Abstract
In budding yeast, exit from the pachytene stage of meiosis requires the mid-meiosis transcription factor Ndt80, which promotes expression of ∼200 genes. Ndt80 is required for meiotic function of polo-like kinase (PLK, Cdc5) and cyclin-dependent kinase (CDK), two cell cycle kinases previously implicated in pachytene exit. We show that ongoing CDK activity is dispensable for two events that accompany exit from pachytene: crossover formation and synaptonemal complex breakdown. In contrast, CDC5 expression in ndt80Δ mutants efficiently promotes both events. Thus, Cdc5 is the only member of the Ndt80 transcriptome required for this critical step in meiotic progression.
Keywords: Saccharomyces cerevisiae, synaptonemal complex, joint molecules, crossover, cyclin-dependent kinase, Holliday junction
During meiosis, a single round of DNA replication is followed by two successive divisions (meiosis I and meiosis II), producing four haploid gametes. This is achieved by segregation of homologs, homologous chromosomes of different parental origin, during meiosis I, followed by segregation of sister chromatids in meiosis II. During the prophase of meiosis I, progressive chromosome condensation is accompanied by homolog pairing and recombination, which culminate at the pachytene stage, where homologs are paired along their entire lengths and connected by the synaptonemal complex (SC). Upon exit from pachytene, the SC disappears and chromosomes become diffuse, only to recondense at diplotene/diakinesis as visible homologs connected by crossovers.
Meiotic recombination initiates in early meiosis I prophase by the formation of DNA double-strand breaks (DSBs), catalyzed by the Spo11 endonuclease (Keeney and Neale 2006). As cells progress toward pachytene, DSBs are repaired to form either noncrossover (NCO) recombinants or joint molecule (JM) intermediates, many of which contain parental homologs connected by double Holliday junctions (Bishop and Zickler 2004). JMs are resolved, primarily as crossover recombinants (COs), at the end of pachytene. Crossovers provide the interhomolog connections that later ensure proper homolog alignment on the metaphase I spindle and thus their accurate segregation at anaphase I.
The exit from pachytene is a crucial transition, since it is associated with the resolution of JMs as COs, SC disassembly, and kinetochore modification so that sister kinetochores orient toward the same spindle pole (mono-orientation), and the separation of duplicated spindle pole bodies (SPBs), the fungal centrosome equivalents (Shuster and Byers 1989; Xu et al. 1995, 1997; Clyne et al. 2003; Lee and Amon 2003b). Failure in any of these processes can result in chromosome nondisjunction and the production of aneuploid progeny. Consequently, checkpoint systems monitor synapsis and recombination and prevent exit from pachytene unless these processes are complete (Roeder and Bailis 2000).
Ndt80, a transcription factor first expressed in late pachytene, is a central target of these checkpoint systems (Xu et al. 1995; Tung et al. 2000). Ndt80 activates expression of more than 200 genes at mid-meiosis, including those required for meiotic divisions and spore formation (Chu and Herskowitz 1998; Chu et al. 1998). Cells lacking Ndt80 fail to exit pachytene, arresting with unresolved JMs, reduced COs, persistent SC, and duplicated but unseparated SPBs (Xu et al. 1995; Allers and Lichten 2001). Ndt80 is extensively phosphorylated during normal meiosis, but in cells that are arrested in pachytene due to defects in synapsis and/or recombination, Ndt80 is less abundant and is hypophosphorylated, and Ndt80-dependent genes are not expressed (Tung et al. 2000; Pak and Segall 2002; Benjamin et al. 2003). Among the targets of Ndt80 regulation are two critical cell cycle kinases: polo-like kinase (PLK) encoded by CDC5, and cyclin-dependent kinase (CDK). Previous studies have suggested roles for both kinases in pachytene exit.
PLKs contain an N-terminal kinase domain and a C-terminal polo-box domain (PBD). PBD is thought to be involved in substrate recognition, often by binding phosphorylated amino acids (Barr et al. 2004; Lowery et al. 2005). PLK is required for many cell division-associated events, including CDK activation, centrosome maturation, spindle formation, cohesin removal from chromosome arms, anaphase promoting complex (APC) activation, CDK inactivation during exit from mitosis, and cytokinesis (Barr et al. 2004). Animal PLKs act late in meiosis during spermatogenesis and oocyte maturation, mainly regulating events associated with the two meiotic divisions (Lee and Amon 2003a; Liu et al. 2007). Animal PLK is also present earlier, during meiosis I prophase, but little is known about its function at this earlier stage (Matsubara et al. 1995; Mirouse et al. 2006).
Budding yeast PLK, Cdc5, plays essential roles in the exit from mitosis and in cytokinesis (Lee et al. 2005). Studies using conditional alleles and alleles that express CDC5 during mitosis but not during meiosis (cdc5-mn, meiotic null) have revealed meiosis I prophase functions. These include promoting kinetochore mono-orientation at metaphase I, removal of cohesin from chromosome arms at anaphase I, and exit from both meiotic divisions (Sharon and Simchen 1990; Clyne et al. 2003; Lee and Amon 2003a, b). Cdc5 also appears to act during pachytene exit, as cdc5-mn mutants accumulate JMs and show reduced COs, consistent with a defect in JM resolution, and SC persists in some cdc5-mn cells (Clyne et al. 2003). These findings raise the possibility that Ndt80 regulates pachytene exit by activating CDC5 expression. However, cdc5 mutant cells undergo SPB separation and establish a metaphase I spindle, indicating that Cdc5 activity is not absolutely required for meiotic progression past pachytene (Clyne et al. 2003; Lee and Amon 2003b).
