Abstract
The enormous dynamic range of human bodily fluid proteomes poses a significant challenge for current MS-based proteomics technologies as it makes it especially difficult to detect low abundance proteins in human biofluids such as blood plasma, which is an essential aspect for successful biomarker discovery efforts. Here we present a novel tandem IgY12-SuperMix immunoaffinity separation system for enhanced detection of low abundance proteins in human plasma. The tandem IgY12-SuperMix system separates ∼60 abundant proteins from the low abundance proteins in plasma, allowing for significant enrichment of low abundance plasma proteins in the SuperMix flow-through fraction. High reproducibility of the tandem separations was observed in terms of both sample processing recovery and LC-MS/MS identification results based on spectral count data. The ability to quantitatively measure differential protein abundances following application of the tandem separations was demonstrated by spiking six non-human standard proteins at three different levels into plasma. A side-by-side comparison between the SuperMix flow-through and IgY12 flow-through samples analyzed by both one- and two-dimensional LC-MS/MS revealed a 60–80% increase in proteome coverage as a result of the SuperMix separations, suggesting significantly enhanced detection of low abundance proteins. A total of 695 plasma proteins were confidently identified in a single analysis (with a minimum of two peptides per protein) by coupling the tandem separation strategy with two-dimensional LC-MS/MS, including 42 proteins with reported normal concentrations of ∼100 pg/ml to 100 ng/ml. The concentrations of two selected proteins, macrophage colony-stimulating factor 1 and matrix metalloproteinase-8, were independently validated by ELISA as 202 pg/ml and 12.4 ng/ml, respectively. Evaluation of binding efficiency revealed that 45 medium abundance proteins were efficiently captured by the SuperMix column with >90% retention. Taken together, these results illustrate the potential broad utilities of this tandem IgY12-SuperMix strategy for proteomics applications involving human biofluids where effectively addressing the dynamic range challenge of the specimen is imperative.
There has been tremendous interest in using advanced proteomics technologies to analyze human bodily fluids such as plasma and serum for the purpose of discovering and verifying new candidate protein biomarkers applicable to different diseases (1, 2). These technologies are challenged to detect low abundance physiologically relevant proteins with extremely wide dynamic ranges in concentrations (i.e., more than 10 orders of magnitude for protein concentrations reported in human plasma) (3, 4). Despite significant recent advances, current proteomics technologies still fall short of being able to reliably detect in blood plasma low ng/ml to sub-ng/ml protein concentrations, a level of detection often required for discovering disease-specific biomarkers (4). Many different fractionation/separation techniques have been developed and applied in a multidimensional fashion to enhance detection of low abundance proteins in human biofluids (5, 6). One of the most commonly applied strategies to potentially alleviate the “masking” effect created by the presence of the highly abundant proteins is to remove them (7). In the human plasma proteome, 22 proteins are responsible for 99% of the bulk mass of the total protein content in human plasma; this leaves perhaps hundreds of thousands of other proteins in only 1% of the protein mass (3). As a result, effective strategies for removing these 22 proteins (and possibly other medium abundance proteins) should greatly enhance detection of low abundance proteins within this important biofluid proteome.
Multicomponent immunoaffinity separation strategies are increasingly being applied in various biomarker discovery applications to remove the abundant proteins and achieve comprehensive surveys of the biofluid proteomes. In such approaches, affinity-purified polyclonal antibodies typically immobilized by cross-linking on either chromatographic matrices or microbeads are used as immunoaffinity reagents to specifically remove abundant proteins. An optimized mixture of different antibody-immobilized beads for targeting a number of proteins within the partitioning column allows for simultaneous and efficient capture of multiple proteins. The principle of multicomponent immunoaffinity separation was initially demonstrated by Pieper et al. (8) when they illustrated a capability for removing 10 high abundance proteins in a single step. At present, several commercially available products are available for simultaneously removing multiple abundant proteins, including the Agilent (Palo Alto, CA) Multiple Affinity Removal System (MARS) (9), GenWay Seppro® IgY12 system (10), and Sigma ProteoPrep® 20 that can separate 7, 12, and 20 human plasma proteins, respectively. These antibody-based separation systems have been demonstrated to be highly efficient for removing the specifically targeted proteins as well as being both reproducible and selective (7, 11, 12). Although the current immunoaffinity partitioning technologies for capturing up to 20 high abundance proteins have provided some promising results, coupling immunoaffinity approaches with 1D1 or 2D LC-MS/MS analysis to identify plasma proteins at concentrations of ng/ml or lower remains a challenge (4). Overcoming this challenge requires an effective fractionation scheme to reduce the dynamic range and enable broader detection of the remaining low abundance proteins of interest.
In this study, we present a new Seppro IgY-SuperMix immunoaffinity separation system for its ability to enhance detection of low abundance proteins in human plasma. The new SuperMix system has been designed to be applied in tandem with the IgY12 system for capturing ∼50 moderately abundant proteins in addition to the 12 most abundant proteins in plasma. Herein we present results from this study that illustrate the potential for enhanced detection of low abundance proteins as well as the reproducibility of the SuperMix partitioning method for LC-MS/MS plasma proteome profiling.
EXPERIMENTAL PROCEDURES
Plasma Sample—
The human blood plasma sample supplied by the Stanford University School of Medicine (Palo Alto, CA) was obtained from a single, healthy volunteer. Approval for the conduct of this study was obtained from the Institutional Review Boards of the Stanford University School of Medicine and the Pacific Northwest National Laboratory in accordance with federal regulations. The initial protein concentration was ∼61 mg/ml as determined by BCA protein assay (Pierce). Unless otherwise noted, all protein sample processing was performed at 4 °C.
Generation of IgY-SuperMix LC2 Column—
To generate an immunoaffinity column with a mixture of antibodies that will bind to those moderately abundant proteins in human plasma, a plasma sample was initially depleted of the 12 highest abundance proteins using an IgY12 column. The flow-through fraction containing medium or low abundance proteins was used as a mixture of antigens for immunizing chickens and generating a mixture of polyclonal IgY antibodies. The IgY12-depleted flow-through fraction was also used as affinity ligands and conjugated to CNBr-activated Sepharose™ 4B (GE Healthcare) for preparing an antigen affinity column, which was used to purify the antibodies from the total IgYs isolated from the chickens immunized with the IgY12-depleted fraction. The mixture of newly purified IgY antibodies from the antigen affinity column was then conjugated to UltraLink Hydrazide Gel (Pierce) and packaged into an immunoaffinity column called the SuperMix LC2 column.
