Abstract
Infection is a leading cause of neonatal morbidity and mortality worldwide. Premature neonates are particularly susceptible to infection because of physiologic immaturity, comorbidity, and extraneous medical interventions. Additionally premature infants are at higher risk of progression to sepsis or severe sepsis, adverse outcomes, and antimicrobial toxicity. Currently initial diagnosis is based upon clinical suspicion accompanied by nonspecific clinical signs and is confirmed upon positive microbiologic culture results several days after institution of empiric therapy. There exists a significant need for rapid, objective, in vitro tests for diagnosis of infection in neonates who are experiencing clinical instability. We used immunoassays multiplexed on microarrays to identify differentially expressed serum proteins in clinically infected and non-infected neonates. Immunoassay arrays were effective for measurement of more than 100 cytokines in small volumes of serum available from neonates. Our analyses revealed significant alterations in levels of eight serum proteins in infected neonates that are associated with inflammation, coagulation, and fibrinolysis. Specifically P- and E-selectins, interleukin 2 soluble receptor α, interleukin 18, neutrophil elastase, urokinase plasminogen activator and its cognate receptor, and C-reactive protein were observed at statistically significant increased levels. Multivariate classifiers based on combinations of serum analytes exhibited better diagnostic specificity and sensitivity than single analytes. Multiplexed immunoassays of serum cytokines may have clinical utility as an adjunct for rapid diagnosis of infection and differentiation of etiologic agent in neonates with clinical decompensation.
Infection, particularly of nosocomial or late onset, is very common in preterm infants (1, 2). The diagnosis of infection in preterm infants can be very difficult. The clinical presentation of neonatal infection is subtle and nonspecific, featuring signs such as jaundice, unstable temperature, difficulty breathing, changes in heart rate, and difficulty in feeding. The diagnostic difficulty is compounded by disease heterogeneity and a lack of reliable, rapid diagnostic tests (3–6). Sources of heterogeneity include etiologic agent, virulence, inoculum, site of primary infection, host genotype, stage of development of host responses, and extraneous clinical interventions. Microbiologic cultures of clinical specimens, the gold standard for diagnosis, have low sensitivity and are not available in time to influence initial therapy. Given the rapid progression and high mortality of sepsis (local infection with evidence of systemic inflammatory response) in preterm infants, broad spectrum antimicrobial chemotherapy is frequently administered at first clinical suspicion of infection (7, 8). Premature infants are at higher risk of drug toxicity because of hepatic and renal organ immaturity, and antimicrobial resistance is an increasing problem in neonatal intensive care settings. Therefore, a reliable and rapid in vitro test is urgently needed for early diagnosis and management of infection in neonates. In addition, the availability of a rapid in vitro test for etiologic agent in neonatal infection would permit early, targeted treatment.
Recently there has been a considerable interest in the application of host biomarkers for diagnostic tests (9). It appears that biological systems are adaptive and that challenges to host homeostasis cause characteristic topological perturbations of molecular networks. A biomarker is a measurable gene, protein, metabolite, or other indicator of network perturbation that correlates with a specific outcome or clinical state (10). Biomarkers are identified through a four-step process of discovery by appropriate multiplex biochemical analysis followed by replication ideally in independent cohorts, validation of diagnostic sensitivity and specificity, and translation into a clinical diagnostic test (9). Numerous candidate biomarkers have been identified in neonatal sepsis: elevated plasma or serum levels of interleukin 6 (IL-6)1 (11, 13), tumor necrosis factor α (TNFα) (11, 13), neutrophil elastase (NE) (14), C-reactive protein (CRP) (12, 15), soluble CD14 (16, 17), granulocyte colony-stimulating factor (G-CSF) (18), soluble intercellular adhesion molecule-1 (ICAM-1) (12), and soluble L-selectin (19) have shown association with infection in neonates. The value of physiological measurements in this context has also been examined recently (20). However, the positive predictive value or negative predictive value (NPV) of individual analytes has not been adequate for routine use in the diagnosis and management of neonatal infection.
In other clinical conditions where individual analytes lack adequate positive predictive value or NPV, the values of several analytes have been combined, together with an algorithm, to provide a biomarker panel with improved clinical performance (9). Multiplexed measurement of plasma proteins is a logical approach because they constitutively function within networks, pathways, complexes, and families (21–23). Indeed the activity of an individual plasma protein is typically dependent not only upon its abundance but also upon the effects of interacting proteins, modifying proteins, and antagonistic and synergistic proteins. This is especially true of cytokines, which are a group of several hundred small, soluble, plasma proteins relevant to sepsis, that are powerful mediators of target cell activities such as migration, activation, phagocytosis, proliferation, and apoptosis.
Several recent studies have explored the utility of measurement of two or more analytes for diagnosis of neonatal infection (11–13, 17). For example, we have shown that CRP > 6 mg/liter and soluble ICAM-1 > 300 ng/ml in combination in plasma samples collected under endotoxin-free conditions (12, 13) were independent predictors of infection and gave both a high sensitivity for clinical infection (95%) and an NPV of 97% (12). We subsequently demonstrated that routinely collected serum samples reproduced a very high NPV for infection (24). We have further examined the diagnostic utility of the combination of low level CRP and soluble immunological markers.2 In a recent review, Ng (25) concluded that a combination of several markers can enhance diagnostic accuracy in neonatal sepsis.
In the present study, we used amplified, multiplexed, immunoassay arrays to measure 142 small serum proteins and assess their utility in differential diagnosis of neonatal infection. In addition, we utilized the most informative of these biomarkers to develop a multiplexed biomarker panel with improved diagnostic performance in neonatal infection.
EXPERIMENTAL PROCEDURES
Patient Population, Samples, and Laboratory Test Results
Serum samples were obtained from acutely ill, preterm (≤36 weeks gestation) infants undergoing intensive care in the neonatal units of the University Hospital, Queen's Medical Centre, Nottingham, UK and the Royal-Jubilee Maternity Service, Royal Hospitals Trust, Belfast, Northern Ireland, UK. These infants were either admitted to the neonatal unit with a suspected diagnosis of infection (early onset and late onset infection) or developed clinical signs suggesting that infection may be complicating their management. As part of their acute management, these infants had blood samples taken for routine laboratory analysis, and an extra aliquot of 0.5 ml of blood was collected for the purposes of this study. At presentation, the culture result, white blood cell count, platelet count, gestational age, primary diagnosis, and clinical signs were also recorded.
Blood samples were kept at ambient temperature until serum was separated (within 24 h), and serum was stored at −20 °C until analysis. No extra venepuncture beyond clinical requirements was necessary as a result of this study for either patients or control subjects.
Clinical diagnosis was established in a prospective manner, blind to the results of the study measurements. Infants were classified as “clinically infected” or “non-infected” based on the initial clinical presentation and results of routine investigations ((a) white cell and platelet counts, (b) blood and other cultures, (c) radiological imaging, and (d) response to antibiotics). The infected group was further subclassified as (a) culture-positive infection or (b) culture-negative infection.
Written informed parental consent was obtained for study subjects. Ethical approval was obtained from the Ethics Committee, University Hospital, Queen's Medical Centre, Nottingham, UK.
Sample Processing
Aliquots of the samples were thawed, centrifuged to remove particulate matter, and diluted 1:5 with 18.75 units/ml heparin (Sigma) in AD1 diluent (Immunochemistry Technologies, Bloomington, MN), and 0.25 mg/ml Heteroblock (Omega Biologicals, Bozeman, MT), 0.25 mg/ml immunoglobulin inhibiting reagent (Bioreclamation, Hicksville, NY), and 0.1% Tween 20 (Sigma) were added prior to assay.
Antibody Microarray Manufacture
Antibody microarray manufacture has been described in detail previously (26). Briefly glass slides were covered with a Teflon mask to provide 16 sample wells and silanized with 3-cyanopropyltriethoxysilane. Each well was printed with quadruplicate spots of each of 64 capture antibodies using a PerkinElmer Life Sciences SpotArray Enterprise piezoelectric arrayer. Six different antibody arrays were prepared, each containing 25–37 unique capture antibodies (Table I).
Table I.