CDKs coordinate progression through the mitotic and meiotic cell cycle (Benjamin et al. 2003; Marston and Amon 2004). Cdc28, the budding yeast CDK catalytic subunit, controls progression through S, G2, and M by associating with regulatory B-type cyclins (Clb1, Clb2, Clb3, Clb4, Clb5, and Clb6). All but Clb2 are produced during meiosis (Chu and Herskowitz 1998). Clb5 (S-phase cyclin) is required for premeiotic DNA replication and meiotic DSB formation (Murakami and Keeney 2008), while Clb1, Clb3, Clb4, and Clb6 (M-phase cyclins), whose expression is Ndt80-dependent, promote meiotic divisions (Chu and Herskowitz 1998; Marston and Amon 2004; Carlile and Amon 2008). Conditional cdc28-ts mutants arrest in meiosis I prophase with unseparated SPBs (Shuster and Byers 1989; Xu et al. 1997), and meiosis I prophase arrest in response to defects in synapsis and/or recombination requires inhibition of CDK by the Swe1 protein (Leu and Roeder 1999). It has thus been inferred that CDK activity is required for exit from pachytene, and it has been suggested that pachytene arrest in ndt80Δ cells is due to the absence of M-phase cyclins (Roeder and Bailis 2000). However, it is possible that partial disruption in cdc28-ts mutants of earlier CDK-dependent events, such as replication or DSB formation, provokes pachytene arrest, and a role for CDK in JM resolution has not been documented.
We show here that, under conditions of high NDT80 expression, ongoing CDK activity is not required for JM resolution or for SC disassembly. In contrast, induced expression of CDC5 in an ndt80Δ strain efficiently promotes both events. We conclude that CDC5 is the only member of the Ndt80 regulon that is required for exit from pachytene.
Results and Discussion
CDK activity is not required for Ndt80-dependent JM resolution and SC breakdown
To examine roles for CDK in exit from pachytene without affecting prior CDK-dependent events, we used an analog-sensitive allele of CDC28, cdc28-as1, which confers sensitivity to the ATP analog 1-NM-PP1 (4-amino-1-tert-butyl-3-[1′-naphthylmethyl] pyrazolo[3,4-d] pyrimidine) (Bishop et al. 2000). We combined cdc28-as1 with an estrogen-inducible NDT80 allele (NDT80-IN) (see Supplemental Material) that confers reversible pachytene arrest (Benjamin et al. 2003; Carlile and Amon 2008). In the absence of inducer (β-estradiol, ED), NDT80 is not expressed, and cells accumulate in pachytene with persistent JMs and full-length SC. ED addition induces NDT80 expression, and cells synchronously exit from pachytene and undergo meiotic nuclear divisions (Fig. 1; Carlile and Amon 2008).
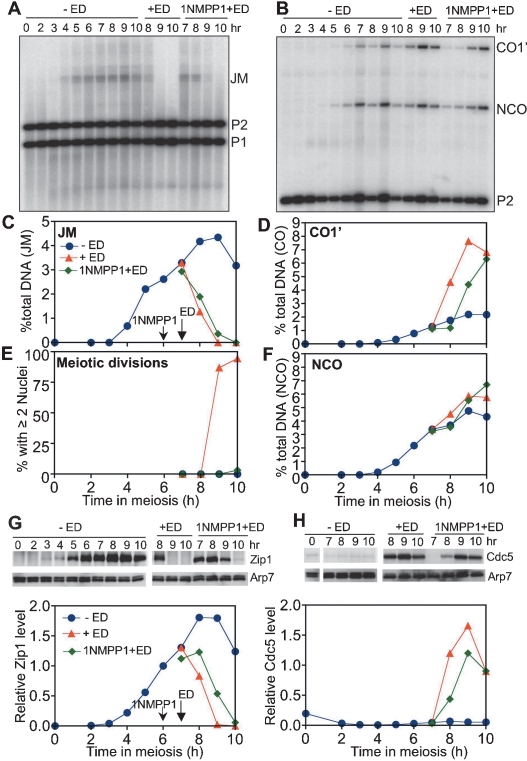
Figure 1.