IgY12 and SuperMix Immunoaffinity Chromatography—
The plasma samples were initially subjected to the separation of 12 high abundance proteins (albumin, IgG, α1-antitrypsin, IgA, IgM, transferrin, haptoglobin, α1-acid glycoprotein, α2-macroglobulin, apolipoprotein A-I, apolipoprotein A-II, and fibrinogen) using a ProteomeLab™ 12.7 × 79.0-mm IgY12 LC10 affinity LC column (Beckman Coulter, Fullerton, CA) with a column capacity of 250 μl of plasma using an Agilent 1100 series HPLC system. The manufacturer's recommendations were followed in these separations, which were similar to those described previously (7). The same three buffers were utilized (dilution/washing: 10 mm Tris-HCl, 150 mm NaCl, pH 7.4 (TBS); stripping/elution buffer: 100 mm glycine, pH 2.5; neutralization buffer: 100 mm Tris-HCl, pH 8.0) in a separation scheme that consisted of sample loading-washing-eluting-neutralization followed by a re-equilibration scheme for a total cycle time of ∼70 min. The flow-through and bound (or eluted) fractions were collected separately.
Following the IgY12 separations, the flow-through fractions were concentrated in Amicon® Ultra-15 (5-kDa nominal molecular mass limit; Millipore, Billerica, MA) concentrators followed by buffer exchange to 50 mm NH4HCO3, pH 8.0. Protein concentration was then determined by BCA protein assay (Pierce).
The concentrated IgY12 flow-through fractions were then partitioned using a Seppro SuperMix LC2 (6.4 × 63.0-mm) immunoaffinity column on the same HPLC system. The separation conditions using the SuperMix column were similar to that described for the IgY12 LC2 separation (7). Both the flow-through and bound fractions from the SuperMix LC2 column were collected and concentrated as described above with buffer exchange to 50 mm NH4HCO3, and protein concentration was measured using the BCA protein assay (Pierce).
To demonstrate the measurements of differential protein abundances, six non-human standard proteins were spiked into three 1-ml human plasma samples at 1, 5, and 25 μg/ml concentrations, respectively. The six proteins spiked into plasma were bovine carbonic anhydrase 2, chicken ovalbumin, horse myoglobin, bovine α-lactalbumin, bovine cytochrome c, and bovine β-lactoglobulin. Each plasma sample with spiked protein was subjected to triplicate tandem IgY12-SuperMix immunoaffinity separations with 250 μl of starting plasma for each replicate. Both the SuperMix flow-through and bound fractions for each of the replicate experiments were concentrated as described above.
Protein Digestion—
The protein samples from IgY12 flow-through, SuperMix flow-through, and SuperMix bound fractions were denatured and reduced in 50 mm NH4HCO3 buffer, pH 8.0, 8 m urea, 10 mm DTT for 1 h at 37 °C. Protein cysteinyl residues were alkylated with 40 mm iodoacetamide for 60 min at room temperature. The resulting protein mixture was diluted 6-fold with 50 mm NH4HCO3, pH 8.0, before sequencing grade modified porcine trypsin (Promega, Madison, WI) was added at a trypsin:protein ratio of 1:50 (w/w). The sample was incubated at 37 °C for 3 h. The tryptically digested sample was then loaded onto a 1-ml SPE C18 column (Supelco, Bellefonte, PA) and washed with 4 ml of 0.1% TFA, 5% acetonitrile. Peptides were eluted from the SPE column with 1 ml of 0.1% TFA, 80% acetonitrile and lyophilized. Final peptide concentration was determined by BCA protein assay (Pierce). Peptide samples were stored at −80 °C until further analysis.
Strong Cation Exchange Fractionation—
∼150 μg of tryptic peptides from the IgY12 flow-through and SuperMix flow-through fractions were resuspended in 150 μl of 10 mm ammonium formate, 25% acetonitrile and fractionated by strong cation exchange chromatography on a 2.1 × 200-mm (5-μm, 300-Å) Polysulfoethyl A LC column (PolyLC, Columbia, MD) preceded by a 2.1 × 10-mm guard column using an 1100 series HPLC system (Agilent) at a flow rate of 200 μl/min. Mobile phases used were 10 mm ammonium formate, pH 3.0, 25% acetonitrile (A) and 500 mm ammonium formate, pH 6.8, 25% acetonitrile (B). Sample was loaded onto the column and run with 100% A for 10 min. Peptides were then separated by a linear gradient to 50% B over 40 min followed by a linear gradient from 50 to 100% B over 10 min. 100% B was then maintained for 20 min. A total of 25 fractions were collected with each fraction being lyophilized prior to reversed-phase LC-MS/MS analysis.
Reversed-phase Capillary LC-MS/MS Analysis—
Peptide samples were analyzed using a custom-built automated four-column high pressure capillary LC system coupled on-line to a linear ion trap mass spectrometer (LTQ; ThermoElectron) via an electrospray ionization interface manufactured in-house. The reversed-phase capillary column was prepared by slurry-packing 3-μm Jupiter C18 bonded particles (Phenomenex, Torrence, CA) into a 65-cm-long, 75-μm-inner diameter fused silica capillary (Polymicro Technologies, Phoenix, AZ). The mobile phase consisted of 0.2% acetic acid, 0.05% TFA in water (A) and 0.1% TFA in 90% acetonitrile, 10% water (B). After loading 5 μl of peptides onto the column, the mobile phase was held at 100% A for 20 min. Exponential gradient elution was performed by increasing the mobile phase composition from 0 to 70% B over 85 min. To identify the eluting peptides, the LTQ mass spectrometer was operated in a data-dependent MS/MS mode (m/z 400–2000) in which a full MS scan was followed by 10 MS/MS scans. The 10 most intensive precursor ions were dynamically selected in the order of highest intensity to lowest intensity and subjected to collision-induced dissociation using a normalized collision energy setting of 35% and a dynamic exclusion duration of 1 min. The heated capillary was maintained at 200 °C; the ESI voltage was kept at 2.2 kV.
Data Analysis—
The LC-MS/MS raw data were converted into a .dta file by Extract_MSn (version 3.0) in Bioworks Cluster 3.2 (Thermo), and the SEQUEST algorithm (version 27, revision 12) was used to independently search all the MS/MS spectra against the human International Protein Index (IPI) database with a total of 61,225 total protein entries (version 3.20, released August 22, 2006). The search parameters used were as follows: 3-Da tolerance for precursor ion masses and 1 Da for fragment ion masses with no enzyme restraint and a maximum of three missed tryptic cleavages. Static carboxamidomethylation of cysteine and dynamic oxidation of methionine were used during the database search.
Because false positive peptide/protein identifications are a common concern in proteomics investigations, we developed and applied a set of criteria based on the reversed database approach for filtering the raw data to limit false positive identifications to <5% at the unique peptide level as described previously (13, 14). The reversed human protein database was created by reversing the order of the amino acid sequences for each protein, and the false positive rate (FPR) for peptide identifications was estimated by dividing the number of unique peptides identified from the reversed database search (NR) by the number of unique peptides identified from the normal database search (NN), i.e. FPR = NR/NN. Table I summarizes the cross-correlation score (Xcorr) and delta correlation (ΔCn) values along with tryptic cleavage states for filtering the SEQUEST raw data. A false positive rate of ∼4% was observed at the unique peptide level following such filtering for this study.
