the overall zinc finger structure (Burke et al. 2001), does not affect Cdt1 interaction and has only a modest effect on HBO1 loading at select origins (Fig. 4G). Thus, the Cdt1-dependent recruitment of HBO1 to origins in vivo is likely to occur via a direct interaction between Cdt1 and HBO1 that is mediated by the MYST domain.
HBO1 enhances Cdt1-dependent rereplication
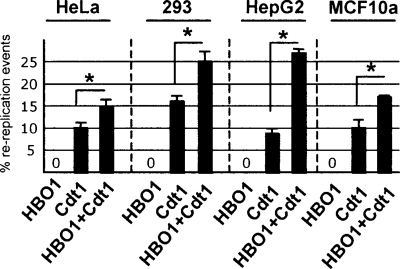
Overexpression of Cdt1 causes cells to accumulate in G2 and in some cases >4N ploidy, indicative of rereplication (Vaziri et al. 2003; Saxena and Dutta 2005; Tatsumi et al. 2006). To demonstrate further the importance of the interaction between Cdt1 and HBO1, we examined whether HBO1 influences Cdt1-dependent rereplication events. In comparison with cells overexpressing Cdt1, cells overexpressing both Cdt1 and HBO1 further show a significantly increased proportion of cells with >4N DNA in all four cell lines tested (Fig. 5). However, overexpression of HBO1 alone does not result in rereplication, indicating that HBO1 is insufficient to license origins, but rather acts as an enhancer of Cdt1 licensing activity. As HBO1 does not affect Cdt1 stability and loading onto origins (Iizuka et al. 2006), even under condition of overexpression (Supplemental Fig. S3), HBO1 regulates licensing activity after it is recruited to origins via its interaction with Cdt1.
Figure 5.
HBO1 enhances Cdt1 licensing activity. Rereplication events in the indicated cells lines overexpressing HBO1, Cdt1, or Cdt1 + HBO1 as defined by the percentage of cells with >4N content as determined by FACS analysis.
HBO1 acts as a coactivator of the licensing factor Cdt1
HBO1 associates with the ORC and MCM complexes in vitro, and it is important for replication licensing, the process that ensures that DNA replication initiates only once per cell cycle. Here, we provide strong evidence that HBO1 functions directly at replication origins during pre-RC formation as a coactivator of the licensing factor Cdt1.
HBO1 associates with replication origins in vivo (Fig. 1), and this association is restricted to G1 (Fig. 2). HBO1 loading on origins occurs after association of ORC and Cdt1, but prior to the assembly of the MCM complex. HBO1 association is affected by conditions that specifically deplete or alter regulation of Cdt1 (Fig. 3), suggesting that Cdt1 is required for HBO1 recruitment. Inhibition of Cdt1 proteolysis (MG132 treatment or the Cdt1Δ32 derivative) permits HBO1 to associate with origins outside of G1 (Fig. 3), and overexpression of GemininL26A does not affect HBO1 association, whereas it inhibits MCM recruitment (Supplemental Fig. S4). Cdt1 directly interacts with HBO1 in vitro and in vivo, and Geminin does not influence this interaction (Fig. 4). This strongly suggests that HBO1 recruitment to origins depends on Cdt1 itself and not on a Geminin-inhibited function of Cdt1.
Both Cdt1 and HBO1 are important for loading of the MCM complex and subsequent DNA replication. Overexpression of HBO1 does not affect MCM complex loading or cell cycle distribution, nor does it cause aberrant rereplication, but it enhances rereplication mediated by Cdt1 in all four cell lines tested (Fig. 5). Given that Cdt1 association occurs prior to, but is required for, HBO1 recruitment, this observation suggests that HBO1 is important for the licensing activity of Cdt1. By analogy with transcriptional coactivators that are recruited to enhancers by DNA-binding proteins, our combined results strongly support a model in which HBO1 is a recruited coactivator of Cdt1.
HBO1, a coactivator for transcriptional regulation and a recently evolved molecule in the licensing process
The proteins comprising the pre-RC (ORC and MCM complexes, Cdt1, Cdc6) are conserved from yeast to human, and regulated proteolysis of Cdt1 is critical for licensing replication origins such that they fire only once per cell cycle. Metazoans have additional mechanisms to control pre-RC formation such as inhibition of Cdt1 licensing activity by Geminin (Wohlschlegel et al. 2000; Tada et al. 2001). Intriguingly, HBO1 does not appear to exist in yeasts or worms (Sanjuan and Marin 2001; Grienenberger et al. 2002), suggesting that HBO1 is a recently evolved molecule in the licensing process and indicating species-specific differences in the mechanism of MCM complex loading. In yeast, a direct interaction with Cdt1 is critical for loading the MCM complex, whereas in human cells the Cdt1-recruited HBO1 is critical. It is unknown to what extent direct interactions between Cdt1 and the MCM complex are important for recruitment of the MCM complex to origins in human cells. However, the fact that Geminin inhibits Cdt1-dependent licensing, but not HBO1 association, suggests a role for a direct Cdt1–MCM complex interaction.
