Abstract
Little is known about the preanalytical fluctuations of phosphoproteins during tissue procurement for molecular profiling. This information is crucial to establish guidelines for the reliable measurement of these analytes. To develop phosphoprotein profiles of tissue subjected to the trauma of excision, we measured the fidelity of 53 signal pathway phosphoproteins over time in tissue specimens procured in a community clinical practice. This information provides strategies for potential surrogate markers of stability and the design of phosphoprotein preservative/fixation solutions. Eleven different specimen collection time course experiments revealed augmentation (±20% from the time 0 sample) of signal pathway phosphoprotein levels as well as decreases over time independent of tissue type, post-translational modification, and protein subcellular location (tissues included breast, colon, lung, ovary, and uterus (endometrium/myometrium) and metastatic melanoma). Comparison across tissue specimens showed an >20% decrease of protein kinase B (AKT) Ser-473 (p < 0.002) and myristoylated alanine-rich C-kinase substrate protein Ser-152/156 (p < 0.0001) within the first 90-min postexcision. Proteins in apoptotic (cleaved caspase-3 Asp-175 (p < 0.001)), proliferation/survival/hypoxia (IRS-1 Ser-612 (p < 0.0003), AMP-activated protein kinase β Ser-108 (p < 0.005), ERK Thr-202/Tyr-204 (p < 0.003), and GSK3αβ Ser-21/9 (p < 0.01)), and transcription factor pathways (STAT1 Tyr-701 (p < 0.005) and cAMP response element-binding protein Ser-133 (p < 0.01)) showed >20% increases within 90-min postprocurement. Endothelial nitric-oxide synthase Ser-1177 did not change over the time period evaluated with breast or leiomyoma tissue. Treatment with phosphatase or kinase inhibitors alone revealed that tissue kinase pathways are active ex vivo. Combinations of kinase and phosphatase inhibitors appeared to stabilize proteins that exhibited increases in the presence of phosphatase inhibitors alone (ATF-2 Thr-71, SAPK/JNK Thr-183/Tyr-185, STAT1 Tyr-701, JAK1 Tyr-1022/1023, and PAK1/PAK2 Ser-199/204/192/197). This time course study 1) establishes the dynamic nature of specific phosphoproteins in excised tissue, 2) demonstrates augmented phosphorylation in the presence of phosphatase inhibitors, 3) shows that kinase inhibitors block the upsurge in phosphorylation of phosphoproteins, 4) provides a rational strategy for room temperature preservation of proteins, and 5) constitutes a foundation for developing evidence-based tissue procurement guidelines.
Elucidation of pathogenic protein signaling networks within tumors offers tremendous promise as a means to individualize molecular targeted cancer therapy. Despite this promise, the data obtained from a diagnostic assay applied to human tissue must be stable, reproducible, monitored, and validated; otherwise a clinical decision may be based on incorrect or biased information.
Modulation of ongoing cellular kinase activity represents one of the most rapidly growing arenas in new drug discovery. Identification of specific phosphoprotein signaling aberrations can be used for the development of targeted therapies for patients with lung, breast, colon, or other cancer (1–6). Profiling the tumor phosphoproteome using human tumor biopsy specimens is an important component of the strategy for individualized cancer therapy (1–6). Nevertheless unless we understand preanalytical variables affecting the fidelity of the phosphoproteome we will fail to utilize this rich repertoire of biological information.
The perishability of tissue molecules may be profoundly influenced by the timing and treatment of the tissue after it is harvested. Tissue is generally snap frozen to perform proteomics research studies. In a busy clinical setting, it may be impossible to immediately preserve procured tissue in liquid nitrogen. Moreover the time delay from patient excision to pathologic examination and molecular analysis is often not recorded and may vary from 30-min to many hours depending on the time of day, length of the procedure, time for pathologic examination, and number of concurrent cases.
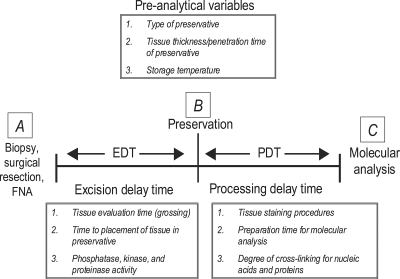
Very little is known about the stability of phosphoproteins in human tissue following surgical procurement, e.g. with regard to the effects of time, method of preservation, organ and tissue of origin, and means of extraction on the yield and fidelity of classes of phosphoproteins. Measurements conducted in tissue may reflect reactive changes in the tissue unrelated to the pathologic state at the time of procurement. Moreover it is unclear whether phosphorylation of certain residues or certain classes of cellular proteins is more labile than that in other classes. The phosphorylated state of a signal protein at any point in time is a function of the balance between kinase and phosphatase enzymes acting on that protein. Because tissue is in a living state following procurement, both kinases and phosphatases remain active. Consequently as the procured tissue incubates in the surgery collection pan or on the pathologist's cutting board, the phosphorylated state of a given phosphoprotein can fluctuate upward or downward, depending on which specific phosphoprotein pathways become activated or suppressed ex vivo (7, 8). Furthermore there exists no means to monitor the quality and yield of phosphoproteins starting from procurement through extraction and quantitative analysis (Fig. 1).
Fig. 1.
Preanalytical variables during tissue acquisition. Categories of preanalytical variables are divided into EDT and PDT. EDT is the time between tissue excision and initiation of specimen preservation (e.g. freezing or fixation); PDT is the time period between sample preservation and molecular analysis (e.g. immunoassay, mass spectrometry, or gene transcript array). FNA, fine needle aspiration.
In the present study we attempted to gather fundamental information about the ex vivo survival of tissue kinase pathways by (a) gathering previously unknown quantitative time course data tracking the stability of tissue phosphoproteins within human surgical tissue collected in the clinical operating room setting, (b) identifying the most labile phosphoproteins and their classes of signal pathways within the first several hours after procurement (Fig. 2), (c) using the addition of phosphatase and kinase inhibitors to study the activity state of kinase pathways in the living tissue postprocurement ex vivo, and (d) interpreting this information to guide strategies for tissue procurement protocols and optimal tissue phosphoprotein stabilization chemistries.
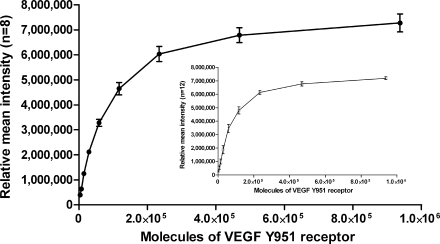
Fig. 2.
Inter- and intraslide precision of reverse phase protein microarrays. Averages of the replicate spot intensities at each dilution were calculated between arrays (n = 8, mean ± S.D.) and within a single array (n = 12, mean ± S.D.). Between-run precision (black circles) of the RPPA ranged from 4.0% (coefficient of variation) in the middle of the linear range to 17.8% at the lower limit of detection with a correlation coefficient of R2 = 0.9761. Within-run precision (inset, white squares) ranged from 5.0 to 18.0% (coefficient of variation) within the linear range with a correlation coefficient of R2 = 0.9693. As a model system, sensitivity of the arrays was 3,660 molecules of phosphorylated VEGFR Tyr-951 (±2 S.D. above background).
EXPERIMENTAL PROCEDURES
Specimens and Patient Data
Surgical tissue specimens were collected from 11 patients under informed consent following an Institutional Review Board-approved protocol in a community hospital (Inova Fairfax Hospital, Fairfax, VA). Tissue was excised in the surgical suite following standard of care guidelines. Tissue was transported at room temperature to the surgical frozen section room. A board-certified pathologist performed gross examination of each tissue sample and provided non-diseased and/or diseased tissue that was not required for diagnosis. Tissue samples were obtained from the following organs: breast, colon, lung, ovary, uterus, and metastatic melanoma (Table I).
Table I.
Clinical sample characteristics and time to cryopreservation data
| Sample 1 | Sample 2 | Sample 3 | Sample 4 | Sample 5 | Sample 6 | Sample 7 | Sample 8 | Sample 9 | Sample 10 | Sample 11 | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Organ | Uterus | Colon | Lung | Lung | Uterus | Ovary | Uterus | Uterus | Breast | Breast | Axillary lymph node |
| Tissue | Endometrium/myometrium | Mucosa, submucosa, muscularis propria, serosa | Alveolar parenchyma | Lung wedge biopsy | Myometrium | Ovary, epithelium | Endometrium/myometrium | Myometrium | Tumor, stroma, and adipose | Mastectomy epithelium, fibrous, and adipose | Lymph node |
| Pathologic diagnosis | Benign polyp | Normal | Normal parenchyma | Squamous cell tumor | Leiomyoma | Mucinous adenocarcinoma | Normal | Leiomyoma | Fibroadenoma | DCIS cribriform variant | Metastatic melanoma |
| Specimen size | 46 × 25 mm | 36 × 25 mm | 10 × 5 mm | 10 × 5 mm | 36 × 25 mm | 15 × 10 mm | 25 × 25-mm wedge | 25-mm diameter × 5 mm thick | 10 × 5 mm, 2 pieces | 5 × 3.5 cm, 4 × 2.5 cm | 9 × 15 mm, 12 × 2.5 mm, 8 × 3 mm, 6 × 3 mm, 5 × 5 mm |
| Elapsed time from excision to initial preservation (min, T0) | 4 | 40 | 10 | 10 | 10 | 30 | 30 | 26 | 18 | 20 | 15 |
| Time course range (min) | 4–91 | 40–70 | 10–20 | 10–20 | 10–190 | 30–120 | 30–120 | 26–949 | 18–138 | 20–320 | 30–90 |
| Temperature (temp; °C) | Room temp | Room temp | Room temp | Room temp | Room temp | Room temp | Room temp and 4 °C | Room temp | Room temp | Room temp | Room temp |
| Time to cryopreservation statistics | |||||||||||
| Average time to cryopreservation: 19.3 min | |||||||||||
| Earliest time: 4 min | |||||||||||
| Longest time: 40 min | |||||||||||
| Mode: 10 |
Specimen 1: Uterus (Endometrium/Myometrium)—
A 46 × 25-mm section of non-diseased uterus from a vaginal hysterectomy was cut with a scalpel into 16 relatively homogeneous pieces (Tables I and II and Fig. 3). Two pieces of tissue, designated as time 0, were immediately processed by embedding one piece in a cryomold with Optimal Cutting Temperature compound (OCT;1 Sakura, Torrence, CA) and then freezing on dry ice; the second piece was placed in a cryovial and snap frozen by immersion in liquid nitrogen (time 0 was 4 min postexcision). The remaining pieces were incubated at room temperature for 9, 16, 24, 34, 48, 60, or 91 min postexcision. Immediately following incubation, one tissue piece was embedded in a cryomold with OCT and frozen on dry ice. A second piece was placed in a cryovial and snap frozen. Tissue blocks/cryovials were stored at −80 °C. Cryosections (8 μm) were prepared from the OCT-embedded tissue blocks using a Tissue Tek (Leica) cryostat. The sections were stored at −80 °C prior to staining with a modified hematoxylin and eosin procedure. Briefly the cryosections were fixed in 70% ethanol, stained in Mayer's hematoxylin, dehydrated in graded ethanol solutions, rinsed in xylene, and air-dried. Tissue sections were immediately solubilized in extraction buffer (tissue protein extraction reagent (T-PER) (Pierce), 2-mercaptoehtanol (Sigma), and 2× SDS-Tris-glycine loading buffer (Invitrogen)) and denatured by heating for 8 min at 100 °C (9–11). Tissue lysates were stored at −80 °C.
Table II.