CDK inhibition does not prevent JM resolution or SC breakdown. NDT80-IN cdc28-as1 (MJL3232) cells were sporulated for 6 h, and the culture was divided into three portions: uninduced (−ED; no ED or 1NMPP1; NDT80off ; blue line), induced (+ED; 1 μM β-estradiol added at 7 h; NDT80on; red line), and CDK-inhibited (+1NMPP1 + ED; 20 μM 1-NM-PP1 added at 6 h followed by 1 μM ED at 7 h; CDKoff; NDT80on; green line). Arrows indicate times of addition. See Supplemental Figure 4 for details of digests. (A) JM recombination intermediates. Blots containing XmnI digests were probed with ARG4 sequences. (P1, P2) Parental fragments containing inserts at LEU2 and HIS4, respectively. Frequencies of JMs are plotted in C as a percentage of total lane signal. (B) CO and NCO products. Blots containing EcoRI/XhoI digests were probed with HIS4 sequences. Frequencies of CO and NCO products are plotted in D and F, respectively, as a percentage of total lane signal. (E) Fraction of cells completing the first meiotic nuclear division. (G,H) Western blots detecting Zip1 (G) and Cdc5 (H). Graphs show normalized Zip1 and Cdc5 levels, which were determined as described in Material and Methods.
To monitor meiotic recombination, we used recombination reporter systems that allow the detection of recombination intermediates and products on Southern blots (Allers and Lichten 2001; Jessop et al. 2005). NDT80-IN cdc28-as1 cells were accumulated at pachytene. To inhibit preexisting Clb5–CDK activity without affecting DSB formation, 1NM-PP1 was added 6 h after initiation of meiosis, a time by which most meiotic DSBs have formed and been repaired (Buhler et al. 2007; data not shown). NDT80 expression was induced 1 h later by ED addition. Induced NDT80 expression restored JM resolution and CO formation, regardless of whether or not CDK was inhibited (Fig. 1A–F). NCO formation was largely unaffected by NDT80 expression or by CDK inhibition (Fig. 1B,F), consistent with previous findings (Allers and Lichten 2001). However, inhibition of CDK blocked meiotic divisions that occurred upon induced NDT80 expression (Fig. 1E). Similar results were obtained when 1-NM-PP1 and ED were added at the same time (Supplemental Fig. 1). Thus, ongoing CDK activity is not required for NDT80-dependent JM resolution as COs.
To determine if CDK is required for SC breakdown, we monitored levels of the SC central element protein Zip1 (Sym et al. 1993). In the absence of NDT80 expression, Zip1 persisted (Fig. 1G). Induction of NDT80 expression led to rapid Zip1 degradation, regardless of whether or not CDK was inhibited (Fig. 1G). Zip1 degradation was accompanied by the disappearance of Zip1 from chromosomes, as monitored in nuclear spreads (data not shown). These results indicate that ongoing CDK activity is not required for SC breakdown. 1NM-PP1 did block meiotic divisions (Fig. 1E) and also impaired the earlier step of SPB separation (data not shown), two events that require active CDK. In a second test, deletion of the four meiotic M-phase cyclin genes—CLB1, CLB3, CLB4, and CLB6—did not affect the timing or extent of DSB repair, JM resolution, CO formation, NCO formation, or SC breakdown, although meiotic divisions and SPB separation were defective (Supplemental Fig. 2; data not shown).
We observed that CDK inhibition delayed JM resolution and SC breakdown by 30–60 min (Fig. 1A,G). To test the possibility that this was due to altered Ndt80 expression or activity, we measured levels of Ndt80 and of three proteins whose production is Ndt80-dependent: Cdc5, Clb1, and Clb3 (Fig. 1H; Supplemental Fig. 3). CDK inhibition caused a 30- to 60-min delay in the accumulation of Ndt80 and the Ndt80 targets Cdc5, Clb1, and Clb3. CDK inhibition also caused a similar delay in Cdc5-dependent Ndt80 modification. Thus, the observed delay in JM resolution and SC breakdown in 1NM-PP1-treated NDT80-IN cdc28-as1 cells is consistent with the suggestion that, while CDK is important for timely Ndt80 protein production and activity, it is not essential for either.
Cdc5 promotes JM resolution as crossovers in ndt80Δ cells
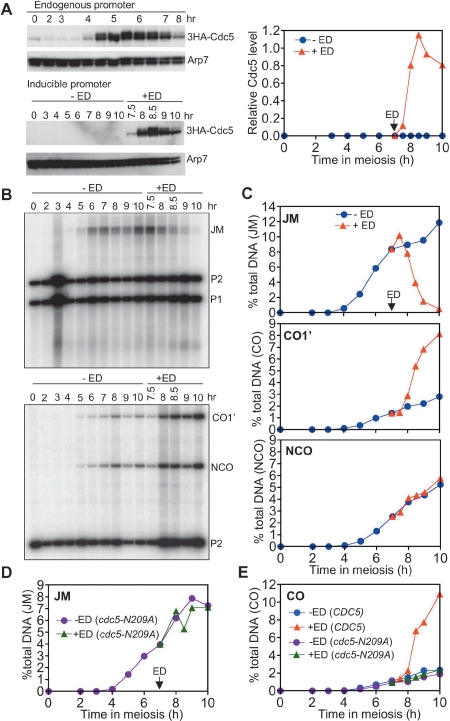
While a previous study had shown that Cdc5 was necessary for efficient JM resolution (Clyne et al. 2003), it remained possible that other proteins encoded by the ∼200 genes of the Ndt80 regulon were also necessary. To determine if Cdc5 was sufficient for JM resolution, we introduced an estrogen-inducible CDC5 allele (CDC5-IN) (Supplemental Material) into ndt80Δ mutant cells, thus enabling Ndt80-independent CDC5 expression during meiosis. Levels of Cdc5p produced using this inducible promoter are similar to Ndt80-dependent levels in wild-type cells (Fig. 2A).