Table I.
SEQUEST filtering criteria used for peptide identifications
| Charge state | ΔCn | Xcorr | Tryptic ends |
|---|---|---|---|
| 1+ | ≥0.05 | ≥1.7 | Fully |
| 1+ | ≥0.1 | ≥1.5 | Fully |
| 1+ | ≥0.05 | ≥3.0 | Partially |
| 1+ | ≥0.16 | ≥2.8 | Partially |
| 2+ | ≥0.05 | ≥2.8 | Fully |
| 2+ | ≥0.1 | ≥2.7 | Fully |
| 2+ | ≥0.16 | ≥2.3 | Fully |
| 2+ | ≥0.05 | ≥3.8 | Partially |
| 2+ | ≥0.1 | ≥3.7 | Partially |
| 2+ | ≥0.16 | ≥3.5 | Partially |
| 3+ | ≥0.05 | ≥3.5 | Fully |
| 3+ | ≥0.1 | ≥3.3 | Fully |
| 3+ | ≥0.16 | ≥3 | Fully |
| 3+ | ≥0.05 | ≥4.6 | Partially |
| 3+ | ≥0.1 | ≥4.5 | Partially |
| 3+ | ≥0.16 | ≥4.3 | Partially |
ProteinProphet™ software was used as a clustering tool to generate a list of non-redundant proteins or protein groups (15). Peptides that passed the filtering criteria were assigned the identical probability score of 1 and entered into the software program (done exclusively for cluster analysis) to generate a final list of non-redundant proteins/protein groups. One protein IPI number was randomly selected to represent each corresponding protein group that consists of a number of database entries. Only those proteins or protein groups with two or more unique peptide identifications were considered as confident protein identifications.
ELISA—
The plasma protein concentrations for macrophage colony-stimulating factor 1 (M-CSF) and matrix metalloproteinase-8 (MMP8) were determined in triplicate using Quantikine ELISA kits (R&D Systems, Minneapolis, MN) following the manufacturer's instructions. Each sample was analyzed at three different dilutions to determine the optimal dilution.
RESULTS
Tandem IgY12-SuperMix Immunoseparation Strategy—
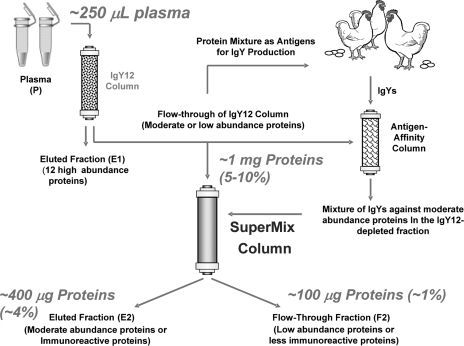
The scheme used to generate the SuperMix column is shown in Fig. 1. The concept behind the SuperMix immunoaffinity separation is that antibodies can be generated against mixed antigens such as the proteins present in human plasma. Because the antibody titers correspond to the abundance and the immunogenicity of the proteins (16) the assumption is that moderately abundant proteins in human plasma may lead to much higher immune response than lower abundance proteins. As a result, a purified mixture of these antibodies can be used to capture a large number of medium abundance proteins especially when applied in tandem with a high abundance protein removal strategy such as IgY12. Because the SuperMix column contains antibodies against those medium abundance proteins from the IgY12-depleted plasma sample, it can be applied in tandem with the IgY12 column as a dual separation strategy (Fig. 1) to significantly enrich low abundance proteins by capturing a relatively large number (>50) of high or medium abundance proteins onto the IgY12 and SuperMix columns. Fig. 1 also indicates typical loading and recoveries for the dual separation strategy; for example, ∼100 μg of total proteins is expected to be recovered from 250 μl of starting plasma sample.
Fig. 1.
The tandem IgY12-SuperMix immunoaffinity separation strategy. The SuperMix column was generated by using the protein mixture from IgY12-depleted human plasma as mixed antigens. For the tandem separations, plasma or other biofluid samples are initially separated by the IgY12 LC-10 column. The flow-through from the IgY12 column is further separated by the SuperMix LC2 column into the flow-through and eluted fractions. The typical recoveries are indicated assuming ∼250 μl of plasma is loaded.
Reproducibility of the Tandem IgY12-SuperMix Separations—
As reproducible separations and sample processing steps are key requirements for quantitative applications in clinical proteomics, we evaluated the reproducibility of tandem IgY12-SuperMix separations Table II shows the reproducibility based on five independent experimental replicates in terms of sample recoveries for each step of separation and sample processing. Recovered amounts of proteins in the IgY12 and SuperMix flow-throughs and SuperMix bound fractions as well as the final digested peptides in the SuperMix flow-through fraction are consistent across the five replicates. This consistency suggests overall good reproducibility of both tandem separations and concentrating, buffer exchange, and protein digestion steps. The variation for finalized recovered peptides in the SuperMix flow-through is slightly higher than the protein level presumably because of contributions from the SPE clean-up step following digestion.
Table II.
Sample recoveries for tandem IgY12 and SuperMix separations
| Replicatea | A | B | C | D | E | Recoveryb |
|---|---|---|---|---|---|---|
| % | ||||||
| IgY12 flow-through (μg)c | 1249 | 1429 | 1281 | 1443 | 1457 | 10.0 ± 0.72 |
| SuperMix bound (μg)d | 485 | 421 | 424 | 438 | 414 | 3.19 ± 0.21 |
| SuperMix flow-through (μg)d | 121 | 128 | 123 | 117 | 122 | 0.89 ± 0.03 |
| SuperMix flow-through postdigestion (μg)d | 68 | 54 | 69 | 78 | 50 | 0.47 ± 0.08 |
For each replicate experiment, 225 μl of plasma was used with the total protein quantity of ∼13.7 mg (plasma concentration, ∼61 mg/ml).
All recoveries are expressed as the percentage of recovered proteins or peptides compared with the 13.7 mg of starting materials along with the standard deviations.
The amount of proteins or peptides recovered for each replicate.
Only nine-tenths of the IgY12 flow-through was used for SuperMix separations.