142 analytes measured by amplified sandwich immunoassays on glass microarrays 1–6
TNFR, TNF receptor; RANTES, regulated on activation normal T cell expressed and secreted.
| No. | Abbreviation | Alternate/full name | Analyte class |
|---|---|---|---|
| 1 | 4-1BB | CD137, TNF receptor superfamily, member 9 | Cytokine receptor |
| 2 | CCL21 | 6Ckine | Cytokine |
| 3 | ACE | Angiotensin I-converting enzyme, CD143 | Enzyme |
| 4 | ACE-2 | Angiotensin I-converting enzyme-2 | Enzyme |
| 5 | AFP | α-Fetoprotein | Oncofetal antigen |
| 6 | AgRP | Agouti-related protein | Cytokine |
| 7 | ALCAM | Activated leukocyte cell adhesion molecule, CD166 | Adhesion molecule |
| 8 | ANG | Angiogenin, ribonuclease 5 | Enzyme |
| 9 | AR | Amphiregulin | Cytokine |
| 10 | BDNF | Brain-derived neurotrophic factor | Cytokine |
| 11 | BLC (BCA-1) | B lymphocyte chemoattractant, CXCL13 | Cytokine |
| 12 | CA125 | Cancer antigen 125, mucin 16, MUC16 | Antigen |
| 13 | CD141 | Thrombomodulin, CD141 | Clotting factor |
| 14 | CD27 | TNF receptor superfamily, member 7 | Cytokine receptor |
| 15 | CD30 | TNF receptor superfamily, member 8 | Cytokine receptor |
| 16 | CD40 | TNF receptor superfamily, member 5 | Cytokine receptor |
| 17 | CNTF | Ciliary neurotrophic factor | Cytokine |
| 18 | CNTF Rα | Ciliary neurotrophic factor receptor α | Cytokine receptor |
| 19 | CRP | C-reactive protein | Pentraxin |
| 20 | CCL27 | Cutaneous T cell-attracting chemokine, CTACK | Cytokine |
| 21 | D-dimer | D dimer | Clotting factor |
| 22 | EGF | Epidermal growth factor | Cytokine |
| 23 | ENA-78, CXCL5 | Epithelial cell-derived neutrophil-activating peptide | Cytokine |
| 24 | Endostatin | Procollagen, type XVIII, α1 | Angiogenesis factor |
| 25 | Endothelin 3 | Endothelin 3 | Cytokine |
| 26 | Eot | Eotaxin, CCL11 | Cytokine |
| 27 | Eot-2 | Eotaxin-2, CCL24 | Cytokine |
| 28 | Eot-3 | Eotaxin-3, CCL26 | Cytokine |
| 29 | ErbB1 | Epidermal growth factor receptor 1, EGFR | Cytokine receptor |
| 30 | ErbB2 | Epidermal growth factor receptor 2, HER2, NEU | Cytokine receptor |
| 31 | E-selectin | E-selectin, CD62E | Adhesion molecule |
| 32 | Fas | CD95, TNF receptor superfamily, member 6 | Cytokine receptor |
| 33 | FGFR3 (IIIc) | Fibroblast growth factor receptor 3 IIIc isoform | Cytokine receptor |
| 34 | FGF2 | Fibroblast growth factor-2 (FGF basic) | Cytokine |
| 35 | FGF4 | Fibroblast growth factor-4 | Cytokine |
| 36 | FGF6 | Fibroblast growth factor-6 | Cytokine |
| 37 | FGF7 | Fibroblast growth factor-7 | Cytokine |
| 38 | FGF9 | Fibroblast growth factor-9 | Cytokine |
| 39 | FGFR3 (IIIb) | Fibroblast growth factor receptor 3 IIIb isoform | Cytokine receptor |
| 40 | FLT3LG | fms-like tyrosine kinase-3 ligand | Cytokine |
| 41 | Follistatin | Follistatin | Cytokine |
| 42 | GCP-2 | Granulocyte chemotactic protein 2, CXCL6 | Cytokine |
| 43 | G-CSF | Granulocyte colony-stimulating factor, CSF3 | Cytokine |
| 44 | GDNF | Glial cell line-derived neurotrophic factor | Cytokine |
| 45 | GM-CSF | Granulocyte macrophage CSF, CSF2 | Cytokine |
| 46 | GRO-γ | Growth-related oncogene γ, CXCL3, MIP2B | Cytokine |
| 47 | HB-EGF | Heparin-binding EGF-like growth factor | Cytokine |
| 48 | HCC4 (NCC4) | Hemofiltrate CC chemokine 4, CCL16 | Cytokine |
| 49 | HCG | Human chorionic gonadotrophin | Hormone |
| 50 | I-309 | I-309, CCL1 | Cytokine |
| 51 | ICAM-1 | Intercellular adhesion molecule 1, CD54 | Adhesion molecule |
| 52 | IFN-α | Interferon α | Cytokine |
| 53 | IFN-β | Interferon γ | Cytokine |
| 54 | IFN-ω | Interferon ω | Cytokine |
| 55 | IGF-1R | Insulin-like growth factor I receptor, CD221 | Cytokine receptor |
| 56 | IGFBP-1 | Insulin-like growth factor-binding protein 1, | Cytokine |
| 57 | IGFBP-3 | Insulin-like growth factor-binding protein 3 | Cytokine |
| 58 | IGFBP-4 | Insulin-like growth factor-binding protein 4 | Cytokine |
| 59 | IGFBP-6 | Insulin-like growth factor-binding protein 6 | Cytokine |
| 60 | IGF2 | Insulin-like growth factor II, somatomedin A | Cytokine |
| 61 | IL-1sR1 | Interleukin 1 soluble receptor I, CD121a | Cytokine receptor |
| 62 | IL-1sRII | Interleukin 1 soluble receptor II, CD121b | Cytokine receptor |
| 63 | IL-10 Rβ | Interleukin 10 receptor β | Cytokine receptor |
| 64 | IL-12b (p40) | Interleukin 12β, p40 | Cytokine |
| 65 | IL-13 | Interleukin 13 | Cytokine |
| 66 | IL-15 | Interleukin 15 | Cytokine |
| 67 | IL-16 | Interleukin 16 | Cytokine |
| 68 | IL-17 | Interleukin 17 | Cytokine |
| 69 | IL-18 | Interleukin 18 | Cytokine |
| 70 | IL-1α | Interleukin 1α | Cytokine |
| 71 | IL-1β | Interleukin 1β | Cytokine |
| 72 | IL-1ra | Interleukin 1 receptor antagonist, IL1RN | Cytokine |
| 73 | IL-2 | Interleukin 2 | Cytokine |
| 74 | IL-2 Rβ | Interleukin 2 receptor β, CD122 | Cytokine receptor |
| 75 | IL-2sRα | Interleukin 2 soluble receptor α, CD25 | Cytokine receptor |
| 76 | IL-3 | Interleukin 3 | Cytokine |
| 77 | IL-4 | Interleukin 4 | Cytokine |
| 78 | IL-5 | Interleukin 5 | Cytokine |
| 79 | IL-6 | Interleukin 6 | Cytokine |
| 80 | IL-7 | Interleukin 7 | Cytokine |
| 81 | IL-8 | Interleukin 8 | Cytokine |
| 82 | I-TAC | IFN-γ-inducible T cell α chemoattractant, CXCL11 | Cytokine |
| 83 | LIF | Leukemia-inhibitory factor | Cytokine |
| 84 | LIF Rα (gp190) | Leukemia-inhibitory factor soluble receptor α | Cytokine receptor |
| 85 | Lptn | Lymphotactin, XCL1 | Cytokine |
| 86 | LT βR | Lymphotoxin-β receptor, TNFR superfamily 3 | Cytokine receptor |
| 87 | MCP-1 | Monocyte chemotactic protein 1, CCL2 | Cytokine |
| 88 | MCP-2 | Monocyte chemotactic protein 2, CCL8 | Cytokine |
| 89 | MCP-3 | Monocyte chemotactic protein 3, CCL7 | Cytokine |
| 90 | M-CSF | Macrophage colony-stimulating factor, CSF1 | Cytokine |
| 91 | M-CSF R | Macrophage colony-stimulating factor receptor CSF1R | Cytokine receptor |
| 92 | MIF | Macrophage migration-inhibitory factor | Cytokine |
| 93 | MIG | Monokine induced by interferon γ, CXCL9 | Cytokine |
| 94 | MIP-1α | Macrophage inflammatory protein 1α, CCL3 | Cytokine |
| 95 | MIP-1β | Macrophage inflammatory protein 1β, CCL4 | Cytokine |
| 96 | MIP-1δ | Macrophage inflammatory protein 1δ, CCL15 | Cytokine |
| 97 | MIP-3α | Macrophage inflammatory protein 3α, CCL20 | Cytokine |
| 98 | MMP-1 | Matrix metalloproteinase 1 | Enzyme |
| 99 | MMP-10 | Matrix metalloproteinase 10 | Enzyme |
| 100 | MMP-2 | Matrix metalloproteinase 2 | Enzyme |
| 101 | MMP-8 (total) | Matrix Metalloproteinase-8 | Enzyme |
| 102 | MPIF | Myeloid progenitor-inhibitory factor 1, CCL23 | Cytokine |
| 103 | NAP-2 | Neutrophil-activating peptide 2, CXCL7 | Cytokine |
| 104 | NE | Neutrophil elastase, elastase 2 | Enzyme |
| 105 | NGF-β | Nerve growth factor-β | Cytokine |
| 106 | NT-3 | Neurotrophin 3, NTF3 | Cytokine |
| 107 | NT-4 | Neurotrophin 5, NTF4 | Cytokine |
| 108 | OSM | Oncostatin M | Cytokine |
| 109 | Osteopontin | SPP1 | Cytokine |
| 110 | PAI-1 | Plasminogen activator inhibitor type 1, serpin E1 | Enzyme inhibitor |
| 111 | PAI-II | Plasminogen activator inhibitor-II, serpin B2 | Enzyme inhibitor |
| 112 | PARC | Pulmonary and activation-regulated chemokine CCL18 | Cytokine |
| 113 | PDGF Rα | Platelet-derived growth factor receptor α CD140A | Cytokine receptor |
| 114 | PDGF Rβ | Platelet-derived growth factor receptor β CD140B | Cytokine receptor |
| 115 | PEDF | Pigment epithelium-derived factor, serpin F1 | Enzyme inhibitor |
| 116 | PF4 | Platelet factor-4, CXCL4 | Cytokine |
| 117 | PlGF | Placental growth factor, PGF | Cytokine |
| 118 | Prolactin | Prolactin | Hormone |
| 119 | Protein C | Protein C | Clotting factor |
| 120 | Protein S | Protein S | Clotting factor |
| 121 | P-selectin | P-selectin, CD62P | Adhesion molecule |
| 122 | RANTES | CCL5 | Cytokine |
| 123 | SCF | Stem cell factor, Kit ligand | Cytokine |
| 124 | sgp130 | Soluble glycoprotein 130, IL6ST, CD130 | Cytokine receptor |
| 125 | sVAP-1 | Soluble vascular adhesion protein-1, AOC3 | Adhesion molecule |
| 126 | sVCAM-1 | Soluble VCAM-1, CD106 | Adhesion molecular |
| 127 | TARC | Thymus and activation-regulated chemokine, CCl17 | Cytokine |
| 128 | TGF-α | Transforming growth factor α | Cytokine |
| 129 | TGF-β RIII | Transforming growth factor β receptor III | Cytokine receptor |
| 130 | Tie-2 | TEK, CD202b | Cytokine receptor |
| 131 | TIMP-2 | Tissue inhibitor of metalloproteinases 2 | Enzyme inhibitor |
| 132 | TNF-α | Tumor necrosis factor α, TNF | Cytokine |
| 133 | TNF-β | Tumor necrosis factor β | Cytokine |
| 134 | TNF-RI | TNF receptor superfamily, member 1A | Cytokine receptor |
| 135 | TRAILR1 | TNF receptor superfamily, member 10A | Cytokine receptor |
| 136 | TSH | Thyroid-stimulating hormone | Hormone |
| 137 | uPA | Urokinase plasminogen activator, PLAU | Clotting factor |
| 138 | uPAR | Urokinase plasminogen activator receptor | Clotting factor |