Figure 2.
Cdc5 expression in ndt80Δ cells promotes JM resolution as crossovers. (A) Cdc5 induction. Western blots detecting Cdc5 protein expressed from its own promoter in a wild-type 3HA-CDC5 strain (MJL3335, top) and from the GAL1 promoter in a CDC5-IN ndt80Δ strain (MJL3267, bottom) in the absence (−ED) or presence (+ED) of 1 μM β-estradiol added at 7 h. The plot shows normalized uninduced (−ED, blue) and induced (+ED, red) levels of Cdc5 protein in the estrogen-inducible strain relative to the maximum level of Cdc5 seen in wild type. (B) JM resolution and CO formation. (Top) JM intermediates. Details are as in Figure1A. (Bottom) CO and NCO products. Details are as in Figure 1B. (C) Quantification of JM, CO, and NCO product frequencies from blots in B. Symbols are as in A. (D,E) Cdc5 kinase activity is required for JM resolution as crossovers. cdc5-N209A-IN ndt80Δ cells (MJL3306) were incubated in sporulation medium for 7 h, at which time expression of kinase-dead cdc5 (cdc5-N209A) was induced by addition of 1 μM estradiol (ED). (−ED, purple line) cdc5-N209A; (+ED, green line) cdc5-N209A; (−ED, blue line) CDC5; (+ED, red line) CDC5. (D) JMs in MJL3306, detected on Southern blots of XmnI-digested DNA samples probed with ARG4 sequences (ArgD); JM levels expressed as a percentage of total insert-containing DNA. (E) Crossovers (CO2) (see Supplemental Figs. 2, 4) in MJL3267 (CDC5-IN) and MJL3306 (cdc5-N209A-IN), detected in Southern blots of XhoI-digested DNA samples probed with ARG4 (ArgD) sequences, expressed as a percentage of total insert-containing chromosomes.
To examine the role of Cdc5 in JM resolution, Cdc5 was produced in pachytene-arrested ndt80Δ cells that had accumulated JMs to relatively high levels (7 h after initiation of meiosis). In the absence of Cdc5 induction, ndt80Δ cells displayed high levels of unresolved JMs and few COs (Fig. 2B,C). Cdc5 production promoted rapid and efficient JM resolution, accompanied by an increase in COs but no net increase in NCOs (Fig. 2B,C). Expression of a kinase-dead cdc5 mutant (cdc5-N209A) (Song et al. 2000) failed to promote JM resolution and CO production (Fig. 2D,E). Thus, Cdc5 kinase promotes JM resolution in the absence of all other members of the Ndt80 regulon.
Cdc5 promotes SC disassembly in ndt80Δ cells
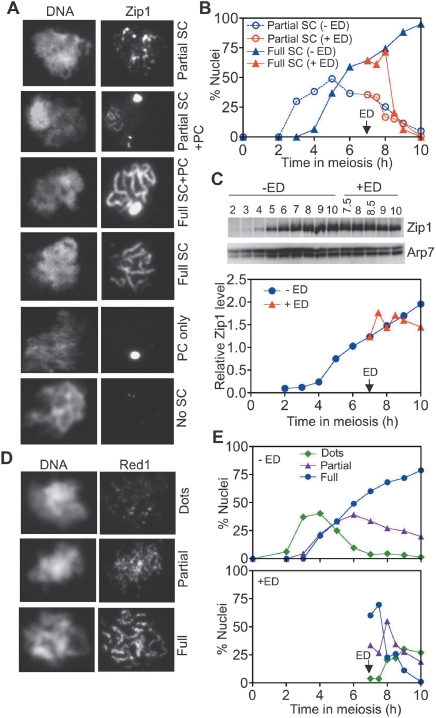
Pachytene-arrested ndt80Δ cells display full-length SC and accumulate polycomplex, an extrachromosomal array of SC proteins (Xu et al. 1995). To monitor the effect of Cdc5 on SC breakdown, nuclear spreads were prepared and stained with antibodies against Zip1. SC persisted in the absence of Cdc5, with most nuclei exhibiting both full-length SC and polycomplex (Fig. 3A,B). Following CDC5 induction, the number of nuclei with full-length SCs dropped rapidly, and most of the SC had disappeared by 3 h after induction, although polycomplex persisted (Fig. 3B; Supplemental Table 1). However, Zip1 protein levels did not decrease after CDC5 induction (Fig. 3C), indicating that Cdc5 mediates disassembly of the SC central element but is insufficient to target Zip1 for proteolysis.
Figure 3.