To further evaluate the reproducibility, three replicates of peptide samples from the IgY12 flow-through, SuperMix flow-through, and SuperMix bound fractions were selected for separate LC-MS/MS analyses. An average of 151 ± 3, 232 ± 6, 104 ± 3 proteins were identified from triplicate analyses of IgY12 flow-through, SuperMix flow-through, and SuperMix bound fractions, respectively. The number of MS/MS spectra that identified a given protein (spectral count) after stringent filtering was used to further evaluate (7) the reproducibility among replicate analyses. Spectral count information has been demonstrated recently as a means for relative quantitation among samples as well as for comparing absolute protein abundances within a sample (17–19). Fig. 2 shows the linear correlations of spectral counts to identified proteins for replicate analyses of IgY12 flow-through samples (A) and SuperMix flow-through samples (B). Comparable reproducibility was observed for both SuperMix and IgY12 flow-through fractions, suggesting that the reproducibility of the tandem IgY12-SuperMix separations is similar to that of the single IgY12 separations.
Fig. 2.
Reproducibility of IgY12 and tandem IgY12-SuperMix separations evaluated by LC-MS/MS. Reproducibility of three replicates of IgY12 separations and tandem IgY12-SuperMix separations was assessed based on spectral count information of identified proteins from individual LC-MS/MS. A, correlation between analyses of IgY12 flow-through samples; B, correlation between SuperMix flow-through samples.
Quantitation of Differential Protein Abundances—
A standard addition experiment using six non-human standard proteins spiked into the original plasma at three different levels, i.e. 1, 5, and 25 μg/ml, was performed to evaluate the ability for relative quantitation of protein abundances applying the tandem separation strategy. Following triplicate immunoaffinity separations, both the SuperMix flow-through and bound fractions from the three plasma samples were analyzed using LC-MS/MS to generate spectral count data for relative quantitation. The spiked proteins were not detected in the SuperMix bound fraction except for β-lactoglobulin, which was observed at the 25 μg/ml spiked level in the bound fraction with an average spectral count of 7 across replicates.
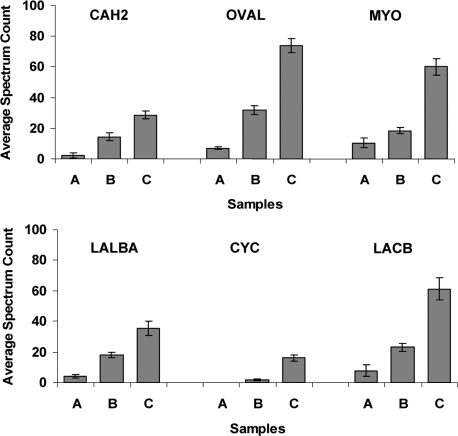
The spectral count data for the six spiked standard proteins as measured in the flow-through samples are plotted in Fig. 3. As shown, reproducible spectral count data within the triplicate experiments were observed for all six proteins at each of the three concentration levels with the sole exception of cytochrome c, which was not detected at the lowest level (1 μg/ml). These results further illustrate the overall reproducibility of the tandem separations combined with LC-MS/MS analyses as well as demonstrate the sensitivity in that five of six proteins were reliably detected at the 1 μg/ml level with single dimensional LC-MS/MS analyses. The ability of this separation to yield relative abundance differences is also clearly demonstrated based on the observed proportional increase in mean spectral counts with the increase of protein concentrations. For all six spiked proteins, the abundance differences between any two spiked levels were observed to be statistically significant (p value <0.02 based on Student's t test analysis of the triplicate data).
Fig. 3.
Measurements of protein concentration differences for six proteins spiked into human plasma at three different levels. Three plasma samples (A, B, and C) were created by spiking six standard proteins at 1, 5, and 25 μg/ml, respectively. Triplicate experiments were performed for each sample by subjecting each sample to independent tandem IgY12-SuperMix immunoaffinity separation, trypsin digestion, and LC-MS/MS analyses of both the SuperMix flow-through and bound fractions. Spectral count data were used to quantify the relative abundance differences between the three samples. The plots were data from the triplicate analyses of the SuperMix flow-through fractions for each sample. The six proteins spiked into plasma are: bovine carbonic anhydrase 2 (CAH2), chicken ovalbumin (OVAL), horse myoglobin (MYO), bovine α-lactalbumin (LALBA), bovine cytochrome c (CYC), and bovine β-lactoglobulin (LACB). Error bars indicate standard deviations of spectral counts among the triplicate experiments.
Enhanced Detection of Low Abundance Proteins—
We next directly compared the tandem IgY12-SuperMix separations with IgY12 separations using both 1D and 2D LC-MS/MS analyses to investigate the potential enhancement in detection of low abundance proteins. For 1D LC-MS/MS analyses, IgY12 and SuperMix flow-through peptide samples from three independent separations experiments were analyzed. For 2D LC-MS/MS, strong cation exchange was used to fractionate approximately the same quantities of peptides from IgY12 and SuperMix flow-through samples into 25 fractions prior to LC-MS/MS. Only those proteins identified with two unique peptides from the entire study set were considered as confident identifications.
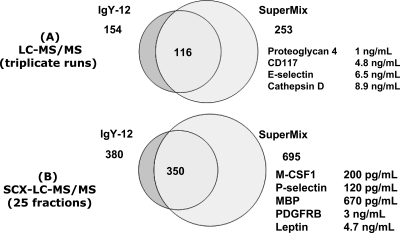
Fig. 4 shows a side-by-side comparison of proteome coverage for IgY12 and SuperMix flow-through peptide samples achieved in 1D and 2D LC-MS/MS analyses. Triplicate LC-MS/MS analyses of SuperMix flow-through indicate an ∼64% improvement in proteome coverage compared with IgY1 (Fig. 4A). Similarly an ∼83% improvement in proteome coverage was obtained using SuperMix for 2D LC-MS/MS compared with IgY12 (Fig. 4B). Note that the SuperMix improvement in proteome coverage was achieved without significantly reducing analytical throughput because only a second separation step was added. In 1D LC-MS/MS analyses, a number of low ng/ml level proteins (based on “normal” concentrations reported in the literature) were identified in the SuperMix flow-through (Fig. 4A), whereas proteins observed in the IgY12 flow-through were in the low μg/ml to high ng/ml range, such as hepatocyte growth factor activator (500 ng/ml) and CD14 (1.4 μg/ml), which is in agreement with our recent report (4). The lists of proteins and peptides identified using SuperMix and IgY12 are available as supplemental Tables 1 and 2, respectively, along with corresponding spectral count information.
Fig. 4.
Comparison of proteome coverage between IgY12 flow-through and SuperMix flow-through as analyzed by 1D or 2D LC-MS/MS. Selected examples of low abundance proteins with reported normal concentrations are listed. MBP, myelin basic protein; SCX, strong cation exchange; PDGFRB, platelet-derived growth factor receptor β.