| 139 | VE-cadherin | Vascular endothelial cadherin, cadherin 5 | Adhesion molecular |
| 140 | VEGF | Vascular endothelial growth factor | Cytokine |
| 141 | VEGF-R3 | VEGF receptor 3, FLT4 | Cytokine receptor |
| 142 | VEGF-D | Vascular endothelial growth factor-D | Cytokine |
Antibody Microarray Assays
Samples were randomized to groups of six, and two groups were randomly assigned to each microarray slide. Each sample was evaluated in duplicate. Quality controls and standards were applied to the four remaining wells on each microarray slide as described previously (26). Assays were performed blinded to the biological identity of the samples. Rolling circle amplification, microarray, sandwich immunoassays were performed on an automated robotic microfluidic station (BioCube, Protedyne Inc., Windsor, CT) as described previously (26, 27). Briefly 15 μl of diluted sample was applied to each well, incubated, and washed. Next a mixture of biotinylated secondary antibodies was applied to each well, incubated, and washed. Then an anti-biotin antibody-DNA conjugate (an anti-biotin antibody with a covalently attached oligonucleotide and an annealed circular nucleotide) was applied to the arrays, incubated, and washed. Rolling circle amplification was then performed for 45 min using T7 native DNA polymerase as described previously (28) and detected with a Cy5-labeled complementary oligonucleotide probe as described previously (26). A Tecan LS200 scanner was used to assay fluorescent signals on the slides, and microarray images were quantified using the image capture software GenePix 4.0.
Data Processing
Data processing procedures were as described previously (26). Briefly data points producing outlier events as a result of missing spots, spots with poor morphology, or printed features demonstrating high pixel outliers were removed. Correlation between sample replicates was performed, and sample replicates with correlation coefficient (R2) values <0.95 were removed. Intensity values were log2-transformed to minimize dependence of variance on intensity and normalized by analysis of variance to reduce variability observed between replicates (26, 29–32). Sample pass rates following data processing procedures are indicated in Table II.
Table II.
Sample pass rate by array
Sample pass rates following the data processing procedures discussed under “Experimental Procedures” are indicated.
| Array | Number of passed samples
|
||
|---|---|---|---|
| One replicate | Two replicates | Total | |
| 1 | 20 | 85 | 105 |
| 2 | 21 | 70 | 91 |
| 4 | 4 | 77 | 81 |
| 5 | 16 | 83 | 99 |
| 6 | 12 | 74 | 86 |
Statistical Analyses
Single Factor Analysis—
Logarithm-transformed mean fluorescence intensities (MFIs) of analyte values in clinically infected and non-infected samples were compared by analysis of variance (ANOVA), Type III, sum of squares (SAS 8.2 general linear model procedure) with a significance level or p value <0.05 as described previously (26). Differences between clinically infected and non-infected samples were stated as effect sizes, which represent a measure of the difference in mean between the two groups, normalized by the within-group standard deviation. This measure is independent of the sample size and is calculated as follows: Estimated effect size = (Mean_Group1 − Mean_Group2)/S.D._Group1_Group2.
The estimated effect size is associated with the predictive ability of a particular analyte. For example, an effect size of 0.3 between two groups is equivalent to a probability of correctly identifying the groups of 0.56. The current study used an effect size ≥0.6 as the cutoff for statistical significance based upon previous studies involving analysis of clinical samples on 142-feature antibody arrays (26, 27).
Pairwise differences in logarithm-transformed MFIs of analyte values in infected neonates with cultures positive for coagulase-negative staphylococci, Escherichia coli, Candida, and group B streptococci were compared by ANOVA, LSMeans (SAS JMP Genomics 3.2) using a false discovery rate correction and a p value <0.05.
Multivariate Analysis—
Because of the rigorous quality controls applied during data processing, the data matrix of samples × analytes contained missing data (Table II). Unfortunately many multivariate analyses are unstable with missing values (33). Model-based methods (34) have long been used to impute values for missing data prior to multivariate analyses, but these methods can introduce bias from incorrectly specified models. To reduce the bias from an incorrectly specified model, we utilized a weighted K-nearest neighbor approach to impute missing values (35). Explicitly we imputed missing values based on corresponding data for the missing analyte from the 20 nearest neighbors irrespective of the clinical diagnosis of the sample. To minimize the impact of imputed data on the discriminant function, we also removed 27 samples and 19 analytes in which more than 25% of the data were imputed. A multivariate discriminant function was developed using the STEPDISC procedure of SAS applied to the average values of log-transformed analyte concentrations for 123 analytes from 80 samples (43 infected and 37 uninfected). The criteria for selection of analytes to enter or remove from the model were based on the significance of the F statistic set to α = 0.1. The ability of the resulting set of analytes to predict infection status was evaluated using the DISC procedure of SAS. Because of unequal and perhaps non-normal multivariate distributions within the infected and non-infected groups, the discriminant function was based on the nearest neighbor method in which K was set to 15. The resulting discriminant function was then tested for predictive value by 80 iterations of “leave-one-out” cross-validation.
RESULTS
Patient Population—
108 serum samples were obtained from 72 infants, comprising 51 samples from 35 clinically infected neonates (early and late onset infection) and 57 samples from 40 non-infected infants. 32 of 51 (63%) clinically infected samples were associated with positive microbiological culture results. Organisms identified in positive cultures were coagulase-negative staphylococci (n = 14), Staphylococcus aureus (n = 2), enterococci (n = 3), group B Streptococcus (n = 3), α hemolytic streptococci (n = 1), E. coli (n = 3), Enterococcus faecium (n = 1), and Candida spp. (n = 4). Univariate statistics revealed the clinically infected samples to be associated with a marginally higher white blood cell count (p < 0.05) and significantly greater C-reactive protein (p = 1.9 × e−25, F-test). Table III shows the demographic characteristics and laboratory values of these groups.
Table III.
Demographics of the study population (mean ± S.E.)
| Clinically infected | Clinically non-infected | |
|---|---|---|
| Number of samples | 51 | 57 |
| Gestational age (weeks) | 28.8 ± 0.6 | 30.2 ± 0.6 |
| White blood cell count | 15.1 ± 1.0 (n = 47) | 11.8 ± 0.8 (n = 42) |
| Platelet count | 241 ± 17 (n = 46) | 251 ± 22 (n = 40) |
| Positive microbiological culture | 32 | 0 |
| Antibiotic therapy | 47 | 35 |
| Duration of antibiotic therapy | 8.5 ± 0.94 (n = 47) | 3.0 ± 0.36 (n = 35) |
| C-reactive protein | 24.4 ± 8.0 (n = 41) | 3.4 ± 1.2 (n = 51) |
Assay Specifications—
Using sandwich immunoassays printed on glass microarrays (26–28), the levels of 142 proteins (Table I) were measured in 108 serum samples from premature neonates. The serum proteins assayed comprised 80 cytokines, 30 soluble cytokine receptors, 12 enzymes or enzyme inhibitors, seven adhesion molecules, six clotting factors, three hormones, and four miscellaneous antigens. The linear dynamic range, sensitivity, specificity, quality metrics, and performance of these antibody microarrays has been described in detail previously (26). Each serum sample was measured in duplicate on 16-well microarray slides following randomization to adjust for location-related bias in values. In addition, the four corner wells in each microarray slide were used to create standard curves for quality control purposes, and slide-to-slide precision was improved with regression-based normalization based upon these standard curves (26). The average slide-to-slide variability (coefficient of variation) was 24, 35, 28, 32, and 20% for Arrays 1, 2, 4, 5, and 6, respectively (Table IV). Analyte levels were expressed as logarithm base 2-transformed MFIs, which show a linear relationship with concentrations (pg/ml) within the linear dynamic range of the assays (26). Because log-transformed analyte concentrations (within the linear range) were of approximately equal scale and variance size, the assumptions underlying classical analysis of variance were approximately satisfied.