Initial time course experiments with human tissue collected in a community hospital
UE, >20% increase within first 60 min postexcision; UM, >20% increase between 60 and 120 min postexcision; UL, >20% increase more than 120 min postexcision; DE, >20% decrease within first 60 min postexcision; DM, >20% decrease between 60 and 120 min postexcision; DL, >20% decrease more than 120 min postexcision; blank, not tested.
| Protein | Function | Significant changes over time course range postexcision
|
|||||
|---|---|---|---|---|---|---|---|
| Sample 1: uterus, benign; 4–91 min | Sample 2: colon, normal; 40–70 min | Sample 3: lung parenchyma; 10–20 min | Sample 4: lung squamous cell carcinoma; 10–20 min | Sample 5: uterine leiomyoma; 10–190 min | Sample 7: uterus, benign; 30–120 min | ||
| BAD (Ser-112) | Apoptosis | UE | a | a | a | a | a |
| Caspase-3, cleaved (Asp-175) | Apoptosis | UE | UE/DM | DE | UE | UE | UE |
| Caspase-7, cleaved (Asp-198) | Apoptosis | UE/DL | |||||
| Caspase-9, cleaved (Asp-330) | Apoptosis | DL | UE | ||||
| CHK1 (Ser-345) | Cell cycle | UM | |||||
| Cyclin A | Cell cycle | UM | |||||
| Acetyl CoA Carboxylase (Ser-79) | Hypoxia/ischemia | a | a | ||||
| eNOS (Ser-1177) | Hypoxia/ischemia | UE | |||||
| HIF-1α | Hypoxia/ischemia | DE/UL | |||||
| p38 MAPK (Thr-180/Tyr-182) | Hypoxia/ischemia | UE | DE | UE | UE | a | DE |
| VEGFR 2 (Tyr-951) | Hypoxia/ischemia | DL | |||||
| AKT (Ser-473) | Proliferation/survival | DE | DE | DE | DE | UE | DE |
| AKT (Thr-308) | Proliferation/survival | UE | DE | a | UE | UE | a |
| EGFR (Tyr-1148) | Proliferation/survival | UE | DE | a | UE | a | |
| eIF4G (Ser-1108) | Proliferation/survival | UE | |||||
| ERK 1/2 (Thr-202/Tyr-204) | Proliferation/survival | UE | DE | a | UE | UE | DE |
| GSK3α (Ser-21/9) | Proliferation/survival | UE | a | UE | UE | a | |
| IRS-1 (Ser-612) | Proliferation/survival | UE | DE | DE | UE | UE | a |
| PDK1 (Ser-241) | Proliferation/survival | UE | |||||
| PKCα/β II (Thr-638/641) | Proliferation/survival | UE | |||||
| ASK1 (Ser-83) | Stress/inflammation | UE | UE/UM | ||||
| ATF-2 (Thr-71) | Stress/inflammation | a | |||||
| IκB-α (Ser-152/156) | Stress/inflammation | a | a | DE | UE | UE | DE/UL |
| JAK1 (Tyr-1022/1023) | Stress/inflammation | ||||||
| MARCKS (Ser-152/156) | Stress/inflammation | DE | DE | DE | DE | DL | DE/DM |
| SAPK/JNK (Thr-183/Tyr-185) | Stress/inflammation | UE | |||||
| Src family (Tyr-416) | Stress/inflammation | DE | |||||
| CREB (Ser-133) | Transcription factor | UE | a | a | a | UE | a |
| STAT1 (Tyr-701) | Transcription factor | UE | DE | UE | UE | UE | a |
| STAT3 (Tyr-705) | Transcription factor | UE | |||||
No change.
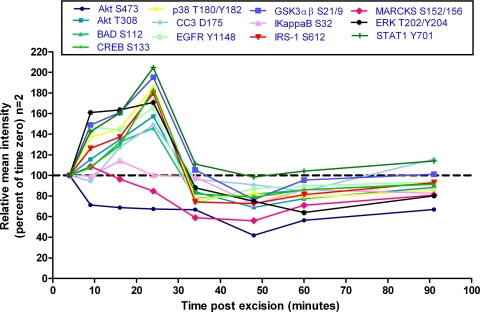
Fig. 3.
Uterine tissue room temperature time course. Uterine tissue was incubated at room temperature and analyzed by reverse phase protein microarray for fluctuations in post-translationally modified proteins. Increases and decreases in specific protein levels are noted over time compared with the initial (time 0) tissue aliquot that was frozen 4 min after excision (represented as 100%). Fluctuations ±20% were considered significant because this coincided with 2 S.D. wherein 1 S.D. is ∼10% of the mean as shown in precision studies (Fig. 2).
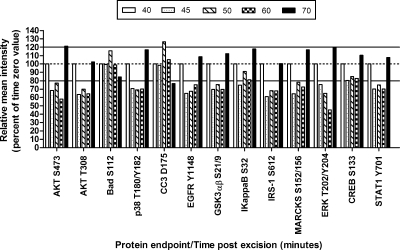
Specimen 2: Colon—
A 36 × 25-mm section of non-diseased colon was cut with a scalpel into 10 relatively homogeneous pieces (Tables I and II and Fig. 4). Tissue was processed as described for specimen 1. Time 0 was 40 min postexcision. The remaining pieces were incubated at room temperature for 45, 50, 60, or 70 min postexcision.
Fig. 4.
Colon tissue room temperature time course. Tissue specimens incubated at room temperature beyond time 0 (40 min postexcision) exhibited fluctuations in specific phosphoprotein analyte levels over time. The initial time 0 aliquot (100%) was frozen immediately, whereas the subsequent tissue aliquots were frozen at various times after room temperature incubation. White, 40 min postexcision; speckled, 45 min; hatched, 50 min; checked, 60 min; black, 70 min postexcision. Solid lines represent ±20% change from the time 0 value.
Specimens 3 and 4: Lung (Non-diseased Lung Alveolar Parenchyma with Patient-matched Squamous Cell Carcinoma)—
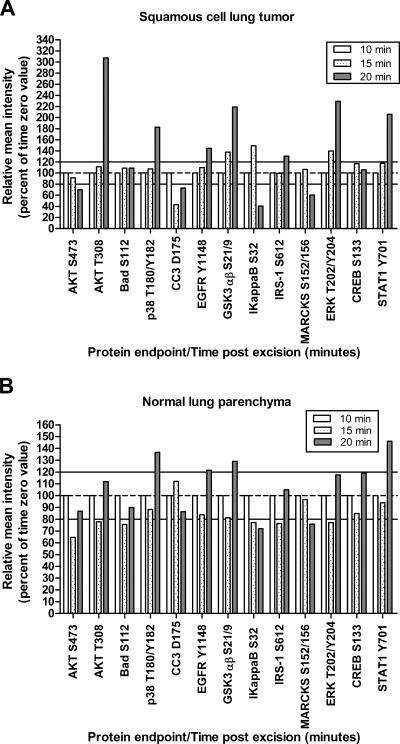
Two pieces of a lung wedge biopsy, each 10 × 5 mm, were provided from the same patient (Tables I and II and Fig. 5). Sample 3 was non-diseased lung parenchyma, and sample 4 was squamous cell carcinoma. Tissue was processed as described for specimen 1. Time 0 was 10 min postexcision. The remaining pieces were incubated at room temperature for 15 and 20 min postexcision.
Fig. 5.
Lung tissue room temperature time course. Lung tissue incubated at room temperature shows fluctuations (±20%) during a short (20-min) time span compared with time 0 (10 min). A, squamous cell lung cancer. B, patient matched normal lung parenchyma. White, 10 min postexcision; speckled, 15 min; gray, 20 min.
Specimen 5: Uterus Leiomyoma (Myometrium)—
A 36 × 25-mm uterine leiomyoma sample was cut with a scalpel into eight relatively homogeneous pieces (Tables I and II). Time 0 was 10 min postexcision. Tissue pieces were incubated at room temperature for 21, 40, 70, 100, 130, 160, and 190 min postexcision prior to embedding in OCT. Cryosectioning, tissue staining, and lysis were performed as described for specimen 1.
Specimen 6: Ovary Tumor—
We collected duplicate samples from a patient diagnosed with an ovarian mucinous adenocarcinoma (Tables I and II and Fig. 6). A 20 × 20-mm section of ovary tissue was cut with a scalpel into 10 relatively homogeneous pieces. Time 0 was 30 min postexcision. Duplicate tissue pieces were incubated at room temperature for 50, 60, 90, or 120 min postexcision prior to snap freezing in liquid nitrogen.
Fig. 6.
Replicate ovarian tissue room temperature time course. A, proteins in stress, adhesion, proliferation/survival, cell cycle, and hypoxia pathways show significant changes over time compared with time 0 (30 min postexcision) n = 2, (mean ± 2 S.D.). ANOVA comparison of means is shown: JAK1 Tyr-1022/1023 (60 min, p < 0.0430; 90 min, p < 0.0156), PAK1 Ser-199/204/PAK2 Ser-192/197 (60 min, p < 0.0418; 90 min, p < 0.0222; 120 min, p < 0.0179), TSC2 Tyr-1571 (90 min, p < 0.0129; 120 min, p < 0.0095), and CHK1 Ser-345 (50 min, p < 0.0238; 60 min, p < 0.0272; 90 min, p < 0.0285; 120 min, p < 0.0172). B, ATF-2 Thr-71 (50 min, p < 0.0503; 60 min, p < 0.0496; 90 min, p < 0.0187; 120 min, p < 0.0075) and eNOS Ser-1177 (50 min, p < 0.0082; 90 min, p < 0.0158; 120 min, p < 0.0149). Statistically significant differences are denoted by asterisks (*). FAK, focal adhesion kinase.
Snap frozen tissue samples were weighed on an analytical balance (Mettler-Toledo) and then pulverized using a Bio-Pulverizer (Research Products International) or a mortal and pestle on dry ice. Protein extraction buffer was added directly to the frozen pulverized tissue. The lysates were denatured by heating at 100 °C for 8 min. Lysates were stored at −80 °C.
Evaluation of Storage Temperature and Preservative Solutions: Specimen 7: Uterus (Endometrium and Myometrium)
To evaluate the effect of storage temperature conditions on post-translationally modified proteins over time, a 25 × 25-mm cross-section of intact uterine tissue (endometrium/myometrium) was cut into 24 relatively homogeneous pieces (Tables I and II). Time 0 was 30 min postexcision. Four pieces of tissue were incubated at 4 °C for 45, 60, 90, or 120 min postexcision (Fig. 7A). An additional four pieces of tissue were incubated at room temperature (Fig. 7B). Tissue was processed as described for specimen 5.
Fig. 7.
4 °C and room temperature time course. A, 4 °C time course for normal uterine tissue. B, room temperature time course for normal uterine tissue. Each analyte listed above the graph is displayed in a different color and symbol as designated. Increases and decreases in specific protein levels are noted over time compared with the initial (time 0) tissue aliquot that was frozen 30 min after excision (represented as 100%). Each time point represents an individual piece of tissue (mean of replicate array spots, n = 2). AcCoA, acetyl-CoA carboxylase.
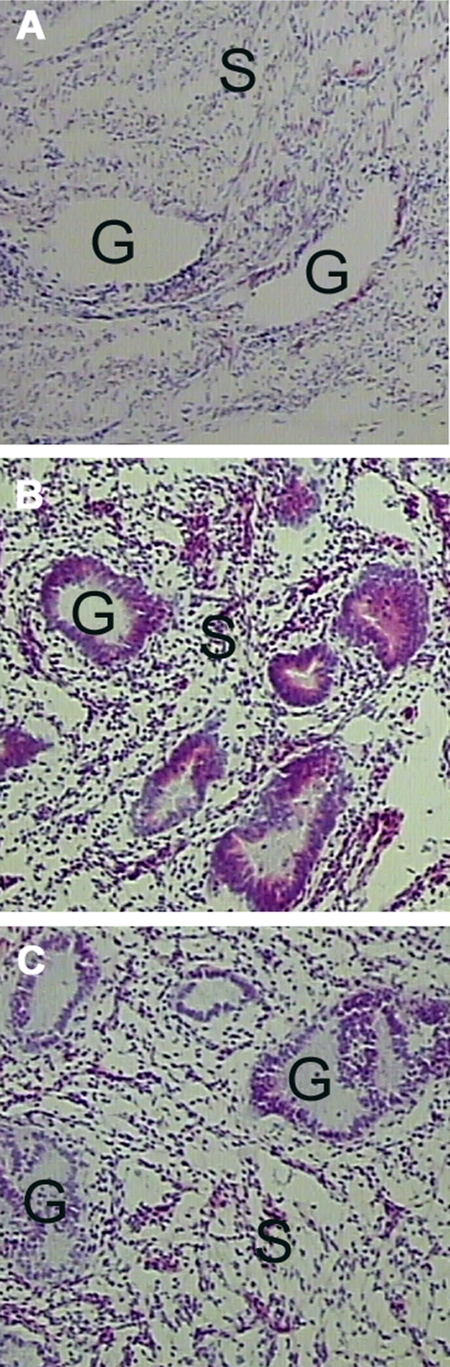
To evaluate the effect of preservative chemistries and phosphatase inhibitors on cellular histomorphology, one of the remaining 15 pieces of tissue was immersed in each of the following solutions 45 min postexcision (see Fig. 13). Base solutions were Cytolyt® (Cytyc Corp., Marlborough, MA), ethanol (Sigma), methanol (Sigma), or Hanks’ balanced salt solution (HBSS) (ThermoFisher, Pittsburgh, PA) with one or more of the following additives: polyethylene glycol (ThermoFisher), Complete™ protease inhibitor tablet (Roche Applied Science), sodium orthovanadate (Sigma), and β-glycerophosphate (EMD Biosciences/Calbiochem). Tissue was removed from solution after 24 h, embedded in OCT, and frozen on dry ice. Tissue was processed as described for specimen 1.