The enhanced detection of low abundance proteins achieved using SuperMix is further illustrated in Fig. 5, which displays protein concentrations in the range of ∼200 pg/ml to 100 ng/ml (based on previous literature data) for 40 low abundance proteins detected in the flow-through fraction analyzed by 2D LC-MS/MS. The concentrations of a cytokine (M-CSF) and MMP8 in the same plasma sample were validated as 202 ± 20 pg/ml and 12.4 ± 0.4 ng/ml, respectively, based on triplicate analyses by ELISAs. Importantly the M-CSF protein was confidently identified by four different peptides as shown in Table III. In comparison only 21 of the 40 proteins were observed in the IgY12 flow-through. The results in Fig. 4 and 5 clearly demonstrate that the use of tandem IgY12-SuperMix separations significantly enhances the detection of low abundance proteins and the overall proteome coverage. Notably this tandem separation strategy coupled with 2D LC-MS/MS analyses has enabled identification of a number of physiologically relevant molecules such as known cytokines and growth factors. Table IV lists 14 known cytokines and growth factors (based on the Ingenuity Knowledgebase) that were identified in this study of which only three were also observed in the IgY12 flow-through.
Fig. 5.
The range of reported normal concentrations for 40 detected low abundance proteins detected in 2D LC-MS/MS analyses of SuperMix flow-through. The protein concentration is plotted on a log scale. All protein concentrations were obtained based on previously reported data for plasma/serum samples from normal human subjects (3, 18, 25). Those proteins highlighted with gray squares were detected in both IgY12 and SuperMix flow-through fractions; the others were detected in SuperMix flow-through only. Protein concentrations based on ELISA results from the present study are marked with # (M-CSF and MMP8). ACE, angiotensin-converting enzyme; TIMP, tissue inhibitor of metalloprotease; VIP36, 36 kDa vesicular integral membrane protein; VCAM, vascular cell adhesion molecule; IGF, insulin-like growth factor; IGFBP, insulin-like growth factor-binding protein; ICAM, intercellular adhesion molecule; PECAM, platelet endothelial cell adhesion molecule; HGF L, hepatocyte growth factor-like; MIF, macrophage migration-inhibitory factor; EGF R, epidermal growth factor receptor; VEGF R, vascular endothelial growth factor receptor; PF4, platelet factor 4; PDGF R, platelet-derived growth factor receptor; MBP, myelin basic protein; MCSF R, M-CSF receptor.
Table III.
Peptide identification information for two selected low abundance proteins reported to be at sub-ng/ml levels
MBP, myelin basic protein.
| Protein | Identified peptide | Charge state | Xcorr | ΔCn | Spectral count |
|---|---|---|---|---|---|
| M-CSF | R↓ SHSSGSVLPLGELEGRR↓ S | 2 | 3.4659 | 0.3521 | 2 |
| M-CSF | K↓ SCFTKDYEEHDKACVR↓ T | 2 | 4.9067 | 0.4207 | 1 |
| M-CSF | K↓ KAFLLVQDIMEDTMR↓ F | 2 | 3.5667 | 0.3052 | 1 |
| M-CSF | R↓ FRDNTPNAIAIVQLQELSLR↓ L | 3 | 4.6686 | 0.2551 | 1 |
| MBP | R↓ TQDENPVVHFFK↓ N | 2 | 3.8375 | 0.3424 | 2 |
| MBP | R↓ HRDTGILDSIGR↓ F | 2 | 3.0247 | 0.3330 | 2 |
Table IV.
A selected list of known cytokines or growth factors identified from 2D LC-MS/MS analyses of IgY12 and SuperMix flow-through fractions
Only proteins observed with two or more spectral counts were considered as confidently detected in individual experiments. The number of unique peptides for a given protein is the combined results for the entire study.
| Protein name | Gene | Unique peptides | Spectral count
|
|
|---|---|---|---|---|
| IgY12 | SuperMix | |||
| Uteroglobin | SCGB1A1 | 3 | 7 | |
| Secreted and transmembrane protein 1 | SECTM1 | 2 | 2 | |
| Macrophage colony-stimulating factor 1 | CSF1 | 5 | 5 | |
| Platelet basic protein | PPBP | 8 | 3 | 36 |
| Platelet factor 4 | PF4 | 3 | 6 | |
| Macrophage migration-inhibitory factor | MIF | 2 | 2 | |
| Neuron-derived neurotrophic factor | NENF | 3 | 3 | |
| Mimecan (osteoglycin) | OGN | 10 | 9 | 11 |
| Inhibin β C chain | INHBC | 5 | 6 | |
| Granulins | GRN | 3 | 3 | |
| Leptin | LEP | 6 | 8 | 5 |
| Insulin-like growth factor I | IGF1 | 7 | 27 | |
| Growth/differentiation factor 8 | GDF8 | 2 | 2 | |
| Insulin-like growth factor II | IGF2 | 7 | 27 | |
Capture Efficiency for Moderately Abundant Proteins by SuperMix Column—
The observed significant improvement in proteome coverage afforded by the SuperMix separation is presumably because of efficient binding of a number of moderately abundant proteins to the column, thus leading to the enrichment of lower abundance proteins. As a result, we examined the binding efficiency of the SuperMix column for moderate abundance proteins. The binding efficiency was estimated on the basis of observed spectral counts from triplicate LC-MS/MS analyses of SuperMix bound and flow-through fractions. Although not quantitative because the bound and flow-through samples are very different in protein/peptide composition, this estimation still allowed us to identify those proteins that are efficiently captured by the column because either zero or a minimum number of spectral counts should be observed in the flow-through.
Table V lists 45 medium abundance proteins that were captured with >90% efficiency by the SuperMix column; ∼35 additional proteins were captured with 20–90% efficiency. Because the SuperMix column contains a mixture of IgY antibodies that were generated on the basis of the immunoreactivity of medium abundance proteins that were present in the IgY12-depleted plasma sample, one might expect that the limited amounts of available antibodies would lead to low binding efficiencies for some of the proteins. To test this hypothesis, we loaded the SuperMix column with twice the amount of sample (IgY12 flow-through from ∼500 μl of plasma). Interestingly overloading of the SuperMix column significantly reduced the total number of proteins identified in the flow-through fraction (188 proteins versus 253 proteins shown in Fig. 4). The number of proteins efficiently captured by the SuperMix column was also reduced from 45 to 29. These results suggest that further optimization of loading the SuperMix column may further enhance detection of low abundance proteins as a result of capturing more moderately abundant proteins.
Table V.