Table IV.
Slide-to-slide variability on a linear scale
| Array | CVa | Number of measurements | S.D. |
|---|---|---|---|
| % | |||
| 1 | 23.6 | 2611 | 17.7 |
| 2 | 34.5 | 2098 | 24.6 |
| 4 | 27.5 | 3208 | 21.3 |
| 5 | 31.6 | 2367 | 23.0 |
| 6 | 19.9 | 2681 | 15.1 |
Coefficient of variation.
Neonatal Infection Biomarker Discovery—
30 of 142 serum analytes showed a significant change (p value <0.05) in expression between clinically infected and non-infected patients based upon ANOVA using logarithm-transformed fluorescence intensities (Table V). 27 of these analytes were in the linear dynamic range of the sandwich immunoassay as evidenced by correlation between concentration (pg/ml) and MFI (data not shown). The measurements of the remaining three analytes (CD40, FGFR3 (IIIb), and IL-12 p40) were below the lower limit of reliable quantitation. For two analytes with statistically significant differences between clinically infected and non-infected samples by microarray immunoassay (CRP and ICAM-1), differences were confirmed by monoplex ELISA retesting (data not shown).
Table V.
Analytes with significantly different levels in clinically infected and non-infected patients
I-TAC, interferon γ-inducible T cell α chemoattractant; OSM, oncostatin M; PAI-II, plasminogen activator inhibitor-II, NGF, nerve growth factor.
| Analyte | Clinically infected
|
Non-infected
|
Effect size | p value | ||||
|---|---|---|---|---|---|---|---|---|
| Mean | S.D. | n | Mean | S.D. | n | |||
| E-selectin | 10.66 | 1.57 | 42 | 9.26 | 1.54 | 51 | 0.90 | 0.00004 |
| IL-18 | 8.38 | 1.55 | 47 | 7.24 | 0.99 | 51 | 0.88 | 0.00004 |
| IL-2sRα | 8.91 | 0.99 | 38 | 8.08 | 0.99 | 52 | 0.84 | 0.0002 |
| CRP | 12.88 | 0.83 | 47 | 12.09 | 1.15 | 51 | 0.77 | 0.0002 |
| uPAR | 9.54 | 0.88 | 45 | 8.95 | 0.61 | 39 | 0.77 | 0.0007 |
| uPA | 7.82 | 0.40 | 45 | 7.53 | 0.47 | 39 | 0.67 | 0.003 |
| P-selectin | 11.65 | 1.19 | 41 | 10.80 | 1.33 | 51 | 0.66 | 0.002 |
| NE | 13.04 | 0.98 | 47 | 12.37 | 1.25 | 49 | 0.60 | 0.004 |
| CA125 | 6.82 | 0.76 | 45 | 6.45 | 0.40 | 40 | 0.59 | 0.008 |
| ICAM-1 | 12.30 | 0.87 | 43 | 11.83 | 0.79 | 37 | 0.57 | 0.01 |
| OSM | 6.75 | 0.49 | 47 | 6.48 | 0.47 | 54 | 0.57 | 0.005 |
| PAI-II | 6.01 | 0.89 | 47 | 5.55 | 0.74 | 50 | 0.57 | 0.006 |
| CCL21 | 8.83 | 0.71 | 44 | 8.43 | 0.71 | 40 | 0.56 | 0.01 |
| ET-3 | 5.81 | 0.57 | 45 | 5.52 | 0.49 | 41 | 0.55 | 0.01 |
| ErbB2 | 9.07 | 0.37 | 43 | 8.87 | 0.39 | 40 | 0.55 | 0.01 |
| FGFR3 (IIIb) | 6.12 | 0.25 | 45 | 5.97 | 0.32 | 41 | 0.55 | 0.01 |
| IL-16 | 7.11 | 0.75 | 43 | 6.73 | 0.63 | 37 | 0.54 | 0.02 |
| MIP-3α | 8.22 | 1.42 | 41 | 7.58 | 0.99 | 38 | 0.51 | 0.03 |
| ErbB1 | 9.98 | 0.32 | 44 | 9.79 | 0.41 | 40 | 0.50 | 0.02 |
| Eot-2 | 9.48 | 1.27 | 36 | 8.90 | 1.09 | 44 | 0.49 | 0.03 |
| NGF-β | 8.49 | 0.75 | 43 | 8.12 | 0.83 | 36 | 0.48 | 0.04 |
| IL-1ra | 7.31 | 1.64 | 45 | 6.68 | 1.00 | 52 | 0.47 | 0.02 |
| I-TAC | 6.95 | 1.01 | 43 | 6.58 | 0.53 | 38 | 0.46 | 0.04 |
| MMP-8 | 12.33 | 1.88 | 41 | 11.41 | 2.06 | 50 | 0.46 | 0.03 |
| MIP-1β | 8.90 | 2.17 | 39 | 8.14 | 1.12 | 46 | 0.45 | 0.04 |
| CD40 | 5.12 | 0.44 | 48 | 4.92 | 0.46 | 51 | 0.44 | 0.03 |
| MPIF-1 | 6.42 | 0.77 | 49 | 6.13 | 0.58 | 55 | 0.44 | 0.03 |
| MCP-2 | 5.97 | 0.78 | 48 | 5.70 | 0.51 | 54 | 0.42 | 0.04 |
| IL-12p40 | 5.68 | 0.59 | 46 | 5.47 | 0.47 | 51 | 0.40 | 0.05 |
| MMP-2 | 13.55 | 0.32 | 44 | 13.70 | 0.30 | 41 | −0.48 | 0.03 |
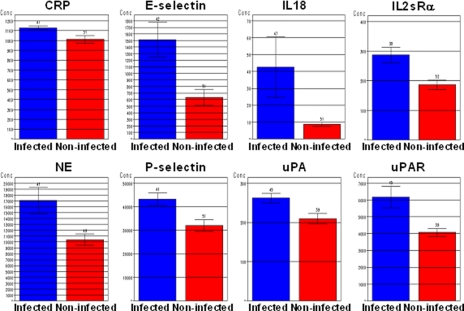
Eight of the 27 analytes (E-selectin, P-selectin, IL-18, IL-2 soluble receptor α chain (IL-2sRα), CRP, urokinase plasminogen activator (uPA), urokinase plasminogen-activated receptor (uPAR), and NE) showed effect sizes greater than 0.6 and mean intensity values in the optimal range of measurement and demonstrated similar trends between fluorescence and derived concentration data. Additionally an interval censored log-linear model of concentration versus group membership resulted in significant difference between infected and non-infected patients for these analytes (data not shown). Fig. 1 shows average concentration (pg/ml) per patient group for each of these eight analytes.
Fig. 1.
Eight serum analytes with greatest differences in levels between clinically infected and non-infected neonates. Each analyte had significant differences between clinically infected and non-infected neonates (p < 0.05 by ANOVA and effect size >0.6 in log-transformed level). Group mean analyte levels are expressed in pg/ml with S.E. (bars).
Multivariate Classification—
Although average values of log-transformed analyte concentrations for several individual analytes showed significant differences between infected and non-infected analytes, single analytes were insufficient for prediction of infection status (data not shown). To develop a predictive model we investigated multivariate functions for their ability to discriminantly predict infection status.
Several multivariate classifier panels were obtained using different parameter settings in discriminant analyses. All contained analytes that exhibited significant group differences in univariate analyses, such as uPAR and uPA. Certain classifiers that predicted infection and non-infection correctly in cross-validation studies were rejected because of overfitting concerns due to featuring relatively large numbers of analytes (data not shown). Others were rejected because they featured well annotated analytes that are established biomarkers in other conditions (e.g. CA125). A single classifier was chosen that featured the smallest number of analytes and greatest number of analytes with significant group differences in univariate analyses (n = 5) and that did not contain analytes lacking biological congruence for neonatal sepsis. This classifier consisted of 13 analytes (analyte, partial R2 value; CD27, 0.06; CD40, 0.05; ciliary neurotrophic factor receptor α, 0.06; FGF6, 0.10; GRO-γ, 0.07; interferon α, 0.05; MCP-3, 0.07; MMP-2, 0.09; P-selectin, 0.09; plasminogen activator inhibitor-II, 0.06; VAP1, 0.08; VEGF-R3, 0.06; uPA, 0.06). This classifier predicted infection in 38 of 43 infected neonates (P(i|i) 0.88) and predicted absence of infection in 31 of 37 uninfected neonates (P(u|u) 0.84). In cross-validation analyses, the classifier predicted infection in 38 of 43 infected neonates (validateP(i|i) 0.88) and predicted absence of infection in 30 of 37 uninfected neonates (validateP(u|u) 0.81).