Cdc5 expression in ndt80Δ cells promotes SC disassembly. A CDC5-IN ndt80Δ strain (MJL3267) was sporulated in the absence (−ED) or presence (+ED) of 1 μM β-estradiol added at 7 h to induce Cdc5 expression. Spread nuclei were stained with anti-Zip1 (A,B) or anti-Red1 (D,E) to detect SC central and lateral elements, respectively. (A) Representative images of nuclei classified as partial SC (discontinuous Zip1), full SC (long, continuous Zip1 lines), and no SC (no Zip1 chromosomal staining). Nuclei with extrachromosomal Zip1 polycomplex (PC; bright staining body) were also scored. (B) SC formation and disassembly. Nuclei (120–152 nuclei for each sample) were scored as illustrated in A. The fraction of nuclei from uninduced cultures (−ED, blue) or cultures with CDC5 expression induced at 7 h (+ED, red) with full SC (triangles) and partial SC (circles) is plotted; nuclei with and without PC were combined. (C) Normalized Zip1 protein levels. (D) Representative images of Red1-stained nuclei scored as having dots, partial (short stretches), and full (long linear stretches) staining patterns. (E) Fraction of nuclei (120–155 nuclei scored for each sample) in each Red1 staining class: (green) dots; (purple) partial; (blue) full. (Top) No CDC5 induction (−ED). (Bottom) CDC5 induced at 7 h (+ED). The number of nuclei in each Zip1 and Red1 category is reported in Supplemental Tables 1 and 2, respectively.
To further characterize the role of Cdc5 in SC disassembly, we examined nuclear spreads for Red1, a component of SC axial elements in cells prior to synapsis and of lateral elements in cells with mature SC (Smith and Roeder 1997). Red1 normally displays a discontinuous distribution along the length of SC (Smith and Roeder 1997). In the absence of CDC5 induction, ndt80Δ nuclei exhibited persistent Red1 with the characteristic discontinuous distribution (Fig. 3D; Supplemental Table 2). Induction of Cdc5 expression resulted in the loss of Red1 from most nuclei, consistent with the suggestion that Cdc5 also mediates disassembly of SC lateral elements (Fig. 3E).
Cdc5 expression in ndt80Δ cells failed to promote the subsequent events of SPB separation and meiotic division (data not shown), consistent with the absence in ndt80Δ cells of M-phase CDK, which is required for both of these events (Shuster and Byers 1989; Xu et al. 1995).
In summary, these data show that, of the ∼200 proteins that are expressed in the NDT80 regulon, Cdc5/PLK is the only protein both necessary and sufficient to trigger two key events of exit from pachytene: JM intermediate resolution and SC disassembly. These events occur earlier in meiosis than do other events in which PLK has been implicated so far. PLK drives monopolin loading on kinetochores (Clyne et al. 2003; Lee and Amon 2003b) late in meiosis I prophase and modifies cohesin on chromosome arms to facilitate its removal at the metaphase I to anaphase I transition (Clyne et al. 2003; Lee and Amon 2003b). PLK is also a component of the FEAR network, which is required for the completion of the first meiotic division (Lee and Amon 2003a). These processes are all involved in either establishing or executing the meiotic division itself. The identification of PLK as being both necessary (Clyne et al. 2003) and sufficient (this study) to trigger exit from pachytene points to its involvement at a much earlier stage, since diffuse, diplotene, and diakinesis stages occur between exit from pachytene and metaphase I.
Evidence for the involvement of PLK in earlier stages of meiosis I prophase in other organisms is limited. Recent studies in Drosophila reveal that inhibition of Polo by the Matrimony protein forms the mechanistic basis for the long arrest of meiotic progression that occurs after pachytene exit in oocytes (Xiang et al. 2007). Matrimony is produced immediately after pachytene, suggesting that Polo is present at this stage, as would be expected if it were involved in promoting exit from pachytene. In addition, localization and mutant studies suggest that PLK-2, a Caenorhabditis elegans Polo homolog, is involved in SC disassembly and chromosome decondensation at the end of pachytene (S. Jover Gil and A. Dernburg, pers. comm.).
How might PLK drive exit from pachytene? Multiple targets for PLK have been identified in animals and budding yeast (Lowery et al. 2005; Lee et al. 2008), including Ime2 (a meiosis-specific CDK-like kinase), Ndt80 (Supplemental Fig. 3), Mam1 (a monopolin component), and Rec8, the meiosis-specific kleisin subunit of cohesin (Clyne et al. 2003; Lee and Amon 2003b). The requirement for persistent arm cohesion through metaphase I for proper homolog disjunction makes cohesin a less likely PLK target in driving exit from pachytene. Consistent with this, CDC5-IN ndt80Δ rec8-17A cells, which lack 17 Cdc5 target sites on Rec8 (Brar et al. 2006), show JM resolution and CO formation similar to those seen in otherwise isogenic REC8 cells (A. Sourirajan and M. Lichten, unpubl.).
One intriguing possibility is that JM resolution is driven by SC disassembly. SC proteins play a critical role in JM formation, by protecting nascent recombination intermediates from the disintegrating activity of the Sgs1/Top3/Rmi1 complex (Jessop et al. 2006; Oh et al. 2007). SC components might play a similar role in protecting JMs from resolving activities until the end of pachytene. In this view, phosphorylation of SC components by PLK, leading to SC disassembly, might expose JMs to resolvases and lead to CO formation.