Fig. 13.
A precipitating fixative with phosphatase and protease inhibitors retains full tissue histomorphology. A, frozen section H&E stain of uterine endometrium and myometrium. B, H&E stain of uterine endometrium and myometrium postincubation in a preservative solution containing methanol, polyethylene glycol, protease inhibitor, sodium orthovanadate, and β-glycerophosphate. C, H&E stain of uterine endometrium and myometrium postincubation in a preservative solution containing ethanol, polyethylene glycol, protease inhibitor, sodium orthovanadate, and β-glycerophosphate. G, endometrial glands; S, stroma.
Precision Studies Using Homogeneous Tissue: Specimen 8: Uterus Leiomyoma (Myometrium)
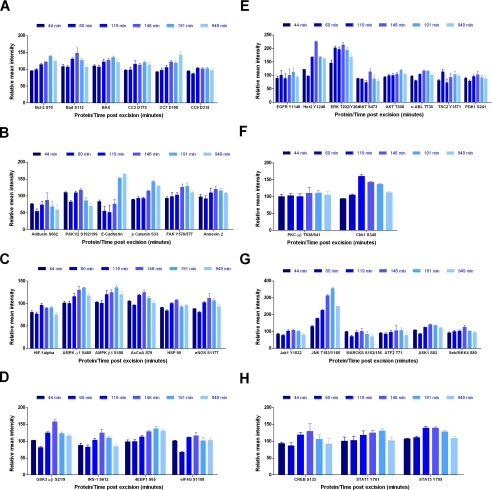
A 25 × 5-mm cross-section of a uterine leiomyoma was cut into 24 uniform, 4 × 4.5-mm cores using a 4-mm Keyes dermal punch (Tables I and III and Fig. 8, A–H). At time 0 (26 min postexcision), three replicate samples were placed in a cryovial and snap frozen in liquid nitrogen. Replicate samples were incubated at room temperature and snap frozen at 44, 60, 119, 146, 191, and 949 min postexcision (Table III). Cryosectioning and tissue lysis was performed as for specimen 6.
Table III.
Evaluation of post-translationally modified proteins over time for uterine leiomyoma tissue
UE, >20% increase within first 60 min postexcision; UM, >20% increase between 60 and 120 min postexcision; UL, >20% increase more than 120 min postexcision; DE, >20% decrease within first 60 min postexcision; DM, >20% decrease between 60 and 120 min postexcision; DL, >20% decrease more than 120 min postexcision.
| Protein | Function | Significant changes over time |
p values for sample 8, uterine leiomyoma (n = 24 tissue pieces), at time postexcision (time 0 = 26 min)
|
|||||
|---|---|---|---|---|---|---|---|---|
| 44 min (n = 3) | 60 min (n = 3) | 119 min (n = 4) | 146 min (n = 4) | 191 min (n = 4) | 949 min (n = 5) | |||
| Adducin (Ser-662) | Adhesion/cytoskeleton | DE/DL | 0.0013 | 0.0437 | 0.0166 | 0.002 | ||
| Annexin II | Adhesion/cytoskeleton | UL | 0.0088 | 0.0408 | ||||
| Catenin (β) (Ser-33/37/Thr-41) | Adhesion/cytoskeleton | UL | 0.0034 | 0.0278 | ||||
| E-cadherin | Adhesion/cytoskeleton | a | a | a | a | a | a | a |
| FAK (Tyr-576/577) | Adhesion/cytoskeleton | UM/JL | 0.0366 | 0.239 | ||||
| PAK1 (Ser-199/204)/PAK2 (Ser-192/197) | Adhesion/cytoskeleton | a | a | a | a | a | a | a |
| BAD (Ser-112) | Apoptosis | UM | 0.0071 | |||||
| BAX | Apoptosis | UM/UL | 0.0052 | 0.0009 | <0.0001 | 0.006 | ||
| BCL-2 (Ser-70) | Apoptosis | UL | 0.0026 | 0.0359 | ||||
| Caspase-3, cleaved (Asp-175) | Apoptosis | a | a | a | a | a | a | a |
| Caspase-7, cleaved (Asp-198) | Apoptosis | UL | 0.0029 | |||||
| Caspase-9, cleaved (Asp-330) | Apoptosis | DE | 0.0462 | 0.0029 | ||||
| CHK1 (Ser-345) | Cell cycle | UM | 0.0141 | 0.0725 | ||||
| Acetyl-CoA carboxylase (Ser-79) | Hypoxia/ischemia | UE/UM | 0.0218 | 0.0037 | ||||
| AMPKα1 (Ser-485) | Hypoxia/ischemia | UM/UL | 0.0064 | 0.002 | ||||
| AMPKβ1 (Ser-108) | Hypoxia/ischemia | UM/UL | 0.0212 | 0.0061 | 0.0002 | 0.0161 | ||
| eNOS (Ser-1177) | Hypoxia/ischemia | a | a | a | a | a | a | a |
| HIF-1α | Hypoxia/ischemia | DE/DL | 0.1043 | 0.0029 | 0.001 | |||
| HSP 90 | Hypoxia/ischemia | a | a | a | a | a | a | a |
| 4EBP1 (Ser-65) | Proliferation/survival | UL | 0.0004 | <0.0001 | 0.0001 | |||
| AKT (Ser-473) | Proliferation/survival | a | a | a | a | a | a | a |
| AKT (Thr-308) | Proliferation/survival | UL | 0.0245 | |||||
| c-ABL (Thr-735) | Proliferation/survival | DE/UM/UL | 0.0029 | 0.0103 | 0.0143 | |||
| EGFR (Tyr-1148) | Proliferation/survival | a | a | a | a | a | a | a |
| eIF4G (Ser-1108) | Proliferation/survival | a | a | a | a | a | a | a |
| ErbB2/HER2 (Tyr-1248) | Proliferation/survival | UL | 0.0015 | |||||
| ERK 1/2 (Thr-202/Tyr-204) | Proliferation/survival | UE/UM/UL | 0.0001 | 0.0002 | <0.0001 | 0.0003 | 0.0035 | |
| GSK3α (Ser-21/9) | Proliferation/survival | UL | 0.0131 | |||||
| IRS-1 (Ser-612) | Proliferation/survival | a | a | a | a | a | a | a |
| PDK1 (Ser-241) | Proliferation/survival | DE | 0.0355 | |||||
| PKCα/β II (Thr-638/641) | Proliferation/survival | a | a | a | a | a | a | a |
| Tuberin/TSC2 (Tyr-1571) | Proliferation/survival | a | a | a | a | a | a | a |
| ASK1 (Ser-83) | Stress/inflammation | UM/UL | 0.0358 | 0.0016 | 0.0044 | |||
| ATF-2 (Thr-71) | Stress/inflammation | a | a | a | a | a | a | a |
| JAK1 (Tyr-1022/1023) | Stress/inflammation | a | a | a | a | a | a | a |
| MARCKS (Ser-152/156) | Stress/inflammation | a | a | a | a | a | a | a |
| NFκB (Ser-536) | Stress/inflammation | a | a | a | a | a | a | a |
| SAP/JNK (Thr-183/Tyr-185) | Stress/inflammation | UE/UM/UL | 0.0437 | 0.0025 | <0.0001 | <0.0001 | 0.0004 | |
| SEK1/MKK4 (Ser-80) | Stress/inflammation | a | a | a | a | a | a | a |
| STAT1 (Tyr-701) | Transcription factor | UM/UL | 0.0371 | 0.0106 | ||||
| STAT3 (Tyr-705) | Transcription factor | UM/UL | <0.0001 | <0.0001 | 0.0002 | |||
No significant change.
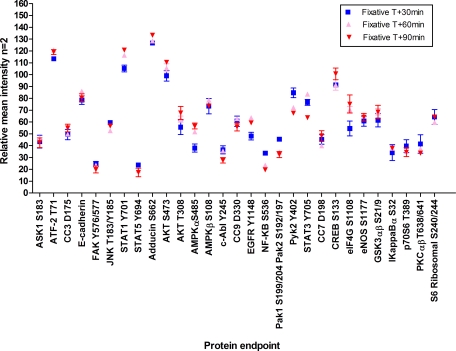
Fig. 8.
Protein fluctuations over time by cell signaling pathway. Leiomyoma tissue replicates incubated at room temperature and analyzed by RPPA reveal pathway- and protein-specific increases and decreases over time (n = 3 for 44 and 60 min; n = 4 for 119, 146, and 191 min; n = 5 for 949 min; ±S.D. as a percentage of time 0 sample). Cell signaling pathways represented are: apoptosis (A), adhesion (B), hypoxia (C), glucose/insulin signaling (D), proliferation/survival (E), cell cycle (F), stress/inflammation (G), and transcription factors (H). FAK, focal adhesion kinase; AcCoA, acetyl-CoA carboxylase.
Effects of Phosphatase and Kinase Inhibitors
Specimen 9: Breast (Fibroadenoma) with Surrounding Stroma/Adipose Tissue—
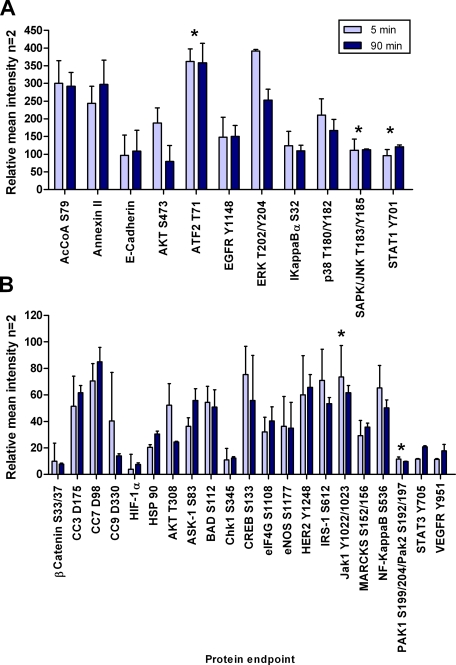
Two 10 × 5-mm breast tissue samples containing tumor, stroma, and adipose tissue were cut into six pieces with a scalpel (Table I and Fig. 11). Six samples were immersed in a solution of ethanol, polyethylene glycol, HBSS, sodium orthovanadate, β-glycerophosphate, staurosporine, and genistein 138 min postexcision. Duplicate samples were removed from solution after 5 min, 90 min, and 24 h; embedded in OCT; and frozen on dry ice. Cryosections were prepared as described for specimen 1.
Fig. 11.
Kinase inhibitors plus phosphatase inhibitors stabilize phosphoproteins over 90 min. Breast fibroadenoma with surrounding stroma tissue was incubated in a solution containing a precipitating fixative, a permeation enhancer, balanced salt solution, and phosphatase and kinase inhibitors. Proteins indicated by an asterisk (*) did not show changes (±20%) over time in the preservative solution indicating attenuation of phosphatase and kinase activity in the tissue samples. Light blue, 5-min incubation; dark blue, 90-min incubation. (n = 2, mean ± S.D.). AcCoA, acetyl-CoA carboxylase.