As might be expected for a recently evolved function, HBO1 appears to affect different steps in DNA replication in a species-specific manner. As shown here for human cells, HBO1 affects Cdt1 licensing activity but not its chromatin loading. However, in Drosophila, targeting of Chameau (the HBO1 homolog) in the vicinity of an artificial chorion origin promotes ORC redistribution on origins and replication initiation (Aggarwal and Calvi 2004), whereas depletion of HBO1 in Xenopus extracts affects Cdt1 association with chromatin but not ORC recruitment (Iizuka et al. 2006). We note, however, that these apparent species-specific differences in the molecular function of HBO1 may reflect the assays used.
HBO1 also functions as a transcriptional coactivator for hormone receptors and AP-1 proteins (Georgiakaki et al. 2006; Miotto and Struhl 2006; Miotto et al. 2006), and we speculate that it may have evolved from this role to become a coactivator for the Cdt1 licensing factor. In addition, the dual role of HBO1 as a coactivator for transcriptional regulation and DNA licensing suggest the possibility that HBO1 might integrate internal and external stimuli to coordinate transcriptional responses with initiation of DNA replication. In particular, DNA damage activated p53 transcription factor regulates HBO1 enzymatic activity and may regulate HBO1 function during DNA licensing (Iizuka et al. 2008).
Materials and methods
The sources of DNAs, antibodies, and other materials are described in the Supplemental Material. Mammalian cell lines were grown as described in the Supplemental Material. To measure growth-dependent regulation of Cdt1, ORC2, MCM2, and HBO1 mRNAs and proteins expression, CCL-156 cells were arrested in the G0 phase by culture in the presence of 0.1% fetal bovine serum for 48 h (i.e., serum deprivation). For cell cycle synchronization in G1, early S phase and M phase, exponentially growing HeLa or CCL-156 cells were treated, respectively, with mimosine (300 μM for 20 h), HU (2 mM for 24 h), or nocodazole (40 ng/mL for 16 h). In all cases, propidium iodide staining and flow cytometry (FACSCalibur system; Becton Dickinson) assays were used to determine the quality of the synchrony. DNA damage was generated by addition of actinomycin D (2 nM for 2 h) in the presence of serum (10%).
Coimmunoprecipitation and in vitro pulldown assays, analysis of RNA levels by RT–PCR, chromatin fractionation, immunofluorescence, and flow cytometry were performed by standard procedures that are detailed in the Supplemental Material. ChIP and sequential ChIP analyses were performed as previously described (Cawley et al. 2004; Geisberg and Struhl 2004). Enrichment for a specific genomic region was defined with respect to a control genomic region (histone H3 exon 2) and expressed as occupancy value (occupancy).
Acknowledgments
We are very grateful to Johannes Walter for insightful comments during the course of this work and on the manuscript. We thank Nathan Lamarre-Vincent, Hideo Nishitani, Stavros Taraviras, Anindya Dutta, Hiroyoshi Ariga, and Marcel Huber for providing reagents, and Benedicte Py for help with flow cytometry procedures and analysis. This work was supported by grants to K.S. from the National Institutes of Health (GM30186).
Footnotes
Supplemental material is available at http://www.genesdev.org.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1674108.