List of moderately abundant proteins that bound to the SuperMix column with relatively high capture efficiency
The spectral count data from triplicate analyses of the flow-through and bound fraction of SuperMix separations are presented for the listed moderately abundant proteins. LMW, low molecular weight; HMW, high molecular weight.
| IPI no. | Protein name | Gene name | Spectral counts
|
Capture efficiencya | |
|---|---|---|---|---|---|
| Flow-through | Bound | ||||
| % | |||||
| IPI00164623.4 | Complement C3 | C3 | 7 | 2060 | >99 |
| IPI00418163.3 | Complement 4B | C4B | 12 | 747 | 98 |
| IPI00032258.4 | Complement C4A | C4A | 12 | 707 | 98 |
| IPI00017601.1 | Ceruloplasmin | CP | 0 | 532 | >99 |
| IPI00022488.1 | Hemopexin | HPX | 3 | 509 | 99 |
| IPI00019591.1 | Complement factor B | CFB | 3 | 425 | 99 |
| IPI00218192.1 | Inter-α-trypsin inhibitor heavy chain H4 | ITIH4 | 4 | 352 | 99 |
| IPI00032291.1 | Complement C5 | C5 | 0 | 306 | >99 |
| IPI00006543.2 | Complement factor H-related 5 | CFHR5 | 0 | 288 | >99 |
| IPI00305461.2 | Inter-α-trypsin inhibitor heavy chain H2 | ITIH2 | 1 | 275 | >99 |
| IPI00515041.2 | Complement factor H | CFH | 0 | 260 | >99 |
| IPI00292530.1 | Inter-α-trypsin inhibitor heavy chain H1 | ITIH1 | 0 | 243 | >99 |
| IPI00019580.1 | Plasminogen | PLG | 0 | 216 | >99 |
| IPI00215894.1 | Isoform LMW of kininogen-1 | KNG1 | 3 | 191 | 98 |
| IPI00032179.2 | Antithrombin III variant | SERPINC1 | 0 | 184 | >99 |
| IPI00032328.1 | Isoform HMW of kininogen-1 | KNG1 | 2 | 178 | 99 |
| IPI00009920.2 | Complement component C6 | C6 | 0 | 151 | >99 |
| IPI00298828.3 | β2-Glycoprotein 1 | APOH | 0 | 125 | >99 |
| IPI00022895.7 | α1B-Glycoprotein | A1BG | 0 | 123 | >99 |
| IPI00022418.1 | Fibronectin | FN1 | 0 | 121 | >99 |
| IPI00294395.1 | Complement C8 β chain | C8B | 0 | 114 | >99 |
| IPI00291866.5 | Plasma protease C1 inhibitor | SERPING1 | 2 | 113 | 98 |
| IPI00022420.3 | Plasma retinol-binding protein | RBP4 | 0 | 100 | >99 |
| IPI00165421.4 | SERPINC1 protein | SERPINC1 | 0 | 98 | >99 |
| IPI00021885.1 | Fibrinogen α chain | FGA | 0 | 90 | >99 |
| IPI00022371.1 | Histidine-rich glycoprotein | HRG | 1 | 88 | 99 |
| IPI00298497.3 | Fibrinogen β chain | FGB | 0 | 83 | >99 |
| IPI00298971.1 | Vitronectin | VTN | 1 | 81 | 99 |
| IPI00021891.5 | Fibrinogen γ chain | FGG | 0 | 68 | >99 |
| IPI00736985.1 | Similar to ceruloplasmin | LOC441368 | 0 | 68 | >99 |
| IPI00021727.1 | C4B-binding protein α chain | C4BPA | 0 | 65 | >99 |
| IPI00296608.6 | Complement component C7 | C7 | 0 | 59 | >99 |
| IPI00011252.1 | Complement C8 α chain | C8A | 0 | 59 | >99 |
| IPI00022391.1 | Serum amyloid P-component | APCS | 1 | 49 | 98 |
| IPI00011261.1 | Complement C8 γ chain | C8G | 0 | 48 | >99 |
| IPI00294004.1 | Vitamin K-dependent protein S | PROS1 | 0 | 36 | >99 |
| IPI00011264.1 | Complement factor H-related protein 1 | CFHR1 | 0 | 32 | >99 |
| IPI00006154.1 | Complement factor H-related protein 2 | CFHR2 | 0 | 13 | >99 |
| IPI00021364.1 | Properdin | CFP | 1 | 13 | 93 |
| IPI00022392.1 | Complement C1Q subunit A | C1QA | 0 | 12 | >99 |
| IPI00022394.2 | Complement C1Q subunit C | C1QC | 0 | 12 | >99 |
| IPI00477992.1 | C1Q subcomponent, B chain | C1QB | 0 | 10 | >99 |
| IPI00027507.1 | Complement factor H-related protein 3 | CFHR3 | 0 | 7 | >99 |
| IPI00025862.1 | C4B-binding protein β chain | C4BPB | 0 | 5 | >99 |
| IPI00029168.1 | Apolipoprotein(a) | LPA | 0 | 5 | >99 |
The capture efficiency is used to estimate the percentage for a given protein to be captured by the SuperMix column; it was estimated for each protein by dividing the spectral count for each protein from the bound fraction (B) by the sum of spectral counts from the bound fraction and flow-through fraction (FT), i.e. B/(B + FT). The estimated capture efficiency will be less accurate for proteins with only a few spectral counts. Even if 0 count was observed in the flow-through, it does not mean a complete capture by the column; therefore, these proteins are represented as >99%.
Several relatively abundant proteins were not bound efficiently by the SuperMix column presumably because of limited immunoreactivity. These proteins included vitamin D-binding proteins, apolipoprotein A-IV, prothrombin, α1-antichymotrypsin, apolipoprotein B-100, and α2-HS-glycoprotein, all of which were captured with <50% efficiency even though their relative abundances based on spectral count ranking were among the top 30 proteins present in the IgY12-depleted plasma. Further details regarding estimated capture efficiency are available in supplemental Table 1.
DISCUSSION
One of the greatest challenges for applying advanced MS-based proteomics to clinical biomarker discovery efforts remains the ability to effectively detect low abundance proteins in complex mixtures. Many different fractionation/separation techniques have been developed and applied in a multidimensional fashion at both the protein and peptide levels to address this dynamic range challenge (5, 20, 21). Essentially all of the chromatography-based separation and fractionation approaches provide some degree of enrichment (or focusing) of a specific subset of low abundance proteins to achieve improved detection. Immunoaffinity chromatography with multiplexed antibodies that target relatively abundant proteins is a commonly used technique for human biofluid proteome profiling (9, 10). In LC format, immunoaffinity separations offer an effective strategy for separating high abundance from relatively low abundance proteins in a highly reproducible fashion (7, 12) thus alleviating the masking effect of high abundance proteins. However, despite advances in recent years, immunoaffinity separation strategies have been limited to the number of antibodies that can be multiplexed on the LC column (currently limited to 20 proteins).