Alternatively, SC disassembly may be triggered by JM resolution. In budding yeast and many other organisms, SC assembly on chromosomes requires recombination, and SC nucleation has been suggested to occur at sites of recombination (Kleckner 2006). It is possible that SC integrity requires the continued presence of JM intermediates and associated late recombination nodules (RNs). In this view, activation of resolvases by PLK would lead to JM resolution, CO formation, and consequent SC disassembly. Determining which of these processes Cdc5 triggers and whether it does so directly or though intermediary regulatory proteins will require the identification of meiotic PLK targets.
In mammals, PLK substrate recognition often occurs when the polo-box domain binds to a portion of the target that is CDK-phosphorylated (Barr et al. 2004; Lowery et al. 2005; Lee et al. 2008). The PLK-dependent, CDK-independent JM resolution and SC breakdown we observe might be due to target recognition by PLK through polo-box-independent or phosphorylation-independent interactions (see Lee et al. 2008 for an example) or by polo-box binding to a CDK-independent phosphomark formed by other kinases that are active during meiosis I prophase. Alternatively, PLK recognition of the relevant target might require a CDK-catalyzed phosphorylation, but one that is established earlier in meiosis and that persists in the absence of ongoing CDK activity.
In summary, our data reveal a novel role for budding yeast PLK Cdc5 in promoting the exit from pachytene and show that ongoing CDK activity is not required for this critical transition. It thus appears that Cdc5 is a major regulator of key prophase I events, in addition to its functions later in meiosis.
Materials and methods
Yeast strains and sporulation
Yeast strains (Supplemental Table 3; construction described in Supplemental Material) are derived from SK1. Sporulation was as described (Allers and Lichten 2001), with 1-NM-PP1 (Toronto Research Chemicals; 10 mM stock in dimethylsulfoxide) and β-estradiol (Sigma; 5 mM stock in ethanol) being added as indicated above.
Recombination intermediate/product analysis
Genomic DNA preparation and analysis on Southern blots were as described (Allers and Lichten 2001; Jessop et al. 2006); see Supplemental Figure 4 for a guide to recombination reporter structures and restriction digests used. XhoI and XmnI digests were probed with ARG4 coding sequences (+165 to +1413). EcoRI/XhoI double digests were probed with HIS4 coding sequences (+538 to +718).
Protein analysis
Total protein was prepared from 2–4 mL of meiotic culture and analyzed on Western blots as described (Jessop and Lichten 2008). After initial probing, blots were stripped and reprobed for Arp7 as a loading control. To quantify protein levels, the band intensity in each lane was divided by the corresponding Arp7 band intensity in the same lane. This ratio is represented in arbitrary units. The primary antisera, sources, and working dilutions were Zip1 and Red1—rabbit polyclonal (a gift from G.S. Roeder), 1:10,000 dilution; Cdc5, Clb1, Clb3, and Arp7—rabbit or goat polyclonal (Santa Cruz Biotechnologies, sc-6733, sc-7647, sc-7167 and sc-8961, respectively), 1:500; influenza hemaglutinin (HA)–mouse monoclonal (Roche Diagnostics, 12CA5), 5 μg/μL, 1:10,000; Ndt80—rabbit polyclonal (a gift from K. Benjamin), 1:10,000.
Meiotic nuclear morphology
Nuclear spreads were formed and stained as described (Bishop 1994) using cells from 5 mL of meiotic culture. Red1 and Zip1 were detected using the primary antisera described above at 1:100 and Alexafluor 488-conjugated goat anti-rabbit IgG (Molecular Probes #A11034) at 1:100 as the secondary antibody. DNA was visualized using DAPI stain present in Vectashield mounting medium (Vector Laboratories). Meiotic spindle formation and nuclear divisions were monitored by staining with anti-Tub2 (a gift from M. Basrai) or DAPI as described (Jessop and Lichten 2008). Sample fluorescence was visualized using a Zeiss Axioplan 2 epifluorescence microscope and a Micromax 1300 CCD camera. Images were acquired using IPlab 3.7 and processed with Adobe Photoshop PS3.
Acknowledgments
We thank Angelika Amon, Munira Basrai, Thomas Carlile, Kyung S. Lee, G. Shirleen Roeder, and Kirsten Benjamin for strains and reagents; Yaron Dayani, Lea Jessop, Jacqueline Segall, Parisha Shah, and Martin Xaver for technical and editorial guidance; and Abby Dernburg for communicating data before publication. We apologize to colleagues whose work was not cited due to space constraints. This research was supported by the Intramural Research Program of the National Cancer Institute, NIH.
Footnotes
Supplemental material is available at http://www.genesdev.org.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1711408.