Specimen 10: Breast (Ductal Carcinoma in Situ) with Surrounding Stroma/Adipose Tissue—
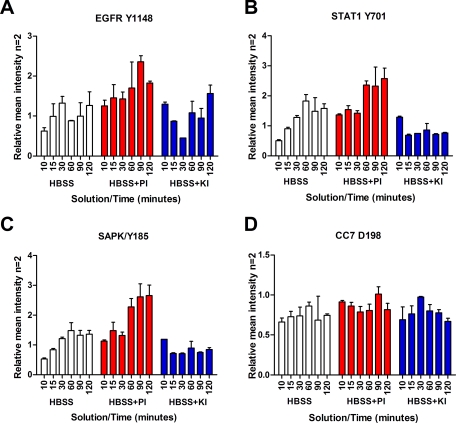
Two breast tissue samples (5 × 3.5 cm and 4 × 2.5 cm) were cut into 40 relatively homogenous pieces. At time 0, 20 min postexcision, duplicate samples were placed in a cryovial and snap frozen in liquid nitrogen. Duplicate samples were incubated at room temperature for 44, 50, 80, 110, 140, 230, 260, and 320 min postexcision and snap frozen in liquid nitrogen (Tables I and IV). The remaining 22 samples were immersed in either HBSS (ThermoFisher), HBSS with sodium orthovanadate (Sigma) and β-glycerophosphate (Calbiochem), or HBSS with staurosporine (Sigma) and genistein (Sigma) 42 min postexcision (Fig. 9). Duplicate tissue samples were removed from each solution after 10, 20, 30, 60, 90, or 120 min and snap frozen in liquid nitrogen. Tissue lysis was performed as for specimen 6.
Table IV.
Time course of breast tissue (DCIS, stroma, and adipose) postexcision
UE, >20% increase within first 60 min postexcision; UM, >20% increase between 60 and 120 min postexcision; UL, >20% increase more than 120 min postexcision; DE, >20% decrease within first 60 min postexcision; DM, >20% decrease between 60 and 120 min postexcision; DL, >20% decrease more than 120 min postexcision.
| Protein | Function | Significant changes over time |
p values for sample 10, breast DCIS, epithelium, and adipose tissue (n = 18 tissue pieces),at time postexcision (time 0 = 20 min)
|
|||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 44 min (n = 2) | 50 min (n = 2) | 80 min (n = 2) | 110 min (n = 2) | 140 min (n = 2) | 230 min (n = 2) | 260 min (n = 2) | 320 min (n = 2) | |||
| Catenin (β) (Ser-33/37/Thr-41) | Adhesion/cytoskeleton | DE/DM/DL | 0.0013 | 0.0083 | 0.0006 | 0.0085 | 0.0063 | 0.0166 | ||
| E-cadherin | Adhesion/cytoskeleton | UE/UM/UL | 0.0005 | 0.0001 | 0.0005 | 0.0319 | 0.0001 | |||
| PAK1 (Ser-199/204)/PAK2 (Ser-192/197) | Adhesion/cytoskeleton | UL | 0.0304 | |||||||
| BAD (Ser-112) | Apoptosis | DL | 0.0132 | 0.0064 | ||||||
| Caspase-3, cleaved (Asp-175) | Apoptosis | UE/UM/UL | <0.0001 | 0.004 | <0.0001 | 0.0018 | 0.0211 | 0.0099 | ||
| Caspase-7, cleaved (Asp-198) | Apoptosis | UE/UM/UL | 0.03 | 0.0139 | 0.0098 | |||||
| Caspase-9, cleaved (Asp-330) | Apoptosis | UE/UM | 0.00333 | 0.0094 | ||||||
| CHK1 (Ser-345) | Cell cycle | a | a | a | a | a | a | a | a | a |
| Acetyl-CoA carboxylase (Ser-79) | Hypoxia/ischemia | UE/UL | 0.0049 | 0.0003 | 0.0484 | |||||
| AMPKα1 (Ser-485) | Hypoxia/ischemia | UE/UL/DL | 0.011 | 0.0328 | 0.011 | 0.0493 | ||||
| AMPKβ1 (Ser-108) | Hypoxia/ischemia | UE/UL/DL | 0.005 | 0.0054 | 0.032 | |||||
| eNOS (Ser-1177) | Hypoxia/ischemia | a | a | a | a | a | a | a | a | a |
| HSP 90 | Hypoxia/ischemia | UL | 0.0341 | |||||||
| VEGF receptor-2 (Tyr-1175) | Hypoxia/ischemia | UE/UM/UL | 0.0188 | 0.004 | 0.0024 | |||||
| AKT (Ser-473) | Proliferation/survival | UE/DL | 0.0288 | 0.0016 | 0.0089 | 0.0021 | 0.0393 | |||
| AKT (Thr-308) | Proliferation/survival | DM/DL | 0.0066 | 0.0076 | 0.0166 | |||||
| c-ABL (Thr-735) | Proliferation/survival | a | a | a | a | a | a | a | a | a |
| EGFR | Proliferation/survival | DM/DL | 0.0382 | 0.0297 | ||||||
| EGFR (Tyr-1148) | Proliferation/survival | DE/DM/DL | 0.0139 | 0.0113 | 0.0132 | 0.004 | 0.0072 | 0.0126 | 0.0034 | 0.016 |
| eIF4G (Ser-1108) | Proliferation/survival | UE/UM/UL | 0.029 | 0.0111 | 0.025 | |||||
| ErbB2 | Proliferation/survival | DM/DL | 0.027 | 0.0471 | 0.0271 | 0.0394 | ||||
| ERK 1/2 (Thr-202/Tyr-204) | Proliferation/survival | DE/DM/DL | 0.0197 | 0.027 | 0.0065 | 0.0129 | 0.0116 | 0.0246 | ||
| GSK3α (Ser-21/9) | Proliferation/survival | DM/DL | 0.0203 | 0.0028 | 0.0091 | 0.0309 | ||||
| HER3 (Tyr-1289) | Proliferation/survival | UL | 0.0241 | |||||||
| IRS-1 (Ser-612) | Proliferation/survival | UE/UM/UL | 0.0221 | 0.0021 | 0.0092 | 0.0003 | ||||
| mTOR (Ser-2481) | Proliferation/survival | a | a | a | a | a | a | a | a | a |
| PKCα/β II (Thr-638/641) | Proliferation/survival | DM/DL | 0.0402 | 0.0451 | 0.0194 | 0.0464 | ||||
| ASK1 (Ser-83) | Stress/inflammation | DE/DM/DL | 0.0055 | 0.0085 | 0.003 | 0.0021 | 0.0055 | |||
| IκB-α (Ser-32/36) | Stress/inflammation | DE/DM/DL | 0.0266 | 0.0107 | 0.0255 | 0.0042 | 0.0026 | 0.0137 | 0.0019 | 0.0093 |
| MARCKS (Ser-152/156) | Stress/inflammation | UE/UM/DM/DL | 0.0013 | 0.0001 | 0.0001 | 0.0087 | 0.0023 | 0.0029 | ||
| SAPK/JNK (Thr-183/Tyr-185) | Stress/inflammation | DM/UL | 0.0347 | 0.0031 | ||||||
| CREB (Ser-133) | Transcription factor | UM/DL | 0.005 | 0.0109 | 0.0023 | 0.0012 | 0.011 | 0.0039 | ||
| STAT1 (Tyr-701) | Transcription factor | UE/UM/UL | 0.0079 | 0.0057 | <0.0001 | 0.0156 | 0.0068 | 0.0002 | ||
| STAT3 (Ser-727) | Transcription factor | a | a | a | a | a | a | a | a | a |
No significant change.
Fig. 9.
Phosphatase inhibitors augment phosphorylation levels in tissue, whereas kinase inhibitors decrease phosphorylation levels. Reverse phase protein microarray analysis of breast tissue incubated for 10–120 min in Hanks’ balanced salt solution only (white), with phosphatase inhibitors (PI) sodium orthovanadate and β-glycerophosphate (red), or with kinase inhibitors (KI) staurosporine and genistein (blue) is shown. A, EGFR Tyr-1148; B, STAT1 Tyr-701; C, SAPK/JNK Thr-183/Tyr-185; D, cleaved caspase-7 Asp-198 (n = 2, mean ± S.D.).
Specimen 11: Metastatic Melanoma to Axillary Lymph Node—
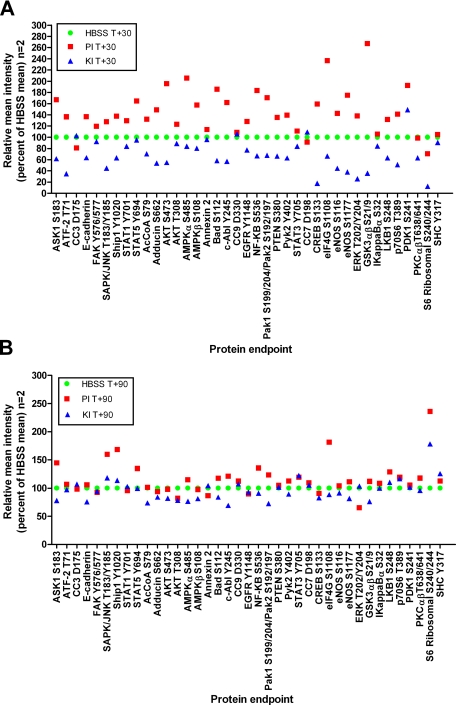
Duplicate tissue samples were embedded in OCT and frozen at time 0 (15 min postexcision). Replicate tissue pieces were immersed in solutions containing either (a) HBSS; (b) HBSS, polyethylene glycol, orthovanadate, and β-glycerophosphate; (c) HBSS, ethanol, polyethylene glycol, genistein, and staurosporine; or (d) HBSS, polyethylene glycol, orthovanadate, β-glycerophosphate, genistein, and staurosporine. Tissue samples were removed 15, 30, and 90 min postimmersion and processed as for specimen 1 (Figs. 10 and 12).
Fig. 10.
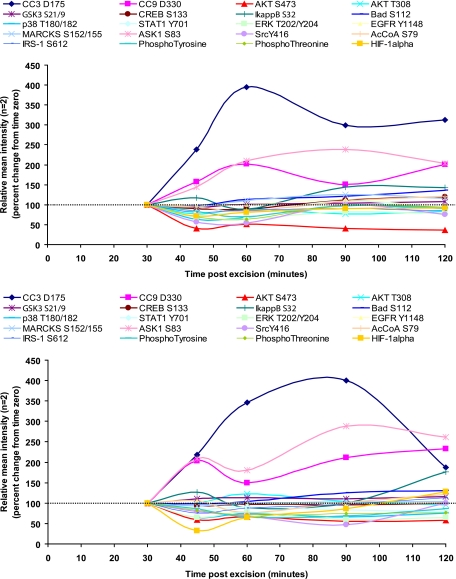
Tissue treated with phosphatase inhibitors exhibits increased phosphorylation of serine, threonine, and tyrosine residues. RPPA analysis of metastatic melanoma tissue incubated for 30 min (A) or 90 min (B) postexcision in Hanks’ balanced salt solution alone (green circles); HBSS with phosphatase inhibitors polyethylene glycol, sodium orthovanadate, and β-glycerophosphate (PI; red squares); and HBSS with kinase inhibitors polyethylene glycol, staurosporine, and genistein (KI; blue triangles) (n = 2, mean as a percentage of the HBSS mean value) is shown. 85% of the end points were elevated with phosphatase inhibitor treatment or decreased with kinase inhibitor treatment by 20% compared with untreated tissue (HBSS) (ANOVA, p < 0.05). Six end points did not change with either inhibitor treatment: CC9 Asp-330, EGFR Tyr-1148, CC7 Asp-198, p70S6 Thr-389, PKCαβ Thr-638/641, and SHC Tyr-317. FAK, focal adhesion kinase; AcCoA, acetyl-CoA carboxylase; PTEN, phosphatase and tensin homologue deleted on chromosome 10.
Fig. 12.
A prototype protein preservative/fixative solution minimizes fluctuations of phosphoproteins in tissue. RPPA analysis of metastatic melanoma tissue immersed in a solution containing a precipitating fixative, a permeation enhancer, balanced salt solution, and phosphatase and kinase inhibitors is shown (blue square, 30 min; pink triangle, 60 min; red triangle, 90 min; n = 2, mean ± range). FAK, focal adhesion kinase.