In this work, we introduced a novel tandem IgY12-SuperMix immunoaffinity separation strategy that allows effective binding of ∼57 high or medium abundance proteins onto the IgY12-SuperMix column. This method of separating >50 abundant proteins from low abundance proteins has led to significant improvements as evidenced by LC-MS/MS results. Improvements in plasma proteome coverage (∼60–80%) (Fig. 4) and detection sensitivity has now allowed identification of a number of low concentration (ng/ml) proteins, including known cytokines and growth factors from a normal human subject (Fig. 5 and Table IV). Importantly one of the cytokines (M-CSF) has been confirmed to be present at ∼200 pg/ml based on ELISA results. These results demonstrate that tandem IgY12-SuperMix immunoaffinity separations with high reproducibility offer an effective means to dig deeper into complex biofluid proteomes for quantitative clinical applications. Unlike other fractionation approaches that typically produce many fractions, the immunoaffinity separations only generate two fractions (the flow-through and bound fractions); thus the improvements are achieved without a significant reduction in analytical throughput. Moreover the separations can be fully automated on an HPLC system with only a slight increase in sample processing because of the addition of concentration and buffer exchange steps. Finally immunoaffinity columns typically allow more than 100 separations per column, alleviating the concern of batch-to-batch variations of affinity columns because a given biological study can be typically completed using a single column.
One of the general concerns in applying immunoaffinity columns is the extent of nonspecific binding. We recently demonstrated that the extent of nonspecific or specific binding to a column is reproducible for a given protein (7). Although we did not observe strong evidence of nonspecific binding on the SuperMix column in this study, we believe the most likely use of the SuperMix column would be as a separation/fractionation tool such that both the bound and flow-through fractions are analyzed to achieve more complete proteome profiling instead of using it as a pure depletion device for removing high and medium abundance proteins. The observed good reproducibility obtained by analyzing both bound and flow-through fractions provides the confidence necessary for quantitative proteomics applications. The ability to determine abundance differences applying the tandem separations was demonstrated in standard protein spiking experiments. The spiking experiments also indicated relatively low nonspecific binding as only one protein of six spiked proteins was detected in the bound fraction at the highest spiked concentration (25 μg/ml).
A potential issue of the SuperMix column is that it contains an undefined mixture of antibodies that are present at various amounts. Based on our data, many proteins could be partially bound to the column; therefore, the SuperMix column should be utilized as a fractionation technique with both the bound and flow-through fractions analyzed for better evaluation of potential abundance differences between conditions. The partial protein binding to the column could amplify the abundance differences depending on the extent of binding without affecting the “directions” of these abundance changes. For example, for a protein present at 1 μg in one sample and at 3 μg in another sample (i.e., a 3-fold difference) and for a column that has an antibody capacity to bind 0.5 μg of the protein for both samples, a 5-fold relative concentration difference between the two samples will be observed (0.5 versus 2.5) following the SuperMix separations. Given the reproducibility of the SuperMix separations, such changes in amplitudes should not have a detrimental effect on our ability to identify statistically significant abundance differences for candidate biomarkers. Nevertheless one does need to keep in mind that observed amplitudes of protein concentration differences may not exactly reflect the true quantitative differences in original samples if there is partial protein binding.
Because generation of the SuperMix column is based on mixed antigens from IgY12-depleted human plasma, the amount of antibodies available for each protein will depend on the immunoreactivity of the proteins in the host system. In fact, we observed several relatively high abundance proteins that were not effectively bound by the SuperMix column. Individual antibodies of these proteins may be developed and added to further enhance the performance of the SuperMix column. If available antibodies are limited, then optimization of the sample loading step may allow more proteins to be effectively captured.
To conclude, the improved proteome coverage/enhanced detection of low abundance proteins, reproducibility, and ability for relative quantitation demonstrate the utility of tandem IgY12-SuperMix immunoaffinity separations for clinical biomarker discovery applications involved with human biofluids. These tandem separations can be coupled with either LC-MS-based or gel-based approaches with or without further fractionation. We anticipate that this tandem IgY12-SuperMix strategy may also significantly enhance the effectiveness of directed MS in the multiple reaction monitoring mode for candidate biomarker verification (22–24).
Acknowledgments
The experimental work described herein was performed in the Environmental Molecular Sciences Laboratory, a national scientific user facility sponsored by the Department of Energy (DOE) and located at Pacific Northwest National Laboratory, which is operated by Battelle Memorial Institute for the DOE under Contract DE-AC05-76RL0 1830.
Footnotes
Published, MCP Papers in Press, July 15, 2008, DOI 10.1074/mcp.M800008-MCP200
The abbreviations used are: 1D, one-dimensional; 2D, two-dimensional; IgY, immunoglobulin yolk; SPE, solid-phase extraction; M-CSF, macrophage colony-stimulating factor 1; LTQ, linear ion trap quadrupole; IPI, International Protein Index; Xcorr, cross-correlation score; ΔCn, delta correlation; MMP8, matrix metalloproteinase-8; α2-HS-glycoprotein, α2-Heremans-Schmid glycoprotein.
This work was supported, in whole or in part, by National Institutes of Health Grants RR018522 from the National Center for Research Resources and U54 GM-62119-02, an NIGMS large scale collaborative research grant. This work was also supported by a grant from the Entertainment Industry Foundation (EIF) and the EIF Women's Cancer Research Fund (to the Breast Cancer Biomarker Discovery Consortium). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.mcponline.org) contains supplemental material.