Differentiation of Etiologic Agent Class by Examination of Host Serum Protein Levels—
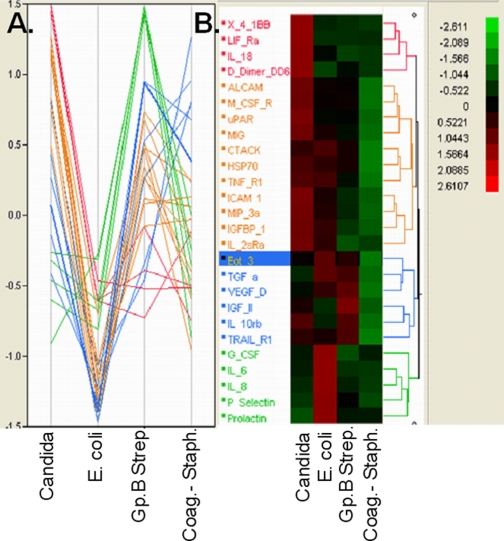
Host biomarkers for early differentiation of etiologic agent in neonates with bacteremia were also sought. Log-transformed levels of serum analytes in three neonatal infections with cultures positive for group B streptococci, four with Candida, 16 with coagulase-negative staphylococci, and three with E. coli were compared. Hierarchical clustering and parallel plots of analytes with largest standardized LSMeans effects are shown in Fig. 2. Host responses differed widely between etiologic agents. Coliform infections evoked little change in the serum analytes examined. In contrast, levels of the proinflammatory cytokines IL-6, IL-8 (CXCL8), MCP1 (CCL2), G-CSF, P-selectin, and prolactin were much higher in group B streptococcal bacteremia than other neonatal infections: average IL-8 levels in group B streptococcal infections were 112,130 pg/ml compared with 40 pg/ml in candidemia and 44 pg/ml in Staphylococcus epidermidis bacteremia. Average IL-6 levels were 270,636 pg/ml in group B streptococcal bacteremia, 18 pg/ml in candidemia, and 45 pg/ml in S. epidermidis. MCP1 differentiated candidemia (177 pg/ml) from group B streptococcal bacteremia or S. epidermidis (216,700 and 109,827 pg/ml, respectively). Fungal infections were associated with elevated levels of the costimulatory receptor 4-1BB, IL-18, IL-2sRα D dimer, uPAR, and chemokines monokine induced by interferon γ (MIG; CXCL9) and MIP3α (CCL23) among others. Cultures positive for coagulase-negative staphylococci were associated with elevated insulin-like growth factor II and TRAILR1.
Fig. 2.
Parallel plot (A) and dendrogram of hierarchical clustering (B) of analytes with largest standardized LSMeans effects in pairwise comparisons of infected neonates with microbiologic cultures positive for coagulase-negative staphylococci (Coag.− Staph.), E. coli, Candida, and group B streptococci (Gp.B Strep.). Eotaxin 3 is highlighted; it is elevated in group B streptococcal infection and depressed in E. coli infection. See Table I for protein abbreviations and full names.
DISCUSSION
A retrospective clinical study was undertaken to identify a serum biomarker panel to diagnose infection in premature neonates. Antibody microarrays were used to measure the levels of 142 small serum proteins in 107 infected and non-infected patients. Amplified antibody microarrays have several potential advantages over conventional monoplex immunoassays or mass spectrometry-based proteomics assays. Sample volumes available from premature neonates preclude use of conventional immunoassays. The small mass and low abundance (pg/ml) of the analytes together with “masking” by more abundant serum proteins diminish the usefulness of mass spectrometry-based proteomics assays. Sensitivity, specificity, linear dynamic range, and coefficients of variation of miniaturized, multiplexed immunoassays can approach that of monoplex sandwich immunoassays (26). However, very limited sample volumes in the present study required sample dilution and performance of assays in duplicate (rather than triplicate), which impaired sample pass rate (Table II) and coefficients of variation (Table IV). Furthermore the present study was undertaken before some recently reported improvements in the standardization of array manufacture and assays were developed (26), and many of the analytes tested have labile immunogenicity. Nevertheless significant differences between groups were observed for 30 of 142 analytes (Table V). Two of these (CRP and ICAM-1) were also measured by conventional, monoplex immunoassays, yielding congruent results. Eight analytes met additional, more stringent conditions for elevation in serum of infected premature neonates (effect sizes greater than 0.6, mean intensity values in the linear range of measurement, and demonstration of similar trends between fluorescence and derived concentration values). They were P-selectin (CD62P), E-selectin (CD62E), IL-2sRα (CD25), IL-18, NE, uPA, uPAR (CD87), and CRP (Fig. 1).
CRP is a critical, phylogenetically ancient component of the innate immune response. Upon CRP recognition and binding to phosphocholine (a phospholipid expressed on the membranes of many bacteria and damaged host cell walls), the acute phase inflammatory response is initiated (36). Accordingly increased levels of CRP are observed early in response to severe bacterial infection (37). Plasma CRP levels were found to be statistically significantly higher in patients with septic syndrome symptoms, and CRP serum concentration correlated with severity of a patient's clinical state (38). In fatal cases high plasma CRP concentration was recorded during hospitalization, whereas in cases of recovery, rapid CRP decrease was noted (38). In our study, CRP was found to be increased in neonatal patients with infection. Therefore, plasma CRP concentration in patients with bacterial sepsis may be helpful in evaluation of clinical state severity and monitoring of the disease course as well as therapy efficacy. Previously we have shown that plasma CRP > 6 mg/liter has a high sensitivity for clinical infection (95%) and NPV of 97% (12). As a classical acute phase reactant, however, CRP elevation alone has insufficient specificity for diagnosis of neonatal infection.
P- and E-selectins are related cell adhesion molecules expressed and secreted by activated endothelial cells (P- and E-selectins) and platelets (P-selectin). Both P- and E-selectins are components of an adhesion cascade that leads to leukocyte and platelet accumulation at sites of inflammation, infection, and/or injury. Uncontrolled adhesiveness in the microcirculatory system leads to tissue hypoxia (cryptic shock) and eventual organ dysfunction, hallmarks of severe sepsis and septic shock (39). Previous studies of P-selectin have demonstrated elevations in serum and platelets in adult sepsis and septic shock and in platelets in neonatal group B Streptococcus sepsis (40–42). However, only elevated levels of E- (but not P-) selectin have been previously documented in neonatal sepsis (43–45).
In part as a result of E-selectin elevation, another early component of the host response to infection is recruitment of neutrophils. Accordingly it was not surprising to find elevated NE in infected neonates. NE is the major serine proteinase secreted by activated neutrophils (or released by damaged neutrophils), and several previous publications have documented NE as an infection parameter for neonatal diagnosis (14, 46–50).
Further evidence of altered endothelial adhesiveness is increased levels of coagulation/fibrinolysis cascade proteins: uPA, an extracellular serine protease, is involved in the catalytic conversion of plasminogen to the active fibrinolytic enzyme plasmin (51). uPA is activated upon binding to its cognate receptor, uPAR, which is a membrane-linked receptor with extracellular protease activity that transduces intracellular signaling pathways. Although in the healthy state, the balance of coagulation and fibrinolysis is tightly regulated, during sepsis coagulation and fibrinolysis are dysregulated as evidenced, for example, by consumption of plasminogen and protein C, elevation of plasminogen activator inhibitor, and occurrence of disseminated intravascular coagulation. Our study, in agreement with others, found increased circulating levels of uPA in the septic state (51). In addition, increased levels of soluble uPAR in response to a variety of infections have been reported and are an indicator of good prognosis in sepsis (52–56). A significant molar excess was observed in the increase in level of uPAR relative to uPA. This raises the possibility that, in neonatal infection, elevated uPAR may act to sequester uPA (despite elevated levels of uPA) and prevent it from binding the membrane-linked uPAR, thus abrogating normal intracellular signaling. Interestingly NE also activates fibrinolysis during sepsis, and its elevation has been suggested as a homeostatic mechanism to overcome impaired fibrinolysis in sepsis (57).
IL-2sRα is a circulating form of one of three components of the high affinity membrane receptor for interleukin 2 present on activated T and B cell thymocyte subsets, pre-B cells and regulatory T cells. Although the biological function of IL-2sR is not completely understood, it is known to indicate T lymphocyte activation during disease. Several previous studies have demonstrated elevated serum levels of IL-2sRα in neonatal sepsis (39–44).
IL-18 is a proinflammatory member of the IL-1 cytokine superfamily. Although IL-18 elevation has not been described previously in neonatal sepsis, recent studies have associated it with other disease morbidity. Neonatal necrotizing enterocolitis (58) and respiratory syncytial virus (59) infection have been associated with genetic polymorphisms in IL-18. In a mouse model, elevated IL-18 levels have been reported in neonatal group B streptococcal infections (60). Furthermore elevated levels of IL-18 have been documented in adult sepsis patients and correlate with adverse outcome and Acute Physiology and Chronic Health Evaluation (APACHE) II score (45, 46).