Reverse Phase Protein Microarrays (RPPAs)
Protein Microarray Construction—
Cellular lysates were printed on glass-backed nitrocellulose array slides (FAST Slides, Whatman, Florham Park, NJ) using an Aushon 2470 arrayer (Aushon BioSystems, Burlington, MA) equipped with 350-μm pins as described previously (9). Cellular lysates prepared from A431 ± epidermal growth factor, HeLa ± pervanadate (BD Biosciences), or Jurkat ± calyculin (Cell Signaling Technology, Danvers, MA) cell lines were printed on each array for quality control assessments. Human endothelial ± pervanadate (BD Biosciences) cellular lysates were printed on arrays for sensitivity and precision comparisons (Fig. 2). Immunostaining was performed as described previously (9) with antibodies listed in Table V. Polyclonal and monoclonal antibodies were purchased from Cell Signaling Technology, BIOSOURCE/Invitrogen, BD Biosciences, and Upstate/Millipore (Billerica, MA). Total protein per microarray spot was determined with a SYPRO Ruby protein stain (Invitrogen/Molecular Probes) according to the manufacturer's directions and imaged with a charge-coupled device camera (NovaRay, Alpha Innotech, San Leandro, CA or Eastman Kodak Co. 4000MM, Carestream Health, New Haven, CT).
Table V.
Proteins evaluated by reverse phase protein microarray in the time course studies
CST, Cell Signaling Technology; BD, BD Biosciences; Upstate, Millipore/Upstate; Biosource, Invitrogen/BIOSOURCE; A, apoptosis; B, transcription factors; C, cell cycle; D, hypoxia/ischemia; E, adhesion/cytoskeletal; F, stress/inflammation; G, proliferation/survival; FAK, focal adhesion kinase; MAPK, mitogen-activated protein kinase.
| Protein/subcellular location | Antibodyvendor/species | Function |
|---|---|---|
| Cytoplasm | ||
| 4EBP1 (Ser-65) | CST/rabbit | G |
| Acetyl-CoA carboxylase (Ser-79) | CST/rabbit | D |
| AKT (Ser-473) | CST/rabbit | G |
| AKT (Thr-308) | CST/rabbit | G |
| AMPKα1 (Ser-485) | CST/rabbit | D |
| AMPKβ (Ser-108) | CST/rabbit | D |
| ASK1 (Ser-83) | CST/rabbit | F |
| c-ABL (Thr-735) | CST/rabbit | G |
| BAD (Ser-112) | CST/rabbit | A |
| Caspase-3, cleaved (Asp-175) | CST/rabbit | A |
| Caspase-7, cleaved (Asp-198) | CST/rabbit | A |
| Caspase-9, cleaved (Asp-330) | CST/rabbit | A |
| E-cadherin | CST/rabbit | E |
| eIF4G (Ser-1108) | CST/rabbit | G |
| eNOS (Ser-1177) | CST/rabbit | D |
| ERK 1/2 (Thr-202/Tyr-204) | CST/rabbit | G |
| FAK (Tyr-576/577) | CST/rabbit | E |
| GSK3α/β (Ser-21/9) | CST/rabbit | G |
| HIF-1α | BD/mouse | D |
| HSP 90 | CST/rabbit | D |
| Iκ B-α (Ser-32/36) | CST/mouse | F |
| IRS-1 (Ser-612) | CST/rabbit | G |
| JAK1 (Tyr-1022/1023) | CST/rabbit | F |
| MARCKS (Ser-152/156) | CST/rabbit | F |
| mTOR (Ser-2481) | CST/rabbit | G |
| NFκB p65 (Ser-536) | CST/rabbit | F |
| p38 MAPK (Thr-180/Tyr-182) | CST/rabbit | D |
| PAK1 (Ser-199/204)/PAK2 (Ser-192/197) | CST/rabbit | E |
| PDK1 (Ser-241) | CST/rabbit | G |
| PKCα/β II (Thr-638/641) | CST/rabbit | G |
| SAPK/JNK (Thr-183/Tyr-183) | CST/rabbit | F |
| SEK1/MKK4 (Ser-80) | CST/rabbit | F |
| Src family (Tyr-146) | CST/rabbit | F |
| STAT1 (Tyr-701) | Upstate/rabbit | B |
| STAT3 (Ser-727) | CST/rabbit | B |
| STAT3 (Tyr-705) | CST/rabbit | B |
| Tuberin/TSC2 (Tyr-1571) | CST/rabbit | G |
| Membrane | ||
| Adducin (Ser-662) | CST/rabbit | E |
| Annexin II | BD/mouse | E |
| EGFR | CST/rabbit | G |
| EGFR (Tyr-1148) | Biosource/rabbit | G |
| ErbB2 | CST/rabbit | G |
| ErbB2/HER2 (Tyr-1248) | CST/rabbit | G |
| HER3 (Tyr-1289) | CST/rabbit | G |
| VEGF receptor-2 (Tyr-1175) | CST/rabbit | D |
| VEGFR 2 (Tyr-951) | CST/rabbit | D |
| Nucleus | ||
| ATF-2 (Thr-71) | CST/rabbit | F |
| β-Catenin (Ser-33/37/Thr-41) | CST/rabbit | E |
| CHK1 (Ser-345) | CST/rabbit | C |
| CREB (Ser-133) | CST/rabbit | B |
| Cyclin A | BD/mouse | C |
| Mitochondria | ||
| BCL-2 (Ser-70) | CST/rabbit | A |
| BAX | CST/rabbit | A |
Statistics—
Analysis of variance (ANOVA) was used to compare the mean values between time 0 samples and all other time points (SAS version 9.1.3, SAS Institute Inc., Cary, NC). p values <0.05 were considered significant. S.D. was calculated for replicate samples.
RESULTS
Reverse Phase Protein Microarray Sensitivity and Precision—
Sensitivity and precision of the RPPA using recombinant prostate-specific antigen molecules, microdissected tissue samples, and fine needle aspirates have been reported previously (3, 10, 11). Because of variation in robotic array printing devices we determined inter- and intraslide precision for the Aushon 2470 arrayer using human endothelial cell lysates treated with pervanadate as a model of phosphorylated VEGF receptor sensitivity and precision. Human endothelial cells are known to express ∼100,000 VEGF receptors/cell. Sensitivity of the arrays using anti-VEGF receptor (VEGFR) Tyr-951 as the primary antibody was found to be 3,660 receptor molecules as indicated by the ability to detect signal greater than 2 S.D. above background (Fig. 2). To determine interslide precision, human endothelial cells treated with pervanadate were printed on RPPAs in 2-fold serial dilutions (undiluted to 1:2,048) in duplicate on eight slides and probed with anti-VEGFR Tyr-951. The mean spot intensity for each dilution was calculated and plotted as a dose-response curve based on the estimated number of VEGF receptors per cell. Excellent dose-response curves were observed between arrays (coefficient of variation, 4.0–17.8%, n = 8, R2 = 0.9761). Within-run variation (n = 12) was found to be within 5.0–18.1% with good linearity (R2 = 0.9693) (Fig. 2, inset).
Post-translationally Modified Cell Signaling Proteins—
53 signal pathway proteins representing phosphorylated, cleaved, or total protein end points from various subcellular compartments (membrane-bound receptors, cytoplasm, mitochondria, and nucleus) were quantitatively measured by RPPA (Table V). Antibodies were extensively validated for specificity (12). End points were selected based on their expected involvement in cellular signaling pathways: hypoxia/ischemia, proliferation/survival, adhesion/cytoskeleton structure, stress/inflammation, or apoptosis. Post-translational modifications, such as caspase cleavage or phosphorylation of specific serine, threonine, or tyrosine residues, were specifically chosen as representative analytes for assessing fluctuations in cellular proteins over time.
Procurement and Time to Preservation Vary in a Hospital Clinical Setting—
The initial specimen collection time course experiments were designed to examine, in a community hospital clinical setting, current specimen collection practices for molecular studies (Table I). The average time we could procure and commence tissue processing was 19.3 min postexcision for uterus, colon, lung (normal and squamous cell carcinoma), ovarian, breast, and metastatic melanoma tissue. The earliest time to process tissue was 4 min (uterus, specimen 1), whereas the longest time was 40 min (colon, specimen 2). Tissue sample size varied from 10 × 5 mm (lung, specimens 3 and 4) to 4 × 3.5 cm (breast specimen 10).
Tissue Is Alive and Reacting to Environmental Stresses Postprocurement—
All tissue specimens evaluated revealed augmentation (±20% from the time 0 sample) of phosphoprotein levels as well as decreases over time independent of tissue type, post-translational modification, and subcellular location (Tables II–IV and Figs. 3–8). Uterine tissue (specimen 1; Fig. 3) showed >20% decreases for AKT Ser-473 and MARCKS Ser-152/156 within the first 60 min postexcision. Proteins in apoptotic (BAD Ser-112 and cleaved caspase-3 Asp-175), hypoxia (p38 Thr-180/Tyr-182), proliferation/survival (AKT Thr-308, EGFR Tyr-1148, ERK Thr-202/Tyr-204, and GSK3αβ Ser-21/9), and transcription factor pathways (STAT1 Tyr-701 and CREB Ser-133) showed >20% increases within the initial 30 min postprocurement. These increases appeared to fluctuate over time, presumably as the tissue reacted to immediate environmental stresses. These findings were not specific to uterus; colon and lung (histologically normal or tumor) tissue samples also showed fluctuations in post-translationally modified protein levels over time (Table II and Figs. 4 and 5). Time 0 for the colon sample (specimen 2) was 40 min; thus early perturbations may have occurred prior to specimen processing. The lung time course (specimens 3 and 4) consisted of three time points because of the small volume of tissue available for analysis. Patient-matched tumor and normal tissue did not show identical responses over time. Squamous cell carcinoma tissue (Fig. 5A) showed decreases in apoptotic proteins (CC3 Asp-175) with concomitant increases in proliferation/survival (AKT Thr-308, EGFR Tyr-1148, GSK3αβ Ser-21/9, ERK Thr-202/Tyr-204, and IRS-1 Ser-612), stress/inflammation (STAT1 Tyr-701 and IκB Ser-32), and hypoxia/ischemia (p38 Thr-180/Tyr-182) proteins. Normal tissue showed increases in proliferation/survival (EGFR Tyr-1148 and GSK3αβ Ser-21/9), hypoxia/ischemia (p38 Thr-180/Tyr-182), and stress/inflammation (STAT1 Tyr-701) with concomitant decreases in IκB Ser-32 (Fig. 5B).
Ovarian tissue (specimen 6) procured, incubated, and analyzed in duplicate exhibited fluctuations over time for a variety of protein end points associated with stress, adhesion, proliferation/survival, cell cycle, and hypoxia pathways (Fig. 6). ANOVA comparison of means for the time 0 sample (30 min postexcision) showed changes over time as compared with tissue incubated at room temperature for the following end points: JAK1 Tyr-1022/1023 (60 min, p < 0.0430; 90 min, p < 0.0156), PAK1 Ser-199/204/PAK2 Ser-192/197 (60 min, p < 0.0418; 90 min, p < 0.0222; 120 min, p < 0.0179), TSC2 Tyr-1571 (90 min, p < 0.0129; 120 min, p < 0.0095), and CHK1 Ser-345 (50 min, p < 0.0238; 60 min, p < 0.0272; 90 min, p < 0.0285; 120 min, p < 0.0172), ATF-2 Thr-71 (50 min, p < 0.0503; 60 min, p < 0.0496; 90 min, p < 0.0187; 120 min, p < 0.0075), and eNOS Ser-1177 (50 min, p < 0.0082; 90 min, p < 0.0158; 120 min, p < 0.0149).
Classes of Phosphoprotein Pathway Fluctuations in Various Tissues—
As summarized in Table I the tissue time course specimens analyzed in this study included a wide variety of tissues and pathologies. An important question was: do classes of phosphoproteins exist that fluctuate in a similar manner independent of tissue type? For specimens 1–5 and specimen 7, general classes of proteins exhibited increases over time compared with the time 0 sample (Table II). Increases were noted for CC3 Asp-175 (apoptosis) in four of six specimens; p38 Thr-180/Tyr-182 (hypoxia/ischemia) in three of six specimens; AKT Thr-308 (three of five specimens), ERK Thr-202/Tyr-204 (three of six specimens), GSK3αβ Ser-21/9 (three of five specimens), and IRS-1 Ser-612 (three of six specimens) (proliferation/survival); and STAT1 Tyr-701 (transcription factor) in four of six specimens. Loss of phosphoprotein levels was noted in five of six specimens for AKT Ser-473 (proliferation/survival) and MARCKS Ser-152/156 (stress/inflammation). Uterine tissue (specimen 7) incubated at either 4 °C (Fig. 7A) or room temperature (Fig. 7B) exhibited similar perturbations, including activation and deactivation, in several classes of kinases over time. Although the magnitude of the changes for CC3 Asp-175, CC9 Asp-330, and ASK1 Ser-83 were somewhat reduced over time for the samples incubated at 4 °C, a marked change from the initial sample (time 0) was observed, similar to the increases noted at room temperature.