References
- Aggarwal B.D., Calvi B.R. Chromatin regulates origin activity in Drosophila follicle cells. Nature. 2004;430:372–376. doi: 10.1038/nature02694. [DOI] [PubMed] [Google Scholar]
- Arias E.E., Walter J.C. Strength in numbers: Preventing rereplication via multiple mechanisms in eukaryotic cells. Genes & Dev. 2007;21:497–518. doi: 10.1101/gad.1508907. [DOI] [PubMed] [Google Scholar]
- Bell S.P., Dutta A. DNA replication in eukaryotic cells. Annu. Rev. Biochem. 2002;71:333–374. doi: 10.1146/annurev.biochem.71.110601.135425. [DOI] [PubMed] [Google Scholar]
- Burke T.W., Cook J.G., Asano M., Nevins J.R. Replication factors MCM2 and ORC1 interact with the histone acetyltransferase HBO1. J. Biol. Chem. 2001;276:15397–15408. doi: 10.1074/jbc.M011556200. [DOI] [PubMed] [Google Scholar]
- Cawley S., Bekiranov S., Ng H.H., Kapranov P., Sekinger E.A., Kampa D., Piccolboni A., Sementchenko V., Cheng J., Williams A.J., et al. Unbiased mapping of transcription factor binding sites along human chromosomes 21 and 22 points to widespread regulation of noncoding RNAs. Cell. 2004;116:499–509. doi: 10.1016/s0092-8674(04)00127-8. [DOI] [PubMed] [Google Scholar]
- Cohen S.M., Hatada S., Brylawski B.P., Smithies O., Kaufman D.G., Cordeiro-Stone M. Complementation of replication origin function in mouse embryonic stem cells by human DNA sequences. Genomics. 2004;84:475–484. doi: 10.1016/j.ygeno.2004.04.008. [DOI] [PubMed] [Google Scholar]
- Danis E., Brodolin K., Menut S., Maiorano D., Girard-Reydet C., Mechali M. Specification of a DNA replication origin by a transcription complex. Nat. Cell Biol. 2004;6:721–730. doi: 10.1038/ncb1149. [DOI] [PubMed] [Google Scholar]
- Doyon Y., Cayrou C., Ullah M., Landry A.J., Cote V., Selleck W., Lane W.S., Tan S., Yang X.J., Cote J. ING tumor suppressor proteins are critical regulators of chromatin acetylation required for genome expression and perpetuation. Mol. Cell. 2006;21:51–64. doi: 10.1016/j.molcel.2005.12.007. [DOI] [PubMed] [Google Scholar]
- Geisberg J.V., Struhl K. Quantitative sequential chromatin immunoprecipitation, a method for analyzing co-occupancy of proteins at genomic regions in vivo. Nucleic Acids Res. 2004;32:e151. doi: 10.1093/nar/gnh148.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgiakaki M., Chabbert-Buffet N., Dasen B., Meduri G., Wenk S., Rajhi L., Amazit L., Chauchereau A., Burger C.W., Blok L.J., et al. Ligand-controlled interaction of histone acetyltransferase binding to ORC-1 (HBO1) with the N-terminal transactivating domain of progesterone receptor induces steroid receptor coactivator 1-dependent coactivation of transcription. Mol. Endocrinol. 2006;20:2122–2140. doi: 10.1210/me.2005-0149. [DOI] [PubMed] [Google Scholar]
- Goren A., Tabib A., Hecht M., Cedar H. DNA replication timing of the human β-globin domain is controlled by histone modification at the origin. Genes & Dev. 2008;22:1319–1324. doi: 10.1101/gad.468308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grienenberger A., Miotto B., Sagnier T., Cavalli G., Schramke V., Geli V., Mariol M.C., Berenger H., Graba Y., Pradel J. The MYST domain acetyltransferase Chameau functions in epigenetic mechanisms of transcriptional repression. Curr. Biol. 2002;12:762–766. doi: 10.1016/s0960-9822(02)00814-x. [DOI] [PubMed] [Google Scholar]
- Hartl T., Boswell C., Orr-Weaver T.L., Bosco G. Developmentally regulated histone modifications in Drosophila follicle cells: Initiation of gene amplification is associated with histone H3 and H4 hyperacetylation and H1 phosphorylation. Chromosoma. 2007;116:197–214. doi: 10.1007/s00412-006-0092-2. [DOI] [PubMed] [Google Scholar]
- Hook S.S., Lin J.J., Dutta A. Mechanisms to control rereplication and implications for cancer. Curr. Opin. Cell Biol. 2007;19:663–671. doi: 10.1016/j.ceb.2007.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iizuka M., Stillman B. Histone acetyltransferase HBO1 interacts with the ORC1 subunit of the human initiator protein. J. Biol. Chem. 1999;274:23027–23034. doi: 10.1074/jbc.274.33.23027. [DOI] [PubMed] [Google Scholar]