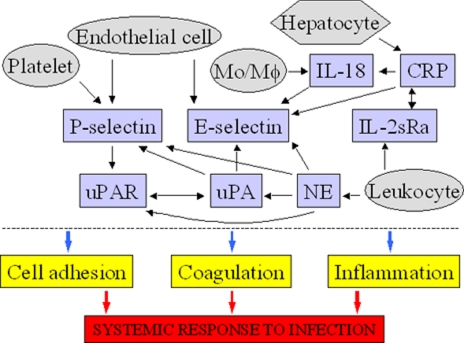
Interestingly it appears that the eight analytes identified as elevated in the infected neonates function in a network that links several processes, which are well known to be important in the response to systemic infection, e.g. coagulation, cell adhesion, and inflammation (Fig. 3). For example, it has been shown that IL-18 induces E-selectin expression on endothelial cells (61); IL-18 release is stimulated by CRP (62). CRP also induces significant E-selectin expression in human endothelial cells (63). Similarly the endothelial expression of E-selectin was increased by human neutrophil elastase (64). In contrast, uPA (urokinase) administration reduces the serum levels of soluble E-selectin in patients with acute myocardial infarction (65, 66). The presence of uPA increases platelet surface P-selectin expression in a concentration-dependent manner (67). It has also been suggested that intravascular fibrinolysis induced by uPA may induce P-selectin (68). uPA itself, as well as uPAR, are cleaved by neutrophil elastase (69, 70). Adhesion of human monocytes to P-selectin via its surface ligand, P-selectin glycoprotein 1, induced uPAR expression (71). Other studies have reported additional links between the above molecules, e.g. colocalization of neutrophil elastase, uPA, and uPAR (72) or colocalization of uPA and IL-2sRα (73). Finally pathogens are known to interact in a specific manner with components of fibrinolytic pathways as well as with CRP (74).
Fig. 3.
Eight serum proteins (P-selectin, E-selectin, IL-2sRα, IL-18, NE, uPA, uPAR, and CRP) that are elevated in infected neonates function in a molecular network that links coagulation/fibrinolysis, cell adhesion, and inflammation, which are critical determinants of the response to systemic infection. Mo/Mφ, monocytes macrophages.
Multivariate analyses were undertaken to derive a multiplexed biomarker panel with improved diagnostic sensitivity and specificity relative to single analyte determinations. Derivation of a multivariate classifier was impeded by the small number of patients studied and by missing values. Several methods for imputation of missing values were utilized but were unsatisfactory. A 13-analyte classifier was developed without missing value imputation whose membership had biological congruence with neonatal infection. In cross-validation analyses, however, the classifier predicted infection in only 38 of 43 infected neonates (validateP(i|i) 0.88) and absence of infection in only 30 of 37 uninfected neonates (validateP(u|u) 0.81). These values provide a proof of concept for the utility of host biomarker measurement in diagnosis of neonatal infection but are insufficient for routine diagnostic use and attest to the extreme heterogeneity of neonatal infection. Additional prospective studies are needed. Future studies should feature larger numbers of patients with matching of acute illness scores and other clinical covariates between groups and/or with synchronization of infection progression through use of a longitudinal study design. In addition, a variety of miniaturized, multiplexed immunoassay formats are now available with improved precision, and a large set of candidate biomarkers are available for testing. Such a study is currently in progress.
Finally host biomarkers for early differentiation of etiologic agent in neonatal infection were sought. Host responses were compared for the four most common species in positive microbiologic cultures: group B streptococci, Candida, E. coli, and coagulase-negative staphylococci. The rationale for differentiation of etiologic agent based on host serum protein levels is that etiologic agents differ predictably in lifestyle and membrane lipid and protein content, evoking characteristic host responses. We found unique signatures of host response to each etiologic agent (Fig. 2). For example, levels of the proinflammatory cytokines IL-6 and IL-8 were more than 1000-fold higher in group B streptococcal bacteremia than in other neonatal infections. These novel findings will require validation in larger case series but offer the exciting prospect of rapid differentiation of at least some etiologic agents in neonatal sepsis, which would enable early institution of targeted antimicrobial therapy. Differences of the magnitude observed with IL-6 and IL-8, if substantiated, would permit etiologic agent differentiation with rapid, point-of-care, semiqualitative immunoassays.
Acknowledgments
We thank the employees of Molecular Staging Inc. who developed amplified antibody microarrays.
Footnotes
Published, MCP Papers in Press, July 13, 2008, DOI 10.1074/mcp.M800175-MCP200
The abbreviations used are: IL, interleukin; TNF, tumor necrosis factor; NE, neutrophil elastase; CRP, C-reactive protein; G-CSF, granulocyte colony-stimulating factor; ICAM-1, intercellular adhesion molecule-1; NPV, negative predictive value; MFI, mean fluorescence intensity; ANOVA, analysis of variance; uPA, urokinase plasminogen activator; uPAR, urokinase plasminogen-activated receptor; IL-2sR, IL-2 soluble receptor.
J. D. M. Edgar, manuscript in preparation.
This work was supported, in whole or in part, by National Institutes of Health Grant AI066569 and Contract HHSN266200400064C. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
REFERENCES
- 1.McGuire, W., Clerihew, L., and Fowlie, P. W. ( 2004) Infection in the preterm infant. BMJ Clin. Res. Ed. 329, 1277–1280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stoll, B. J., Hansen, N., Fanaroff, A. A., Wright, L. L., Carlo, W. A., Ehrenkranz, R. A., Lemons, J. A., Donovan, E. F., Stark, A. R., Tyson, J. E., Oh, W., Bauer, C. R., Korones, S. B., Shankaran, S., Laptook, A. R., Stevenson, D. K., Papile, L. A., and Poole, W. K. ( 2002) Late-onset sepsis in very low birth weight neonates: the experience of the NICHD Neonatal Research Network. Pediatrics 110, 285–291 [DOI] [PubMed] [Google Scholar]
- 3.Baltimore, R. S. ( 2003) Neonatal sepsis: epidemiology and management. Paediatr. Drugs 5, 723–740 [DOI] [PubMed] [Google Scholar]
- 4.Gerdes, J. S. ( 1991) Clinicopathologic approach to the diagnosis of neonatal sepsis. Clin. Perinatol. 18, 361–381 [PubMed] [Google Scholar]
- 5.Malik, A., Hui, C. P., Pennie, R. A., and Kirpalani, H. ( 2003) Beyond the complete blood cell count and C-reactive protein: a systematic review of modern diagnostic tests for neonatal sepsis. Arch. Pediatr. Adolesc. Med. 157, 511–516 [DOI] [PubMed] [Google Scholar]
- 6.Polin, R. A. ( 2003) The “ins and outs” of neonatal sepsis. J. Pediatr. 143, 3–4 [DOI] [PubMed] [Google Scholar]
- 7.McCracken, G. J., and Freij, B. ( 1987) Perinatal bacterial diseases, in Textbook of Pediatric Infectious Diseases (Feigin, R., and Cherry, J., eds) 2nd Ed., pp. 940–966, W. B. Saunders, Philadelphia
- 8.Philip, A. G., and Hewitt, J. R. ( 1980) Early diagnosis of neonatal sepsis. Pediatrics 65, 1036–1041 [PubMed] [Google Scholar]
- 9.Kingsmore, S. F. ( 2006) Multiplexed protein measurement: technologies and applications of protein and antibody arrays. Nat. Rev. 5, 310–320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Frank, R., and Hargreaves, R. ( 2003) Clinical biomarkers in drug discovery and development. Nat. Rev. 2, 566–580 [DOI] [PubMed] [Google Scholar]
- 11.de Bont, E. S., Martens, A., van Raan, J., Samson, G., Fetter, W. P., Okken, A., de Leij, L. H., and Kimpen, J. L. ( 1994) Diagnostic value of plasma levels of tumor necrosis factor alpha (TNF alpha) and interleukin-6 (IL-6) in newborns with sepsis. Acta Paediatr. 83, 696–699 [DOI] [PubMed] [Google Scholar]
- 12.Edgar, J. D., Wilson, D. C., McMillan, S. A., Crockard, A. D., Halliday, M. I., Gardiner, K. R., Rowlands, B. J., Halliday, H. L., and McNeill, T. A. ( 1994) Predictive value of soluble immunological mediators in neonatal infection. Clin. Sci. (Lond.) 87, 165–171 [DOI] [PubMed] [Google Scholar]