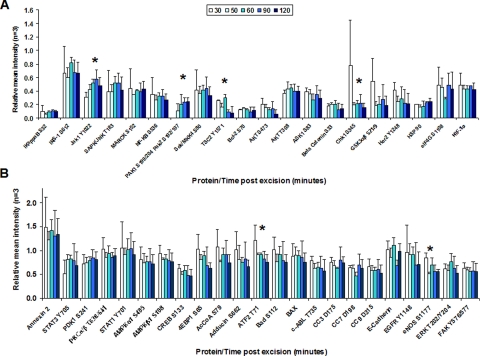
Leiomyoma tissue (specimen 8; Table III and Fig. 8, A–H) provided homogeneous tissue for precision studies with biological and experimental replicates. ANOVA comparison of means between the initial time 0 sample and every other time point revealed significant decreases (p < 0.05) over time for Adducin Ser-662 in four of six time points and HIF-1α in three of six time points. Significant increases (individual p values are listed in Table III and range from p <0.04 to <0.001) over time compared with time 0 were seen with BAX in four of six time points, AMPKβ1 Ser-108 in four of six time points, 4EBP1 Ser-65 in three of six time points, ERK Thr-202/Tyr-204 in five of six time points, ASK1 Ser-83 in three of six time points, SAPK/JNK Thr-183/Tyr-185 in five of six time points, and STAT3 Tyr-705 in three of six time points. No significant differences were noted for E-cadherin, PAK1 Ser-199/204/PAK2 Ser-192/197, CC3 Asp-175, eNOS Ser-1177, HSP 90, AKT Ser-473, EGFR Tyr-1148, eIF4G Ser-1108, IRS-1 Ser-612, PKCαβ II Thr-638/641, TSC2 Tyr-1571, ATF-2 Thr-71, JAK1 Tyr-1022/1023, MARCKS Ser-152/156, NFκB Ser-536, and SEK1/MKK4 Ser-80 over the evaluated time period.
Heterogeneous breast tissue (specimen 10; Table IV) exhibited a different activation/deactivation protein profile over time compared with the leiomyoma tissue. Decreases (p < 0.05, individual p values are listed in Table IV) were noted in β-catenin Ser-33/37/Thr-41 in six of eight time points, EGFR Tyr-1148 in eight of eight time points, ERK Thr-202/Tyr-204 in six of eight time points, ErbB2 in four of eight time points, GSK3αβ Ser-21/9 in four of eight time points, ASK1 Ser-83 in five of eight time points, PKCαβ II Thr-638/641 in four of eight time points, and IκB Ser-32 in eight of eight time points. Increases were seen in E-cadherin in five of eight time points, CC3 Asp-175 in six of eight time points, AMPKα1 Ser-485 in four of eight time points, IRS-1 Ser-612 in four of eight time points, and STAT1 Tyr-701 in six of eight time points. MARCKS Ser-152/156 and CREB Ser-133 exhibited increases in the early/middle time points with decreases noted at later time points (more than 230 min postexcision). No significant differences were noted for CHK1 Ser-345, eNOS Ser-1177, c-ABL Thr-735, mTOR Ser-2481, or STAT3 Ser-727.
Phosphatase and Kinase Inhibitor Treatment of Tissue Reveals Active and Reactive Kinase Pathways Postprocurement—
As a model for evaluating the effects of cell-permeable phosphatase inhibitors on cell signaling kinases we treated A549 wild type EGFR lung cancer cells and H1975 EGFR L858R mutant cells with 1 mm pervanadate solution. Sustained increases in serine and tyrosine phosphorylation in the cell line models were noted (data not shown).
To assess the effects of phosphatase inhibitors within intact tissue in a liquid-based preservative solution, we immersed fresh tissue in HBSS with or without phosphatase inhibitors (sodium orthovanadate and β-glycerophosphate) or kinase inhibitors (genistein and staurosporine). Duplicate breast tissue (epithelium, tumor, and adipose tissue) (specimen 10) samples incubated in HBSS plus phosphatase inhibitors confirmed the presence of elevated levels of phosphoproteins over time compared with untreated tissue (HBSS only) because of the continued activity of kinases (Fig. 9). EGFR Tyr-1148, STAT1 Tyr-701, and SAPK/JNK Thr-183/Tyr-185 exhibited elevated phosphorylation in the presence of phosphatase inhibitors over time compared with untreated tissue. The same end points showed a lack of elevation over time in the presence of kinase inhibitors. Phosphatase and kinase inhibitors did not show a notable effect on CC7 Asp-198 compared with incubation in HBSS only perhaps because of the lack of a direct phosphorylation event required for caspase activation (Fig. 9D).
We repeated this experiment with metastatic melanoma tissue (specimen 11). As shown in Fig. 10, for 39 phosphoprotein end points measured, 85% were elevated following phosphatase inhibitor treatment, whereas 85% were suppressed following kinase inhibitor treatment (Fig. 10). This treatment effect was most prominent at early time points in keeping with the early elevation of phosphoprotein end points shown in Fig. 9. At 90 min, phosphatase inhibitor treatment continued to augment the levels of a subset of proteins compared with untreated tissue. A subset, including JNK Thr-183/Tyr-185, ASK1 Ser-183, SHIP1 Tyr-1020, eIF4G Ser-1108, and S6 ribosomal protein Ser-240/244, remained elevated after 90 min compared with the untreated tissue. In contrast, the levels of cleaved caspases were not altered by the kinase or phosphatase inhibitor treatment of the tissue (Fig. 10). These data strongly support the conclusion that kinase pathways containing the phosphoproteins being measured are active and reactive in the tissue for at least 90 min following tissue procurement.
Phosphatase plus Kinase Inhibitors Stabilize Phosphoprotein Fluctuations over 90 Min—
Because the experiments above demonstrated that kinase pathways remained reactive ex vivo we tested the hypothesis that a combination of phosphatase and kinase inhibitors would suppress phosphoprotein fluctuations by blocking both kinase and phosphatase activities on the phosphoprotein substrates. Six breast fibroadenoma tissue pieces (specimen 9) were immersed in a solution of ethanol, polyethylene glycol, HBSS, sodium orthovanadate, β-glycerophosphate, staurosporine, and genistein 138 min postexcision. Morphological examination of the tissue samples at 5 min, 90 min, and 24 h postincubation in the fixative solution containing phosphatase and kinase inhibitors revealed the presence of mainly adipose tissue in the 24-h sample (hematoxylin and eosin (H&E) stain; data not shown). Therefore, data for the 24-h time point was not used for comparison because of lack of tissue homogeneity with the samples fixed for 5 and 90 min. Proteins that we expected to increase ±20% (ATF-2 Thr-71, SAPK/JNK Thr-183/Tyr-185, STAT1 Tyr-701, JAK1 Tyr-1022/1023, and PAK1 Ser-199/204/PAK2 Ser-192/197) based on observations from specimens 10 and 11 appeared to be stabilized over the 90-min time period in the presence of kinase and phosphatase inhibitors (Fig. 11). In contrast, these same phosphoproteins increased in the presence of a phosphatase inhibitor alone (Fig. 9). This stabilization of labile phosphoproteins was confirmed in a separate tissue sample examined at 30, 60, and 90 min postprocurement (metastatic melanoma) (Fig. 12).
Histomorphology and Phosphoprotein Preservation in the Presence of Kinase and/or Phosphatase Inhibitors—
Histology preservation and morphology was compared for uterine endometrium/myometrium (specimen 7) tissue incubated in candidate preservative solutions containing phosphatase and protease inhibitors with methanol or ethanol with a standard frozen section preparation (Fig. 13). Adequate cellular morphology was seen with an H&E stain for the frozen section (Fig. 13A). Tissue incubated in either the methanol-based or ethanol-based solution containing phosphatase and protease inhibitors revealed excellent cellular color and nuclear detail with minimal cell shrinkage (Fig. 13, B and C).
DISCUSSION
This study provides previously unknown quantitative information about the stability of 53 tissue phosphoproteins and signal pathway proteins at room temperature following collection of surgical specimens in the setting of a community hospital. The results support the conclusion that kinase pathways remain active and reactive during the immediate ex vivo period. These data have profound implications for tissue collection protocols, tissue banking, and development of preservative chemistries for molecular profiling of proteins.
Tissue Acquisition and Accessibility—
Tissue is generally procured for pathologic examination in three main settings: (a) surgery in a hospital-based operating room, (b) biopsy conducted in an outpatient clinic, and (c) image-directed needle biopsies or needle aspirates conducted in a radiologic suite. The first questions we needed to address in this study were: 1) what is the accessibility of tissue for research in a community hospital setting? and 2) how long are tissue proteins, particularly phosphoproteins, stable?
Fig. 1 depicts the two categories of variable time periods that define the stability intervals for tissue procurement. Time point A is defined as the moment tissue is excised from the patient and becomes available ex vivo for analysis and processing. The postexcision delay time (EDT) is the time from time point A to the time that the specimen is placed in a stabilized state, e.g. immersed in fixative or snap frozen in liquid nitrogen, time point B. Given the complexity of patient care settings, during the EDT the tissue may reside at room temperature in the operating room or on the pathologist's cutting board, or it may be refrigerated in a specimen container. The second variable time period is the processing delay time (PDT). At the beginning of this interval the tissue is immersed in a preservative solution or stored in a freezer. At the end of this interval, time point C, the tissue is subjected to processing for molecular analysis. In addition to the uncertainty about the length of these two time intervals, a host of known and unknown variables can influence the stability of tissue molecules during these time periods. These include 1) temperature fluctuations prior to fixation or freezing; 2) preservative chemistry and rate of tissue penetration; 3) size of the tissue specimen; 4) extent of handling, cutting, and crushing of the tissue; 5) fixation and staining prior to microdissection or histologic processing; and 6) tissue hydration and dehydration.
Tissue was selected for study based on 1) availability (excess tissue not required to render diagnosis), 2) organ/tissue type (epithelial/fibrous), and 3) disease status (normal/benign/diseased). The majority of the diseased tissue sample (squamous cell carcinoma, breast fibroadenoma, and metastatic melanoma) was required for diagnosis. Therefore the sample size available for research for the diseased tissue (cancer) was much smaller compared with normal/benign tissue samples (uterus/leiomyoma and colon). Laser microdissection was not performed on these samples in order to analyze the total repertoire of all cell signaling classes in a variety of cellular phenotypes. H&E-stained tissue sections were evaluated for confirmation of tissue type (data not shown), and contiguous tissue cryosections were analyzed as a means of maintaining tissue block homogeneity.
The average time from specimen excision to preservation was 19.3 min with a range of 4–40 min. Factors affecting these times were (a) proximity of the surgery suite to the pathology area, (b) number and size of specimens submitted, (c) request for pathology frozen section evaluation, (d) number of concurrent surgical cases, and (e) efficiency of sample collection coordination between surgical, pathology, and research staff.
Collection of research samples should not interfere with standard of care in a clinical setting. As such, routine specimen collection protocols for diagnosis were followed in all cases. This requirement profoundly limited the volume of clinical tissue that could be released for research purposes. Samples were routinely transported in closed polypropylene or polystyrene containers without additional fluids. We observed that a high degree of coordination was needed between surgical, pathology, and research teams to effectively communicate research project requirements such as specimen type and handling. Community hospitals and clinics often rotate staff assignments, unlike research entities in which dedicated staff oversee a project, necessitating timely communication between the groups. We noted some common pathology gross examination practices that should, at the very least, be noted as part of the research record because of their potential impact on molecular analyses. One practice we observed was the rinsing of tissue in running tap water to remove excess blood prior to the initial dissection of the specimen (specimen 6, ovarian mucinous adenocarcinoma). This practice may cause tissue osmotic shock (rapid hydration and changes in temperature). Depending on the sensitivity of the research assay, this may confound the molecular analysis.