- Iizuka M., Matsui T., Takisawa H., Smith M.M. Regulation of replication licensing by acetyltransferase Hbo1. Mol. Cell. Biol. 2006;26:1098–1108. doi: 10.1128/MCB.26.3.1098-1108.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iizuka M., Sarmento O.F., Sekiya T., Scrable H., Allis C.D., Smith M.M. Hbo1 Links p53-dependent stress signaling to DNA replication licensing. Mol. Cell. Biol. 2008;28:140–153. doi: 10.1128/MCB.00662-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karnani N., Taylor C., Malhotra A., Dutta A. Pan-S replication patterns and chromosomal domains defined by genome-tiling arrays of ENCODE genomic areas. Genome Res. 2007;17:865–876. doi: 10.1101/gr.5427007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemp M.G., Ghosh M., Liu G., Leffak M. The histone deacetylase inhibitor trichostatin A alters the pattern of DNA replication origin activity in human cells. Nucleic Acids Res. 2005;33:325–336. doi: 10.1093/nar/gki177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucas I., Palakodeti A., Jiang Y., Young D.J., Jiang N., Fernald A.A., Le Beau M.M. High-throughput mapping of origins of replication in human cells. EMBO Rep. 2007;8:770–777. doi: 10.1038/sj.embor.7401026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendez J., Zou-Yang X.H., Kim S.Y., Hidaka M., Tansey W.P., Stillman B. Human origin recognition complex large subunit is degraded by ubiquitin-mediated proteolysis after initiation of DNA replication. Mol. Cell. 2002;9:481–491. doi: 10.1016/s1097-2765(02)00467-7. [DOI] [PubMed] [Google Scholar]
- Miotto B., Struhl K. Differential gene regulation by selective association of transcriptional coactivators and bZIP DNA-binding domains. Mol. Cell. Biol. 2006;26:5969–5982. doi: 10.1128/MCB.00696-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miotto B., Sagnier T., Berenger H., Bohmann D., Pradel J., Graba Y. Chameau HAT and DRpd3 HDAC function as antagonistic cofactors of JNK/AP-1-dependent transcription during Drosophila metamorphosis. Genes & Dev. 2006;20:101–112. doi: 10.1101/gad.359506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niida H., Nakanishi M. DNA damage checkpoints in mammals. Mutagenesis. 2006;21:3–9. doi: 10.1093/mutage/gei063. [DOI] [PubMed] [Google Scholar]
- Sanjuan R., Marin I. Tracing the origin of the compensasome: Evolutionary history of DEAH helicase and MYST acetyltransferase gene families. Mol. Biol. Evol. 2001;18:330–343. doi: 10.1093/oxfordjournals.molbev.a003809. [DOI] [PubMed] [Google Scholar]
- Saxena S., Dutta A. Geminin-Cdt1 balance is critical for genetic stability. Mutat. Res. 2005;569:111–121. doi: 10.1016/j.mrfmmm.2004.05.026. [DOI] [PubMed] [Google Scholar]
- Tada S., Li A., Maiorano D., Mechali M., Blow J.J. Repression of origin assembly in metaphase depends on inhibition of RLF-B/Cdt1 by geminin. Nat. Cell Biol. 2001;3:107–113. doi: 10.1038/35055000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda D.Y., Parvin J.D., Dutta A. Degradation of Cdt1 during S phase is Skp2-independent and is required for efficient progression of mammalian cells through S phase. J. Biol. Chem. 2005;280:23416–23423. doi: 10.1074/jbc.M501208200. [DOI] [PubMed] [Google Scholar]
- Tatsumi Y., Sugimoto N., Yugawa T., Narisawa-Saito M., Kiyono T., Fujita M. Deregulation of Cdt1 induces chromosomal damage without rereplication and leads to chromosomal instability. J. Cell Sci. 2006;119:3128–3140. doi: 10.1242/jcs.03031. [DOI] [PubMed] [Google Scholar]
- Thommes P., Blow J.J. The DNA replication licensing system. Cancer Surv. 1997;29:75–90. [PubMed] [Google Scholar]
- Vaziri C., Saxena S., Jeon Y., Lee C., Murata K., Machida Y., Wagle N., Hwang D.S., Dutta A. A p53-dependent checkpoint pathway prevents rereplication. Mol. Cell. 2003;11:997–1008. doi: 10.1016/s1097-2765(03)00099-6. [DOI] [PubMed] [Google Scholar]
- Wohlschlegel J.A., Dwyer B.T., Dhar S.K., Cvetic C., Walter J.C., Dutta A. Inhibition of eukaryotic DNA replication by geminin binding to Cdt1. Science. 2000;290:2309–2312. doi: 10.1126/science.290.5500.2309. [DOI] [PubMed] [Google Scholar]
- Wohlschlegel J.A., Kutok J.L., Weng A.P., Dutta A. Expression of geminin as a marker of cell proliferation in normal tissues and malignancies. Am. J. Pathol. 2002;161:267–273. doi: 10.1016/S0002-9440(10)64178-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J., Chau C., Deng Z., Stedman W., Lieberman P.M. Epigenetic control of replication origins. Cell Cycle. 2005;4:889–892. doi: 10.4161/cc.4.7.1823. [DOI] [PubMed] [Google Scholar]