- 13.Wilson, D., and Edgar, J. ( 1997) Predictors of bacterial infection in neonates. J. Paediatr. 130, 166. [DOI] [PubMed] [Google Scholar]
- 14.Philip, A. G., Tito, A. M., Gefeller, O., and Speer, C. P. ( 1994) Neutrophil elastase in the diagnosis of neonatal infection. Pediatr. Infect. Dis. J. 13, 323–326 [DOI] [PubMed] [Google Scholar]
- 15.Wasunna, A., Whitelaw, A., Gallimore, R., Hawkins, P. N., and Pepys, M. B. ( 1990) C-reactive protein and bacterial infection in preterm infants. Eur. J. Pediatr. 149, 424–427 [DOI] [PubMed] [Google Scholar]
- 16.Blanco, A., Solis, G., Arranz, E., Coto, G. D., Ramos, A., and Telleria, J. ( 1996) Serum levels of CD14 in neonatal sepsis by Gram-positive and Gram-negative bacteria. Acta Paediatr. 85, 728–732 [DOI] [PubMed] [Google Scholar]
- 17.Lehrnbecher, T., Schrod, L., Rutsch, P., Roos, T., Martius, J., and von Stockhausen, H. B. ( 1996) Immunologic parameters in cord blood indicating early-onset sepsis. Biol. Neonate 70, 206–212 [DOI] [PubMed] [Google Scholar]
- 18.Kennon, C., Overturf, G., Bessman, S., Sierra, E., Smith, K. J., and Brann, B. ( 1996) Granulocyte colony-stimulating factor as a marker for bacterial infection in neonates. J. Pediatr. 128, 765–769 [DOI] [PubMed] [Google Scholar]
- 19.Kourtis, A. P., Lee, F. K., and Stoll, B. J. ( 2003) Soluble L-selectin, a marker of immune activation, in neonatal infection. Clin. Immunol. (Orlando) 109, 224–228 [DOI] [PubMed] [Google Scholar]
- 20.Griffin, M. P., Lake, D. E., Bissonette, E. A., Harrell, F. E., Jr., O'Shea, T. M., and Moorman, J. R. ( 2005) Heart rate characteristics: novel physiomarkers to predict neonatal infection and death. Pediatrics 116, 1070–1074 [DOI] [PubMed] [Google Scholar]
- 21.Kingsmore, S. F., and Patel, D. D. ( 2003) Multiplexed protein profiling on antibody-based microarrays by rolling circle amplification. Curr. Opin. Biotechnol. 14, 74–81 [DOI] [PubMed] [Google Scholar]
- 22.Petricoin, E. F., III, Bichsel, V. E., Calvert, V. S., Espina, V., Winters, M., Young, L., Belluco, C., Trock, B. J., Lippman, M., Fishman, D. A., Sgroi, D. C., Munson, P. J., Esserman, L. J., and Liotta, L. A. ( 2005) Mapping molecular networks using proteomics: a vision for patient-tailored combination therapy. J. Clin. Oncol. 23, 3614–3621 [DOI] [PubMed] [Google Scholar]
- 23.Schweitzer, B., and Kingsmore, S. F. ( 2002) Measuring proteins on microarrays. Curr. Opin. Biotechnol. 13, 14–19 [DOI] [PubMed] [Google Scholar]
- 24.Edgar, D., Gabriel, V., Craig, A., Wheeler, D., Thomas, M., and Grant, J. ( 2002) A low serum sICAM-1 level may assist in the exclusion of neonatal infection. Biol. Neonate 81, 105–108 [DOI] [PubMed] [Google Scholar]
- 25.Ng, P. C. ( 2004) Diagnostic markers of infection in neonates. Arch. Dis. Child. Fetal Neonatal Ed. 89, F229–F235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Perlee, L., Christiansen, J., Dondero, R., Grimwade, B., Lejnine, S., Mullenix, M., Shao, W., Sorette, M., Tchernev, V., Patel, D., and Kingsmore, S. ( 2004) Development and standardization of multiplexed antibody microarrays for use in quantitative proteomics. Proteome Sci. 2, 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schweitzer, B., Roberts, S., Grimwade, B., Shao, W., Wang, M., Fu, Q., Shu, Q., Laroche, I., Zhou, Z., Tchernev, V. T., Christiansen, J., Velleca, M., and Kingsmore, S. F. ( 2002) Multiplexed protein profiling on microarrays by rolling-circle amplification. Nat. Biotechnol. 20, 359–365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schweitzer, B., Wiltshire, S., Lambert, J., O'Malley, S., Kukanskis, K., Zhu, Z., Kingsmore, S. F., Lizardi, P. M., and Ward, D. C. ( 2000) Inaugural article: immunoassays with rolling circle DNA amplification: a versatile platform for ultrasensitive antigen detection. Proc. Natl. Acad. Sci. U. S. A. 97, 10113–10119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Geller, S. C., Gregg, J. P., Hagerman, P., and Rocke, D. M. ( 2003) Transformation and normalization of oligonucleotide microarray data. Bioinformatics (Oxf.) 19, 1817–1823 [DOI] [PubMed] [Google Scholar]
- 30.Huber, W., von Heydebreck, A., Sultmann, H., Poustka, A., and Vingron, M. ( 2002) Variance stabilization applied to microarray data calibration and to the quantification of differential expression. Bioinformatics (Oxf.) 18, Suppl. 1, S96–S104 [DOI] [PubMed] [Google Scholar]
- 31.Kerr, M. K., Martin, M., and Churchill, G. A. ( 2000) Analysis of variance for gene expression microarray data. J. Comput. Biol. 7, 819–837 [DOI] [PubMed] [Google Scholar]
- 32.Rocke, D. M., and Durbin, B. ( 2003) Approximate variance-stabilizing transformations for gene-expression microarray data. Bioinformatics (Oxf.) 19, 966–972 [DOI] [PubMed] [Google Scholar]
- 33.Liu, L., Hawkins, D. M., Ghosh, S., and Young, S. S. ( 2003) Robust singular value decomposition analysis of microarray data. Proc. Natl. Acad. Sci. U. S. A. 100, 13167–13172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wilkinson, G. N. ( 1958) Estimation of missing values for the analysis of incomplete data. Biometrics 14, 257–286 [Google Scholar]
- 35.Troyanskaya, O., Cantor, M., Sherlock, G., Brown, P., Hastie, T., Tibshirani, R., Botstein, D., and Altman, R. B. ( 2001) Missing value estimation methods for DNA microarrays. Bioinformatics (Oxf.) 17, 520–525 [DOI] [PubMed] [Google Scholar]
- 36.Gewurz, H., Mold, C., Siegel, J., and Fiedel, B. ( 1982) C-reactive protein and the acute phase response. Adv. Int. Med. 27, 345–372 [PubMed] [Google Scholar]
- 37.Kanda, T. ( 2001) C-reactive protein (CRP) in the cardiovascular system. Rinsho Byori 49, 395–401 [PubMed] [Google Scholar]
- 38.Kepa, L., and Oczko-Grzesik, B. ( 2001) Usefulness of plasma C-reactive protein (CRP) estimation in patients with bacterial sepsis. Przegl. Epidemiol. 55, Suppl. 3, 63–67 [PubMed] [Google Scholar]
- 39.Parent, C., and Eichacker, P. Q. ( 1999) Neutrophil and endothelial cell interactions in sepsis. The role of adhesion molecules. Infect. Dis. Clin. N. Am. 13, 427–447 [DOI] [PubMed] [Google Scholar]
- 40.Fijnheer, R., Frijns, C. J., Korteweg, J., Rommes, H., Peters, J. H., Sixma, J. J., and Nieuwenhuis, H. K. ( 1997) The origin of P-selectin as a circulating plasma protein. Thromb. Haemostasis 77, 1081–1085 [PubMed] [Google Scholar]
- 41.Nakamura, T., Ebihara, I., Shoji, H., Ushiyama, C., Suzuki, S., and Koide, H. ( 1999) Treatment with polymyxin B-immobilized fiber reduces platelet activation in septic shock patients: decrease in plasma levels of soluble P-selectin, platelet factor 4 and beta-thromboglobulin. Inflamm. Res. 48, 171–175 [DOI] [PubMed] [Google Scholar]
- 42.Siauw, C., Kobsar, A., Dornieden, C., Beyrich, C., Schinke, B., Schubert-Unkmeir, A., Abele-Horn, M., Speer, C. P., and Eigenthaler, M. ( 2006) Group B streptococcus isolates from septic patients and healthy carriers differentially activate platelet signaling cascades. Thromb. Haemostasis 95, 836–849 [DOI] [PubMed] [Google Scholar]
- 43.Austgulen, R., Arntzen, K. J., Haereid, P. E., Aag, S., and Dollner, H. ( 1997) Infections in neonates delivered at term are associated with increased serum levels of ICAM-1 and E-selectin. Acta Paediatr. 86, 274–280 [DOI] [PubMed] [Google Scholar]
- 44.Dollner, H., Vatten, L., and Austgulen, R. ( 2001) Early diagnostic markers for neonatal sepsis: comparing C-reactive protein, interleukin-6, soluble tumour necrosis factor receptors and soluble adhesion molecules. J. Clin. Epidemiol. 54, 1251–1257 [DOI] [PubMed] [Google Scholar]
- 45.Ng, P. C., Cheng, S. H., Chui, K. M., Fok, T. F., Wong, M. Y., Wong, W., Wong, R. P., and Cheung, K. L. ( 1997) Diagnosis of late onset neonatal sepsis with cytokines, adhesion molecule, and C-reactive protein in preterm very low birthweight infants. Arch. Dis. Child. Fetal Neonatal. Ed. 77, F221–F227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fischer, J. E., Brunner, A., Janousek, M., Nadal, D., Blau, N., and Fanconi, S. ( 2000) Diagnostic potential of neutrophil elastase inhibitor complex in the routine care of critically ill newborn infants. Eur. J. Pediatr. 159, 659–662 [DOI] [PubMed] [Google Scholar]