Tissue Signal Pathway Phosphoproteins Fluctuate ex Vivo at Room Temperature—
Previously quantitative data did not exist for the effects of time, method of preservation, organ and tissue of origin, and means of surgical extraction on the yield and fidelity of classes of phosphoproteins in human tissue procured in a typical surgical environment. To date, clinical preservation practices (e.g. to preserve proteins) routinely rely on protocols that are decades old, such as formalin fixation, and are designed to preserve specimens for histologic examination (13, 14). Although it is now possible to extract proteins from formalin-fixed tissue, formalin penetrates tissue at a rate of less than 1 mm/h (13). During this time delay period the portion of living tissue deeper than several millimeters would be expected to undergo significant fluctuations (Figs. 3–8) with respect to phosphoprotein analytes (15). Because the dimensions of the tissue and the depth of the block for which the samples are obtained are unknown variables, formalin fixation would be expected to cause pronounced variability in phosphoprotein levels for molecular diagnostics.
For the few studies that have been conducted in the past concerning the stability of proteins and phosphoproteins in tissue, the main analytical tool has been immunohistochemistry with incidental use of Western blots (7, 8). Baker et al. (7) reported disparities of AKT Ser-473 staining in human surgical sections compared with biopsy samples, all of which were fixed in 10% buffered formalin. One of the uncontrolled aspects of this prior study was the standardization of tissue sample size such that the formalin penetration rate and thus the rate of protein fixation were not similar in both study sets. Thus conclusions regarding the rate of decline in AKT Ser-473 activity should be judged in the context of tissue size, formalin penetration rate, and the depth of the tissue that was used for analysis in relation to overall size of the tissue block.
Immunohistochemistry is valuable for cellular localization information but cannot be considered a true quantitative tool at least because immunohistochemistry lacks sensitivity and quantitation for phosphoproteins (16). This reflects the fact that antibody reactivity is a complex product of antigen availability within the permeabilized cell, the chemistry and timing of fixation, level of cross-linking, and antigen retrieval. Immunohistochemistry is thousands of times less sensitive than alternative molecular procedures, such as direct extraction and solubilization for protein microarray analysis (10, 11). Western blotting is non-quantitative and is 100–1000 times less sensitive compared with quantitative protein microarray analysis.
Measurement of protein signaling molecules may provide key information for individualized therapy because the protein signaling molecules constitute the actual drug targets of molecular inhibitors. Despite this promise, molecular profiling can never be translated to patient benefit unless the fundamental problem of how to minimize preanalytical fluctuations of proteins, particularly post-translationally modified proteins, in tissue samples is solved. The instant a tissue biopsy is removed from a patient the cells within the tissue react and adapt to the absence of vascular perfusion, ischemia, hypoxia, acidosis, accumulation of cellular waste, absence of electrolytes, and temperature changes. In as little as 30 min postexcision (Figs. 3–5), drastic changes can occur in the protein signaling pathways of the biopsy tissue as the tissue remains in the operating room suite or on the pathologist's cutting board. To address the uncertainties of molecular preservation, freezing the tissue has been the major conventional method to preserve RNA and protein molecules (17, 18). The tissue is placed in liquid nitrogen or embedded in cryoprotectant medium and transported on dry ice. Although freezing may preserve RNA and protein pathway signatures, it is impractical for routine use in the clinic as many physician offices and operating rooms are not equipped with these reagents. Moreover frozen material is inadequate for routine pathological diagnosis, which requires chemical fixation of the tissue to maintain cellular morphology. In addition, freezing does not solve the problem of the unknown or unknowable time delay following removal of the tissue from the patient during surgery.
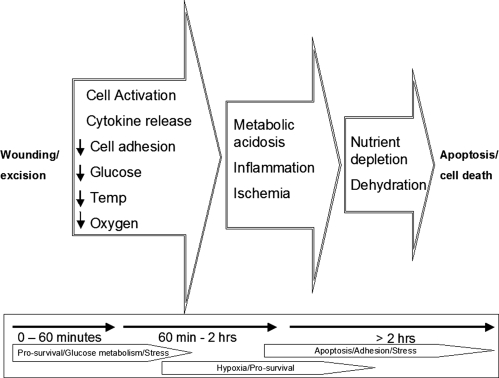
We have shown, with biological and experimental replicates, the dynamic nature of cell signaling pathways postexcision (Figs. 6, 8, and 9). Tissue biopsies can no longer be viewed as non-reactive or unchanging after procurement. The excised living tissue is likely reacting to environmental stress and the wounding insult. Cascades of molecular events due to cytokine release, hypotension, vascular stasis, decreases in glucose concentration, and temperature and oxygen saturation potentially contribute to the previously unrecognized activation and/or deactivation of stress, hypoxia, survival, and apoptotic pathways within excised tissue (Fig. 14).
Fig. 14.
Molecular stages and time line of tissue cell death. Post-tissue excision, cascades of cellular kinases are activated and deactivated as tissue reacts to wounding, ischemia, inflammation, environmental stresses, hypoxia, and nutrient depletion. temp, temperature.
Conventional wisdom dictates that tissue should be stored at 4 °C if a processing delay is expected or likely. Based solely on classical enzyme kinetics one would surmise that this practice would delay or suppress cellular kinase and phosphatase activity in tissue samples (19). In the case of replicate uterine tissue samples incubated at 4 °C and room temperature (Fig. 7) fluctuations in kinase substrates were exhibited at both temperatures, although there was apparent reduction of kinase activity over time for the refrigerated samples. In the absence of preservative chemistries or immediate processing (freezing), the recommendation is to freeze tissue within 20 min of excision to preserve the state of phosphoproteins. The effect of refrigeration in comparison with room temperature storage is the subject of an on-going, in-depth study.
Each tissue specimen behaved somewhat differently over time with regard to phosphoprotein fluctuations (Tables II–IV). Presumably this is a reflection of the survival struggle of the tissue overlaid by the pathological state and the cellular composition of the individual tissue. As expected, this time course study revealed tissue phosphoprotein fluctuations that were potentially influenced by neoplastic state or tissue of origin and other factors that appear independent of tissue histology. Those tissue-specific classes of proteins that changed over time may reflect the predominant pathways normally in use in that particular tissue. Interestingly in both leiomyoma and breast tissue (DCIS with surrounding adipose and stromal tissue) eNOS Ser-1177 was the only protein in common of those studied that did not change over time.
These data constitute the first systematic attempt to quantitatively explore the stability of disease-relevant phosphoproteins in freshly obtained human tissue specimens. As such a variety of specimen tissue types and pathologic states were investigated to answer basic questions such as (a) are the phosphoproteins changing at all?, (b) what is the direction of change?, (c) what is the reason for the phosphoprotein changes?, and (d) how can this guide the design of improved room temperature preservative chemistries? We did obtain answers to these fundamental questions. Nevertheless the data generated are limited to the specimen types available. Because we did not evaluate all classes of tissue and organs, nor a wide variety of pathologies, nor a series of matched normal and cancer specimens from the same patient, these conclusions cannot be generalized with regard to organ- or cancer-specific phosphoprotein differences. These findings support the important need for future in-depth investigation of the fluctuations of protein signaling and post-translational modifications ex vivo following tissue procurement. Gathering these data are a necessary step before a phosphoprotein end point is selected for use as a clinical predictor (see guidelines for tissue procurement in Table VI). Moreover the ex vivo fluctuations in the reactive and reacting signal pathways of cancer cells may be different compared with their normal counterpart. This knowledge could provide insights into the adaptation of cancer cells in the native tissue to hypoxic and metabolic stress.
Table VI.
Concepts in tissue procurement and preservation
| Tissue protein biomarkers: preanalytical procurement validation and guidelines for minimizing preanalytical variables during clinical tissue procurement |
|---|
| 1. Tissue procurement protocols must be based on the recognition that the excised tissue is alive and reactive to ex vivo stress. Kinase pathways are active and reactive until the tissue cells are stabilized. |
| 2. Based on time course analysis of phosphoproteins, tissue should be stabilized as soon as possible after excision. Taking into consideration the average time for procurement in a community hospital, the recommended maximum elapsed time is 20 min from excision to stabilization (e.g. flash freezing, thermal denaturation, or chemical stabilization). |
| 3. For preservation of kinase pathway proteins, stabilization methods should be designed to block both sides of the kinase/phosphatase kinetics. Blocking the phosphatases only can cause false elevation of the phosphorylation level of the analyte. |
| 4. Reactive changes occurring in tissue postexcision can generate false elevation as well as false decline in protein (or molecular) analytes. This is a significant source of bias for clinical biomarker qualification and must be known before a tissue biomarker is selected for clinical use. |
Phosphoprotein Stability: the Balance between Kinases and Phosphatases: Implications for Preservation Chemistry—
Phosphorylation and dephosphorylation of structural and regulatory proteins are major intracellular control mechanisms (20). Protein-tyrosine phosphatases remove phosphate groups from phosphorylated tyrosine residues of proteins. Protein-tyrosine phosphatases display diverse structural features and play important roles in the regulation of cell proliferation, differentiation, cell adhesion and motility, and cytoskeletal function (21). At any point in time within the cellular microenvironment, the phosphorylated state of a protein is a function of the local stoichiometry of associated kinases and phosphatases specific for the phosphorylated residue. During the ex vivo time period, if the cell remains alive, phosphorylation of certain kinase substrates may transiently increase because of the persistence of functional signaling, activation by hypoxia, or some other stress response signal. On the other hand, the availability of ubiquitous cellular phosphatases would be expected to ultimately destroy phosphorylation sites given enough time. This conclusion is in keeping with the time course data that generally revealed a surge of phosphorylation in early times, consistent with reactive kinase pathways, followed by a general long term loss of all phosphoprotein analytes as the tissue died and the kinase pathways ceased to function while the phosphatases remained active. Cultured cell lines and human tissue exhibited enhanced phosphorylation in the presence of phosphatase inhibitors compared with untreated tissue, whereas the same proteins incubated with kinase inhibitors did not increase over time, indicating the explicit need for both kinase and phosphatase inhibitors in a tissue preservative (Figs. 9 and 10).
These data do not support the use of phosphatase inhibitors alone for the stabilization of tissue phosphoproteins. Several studies have been conducted concerning the stability of RNA in tissue ex vivo (22–26). These studies indicate that refrigeration is superior to room temperature storage and that the addition of RNase inhibitors may be useful as RNA preservatives. Although this information is applicable to gene array profiling, it has little if any bearing on protein stability in general or phosphoprotein stability specifically at least because gene transcript array data cannot reflect the post-translational state of a protein. For the case of phosphoprotein preservation, because the tissue kinase pathways remain active, as shown in Figs. 9 and 10, phosphatase inhibitors alone will cause a non-physiologic false elevation of phosphorylation that is not representative of the in vivo state prior to procurement. In contrast, simultaneous inhibition of kinases and phosphatases (Figs. 11 and 12) stabilizes phosphoproteins to a greater degree compared with phosphatases alone.
An important long term goal of clinical proteomics for cancer diagnostics is to use the chemistry of kinases and phosphatases to design rational stabilizers for the preservation of phosphoprotein stability without freezing. There is a need for methods to preserve post-translationally modified proteins, such as phosphoproteins (e.g. within about 20 min of corporal extraction), and for methods to monitor the status (e.g. phosphorylation state) of proteins during the EDT and PDT periods (Fig. 1). A variety of chemical- and protein-based inhibitors of phosphatases exist (27, 28). Our data support a room temperature preservative chemistry that incorporates both phosphatase and kinase inhibitors to stabilize phosphoproteins at room temperature. Initial evaluation of this solution shows that it is compatible with histological examination of tissue (Fig. 13) and fully stabilizes end points that we expected to increase dramatically over the immediate postprocurement 90-min period (Figs. 11 and 12). Additionally this prototype stabilization chemistry did not appreciably alter the total yield of candidate phosphoprotein identifications by mass spectrometry applied to peripheral blood mononuclear cells compared with untreated peripheral blood mononuclear cells (data not shown). If we are to develop a set of surrogate markers for tissue quality, it would be interesting to know what role other markers, or “alarmins,” play in signal transduction of excised tissue (29). From a cellular viewpoint, excision is traumatic injury. We are currently restricted to evaluating known cell signaling markers, although a host of as yet unknown or unrecognized molecules may be contributing to cellular response post-trauma. Alarmins, as a class, will undoubtedly provide a source of molecules to study in the future for developing a definitive portrait of the forensic time course of tissue stability.