References
- Allers T., Lichten M. Differential timing and control of noncrossover and crossover recombination during meiosis. Cell. 2001;106:47–57. doi: 10.1016/s0092-8674(01)00416-0. [DOI] [PubMed] [Google Scholar]
- Barr F.A., Sillje H.H., Nigg E.A. Polo-like kinases and the orchestration of cell division. Nat. Rev. Mol. Cell Biol. 2004;5:429–440. doi: 10.1038/nrm1401. [DOI] [PubMed] [Google Scholar]
- Benjamin K.R., Zhang C., Shokat K.M., Herskowitz I. Control of landmark events in meiosis by the CDK Cdc28 and the meiosis-specific kinase Ime2. Genes & Dev. 2003;17:1524–1539. doi: 10.1101/gad.1101503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop D.K. RecA homologs Dmc1 and Rad51 interact to form multiple nuclear complexes prior to meiotic chromosome synapsis. Cell. 1994;79:1081–1092. doi: 10.1016/0092-8674(94)90038-8. [DOI] [PubMed] [Google Scholar]
- Bishop D.K., Zickler D. Early decision; meiotic crossover interference prior to stable strand exchange and synapsis. Cell. 2004;117:9–15. doi: 10.1016/s0092-8674(04)00297-1. [DOI] [PubMed] [Google Scholar]
- Bishop A.C., Ubersax J.A., Petsch D.T., Matheos D.P., Gray N.S., Blethrow J., Shimizu E., Tsien J.Z., Schultz P.G., Rose M.D., et al. A chemical switch for inhibitor-sensitive alleles of any protein kinase. Nature. 2000;407:395–401. doi: 10.1038/35030148. [DOI] [PubMed] [Google Scholar]
- Brar G.A., Kiburz B.M., Zhang Y., Kim J.E., White F., Amon A. Rec8 phosphorylation and recombination promote the step-wise loss of cohesins in meiosis. Nature. 2006;441:532–536. doi: 10.1038/nature04794. [DOI] [PubMed] [Google Scholar]
- Buhler C., Borde V., Lichten M. Mapping meiotic single-strand DNA reveals a new landscape of DNA double-strand breaks in Saccharomyces cerevisiae. PLoS Biol. 2007;5:e324. doi: 10.1371/journal. pbio.0050324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlile T.M., Amon A. Meiosis I is established through division-specific translational control of a cyclin. Cell. 2008;133:280–291. doi: 10.1016/j.cell.2008.02.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu S., Herskowitz I. Gametogenesis in yeast is regulated by a transcriptional cascade dependent on Ndt80. Mol. Cell. 1998;1:685–696. doi: 10.1016/s1097-2765(00)80068-4. [DOI] [PubMed] [Google Scholar]
- Chu S., DeRisi J., Eisen M., Mulholland J., Botstein D., Brown P.O., Herskowitz I. The transcriptional program of sporulation in budding yeast. Science. 1998;282:699–705. doi: 10.1126/science.282.5389.699. [DOI] [PubMed] [Google Scholar]
- Clyne R.K., Katis V.L., Jessop L., Benjamin K.R., Herskowitz I., Lichten M., Nasmyth K. Polo-like kinase Cdc5 promotes chiasmata formation and cosegregation of sister centromeres at meiosis I. Nat. Cell Biol. 2003;5:480–485. doi: 10.1038/ncb977. [DOI] [PubMed] [Google Scholar]
- Jessop L., Lichten M. Mus81/Mms4 endonuclease and Sgs1 helicase collaborate to ensure proper recombination intermediate metabolism during meiosis. Mol. Cell. 2008;31:313–323. doi: 10.1016/j.molcel.2008.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jessop L., Allers T., Lichten M. Infrequent co-conversion of markers flanking a meiotic recombination initiation site in Saccharomyces cerevisiae. Genetics. 2005;169:1353–1367. doi: 10.1534/genetics.104.036509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jessop L., Rockmill B., Roeder G.S., Lichten M. Meiotic chromosome synapsis-promoting proteins antagonize the anti-crossover activity of Sgs1. PLoS Genet. 2006;2:e155. doi: 10.1371/journal. pgen.0020155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeney S., Neale M.J. Initiation of meiotic recombination by formation of DNA double-strand breaks: Mechanism and regulation. Biochem. Soc. Trans. 2006;34:523–525. doi: 10.1042/BST0340523. [DOI] [PubMed] [Google Scholar]
- Kleckner N. Chiasma formation: Chromatin/axis interplay and the role(s) of the synaptonemal complex. Chromosoma. 2006;115:175–194. doi: 10.1007/s00412-006-0055-7. [DOI] [PubMed] [Google Scholar]
- Lee B.H., Amon A. Polo kinase–meiotic cell cycle coordinator. Cell Cycle. 2003a;2:400–402. [PubMed] [Google Scholar]
- Lee B.H., Amon A. Role of Polo-like kinase CDC5 in programming meiosis I chromosome segregation. Science. 2003b;300:482–486. doi: 10.1126/science.1081846. [DOI] [PubMed] [Google Scholar]
- Lee K.S., Park J.E., Asano S., Park C.J. Yeast polo-like kinases: Functionally conserved multitask mitotic regulators. Oncogene. 2005;24:217–229. doi: 10.1038/sj.onc.1208271. [DOI] [PubMed] [Google Scholar]