REFERENCES
- 1.Hu, S., Loo, J. A., and Wong, D. T. ( 2006) Human body fluid proteome analysis. Proteomics 6, 6326–6353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rifai, N., Gillette, M. A., and Carr, S. A. ( 2006) Protein biomarker discovery and validation: the long and uncertain path to clinical utility. Nat. Biotechnol. 24, 971–983 [DOI] [PubMed] [Google Scholar]
- 3.Anderson, N. L., and Anderson, N. G. ( 2002) The human plasma proteome: history, character, and diagnostic prospects. Mol. Cell. Proteomics 1, 845–867 [DOI] [PubMed] [Google Scholar]
- 4.Qian, W. J., Jacobs, J. M., Liu, T., Camp, D. G., II, and Smith, R. D. ( 2006) Advances and challenges in liquid chromatography-mass spectrometry-based proteomics profiling for clinical applications. Mol. Cell. Proteomics 5, 1727–1744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lee, H. J., Lee, E. Y., Kwon, M. S., and Paik, Y. K. ( 2006) Biomarker discovery from the plasma proteome using multidimensional fractionation proteomics. Curr. Opin. Chem. Biol. 10, 42–49 [DOI] [PubMed] [Google Scholar]
- 6.Wang, H., Clouthier, S. G., Galchev, V., Misek, D. E., Duffner, U., Min, C. K., Zhao, R., Tra, J., Omenn, G. S., Ferrara, J. L., and Hanash, S. M. ( 2005) Intact-protein-based high-resolution three-dimensional quantitative analysis system for proteome profiling of biological fluids. Mol. Cell. Proteomics 4, 618–625 [DOI] [PubMed] [Google Scholar]
- 7.Liu, T., Qian, W. J., Mottaz, H. M., Gritsenko, M. A., Norbeck, A. D., Moore, R. J., Purvine, S. O., Camp, D. G., II, and Smith, R. D. ( 2006) Evaluation of multiprotein immunoaffinity subtraction for plasma proteomics and candidate biomarker discovery using mass spectrometry. Mol. Cell. Proteomics 5, 2167–2174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pieper, R., Su, Q., Gatlin, C. L., Huang, S. T., Anderson, N. L., and Steiner, S. ( 2003) Multi-component immunoaffinity subtraction chromatography: an innovative step towards a comprehensive survey of the human plasma proteome. Proteomics 3, 422–432 [DOI] [PubMed] [Google Scholar]
- 9.Zolotarjova, N., Martosella, J., Nicol, G., Bailey, J., Boyes, B. E., and Barrett, W. C. ( 2005) Differences among techniques for high-abundant protein depletion. Proteomics 5, 3304–3313 [DOI] [PubMed] [Google Scholar]
- 10.Huang, L., Harvie, G., Feitelson, J. S., Gramatikoff, K., Herold, D. A., Allen, D. L., Amunngama, R., Hagler, R. A., Pisano, M. R., Zhang, W. W., and Fang, X. ( 2005) Immunoaffinity separation of plasma proteins by IgY microbeads: meeting the needs of proteomic sample preparation and analysis. Proteomics 5, 3314–3328 [DOI] [PubMed] [Google Scholar]
- 11.Yocum, A. K., Yu, K., Oe, T., and Blair, I. A. ( 2005) Effect of immunoaffinity depletion of human serum during proteomic investigations. J. Proteome Res. 4, 1722–1731 [DOI] [PubMed] [Google Scholar]
- 12.Brand, J., Haslberger, T., Zolg, W., Pestlin, G., and Palme, S. ( 2006) Depletion efficiency and recovery of trace markers from a multiparameter immunodepletion column. Proteomics 6, 3236–3242 [DOI] [PubMed] [Google Scholar]
- 13.Qian, W. J., Liu, T., Monroe, M. E., Strittmatter, E. F., Jacobs, J. M., Kangas, L. J., Petritis, K., Camp, D. G., and Smith, R. D. ( 2005) Probability-based evaluation of peptide and protein identifications from tandem mass spectrometry and SEQUEST analysis: the human proteome. J. Proteome Res. 4, 53–62 [DOI] [PubMed] [Google Scholar]
- 14.Liu, T., Qian, W. J., Gritsenko, M. A., Xiao, W., Moldawer, L. L., Kaushal, A., Monroe, M. E., Varnum, S. M., Moore, R. J., Purvine, S. O., Maier, R. V., Davis, R. W., Tompkins, R. G., Camp, D. G., II, and Smith, R. D. ( 2006) High dynamic range characterization of the trauma patient plasma proteome. Mol. Cell. Proteomics 5, 1899–1913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nesvizhskii, A. I., Keller, A., Kolker, E., and Aebersold, R. ( 2003) A statistical model for identifying proteins by tandem mass spectrometry. Anal. Chem. 75, 4646–4658 [DOI] [PubMed] [Google Scholar]
- 16.Anderson, N. G., Willis, D. D., Holladay, D. W., Caton, J. E., Holleman, J. W., Eveleigh, J. W., Attrill, J. E., Ball, F. L., and Anderson, N. L. ( 1975) Analytical techniques for cell fractions. XX. Cyclic affinity chromatography: principles and applications. Anal. Biochem. 68, 371–393 [DOI] [PubMed] [Google Scholar]
- 17.Liu, H., Sadygov, R. G., and Yates, J. R., III ( 2004) A model for random sampling and estimation of relative protein abundance in shotgun proteomics. Anal. Chem. 76, 4193–4201 [DOI] [PubMed] [Google Scholar]
- 18.Qian, W. J., Jacobs, J. M., Camp, II, D. G., Monroe, M. E., Moore, R. J., Gritsenko, M. A., Calvano, S. E., Lowry, S. F., Xiao, W., Moldawer, L. L., Davis, R. W., Tompkins, R. G., and Smith, R. D. ( 2005) Comparative proteome analyses of human plasma following in vivo lipopolysaccharide administration using multidimensional separations coupled with tandem mass spectrometry. Proteomics 5, 572–584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zybailov, B., Coleman, M. K., Florens, L., and Washburn, M. P. ( 2005) Correlation of relative abundance ratios derived from peptide ion chromatograms and spectrum counting for quantitative proteomic analysis using stable isotope labeling. Anal. Chem. 77, 6218–6224 [DOI] [PubMed] [Google Scholar]
- 20.Wang, H., and Hanash, S. ( 2005) Intact-protein based sample preparation strategies for proteome analysis in combination with mass spectrometry. Mass Spectrom. Rev. 24, 413–426 [DOI] [PubMed] [Google Scholar]
- 21.Sheng, S., Chen, D., and Van Eyk, J. E. ( 2006) Multidimensional liquid chromatography separation of intact proteins by chromatographic focusing and reversed phase of the human serum proteome: optimization and protein database. Mol. Cell. Proteomics 5, 26–34 [DOI] [PubMed] [Google Scholar]
- 22.Keshishian, H., Addona, T., Burgess, M., Kuhn, E., and Carr, S. A. ( 2007) Quantitative, multiplexed assays for low abundance proteins in plasma by targeted mass spectrometry and stable isotope dilution. Mol. Cell. Proteomics 6, 2212–2229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Anderson, L., and Hunter, C. L. ( 2006) Quantitative mass spectrometric multiple reaction monitoring assays for major plasma proteins. Mol. Cell. Proteomics 5, 573–588 [DOI] [PubMed] [Google Scholar]
- 24.Stahl-Zeng, J., Lange, V., Ossola, R., Eckhardt, K., Krek, W., Aebersold, R., and Domon, B. ( 2007) High sensitivity detection of plasma proteins by multiple reaction monitoring of N-glycosites. Mol. Cell. Proteomics 6, 1809–1817 [DOI] [PubMed] [Google Scholar]
- 25.Haab, B. B., Geierstanger, B. H., Michailidis, G., Vitzthum, F., Forrester, S., Okon, R., Saviranta, P., Brinker, A., Sorette, M., Perlee, L., Suresh, S., Drwal, G., Adkins, J. N., and Omenn, G. S. ( 2005) Immunoassay and antibody microarray analysis of the HUPO Plasma Proteome Project reference specimens: systematic variation between sample types and calibration of mass spectrometry data. Proteomics 5, 3278–3291 [DOI] [PubMed] [Google Scholar]