A desired standard operating procedure would be to place the tissue directly in the preservative chemistry solution at the time of excision. The ideal chemistry would (a) fully preserve histology for pathologic diagnosis, (b) permit cryosection preparation, and (c) remain compatible with subsequent formalin fixation and paraffin embedding if desired. Thus no further manipulation and no subdivision of tissue for molecular analysis would be necessary compared with the current standard of practice. Continued studies of tissue phosphoprotein preservative chemistries to achieve this standard operating procedure are ongoing in our laboratory.
Footnotes
Published, MCP Papers in Press, July 30, 2008, DOI 10.1074/mcp.M700596-MCP200
The abbreviations used are: OCT, Optimal Cutting Temperature compound; AKT, protein kinase B; ATF-2, cyclic AMP-dependent transcription factor protein; AMPK, AMP-activated protein kinase; ANOVA, analysis of variance; ASK1, apoptosis signal-regulating kinase 1; CC, cleaved caspase; CREB, cAMP response element-binding protein; DCIS, ductal carcinoma in situ; EDT, excision delay time; EGFR, epidermal growth factor receptor; eIF4G, eukaryotic initiation factor 4G; eNOS, endothelial nitric-oxide synthase; ErbB, family of epidermal growth factor receptors; ERK, extracellular signal-regulated kinase; HSP, heat shock protein; GSK3αβ, glycogen synthase kinase 3 protein; IκB, inhibitor κB, IRS-1, insulin receptor substrate; HBSS, Hanks’ balanced salt solution; H&E, hematoxylin and eosin; MARCKS, myristoylated alanine-rich C-kinase substrate protein; mTOR, mammalian target of rapamycin; NFκB, nuclear factor κB protein; PAK1/2, p21-activated kinases 1 and 2; PDT, processing delay time; PKC, protein kinase C; p70S6, ribosomal protein S6 kinase; RPPA, reverse phase protein microarray; SAPK/JNK, stress-activated protein kinase/c-Jun NH2-terminal kinase; SEK1/MKK4, SAPK/ERK kinase 1/mitogen-activate protein kinase kinase 4; STAT, signal transducers and activator of transcription; TSC2, tuberous sclerosis complex 2; VEGF, vascular endothelial cell growth factor; VEGFR, vascular endothelial cell growth factor receptor.
This work was supported, in whole or in part, by a National Institutes of Health grant from the National Cancer Institute program “Innovations in Cancer Sample Preparation” (to L. A. L.). This work was also supported by George Mason University, Inova Fairfax Hospital, and the Istituto Superiore di Sanita, Italy. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
REFERENCES
- 1.Araujo, R. P., Liotta, L. A., and Petricoin, E. F. ( 2007) Proteins, drug targets and the mechanisms they control: the simple truth about complex networks. Nat. Rev. Drug Discov. 6, 871–880 [DOI] [PubMed] [Google Scholar]
- 2.Collins, M. O., Yu, L., and Choudhary, J. S. ( 2007) Analysis of protein phosphorylation on a proteome-scale. Proteomics 7, 2751–2768 [DOI] [PubMed] [Google Scholar]
- 3.Gulmann, C., Espina, V., Petricoin, E., III, Longo, D. L., Santi, M., Knutsen, T., Raffeld, M., Jaffe, E. S., Liotta, L. A., and Feldman, A. L. ( 2005) Proteomic analysis of apoptotic pathways reveals prognostic factors in follicular lymphoma. Clin. Cancer Res. 11, 5847–5855 [DOI] [PubMed] [Google Scholar]
- 4.Haab, B. B. ( 2005) Antibody arrays in cancer research. Mol. Cell. Proteomics 4, 377–383 [DOI] [PubMed] [Google Scholar]
- 5.Petricoin, E. F., III, Bichsel, V. E., Calvert, V. S., Espina, V., Winters, M., Young, L., Belluco, C., Trock, B. J., Lippman, M., Fishman, D. A., Sgroi, D. C., Munson, P. J., Esserman, L. J., and Liotta, L. A. ( 2005) Mapping molecular networks using proteomics: a vision for patient-tailored combination therapy. J. Clin. Oncol. 23, 3614–3621 [DOI] [PubMed] [Google Scholar]
- 6.Schmelzle, K., and White, F. M. ( 2006) Phosphoproteomic approaches to elucidate cellular signaling networks. Curr. Opin. Biotechnol. 17, 406–414 [DOI] [PubMed] [Google Scholar]
- 7.Baker, A. F., Dragovich, T., Ihle, N. T., Williams, R., Fenoglio-Preiser, C., and Powis, G. ( 2005) Stability of phosphoprotein as a biological marker of tumor signaling. Clin. Cancer Res. 11, 4338–4340 [DOI] [PubMed] [Google Scholar]
- 8.De Marzo, A. M., Fedor, H. H., Gage, W. R., and Rubin, M. A. ( 2002) Inadequate formalin fixation decreases reliability of p27 immunohistochemical staining: probing optimal fixation time using high-density tissue microarrays. Hum. Pathol. 33, 756–760 [DOI] [PubMed] [Google Scholar]
- 9.Petricoin, E. F., III, Espina, V., Araujo, R. P., Midura, B., Yeung, C., Wan, X., Eichler, G. S., Johann, D. J., Jr., Qualman, S., Tsokos, M., Krishnan, K., Helman, L. J., and Liotta, L. A. ( 2007) Phosphoprotein pathway mapping: Akt/mammalian target of rapamycin activation is negatively associated with childhood rhabdomyosarcoma survival. Cancer Res. 67, 3431–3440 [DOI] [PubMed] [Google Scholar]
- 10.Rapkiewicz, A., Espina, V., Zujewski, J. A., Lebowitz, P. F., Filie, A., Wulfkuhle, J., Camphausen, K., Petricoin, E. F., III, Liotta, L. A., and Abati, A. ( 2007) The needle in the haystack: application of breast fine-needle aspirate samples to quantitative protein microarray technology. Cancer 111, 173–184 [DOI] [PubMed] [Google Scholar]
- 11.Paweletz, C. P., Charboneau, L., Bichsel, V. E., Simone, N. L., Chen, T., Gillespie, J. W., Emmert-Buck, M. R., Roth, M. J., Petricoin, I. E., and Liotta, L. A. ( 2001) Reverse phase protein microarrays which capture disease progression show activation of pro-survival pathways at the cancer invasion front. Oncogene 20, 1981–1989 [DOI] [PubMed] [Google Scholar]
- 12.Major, S. M., Nishizuka, S., Morita, D., Rowland, R., Sunshine, M., Shankavaram, U., Washburn, F., Asin, D., Kouros-Mehr, H., Kane, D., and Weinstein, J. N. ( 2006) AbMiner: a bioinformatic resource on available monoclonal antibodies and corresponding gene identifiers for genomic, proteomic, and immunologic studies. BMC Bioinformatics 7, 192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kiernan, J. A. ( 2000) Formaldehyde, formalin, paraformaldehyde and glutaraldehyde: what they are and what they do. Microsc. Today 00, 8–12
- 14.Titford, M. E., and Horenstein, M. G. ( 2005) Histomorphologic assessment of formalin substitute fixatives for diagnostic surgical pathology. Arch. Pathol. Lab. Med. 129, 502–506 [DOI] [PubMed] [Google Scholar]
- 15.Becker, K. F., Schott, C., Hipp, S., Metzger, V., Porschewski, P., Beck, R., Nahrig, J., Becker, I., and Hofler, H. ( 2007) Quantitative protein analysis from formalin-fixed tissues: implications for translational clinical research and nanoscale molecular diagnosis. J. Pathol. 211, 370–378 [DOI] [PubMed] [Google Scholar]
- 16.Wong, S. C., Chan, J. K., Lo, E. S., Chan, A. K., Wong, M. C., Chan, C. M., Lam, M. Y., and Chan, A. T. ( 2007) The contribution of bifunctional SkipDewax pretreatment solution, rabbit monoclonal antibodies, and polymer detection systems in immunohistochemistry. Arch. Pathol. Lab. Med. 131, 1047–1055 [DOI] [PubMed] [Google Scholar]
- 17.Gillespie, J. W., Best, C. J., Bichsel, V. E., Cole, K. A., Greenhut, S. F., Hewitt, S. M., Ahram, M., Gathright, Y. B., Merino, M. J., Strausberg, R. L., Epstein, J. I., Hamilton, S. R., Gannot, G., Baibakova, G. V., Calvert, V. S., Flaig, M. J., Chuaqui, R. F., Herring, J. C., Pfeifer, J., Petricoin, E. F., Linehan, W. M., Duray, P. H., Bova, G. S., and Emmert-Buck, M. R. ( 2002) Evaluation of non-formalin tissue fixation for molecular profiling studies. Am. J. Pathol. 160, 449–457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mutter, G. L., Zahrieh, D., Liu, C., Neuberg, D., Finkelstein, D., Baker, H. E., and Warrington, J. A. ( 2004) Comparison of frozen and RNALater solid tissue storage methods for use in RNA expression microarrays. BMC Genomics 5, 88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Voet, D., and Voet, J. G. ( 1995) Biochemistry, 2nd Ed., pp. 331–410, John Wiley & Sons, New York
- 20.Khan, I. H., Mendoza, S., Rhyne, P., Ziman, M., Tuscano, J., Eisinger, D., Kung, H. J., and Luciw, P. A. ( 2006) Multiplex analysis of intracellular signaling pathways in lymphoid cells by microbead suspension arrays. Mol. Cell. Proteomics 5, 758–768 [DOI] [PubMed] [Google Scholar]
- 21.Stone, R. L., and Dixon, J. E. ( 1994) Protein-tyrosine phosphatases. J. Biol. Chem. 269, 31323–31326 [PubMed] [Google Scholar]
- 22.Ohashi, Y., Creek, K. E., Pirisi, L., Kalus, R., and Young, S. R. ( 2004) RNA degradation in human breast tissue after surgical removal: a time-course study. Exp. Mol. Pathol. 77, 98–103 [DOI] [PubMed] [Google Scholar]
- 23.Paska, C., Bogi, K., Szilak, L., Tokes, A., Szabo, E., Sziller, I., Rigo, J., Jr., Sobel, G., Szabo, I., Kaposi-Novak, P., Kiss, A., and Schaff, Z. ( 2004) Effect of formalin, acetone, and RNAlater fixatives on tissue preservation and different size amplicons by real-time PCR from paraffin-embedded tissue. Diagn. Mol. Pathol. 13, 234–240 [DOI] [PubMed] [Google Scholar]
- 24.Imbeaud, S., Graudens, E., Boulanger, V., Barlet, X., Zaborski, P., Eveno, E., Mueller, O., Schroeder, A., and Auffray, C. ( 2005) Towards standardization of RNA quality assessment using user-independent classifiers of microcapillary electrophoresis traces. Nucleic Acids Res. 33, e56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Srinivasan, M., Sedmak, D., and Jewell, S. ( 2002) Effect of fixatives and tissue processing on the content and integrity of nucleic acids. Am. J. Pathol. 161, 1961–1971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chu, W. S., Furusato, B., Wong, K., Sesterhenn, I. A., Mostofi, F. K., Wei, M. Q., Zhu, Z., Abbondanzo, S. L., and Liang, Q. ( 2005) Ultrasound-accelerated formalin fixation of tissue improves morphology, antigen and mRNA preservation. Mod. Pathol. 18, 850–863 [DOI] [PubMed] [Google Scholar]
- 27.Neel, B. G., and Tonks, N. K. ( 1997) Protein tyrosine phosphatases in signal transduction. Curr. Opin. Cell Biol. 9, 193–204 [DOI] [PubMed] [Google Scholar]
- 28.Theodosiou, A., and Ashworth, A. ( 2002) MAP kinase phosphatases. Genome Biol. 3, REVIEWS3009 [DOI] [PMC free article] [PubMed]
- 29.Bianchi, M. ( 2007) DAMPs, PAMPs and alarmins: all we need to know about danger. J. Leukoc. Biol. 81, 1–5 [DOI] [PubMed] [Google Scholar]