- Lee K.S., Park J.E., Kang Y.H., Zimmerman W., Soung N.K., Seong Y.S., Kwak S.J., Erikson R.L. Mechanisms of mammalian polo-like kinase 1 (Plk1) localization: Self- versus non-self-priming. Cell Cycle. 2008;7:141–145. doi: 10.4161/cc.7.2.5272. [DOI] [PubMed] [Google Scholar]
- Leu J.Y., Roeder G.S. The pachytene checkpoint in S. cerevisiae depends on Swe1-mediated phosphorylation of the cyclin-dependent kinase Cdc28. Mol. Cell. 1999;4:805–814. doi: 10.1016/s1097-2765(00)80390-1. [DOI] [PubMed] [Google Scholar]
- Liu J., Grimison B., Maller J.L. New insight into metaphase arrest by cytostatic factor: From establishment to release. Oncogene. 2007;26:1286–1289. doi: 10.1038/sj.onc.1210203. [DOI] [PubMed] [Google Scholar]
- Lowery D.M., Lim D., Yaffe M.B. Structure and function of Polo-like kinases. Oncogene. 2005;24:248–259. doi: 10.1038/sj.onc.1208280. [DOI] [PubMed] [Google Scholar]
- Marston A.L., Amon A. Meiosis: Cell-cycle controls shuffle and deal. Nat. Rev. Mol. Cell Biol. 2004;5:983–997. doi: 10.1038/nrm1526. [DOI] [PubMed] [Google Scholar]
- Matsubara N., Yanagisawa M., Nishimune Y., Obinata M., Matsui Y. Murine polo like kinase 1 gene is expressed in meiotic testicular germ cells and oocytes. Mol. Reprod. Dev. 1995;41:407–415. doi: 10.1002/mrd.1080410403. [DOI] [PubMed] [Google Scholar]
- Mirouse V., Formstecher E., Couderc J.L. Interaction between Polo and BicD proteins links oocyte determination and meiosis control in Drosophila. Development. 2006;133:4005–4013. doi: 10.1242/dev.02565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami H., Keeney S. Regulating the formation of DNA double-strand breaks in meiosis. Genes & Dev. 2008;22:286–292. doi: 10.1101/gad.1642308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh S.D., Lao J.P., Hwang P.Y., Taylor A.F., Smith G.R., Hunter N. BLM ortholog, Sgs1, prevents aberrant crossing-over by suppressing formation of multichromatid joint molecules. Cell. 2007;130:259–272. doi: 10.1016/j.cell.2007.05.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pak J., Segall J. Role of Ndt80, Sum1, and Swe1 as targets of the meiotic recombination checkpoint that control exit from pachytene and spore formation in Saccharomyces cerevisiae. Mol. Cell. Biol. 2002;22:6430–6440. doi: 10.1128/MCB.22.18.6430-6440.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roeder G.S., Bailis J.M. The pachytene checkpoint. Trends Genet. 2000;16:395–403. doi: 10.1016/s0168-9525(00)02080-1. [DOI] [PubMed] [Google Scholar]
- Sharon G., Simchen G. Mixed segregation of chromosomes during single-division meiosis of Saccharomyces cerevisiae. Genetics. 1990;125:475–485. doi: 10.1093/genetics/125.3.475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shuster E.O., Byers B. Pachytene arrest and other meiotic effects of the start mutations in Saccharomyces cerevisiae. Genetics. 1989;123:29–43. doi: 10.1093/genetics/123.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith A.V., Roeder G.S. The yeast Red1 protein localizes to the cores of meiotic chromosomes. J. Cell Biol. 1997;136:957–967. doi: 10.1083/jcb.136.5.957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song S., Grenfell T.Z., Garfield S., Erikson R.L., Lee K.S. Essential function of the polo box of Cdc5 in subcellular localization and induction of cytokinetic structures. Mol. Cell. Biol. 2000;20:286–298. doi: 10.1128/mcb.20.1.286-298.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sym M., Engebrecht J.A., Roeder G.S. ZIP1 is a synaptonemal complex protein required for meiotic chromosome synapsis. Cell. 1993;72:365–378. doi: 10.1016/0092-8674(93)90114-6. [DOI] [PubMed] [Google Scholar]
- Tung K.S., Hong E.J., Roeder G.S. The pachytene checkpoint prevents accumulation and phosphorylation of the meiosis-specific transcription factor Ndt80. Proc. Natl. Acad. Sci. 2000;97:12187–12192. doi: 10.1073/pnas.220464597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang Y., Takeo S., Florens L., Hughes S.E., Huo L.J., Gilliland W.D., Swanson S.K., Teeter K., Schwartz J.W., Washburn M.P., et al. The inhibition of polo kinase by Matrimony maintains G2 arrest in the meiotic cell cycle. PLoS Biol. 2007;5:e323. doi: 10.1371/ journal.pbio.0050323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L., Ajimura M., Padmore R., Klein C., Kleckner N. NDT80, a meiosis-specific gene required for exit from pachytene in Saccharomyces cerevisiae. Mol. Cell. Biol. 1995;15:6572–6581. doi: 10.1128/mcb.15.12.6572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L., Weiner B.M., Kleckner N. Meiotic cells monitor the status of the interhomolog recombination complex. Genes & Dev. 1997;11:106–118. doi: 10.1101/gad.11.1.106. [DOI] [PubMed] [Google Scholar]