- 47.Jensen, J. G., Madsen, P., Rix, M., Rosthoj, S., and Ebbesen, F. ( 1996) Capillary plasma neutrophil elastase alpha-1-proteinase inhibitor as infection parameter in neonates. Scand. J. Clin. Lab. Investig. 56, 37–40 [DOI] [PubMed] [Google Scholar]
- 48.Ksssler, A., Grunert, C., and Wood, W. G. ( 1994) The limitations and usefulness of C-reactive protein and elastase-alpha 1-proteinase inhibitor complexes as analytes in the diagnosis and follow-up of sepsis in newborns and adults. Eur. J. Clin. Chem. Clin. Biochem. 32, 365–368 [DOI] [PubMed] [Google Scholar]
- 49.Salzer, H. R., Pollak, A., Herkner, K., Weninger, M., and Schemper, W. ( 1993) Value of measurement of neutrophil elastase-alpha 1 proteinase inhibitor levels in the early diagnosis of neonatal infection. Eur. J. Clin. Microbiol. Infect. Dis. 12, 553–556 [DOI] [PubMed] [Google Scholar]
- 50.Wunderer, G., Rohrig, E., Suschke, H., and Walter, I. ( 1989) Leukocyte elastase as an indicator for early detection of neonatal infection. Gynakol. Rundsch. 29, Suppl. 2, 161–162 [PubMed] [Google Scholar]
- 51.Abraham, E., Gyetko, M. R., Kuhn, K., Arcaroli, J., Strassheim, D., Park, J. S., Shetty, S., and Idell, S. ( 2003) Urokinase-type plasminogen activator potentiates lipopolysaccharide-induced neutrophil activation. J. Immunol. 170, 5644–5651 [DOI] [PubMed] [Google Scholar]
- 52.Dekkers, P. E., ten Hove, T., te Velde, A. A., van Deventer, S. J., and van Der Poll, T. ( 2000) Upregulation of monocyte urokinase plasminogen activator receptor during human endotoxemia. Infect. Immun. 68, 2156–2160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ostrowski, S. R., Plomgaard, P., Fischer, C. P., Steensberg, A. S., Moller, K., Hoyer-Hansen, G., Pedersen, B. K., and Ullum, H. ( 2005) Interleukin-6 infusion during human endotoxaemia inhibits in vitro release of the urokinase receptor from peripheral blood mononuclear cells. Scand. J. Immunol. 61, 197–206 [DOI] [PubMed] [Google Scholar]
- 54.Eugen-Olsen, J., Gustafson, P., Sidenius, N., Fischer, T. K., Parner, J., Aaby, P., Gomes, V. F., and Lisse, I. ( 2002) The serum level of soluble urokinase receptor is elevated in tuberculosis patients and predicts mortality during treatment: a community study from Guinea-Bissau. Int. J. Tuberc. Lung Dis. 6, 686–692 [PubMed] [Google Scholar]
- 55.Ostergaard, C., Benfield, T., Lundgren, J. D., and Eugen-Olsen, J. ( 2004) Soluble urokinase receptor is elevated in cerebrospinal fluid from patients with purulent meningitis and is associated with fatal outcome. Scand. J. Infect. Dis. 36, 14–19 [DOI] [PubMed] [Google Scholar]
- 56.Sidenius, N., Sier, C. F., Ullum, H., Pedersen, B. K., Lepri, A. C., Blasi, F., and Eugen-Olsen, J. ( 2000) Serum level of soluble urokinase-type plasminogen activator receptor is a strong and independent predictor of survival in human immunodeficiency virus infection. Blood 96, 4091–4095 [PubMed] [Google Scholar]
- 57.Gando, S., Hayakawa, M., Sawamura, A., Hoshino, H., Oshiro, A., Kubota, N., and Jesmin, S. ( 2007) The activation of neutrophil elastase-mediated fibrinolysis is not sufficient to overcome the fibrinolytic shutdown of disseminated intravascular coagulation associated with systemic inflammation. Thromb. Res. 121, 67–73 [DOI] [PubMed] [Google Scholar]
- 58.Treszl, A., Tulassay, T., and Vasarhelyi, B. ( 2006) Genetic basis for necrotizing enterocolitis—risk factors and their relations to genetic polymorphisms. Front. Biosci. 11, 570–580 [DOI] [PubMed] [Google Scholar]
- 59.Puthothu, B., Krueger, M., Forster, J., Heinze, J., Weckmann, M., and Heinzmann, A. ( 2007) Interleukin (IL)-18 polymorphism 133C/G is associated with severe respiratory syncytial virus infection. Pediatr. Infect. Dis. J. 26, 1094–1098 [DOI] [PubMed] [Google Scholar]
- 60.Cusumano, V., Midiri, A., Cusumano, V. V., Bellantoni, A., De Sossi, G., Teti, G., Beninati, C., and Mancuso, G. ( 2004) Interleukin-18 is an essential element in host resistance to experimental group B streptococcal disease in neonates. Infect. Immun. 72, 295–300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Morel, J. C., Park, C. C., Woods, J. M., and Koch, A. E. ( 2001) A novel role for interleukin-18 in adhesion molecule induction through NFκB and phosphatidylinositol (PI) 3-kinase-dependent signal transduction pathways. J. Biol. Chem. 276, 37069–37075 [DOI] [PubMed] [Google Scholar]
- 62.Yamaoka-Tojo, M., Tojo, T., Masuda, T., Machida, Y., Kitano, Y., Kurosawa, T., and Izumi, T. ( 2003) C-reactive protein-induced production of interleukin-18 in human endothelial cells: a mechanism of orchestrating cytokine cascade in acute coronary syndrome. Heart Vessels 18, 183–187 [DOI] [PubMed] [Google Scholar]
- 63.Pasceri, V., Willerson, J. T., and Yeh, E. T. ( 2000) Direct proinflammatory effect of C-reactive protein on human endothelial cells. Circulation 102, 2165–2168 [DOI] [PubMed] [Google Scholar]
- 64.Nozawa, F., Hirota, M., Okabe, A., Shibata, M., Iwamura, T., Haga, Y., and Ogawa, M. ( 2000) Elastase activity enhances the adhesion of neutrophil and cancer cells to vascular endothelial cells. J. Surg. Res. 94, 153–158 [DOI] [PubMed] [Google Scholar]
- 65.Squadrito, F., Saitta, A., Altavilla, D., Ioculano, M., Canale, P., Campo, G. M., Squadrito, G., Di Tano, G., Mazzu, A., and Caputi, A. P. ( 1996) Thrombolytic therapy with urokinase reduces increased circulating endothelial adhesion molecules in acute myocardial infarction. Inflamm. Res. 45, 14–19 [DOI] [PubMed] [Google Scholar]
- 66.Squadrito, F., Altavilla, D., Ioculano, M., Canale, P., Campo, G. M., Squadrito, G., Ditano, G., Freni, F., Saitta, A., and Caputi, A. P. ( 1995) Soluble E-selectin levels in acute human myocardial infarction. Int. J. Microcirc. Clin. Exp. 15, 80–84 [DOI] [PubMed] [Google Scholar]
- 67.Kawano, K., Aoki, I., Aoki, N., Homori, M., Maki, A., Hioki, Y., Hasumura, Y., Terano, A., Arai, T., Mizuno, H., and Ishikawa, K. ( 1998) Human platelet activation by thrombolytic agents: effects of tissue-type plasminogen activator and urokinase on platelet surface P-selectin expression. Am. Heart J. 135, 268–271 [DOI] [PubMed] [Google Scholar]
- 68.Holvoet, P., and Collen, D. ( 1997) Thrombosis and atherosclerosis. Curr. Opin. Lipidol. 8, 320–328 [DOI] [PubMed] [Google Scholar]
- 69.Beaufort, N., Leduc, D., Rousselle, J. C., Magdolen, V., Luther, T., Namane, A., Chignard, M., and Pidard, D. ( 2004) Proteolytic regulation of the urokinase receptor/CD87 on monocytic cells by neutrophil elastase and cathepsin G. J. Immunol. 172, 540–549 [DOI] [PubMed] [Google Scholar]
- 70.Moir, E., Robbie, L. A., Bennett, B., and Booth, N. A. ( 2002) Polymorphonuclear leucocytes have two opposing roles in fibrinolysis. Thromb. Haemostasis 87, 1006–1010 [PubMed] [Google Scholar]
- 71.Mahoney, T. S., Weyrich, A. S., Dixon, D. A., McIntyre, T., Prescott, S. M., and Zimmerman, G. A. ( 2001) Cell adhesion regulates gene expression at translational checkpoints in human myeloid leukocytes. Proc. Natl. Acad. Sci. U. S. A., 98, 10284–10289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Pedersen, T. L., Plesner, T., Horn, T., Hoyer-Hansen, G., Sorensen, S., and Hansen, N. E. ( 2000) Subcellular distribution of urokinase and urokinase receptor in human neutrophils determined by immunoelectron microscopy. Ultrastruct. Pathol. 24, 175–182 [DOI] [PubMed] [Google Scholar]
- 73.Falkenberg, M., Giglio, D., Bjornheden, T., Nygren, H., and Risberg, B. ( 1998) Urokinase plasminogen activator colocalizes with CD25+ cells in atherosclerotic vessels. J. Vasc. Res. 35, 318–324 [DOI] [PubMed] [Google Scholar]
- 74.Bergmann, S., and Hammerschmidt, S. ( 2007) Fibrinolysis and host response in bacterial infections. Thromb. Haemostasis 98, 512–520 [PubMed] [Google Scholar]